1. Introduction
The human body is colonised by an abundance of microbial cells which have a significant role in health and disease. Next-generation sequencing techniques allowed to characterise the microbiome of various body sites including skin and oral, nasal, conjunctival, vaginal and gut mucosa in health as well as in distinct disease states, e.g. in inflammatory bowel disease, pre-diabetes or preterm birth [
1,
2].
Conflicting results have been reported in samples from body sites commonly considered free of bacteria, such as cerebrospinal fluid or placental tissue. A placental microbiome has been described [
3], whereas another study argued against a viable microbial colonisation of the human placenta [
4].
Using 16S-PCR and short read sequencing, bacterial DNA can be detected with high sensitivity and specificity. This technique also allows to detect microbes not amenable to cultivation. However, the presence of bacterial DNA does not necessarily indicate the existence of living bacteria as bacterial remnants can be stable for longer periods and phagocytic or infected cells may carry bacterial DNA to distant sites [
5]. Furthermore, the ubiquitous presence of microbes in the environment and the extremely high sensitivity of PCR raises the risk to detect contaminating DNA and mistake the result as indicating resident microbial colonisation [
5]. This issue is of particular importance when body sites with very low bacterial load, such as the ocular surface, are examined. Thus, particular care must be taken to control for the presence of background bacterial DNA in these analyses.
The microbial load of skin or oral mucosa (12 bacteria per epithelial cell) is about 200 times higher than that of conjunctival mucosa (0.06 bacteria per epithelial cell, Doan et al. 2016). However, the microbiome of the healthy ocular surface has been characterised [
7]. The current literature suggests that several factors influence the microbial patterns detected on the ocular surface, namely the sampling method [
8,
9], the timing of sampling, as well as ethnicity, age and sex of the sampled individual [
10,
11] and the sampling location (upper or lower fornix, limbus, cornea) [
12,
13].
Today, 16S rDNA gene sequencing facilitates insights not only into the healthy, but also the diseased ocular surface that would never have been possible with bacterial culture methods [
14].
Under healthy conditions, living bacteria should be prevented from passing major barriers such as the blood-brain, blood-placenta [
15,
16], or the blood-retina barrier [
17]. However, it has been reported that these barriers may be breached under certain conditions [
18,
19]. Currently, little is known about a possible physiological presence of microbes in healthy eyes. In one study comparing vitreal samples from patients with or without bacterial endophthalmitis, no bacterial DNA was retrieved from healthy eyes by whole genome sequencing [
19], but so far no comparable study was performed by the more sensitive 16S rDNA-sequencing. Another study did not find an intraocular microbiome in the aqueous humour of pseudophakic donor eyes using shotgun sequencing but detected contaminating environmental DNA [
20]. If healthy eyes harbored resident microbes, it would be important to understand their role in immunological pathomechanisms and the anterior chamber associated immunodeficiency (ACAID) [
21,
22]. To further explore this issue, we used 16S rDNA gene sequencing and employed several controls to detect potential contaminants.
2. Materials and Methods
This study followed the rules of the Declaration of Helsinki of 1975, revised in 2013. It was approved by the local ethics committee (23/20) and is listed in “Deutsches Register Klinischer Studien” (DRKS00020512). All subjects gave their informed consent for inclusion before they participated in the study.
2.1. Patients
In total, three Caucasian patients undergoing cataract surgery by the same surgeon on the same day in the same operation theatre were included. There were two inclusion criteria: informed consent and the absence of acute or chronic eye disease besides cataract at the time of surgery. Exclusion criteria were: local (eye drops/ointment) or systemic antibiotic therapy during the last 6 months, contact lens wearing, history of infectious or noninfectious inflammatory eye disease in the past, known allergy against any of the drugs for surgery (see ‘sampling’ below), diabetes mellitus and chronic inflammatory bowel disease. Two other Caucasian individuals who met the inclusion and exclusion criteria but without cataract and planned cataract surgery served as conjunctival and air sampling controls. These two samples were taken together on the same day in one operation theatre, but at a different date than the three cataract patients’ samples.
2.2. Sampling
All three cataract patients underwent routine cataract surgery. For topical anesthesia, xylocaine 2% and proparacaine 0.5% were applied to the ocular surface three times every five minutes. Subsequently, 5% povidone-iodine was applied to the ocular surface, superior and inferior fornices and the periocular lid region. After 3 minutes, a sterile cover was placed on the face and eyelids, a lid retractor was inserted and the povidone-iodine was flushed from the conjunctiva by balanced salt solution (BSS, Alcon, Germany). From each patient, three samples were taken by one surgeon wearing sterile gloves and clothes: first, a dry swab was wagged in the air above the ocular surface and was put into a 2ml sterile microtube (Sarstedt, Germany). Second, another dry swab was used to wipe the limbal conjunctiva at the site of the future incision and was also placed in a microtube. Third, just after the initial corneal incision, 60-100µL of anterior chamber fluid were aspirated with a 1ml syringe and injection needle and the collected fluid was placed in a microtube. The samples were put on dry ice immediately and prepared for shipment. For the two conjunctival swab controls, sampling was performed using a first dry swab wagged in ambient air and a second swab to wipe the inferior fornix (without any pretreatment). The swabs were transferred to microtubes, placed on dry ice and prepared for shipment. Individual sterile gloves were used for each sample. All samples were sent to the sequencing service provider on dry ice (Microsynth AG, Balgach, Switzerland).
2.3. Sample processing
Extraction, lysis and DNA isolation were done according to manufacturer's recommendations (ZymoBIOMICS DNA Mini Kit). Bead beating was run on a FastPrep-24 instrument (MPBiomedicals; 4 cycles of 45s at speed 4 followed by 1 cycle of 45s at speed 6.5). 400µl of raw extract was prepared for DNA-isolation. Concentration of the isolated DNA was assessed with PicoGreen measurement (Quant-iT™ PicoGreen™ dsDNA Assay Kit, Thermo Fisher).
To sequence the V3 and V4 regions of the bacterial 16S rDNA gene, two-step, Nextera barcoded PCR libraries using the locus specific primer pair 341F (5′- CCT ACG GGN GGC WGC AG -3′) and 805R (5′- GAC TAC HVG GGT ATC TAA TCC -3′) with 20 PCR cycles for the first step and 20 PCR cycles for the second step were created. Subsequently the libraries were sequenced on an Illumina MiSeq platform using a v2 500 cycles kit.
2.4. Biostatistics
The produced paired-end reads which passed Illumina’s chastity filter were subject to de-multiplexing and trimming of Illumina adaptor residuals using Illumina’s bcl2fastq software version v2.20.0.422. The quality of the reads was checked with the software FastQC version 0.11.8 and sequencing reads that fell below an average Q-score of 20 or had any uncalled bases (N) were removed from further analysis. The locus specific V3 and V4 primers were trimmed from the sequencing reads with the software cutadapt v3.2. Paired-end reads were discarded if the primer could not be trimmed. Trimmed forward and reverse reads of each paired-end read were merged to reform the sequenced molecule
in-silico considering a minimum overlap of 15 bases using the software USEARCH version 11.0.667. Merged reads that contained ambiguous bases or were outliers regarding the expected amplicon size distribution were also discarded. The remaining reads were denoised using the UNOISE algorithm implemented in USEARCH to form ASVs (Amplicon Sequence Variants) discarding singletons and chimeras in the process. The resulting ASV abundance table was then filtered for possible barcode bleed-in contaminations using the UNCROSS algorithm. ASV sequences were compared to the reference sequences of the RDP 16S database provided by
https://www.drive5.com/usearch/manual/sintax_downloads.html and taxonomies were predicted considering a minimum confidence threshold of 0.5 using the SINTAX algorithm implemented in USEARCH. The metagenome was visualized via krona charts. Alpha diversity was estimated using the Richness (Observed), Simpson and Shannon indices. Rarefaction analysis was carried out to estimate the coverage of the captured metagenome in contrast to the potential metagenome. Beta diversity was calculated using the weighted Unifrac distance method on the basis of rarefied ASV abundance counts per sample. These sample distances were then used in a detrended correspondence analysis (DCA) to reveal possible patterns of inter-sample relations. Alpha and beta diversity calculations and the rarefaction analysis were performed with the R software packages phyloseq v1.26.1 and vegan v2.5-5. To detect differentially abundant ASVs depending on collected sample metadata (e.g. sample category – air, control, etc.) differential ASV analysis using normalized abundance counts was performed with the R software package DESeq2 v1.26.0.
3. Results
3.1. The patient’s and control’s demographic data are displayed in Table 1.
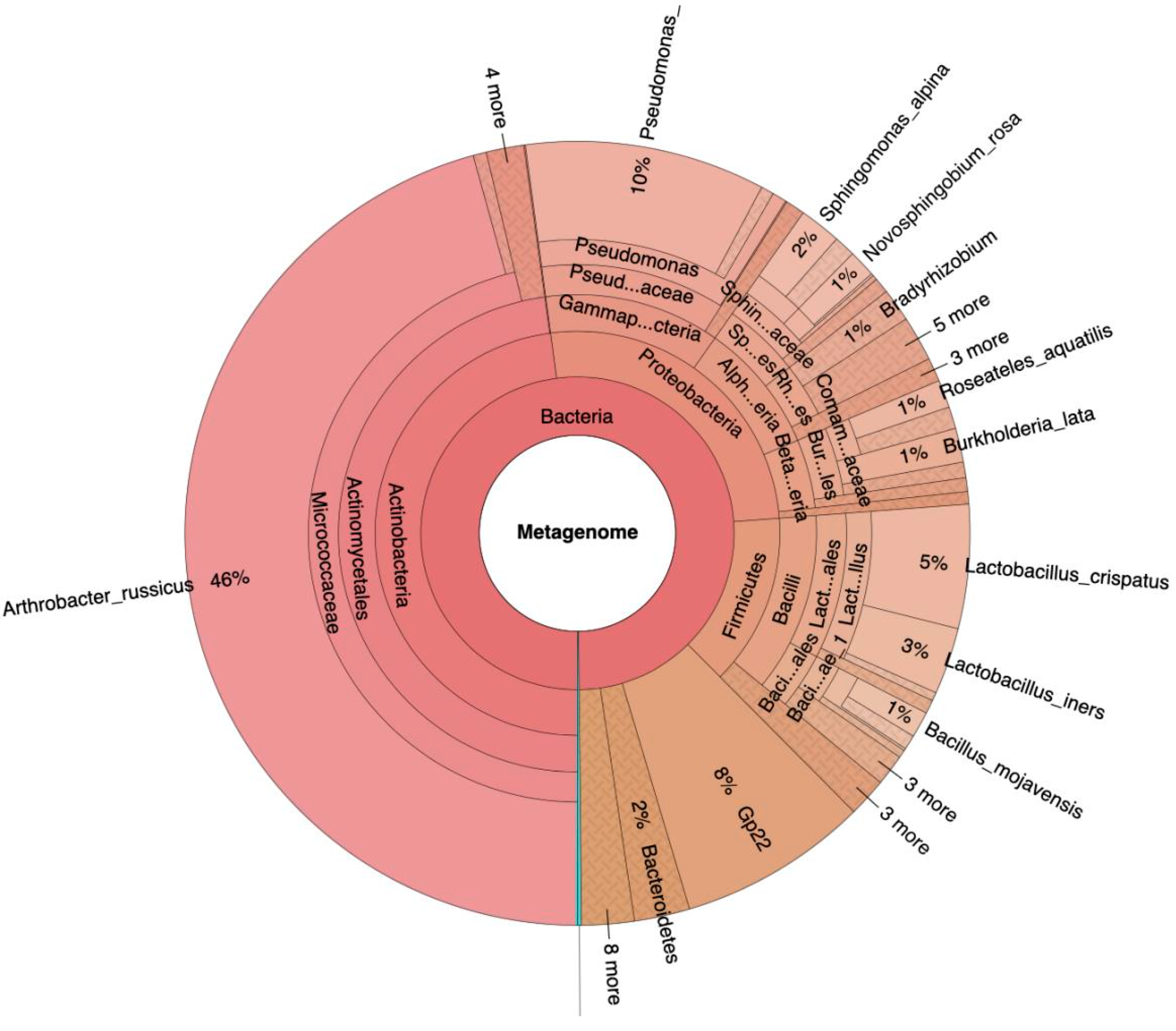
Following isolation, amplification and filtering of the DNA, 1107324 reads were obtained in total (median 26787 +/-7315 per sample). Mean read length per sample was 236 +/-12 base pairs. Description of the analysis quality via FastQC (version 0.11.8) revealed acceptable parameters despite the little content of DNA per sample. Classification of the bacterial DNA showed Actinobacteria (48%), Proteobacteria (26%), Firmicutes (14%), Acidobacteria (8%) and Bacteroidetes (2%) as main phyla in all samples (
Figure 1).
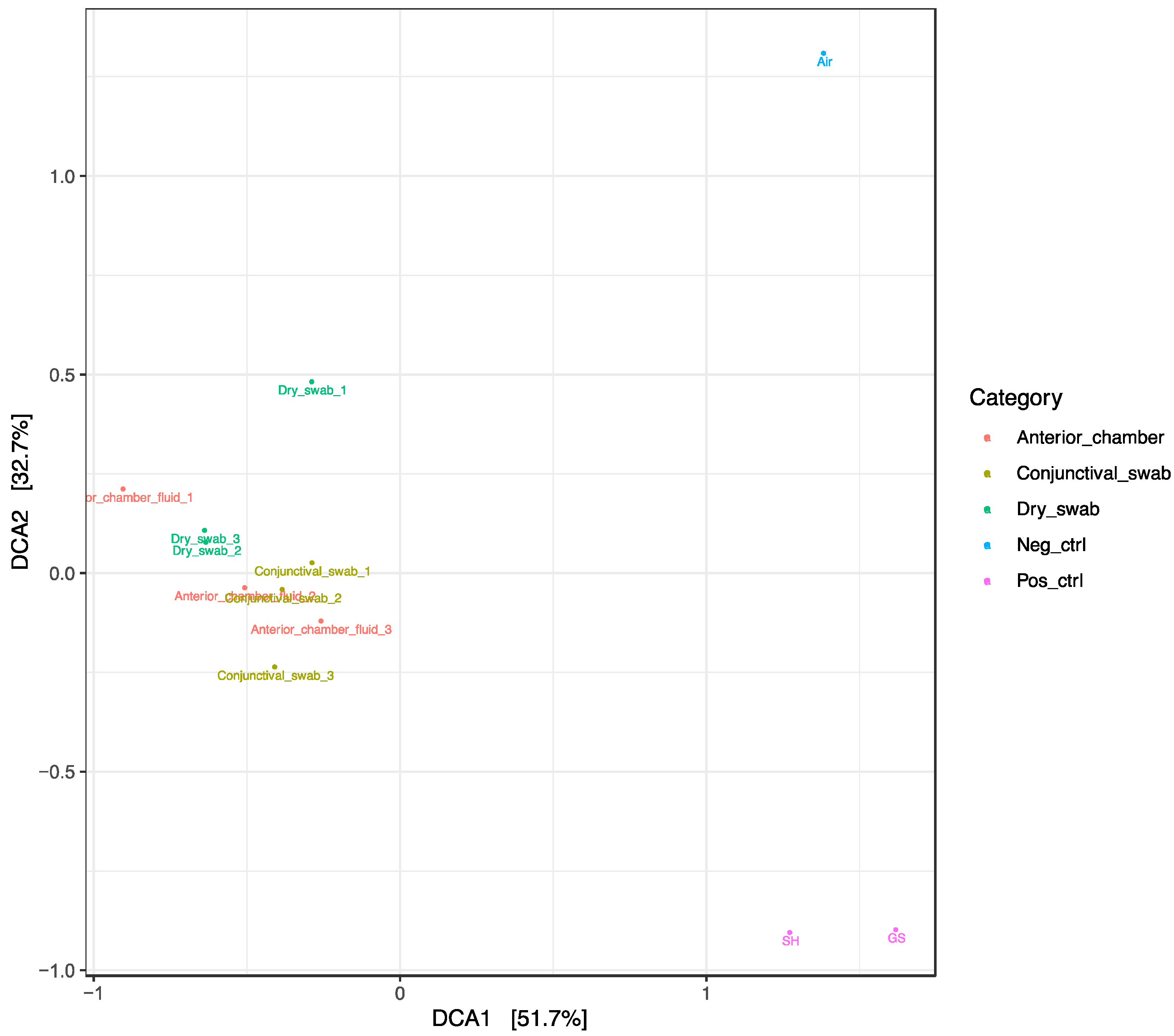
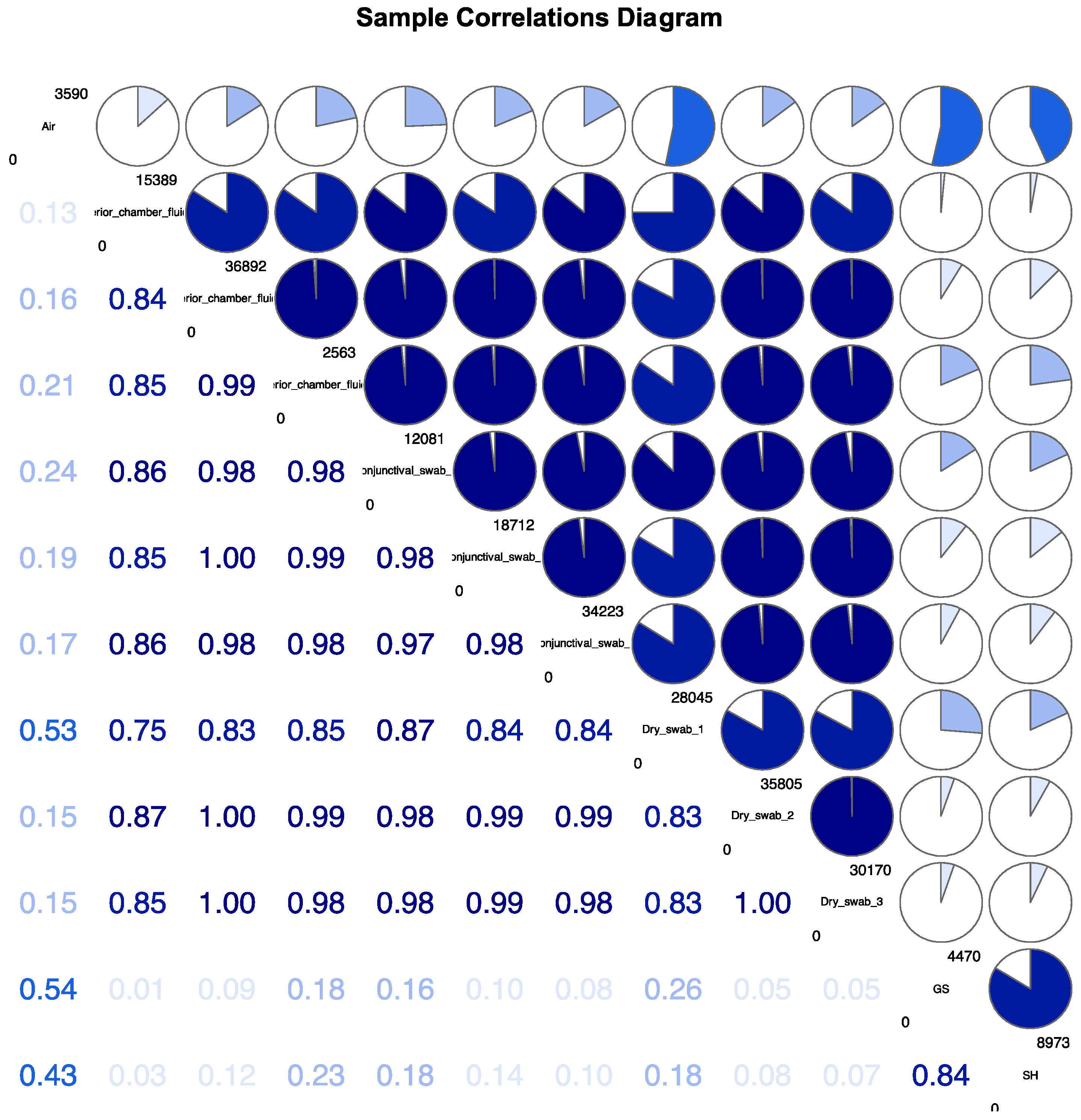
For displaying differences between the samples, a Detrended Correspondence Analysis (DCA) and a sample correlations diagram were used (
Figure 2a,b).
Figure 2.
a. Detrended correspondence analysis (DCA) using the normalized ASV abundances of the sequenced samples. Most of the observed abundance differences in this study is explained by differences between cataract patient samples and untreated controls (DCA1), and between negative and positive untreated controls (DCA2), but not by differences between the different types of cataract patient samples.
Figure 2.
a. Detrended correspondence analysis (DCA) using the normalized ASV abundances of the sequenced samples. Most of the observed abundance differences in this study is explained by differences between cataract patient samples and untreated controls (DCA1), and between negative and positive untreated controls (DCA2), but not by differences between the different types of cataract patient samples.
Figure 2.
b. Sample Correlations Diagram.
Figure 2.
b. Sample Correlations Diagram.
DCA reveals clustering of all samples related to the surgical patients, including the corresponding dry swab negative controls. These “surgical” samples contained mainly Acinetobacteria, classified as Arthrobacter russicus (see
Figure 1 left side), and waterborne bacteria (e.g. Pseudomonas and Sphingomonas species) which are typically not dominant in published reports on the conjunctival microbiome. The positive controls of two healthy eyes (“native” samples) not undergoing cataract surgery were clearly distinct from a corresponding air swab negative control. These positive and negative controls strongly differed from the cluster of “surgical” samples (
Figure 2a,b). The sample correlations diagram shows correlation coefficients of the samples (
Figure 2b).
3.2. Figures, Tables and Schemes
Table 1.
Demographics of the patients and control subjects.
Table 1.
Demographics of the patients and control subjects.
| |
Age (years) |
Sex |
Ethnicity |
Eye |
| Patient 1 |
70 |
Female |
Caucasian |
Left |
| Patient 2 |
73 |
Male |
Caucasian |
Right |
| Patient 3 |
83 |
Female |
Caucasian |
Right |
| Control 1 |
56 |
Male |
Caucasian |
Right |
| Control 2 |
43 |
Female |
Caucasian |
Right |
Correlations of ASV abundances for all 12 samples: “Air”, “GS” and “SH” show the least correlation in comparison to the intraoperatively obtained samples (dry swab, conjunctival swab and anterior chamber fluid, 1-3 each, numbered top down). Inbetween the intraoperative categories, specific correlation is detectable as well: the probes from different patients differ among each other. Anterior chamber fluid 1 and dry swab 1 show the most different bacterial DNA of all “intraoperative” samples.
4. Discussion
This study aims to explore a possible microbiomic signature of human aqueous humor by Next Generation 16S rDNA gene sequencing.
As this method is highly sensitive, care was taken to characterize environmental background DNA using several controls. The two positive control conjunctival swabs from untreated conjunctiva clearly differ from a corresponding air swab pointing to the presence of a local microbiome on an untreated ocular surface as reported earlier [
8,
13,
23]. Unexpected was the clustering of all surgery-related samples including intraocular aspirates, conjunctival swabs and air swab negative controls (
Figure 2) which were in marked contrast to the positive and negative controls unrelated to surgery mentioned above. This strongly suggests an effect of preoperative disinfection and irrigation, which appear to clear bacterial DNA from the ocular surface (see also [
24]). This could explain the similarity of conjunctival samples from surgical patients with the corresponding dry air swabs. A clustering of aqueous humor samples with the former two groups is consistent with the absence of a distinct intraocular microbiome. Arthrobacter species detected in great abundance in all samples related to surgical patients are frequently isolated from environmental sources including the air of ventilation systems [
25]. To date cultivation of Arthrobacter russicus from the ocular surface has not been reported. Together this makes contamination from the environment the most plausible explanation for this finding. Intriguingly, the difference in the surgery-related air swabs and the positive control-related air swab obtained at a different date, but in the same operating theatre, indicates that the method is sensitive enough to detect differences in background contamination which were more pronounced than the differences detected between the surgery-related samples (air swab, cleaned conjunctiva, aqueous humor) on a single day.
In general, detection of bacterial DNA by 16S rDNA gene sequencing does not allow a statement about living bacteria. It is possible that sterile surgical instruments and flushing solutions like basal salt solution contain bacterial DNA introduced in the production and sterilisation processes, typically of waterborne species like in our surgery-related samples. If the DNA content of the sampled material of interest is very low, this background DNA may become dominant due to the high sensitivity of the detection system, rendering a poor signal-to-noise ratio.
5. Conclusions
In conclusion, these data are in line with other observations in ocular [
19,
20], placental [
26] and cerebrospinal [
27] material. The findings strongly suggest that there usually are no significant amounts of living bacteria in body sites shielded by blood-tissue barriers. With the next generation sequencing methods currently available, no intraocular microbiome has been detected.
Author Contributions
Conceptualization, S.H, T.R. and G.S.; methodology, S.S., W.M., S.H., G.S..; software, S.S., W.M.; validation, B.M., W.M., G.S., P.M. and S.H.; formal analysis, S.S.; investigation, S.H., P.M., T.R.; resources, T.R..; data curation, S.S.; writing—original draft preparation, S.H.; writing—review and editing, S.H., G.S., T.R.; visualization, S.S., W.M.; supervision, B.M., S.H., G.S.; project administration, S.H; funding acquisition, T.R.. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Ethics Committee of Albert-Ludwigs-Universität Freiburg (23/20) and is listed in “Deutsches Register Klinischer Studien” (DRKS00020512).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Due to ethical and safety reasons, original data is unavailable for public.
Acknowledgments
These data have not been presented elsewhere in oral or written form. The study was financed by institutional research funds.
Conflicts of Interest
Strempel is employee of Microsynth AG. None of the remaining authors has any potential sources of conflict of interest regarding this study.
References
- The Integrative HMP (iHMP) Research Network Consortium The Integrative Human Microbiome Project. Nature 2019, 569, 641–648. [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The Human Microbiome Project. Nature 2007, 449, 804–810. [CrossRef]
- Aagaard, K.; Ma, J.; Antony, K.M.; Ganu, R.; Petrosino, J.; Versalovic, J. The Placenta Harbors a Unique Microbiome. Sci. Transl. Med. 2014, 6. [CrossRef]
- de Goffau, M.C.; Lager, S.; Sovio, U.; Gaccioli, F.; Cook, E.; Peacock, S.J.; Parkhill, J.; Charnock-Jones, D.S.; Smith, G.C.S. Human Placenta Has No Microbiome but Can Contain Potential Pathogens. Nature 2019, 572, 329–334. [CrossRef]
- Stinson, L.F.; Keelan, J.A.; Payne, M.S. Characterization of the Bacterial Microbiome in First-Pass Meconium Using Propidium Monoazide (PMA) to Exclude Nonviable Bacterial DNA. Lett. Appl. Microbiol. 2019, 68, 378–385. [CrossRef]
- Doan, T.; Akileswaran, L.; Andersen, D.; Johnson, B.; Ko, N.; Shrestha, A.; Shestopalov, V.; Lee, C.S.; Lee, A.Y.; Van Gelder, R.N. Paucibacterial Microbiome and Resident DNA Virome of the Healthy Conjunctiva. Investigative Opthalmology & Visual Science 2016, 57, 5116. [CrossRef]
- Delbeke, H.; Younas, S.; Casteels, I.; Joossens, M. Current Knowledge on the Human Eye Microbiome: A Systematic Review of Available Amplicon and Metagenomic Sequencing Data. Acta Ophthalmol 2021, 99, 16–25. [CrossRef]
- Dong, Q.; Brulc, J.M.; Iovieno, A.; Bates, B.; Garoutte, A.; Miller, D.; Revanna, K.V.; Gao, X.; Antonopoulos, D.A.; Slepak, V.Z.; et al. Diversity of Bacteria at Healthy Human Conjunctiva. Investigative Opthalmology & Visual Science 2011, 52, 5408. [CrossRef]
- Katzka, W.; Dong, T.S.; Luu, K.; Lagishetty, V.; Sedighian, F.; Arias-Jayo, N.; Jacobs, J.P.; Hsu, H.Y. The Ocular Microbiome Is Altered by Sampling Modality and Age. Trans. Vis. Sci. Tech. 2021, 10, 24. [CrossRef]
- Ozkan, J.; Nielsen, S.; Diez-Vives, C.; Coroneo, M.; Thomas, T.; Willcox, M. Temporal Stability and Composition of the Ocular Surface Microbiome. Sci Rep 2017, 7, 9880. [CrossRef]
- Wen, X.; Miao, L.; Deng, Y.; Bible, P.W.; Hu, X.; Zou, Y.; Liu, Y.; Guo, S.; Liang, J.; Chen, T.; et al. The Influence of Age and Sex on Ocular Surface Microbiota in Healthy Adults. Invest. Ophthalmol. Vis. Sci. 2017, 58, 6030–6037. [CrossRef]
- Matysiak, A.; Kabza, M.; Karolak, J.A.; Jaworska, M.M.; Rydzanicz, M.; Ploski, R.; Szaflik, J.P.; Gajecka, M. Characterization of Ocular Surface Microbial Profiles Revealed Discrepancies between Conjunctival and Corneal Microbiota. Pathogens 2021, 10, 405. [CrossRef]
- Ozkan, J.; Coroneo, M.; Willcox, M.; Wemheuer, B.; Thomas, T. Identification and Visualization of a Distinct Microbiome in Ocular Surface Conjunctival Tissue. Invest. Ophthalmol. Vis. Sci. 2018, 59, 4268–4276. [CrossRef]
- Borroni, D.; Romano, V.; Kaye, S.B.; Somerville, T.; Napoli, L.; Fasolo, A.; Gallon, P.; Ponzin, D.; Esposito, A.; Ferrari, S. Metagenomics in Ophthalmology: Current Findings and Future Prospectives. BMJ Open Ophthalmology 2019, 4, e000248. [CrossRef]
- Ander, S.E.; Diamond, M.S.; Coyne, C.B. Immune Responses at the Maternal-Fetal Interface. Sci Immunol 2019, 4, eaat6114. [CrossRef]
- Kniesel, U.; Wolburg, H. [No Title Found]. Cellular and Molecular Neurobiology 2000, 20, 57–76. [CrossRef]
- Mölzer, C.; Heissigerova, J.; Wilson, H.M.; Kuffova, L.; Forrester, J.V. Immune Privilege: The Microbiome and Uveitis. Front Immunol 2020, 11, 608377. [CrossRef]
- Al-Obaidi, M.M.J.; Desa, M.N.M. Mechanisms of Blood Brain Barrier Disruption by Different Types of Bacteria, and Bacterial–Host Interactions Facilitate the Bacterial Pathogen Invading the Brain. Cell Mol Neurobiol 2018, 38, 1349–1368. [CrossRef]
- Kirstahler, P.; Bjerrum, S.S.; Friis-Møller, A.; la Cour, M.; Aarestrup, F.M.; Westh, H.; Pamp, S.J. Genomics-Based Identification of Microorganisms in Human Ocular Body Fluid. Sci Rep 2018, 8, 4126. [CrossRef]
- Mazoteras, P.; Quiles, M.G.; Martins Bispo, P.J.; Höfling-Lima, A.L.; Pignatari, A.C.; Casaroli-Marano, R.P. Analysis of Intraocular Lens Biofilms and Fluids After Long-Term Uncomplicated Cataract Surgery. Am J Ophthalmol 2016, 169, 46–57. [CrossRef]
- Rosenbaum, J.T.; Asquith, M. The Microbiome and HLA-B27-Associated Acute Anterior Uveitis. Nat Rev Rheumatol 2018, 14, 704–713. [CrossRef]
- Vendomèle, J.; Khebizi, Q.; Fisson, S. Cellular and Molecular Mechanisms of Anterior Chamber-Associated Immune Deviation (ACAID): What We Have Learned from Knockout Mice. Front Immunol 2017, 8, 1686. [CrossRef]
- Huang, Y.; Yang, B.; Li, W. Defining the Normal Core Microbiome of Conjunctival Microbial Communities. Clinical Microbiology and Infection 2016, 22, 643.e7-643.e12. [CrossRef]
- Fan, C.; Yang, B.; Huang, Y. Efficacy of 0.5% Levofloxacin and 5.0% Povidone-Iodine Eyedrops in Reducing Conjunctival Bacterial Flora: Metagenomic Analysis. Journal of Ophthalmology 2020, 2020, 1–9. [CrossRef]
- Li, Y.; Kawamura, Y.; Fujiwara, N.; Naka, T.; Liu, H.; Huang, X.; Kobayashi, K.; Ezaki, T. Rothia Aeria Sp. Nov., Rhodococcus Baikonurensis Sp. Nov. and Arthrobacter Russicus Sp. Nov., Isolated from Air in the Russian Space Laboratory Mir. International Journal of Systematic and Evolutionary Microbiology 2004, 54, 827–835. [CrossRef]
- Segata, N. No Bacteria Found in Healthy Placentas. Nature 2019, 572, 317–318. [CrossRef]
- Perlejewski, K.; Bukowska-Ośko, I.; Nakamura, S.; Motooka, D.; Stokowy, T.; Płoski, R.; Rydzanicz, M.; Zakrzewska-Pniewska, B.; Podlecka-Piętowska, A.; Nojszewska, M.; et al. Metagenomic Analysis of Cerebrospinal Fluid from Patients with Multiple Sclerosis. Adv. Exp. Med. Biol. 2016, 935, 89–98. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).