Patients and Methods
Ethical statement:
All relevant institutional, national, and/or worldwide guidelines for the care and use of humans and animals were prioritized in the current survey. The Ethical Committee for Human and Animal Handling at Cairo University( ECAHCU), at the Faculty of Pharmacy, Cairo University, Egypt, approved all study procedures involving humans and animals by the recommendations of the Weatherall Report( approval number T-11-3-2023). The number of humans and animals included in the study, as well as their suffering, was minimized at all costs. The randomized human clinical trials phase 1/2 registration number was NCT00000714/ 2023.
Type of the study:
Screening experimental study.
Place and date of the study:
This study was finished at faculty of pharmacy, Cairo university, Egypt between March 2023 and January 2024.
Source of animal models:
They were obtained and legalized from the pharmacology and toxicology department of the faculty of pharmacy, Cairo university, Egypt.
Inclusion criteria for animal models:
Adult male obese rabbit animal models weighing about 2 kg; can be inoculated with different bacterial infections. Rabbits were acclimatized for one week before the experiment. At a humidity( 50 % ±5), light-dark cycle( 12/ 12 h), and a controlled temperature( 25±2 0C). The rabbits were fed with fresh grass.
Exclusion criteria for animal models:
Young and female rabbits; Non-obese rabbits weighing less than 2 kg.
Collection of 100 soil samples:
The samples were randomly selected grassland soils taken from various soil settings in Egypt at a depth of 30 cm. Before being processed, samples were kept at 4 ℃ in sterile containers. Each soil sample was weighed at one gram, and each 250 ml Erlenmeyer flask had 99 ml of sterile distilled water. The flasks were shaken at 400 rpm using a gyrator shaker for five minutes. Following dilutions from 10-1 to 10-6 in sterile distilled water, the soil suspensions were plated on selective Casein yeast peptone agar( CYP) medium( bought from Sigma-Aldrich, USA). 50 cc of nutrient broth liquid at PH 7 was added to 250 ml Erlenmeyer flasks to create the inoculum for the bacterial isolate under investigation.
The medium was autoclaved and then infected with a loopful of culture from a nutritional agar slant that had been left overnight. The inoculum was the inoculated flasks, which were shaken for a whole day at 150 rpm.
Instruments
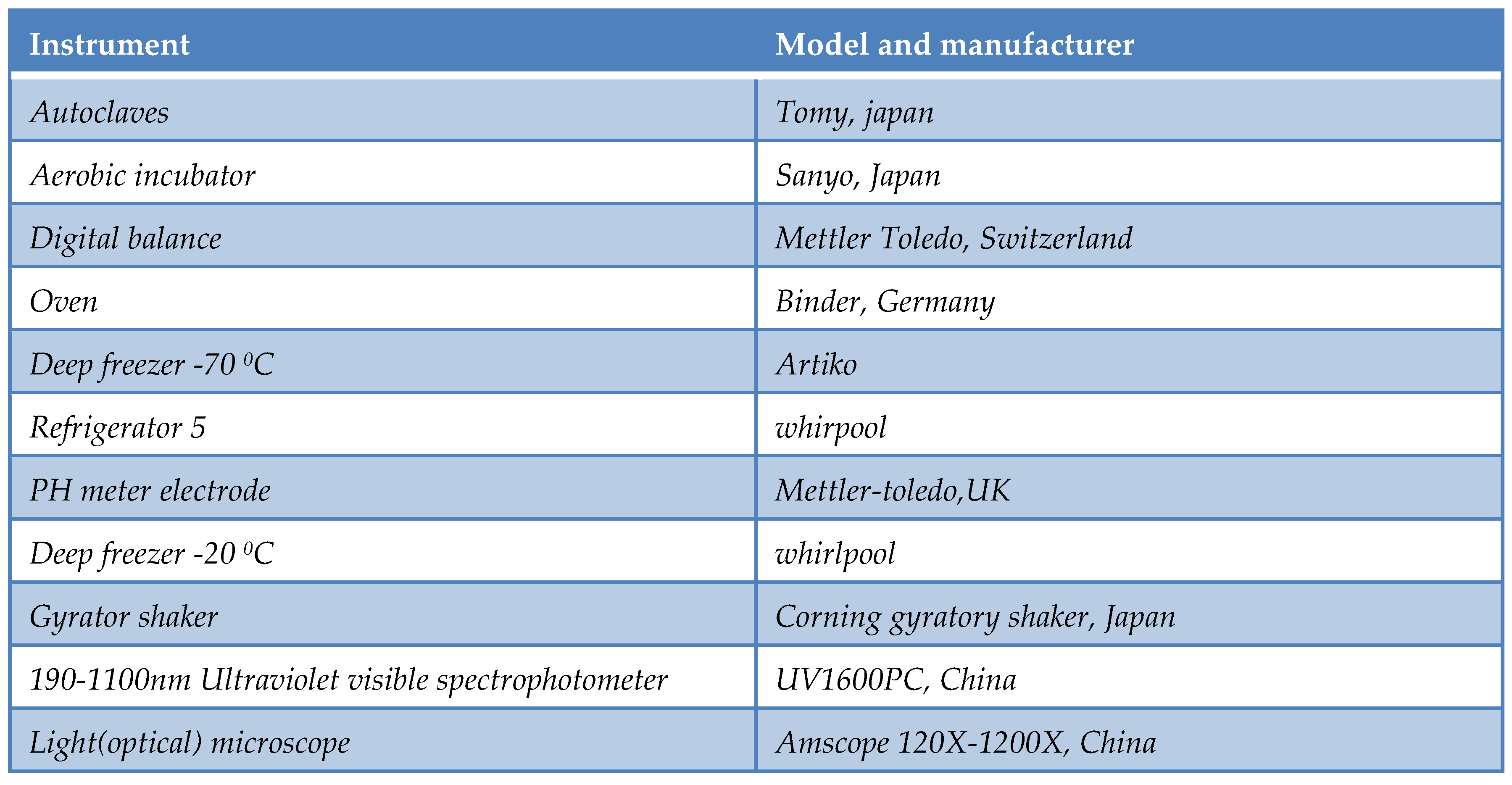
Table 1.
List of instruments.
Table 1.
List of instruments.
Material:
All chemical and biochemical substances were purchased from Algomhuria pharmaceutical company and Alnasr chemical company, Egypt. All chemical reagents used were of analytical grade.
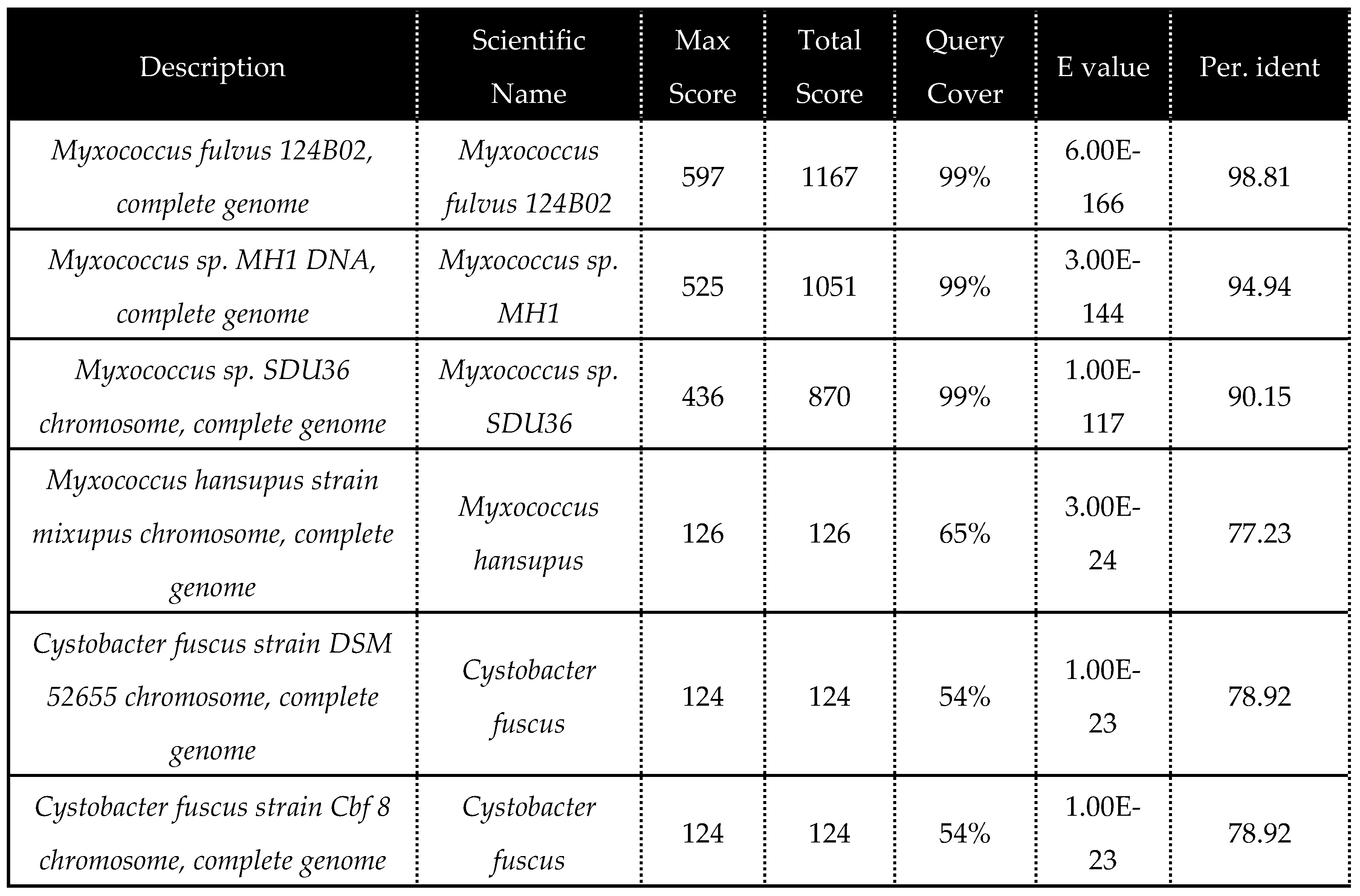
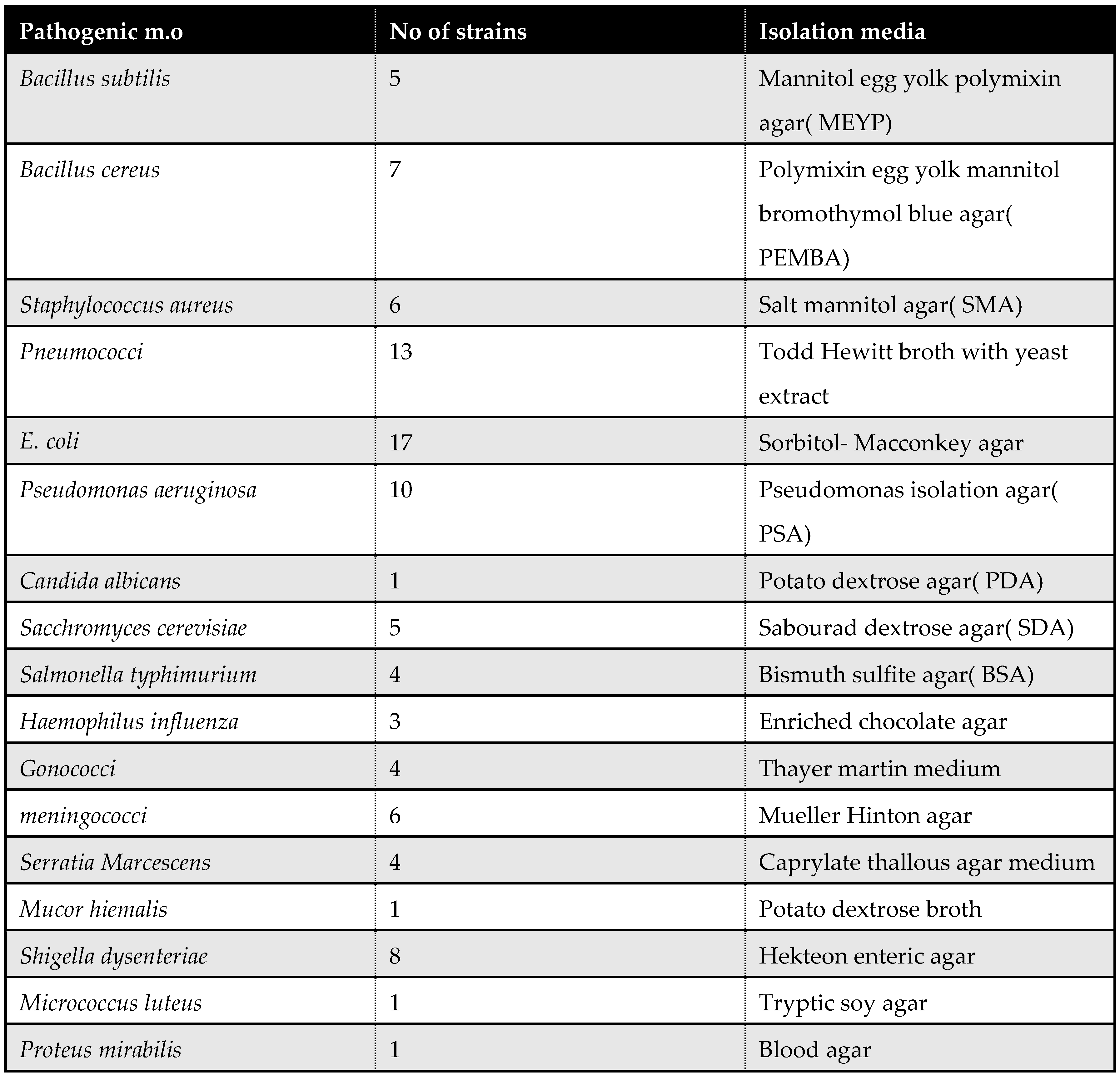
Isolation of Myxococcus fulvus 124B02 producing Myxopyronin antibiotics:
The selective isolation of species of
Myxococcus fulvus 124B02 from different soil samples was directly achieved using dilution plating. The technique comprised the suppression of competing bacteria exploiting antibiotics such as
10 mcg/ ml Vancomycin and/ or
10 mcg/ ml Chloramphenicol combined with wet heat treatment of soils and air drying. Fungi were eliminated via supplementing the plating medium with
2 mcg/ ml Terbinafine HCl. Swarming of
Myxococcus fulvus 124B02 colonies was controlled with
Casein Yeast Peptone(
CYP) plates incubated at
30℃ and
PH 7.2 for
5 days. The composition of
CYP plate included
0.4 % Peptone from
Casein, tryptically digested,
0.3 % CaCl2.2H2O,
0.1 % MgSO4.7H2O,
PH 7.2. The potent bacterial isolate producing Myxopyronin was performed utilizing
16 S rRNA sequencing technique. The predominant bacterial isolate with high antibacterial activity was identified using
16S rRNA sequencing and other biochemical tests. Nucleic acid was extracted from a swab by bead-beating in a buffered solution containing Phenol, Chloroform and Isoamyl alcohol. Variable region of
16S rRNA gene was then amplified from the resulting nucleic acid using
PCR. The genomic
DNA was extracted from
120 hours cultured cells using a
DNA purification kit[
PurreLinkTM Genomic DNA Mini Kit with Catalog number: K182002 was purchased from Invitrogen, USA] according to the protocol provided by the manufacturer of
DNA purification kit.
The 16S rRNA gene was amplified by
PCR[
PCR SuperMix kit was purchased from Invitrogen,
USA] using forward[
5-AGAGTTTGATCCTGGCTCAG-3-] and reverse[
5-GGTTACCTTGTTACGACTT-3-]
primers.
PCR amplicons from up to hundreds of samples were then combined and sequenced on a single run. The resulting sequences were matched to a reference database to determine relative bacterial abundances. Polymerase Chain Reaction (
PCR) was a powerful method for amplifying particular segments of
DNA.
PCR used the enzyme
PlatinumTM Taq DNA polymerase with catalog number
10966018[ purchased from Invitrogen,
USA] that directed the synthesis of
DNA from deoxynucleotide substrates on a single-stranded
DNA template.
DNA polymerase added nucleotides to the 3` end of a custom-designed oligonucleotide when it was annealed to a longer template
DNA. Thus, if a synthetic oligonucleotide was annealed to a single-stranded template that contained a region complementary to the oligonucleotide,
DNA polymerase could use the oligonucleotide as a primer and elongate its 3` end to generate an extended region of double stranded
DNA. Denaturation was the initial
PCR cycle stage The
DNA template was heated to
94° C. This broke down the weak hydrogen bonds that held
DNA strands together in a helix, allowing the strands to separate creating single stranded
DNA. Annealing was the second
PCR cycle. The mixture was cooled to anywhere from
50-70° C. This allowed the primers to bind (anneal) to their complementary sequence in the template
DNA. Extension was the final step of
PCR cycle. The reaction was ; then heated up to
72° C, the optimal temperature for
DNA polymerase to act.
DNA polymerase extended the primers, adding nucleotides onto the primer in a sequential manner, using the target
DNA as a template.
With one cycle, a single segment of double-stranded DNA template was amplified into two separate pieces of double-stranded DNA. These two pieces were then available for amplification in the next cycle. As the cycles were repeated, more and more copies were generated and the number of copies of the template was increased exponentially. The amplified
PCR product was sequenced using a genetic analyzer
3130XL[ purchased from Applied biosystems,
USA].
DNA sequence homology search analysis of the predominant bacterial isolate was achieved using Blastn algorithm at
NCBI website. Fruiting bodies were examined using a
Stereomicroscope(
dissecting microscope) MSC-ST45T( purchased from Infetik, China). Wet mounts from crushed fruiting bodies were prepared. The refaractility, shape and the size of Myxospores were determined victimizing phase contrast microscopy. On the other hand the plates were exposed to
360 nm wavelength ultraviolet light to assess the fruiting bodies fluoresced [
16].
Identification Myxopyronin A producing bacterial isolates:
Gram stain:
It classified bacteria into two categories based on the makeup of their cell walls. The bacterial cells became purple after being treated with a solution of crystal violet and subsequently iodine on a microscope slide. When colored cells were treated with a solvent such as alcohol or acetone, gram-positive organisms kept the stain whereas gram-negative organisms lost the stain and turned colorless. With the addition of the counter-stain safranin, the clear, gram-negative bacteria became pink [
17].
Spore shape:
This was discovered using the spore staining method. To get rid of any fingerprints, the slide was wiped with alcohol and a Kim-wipe. On the bottom of the slide, a Sharpie was used to create two circles. Each circle was filled with two tiny droplets of water using an inoculation loop. A very small amount of germs was taken out of the culture tube using an aseptic method. The water droplet on the slide had microorganisms on it. The slide was thoroughly dried by air. Bypassing the slide through the flame three to four times with the smear side up, the slide was heat-fixed. It took a while for the slide to completely cool. A piece of paper towel placed inside the slide’s border was used to hide the streaks. A beaker of heating water was situated over the slide. The slide was allowed to steam for three to five minutes; while the paper towel was covered with a malachite green liquid. Removed and thrown away was the discolored paper towel. To get rid of any stray paper towel bits, the slide was gently cleaned with water. The counter-stain was safranin for 1 minute. Before putting the slide on the microscope’s stage and seeing it via the oil immersion lens, the slide’s bottom was dried [
18].
Spore site:
During the Gram stain test, the spore location was established [
19].
Cell shape:
During the Gram stain test, the cell shape was assessed [
20].
Blood haemolysis:
On blood agar media, the test antibiotic capacity to haemolyze the blood was tested [
21].
Motility test:
It discriminated between motile bacteria and non-motile bacteria.
A sterile needle was used to penetrate the medium to within 1 cm of the tube’s bottom to select a well-isolated colony and test for motility. The needle was certainly retained in the same position as it was inserted and removed from the medium. It took
18 hours of incubation at
35°C, or until noticeable growth appeared [
22].
Nitrate reduction test:
0.5 ml of nitrate broth was added in a clean test tube, was autoclaved for 15 minutes at
15 lbs pressure and
121°C, and was let to cool to room temperature. The tube was inoculated with a heavy inoculum of fresh bacterial culture and was incubated at
35°C for
2 hours.
2 drops of reagent A and
2 drops of reagent B were added and mixed well. The development of red color within
2 minutes was observed for. If no red color was developed, a small amount of
zinc dust was added and observed for the development of the red color within
5 minutes [
23].
Methyl red test:
In the Methyl Red test, an infected tube of
MR broth was used before adding the methyl red
PH indicator. The buffers in the medium were overcome by the acids when an organism used the mixed acid fermentation pathway and produced stable acidic end products, resulting in an acidic environment [
24].
Catalase test:
A little inoculum of a specific bacterial strain was introduced to a
3% hydrogen peroxide solution to see if it might produce catalase. It was observed for the rapid emergence of oxygen bubbles [
25].
Oxidase test:
The 1% Kovács oxidase reagent was applied to a tiny piece of filter paper, which was then allowed to air dry. A well-isolated colony was taken from a fresh (
18 to 24-hour culture) bacterial plate using a sterile loop, and it was then rubbed onto prepared filter paper. Color alterations were noticed [
26].
Citrate utilization:
Five milliliters of a Simmon Koser’s citrate medium were taken after it had been autoclaved at
15 pounds for
15 minutes. To create a clear slant and butt, the test tube containing melted citrate medium was slanted. Using sterilized wire and labeled tubes, the specified samples of microbe were injected on the media’s incline. For
24 hours, the tubes were incubated at
37°C. The medium’s color shift was watched for [
27].
Starch hydrolysis:
For
48 hours at
37°C, the bacterium plates were injected. After incubation, a dropper was used to saturate the surface of the plates with an iodine solution for
30 seconds. Iodine that was in excess was afterward poured out. The area surrounding the bacterial growth line was looked at [
28].
Tween 80 hydrolysis:
1% Tween 80 was used to create agar media. The supplied microorganism was added to the
Tween 80 agar plates by utilizing an inoculating loop to create a single center streak in the plate. The plates were incubated for
24 hours at
37 °C.
HgCl2 solution was poured over the plates. After a short while, the plates were examined. Positive test result; distinct halo-zone surrounding the injected region showed
Tween 80 hydrolysis [
29].
Growth at 10-45 0C:
On nutrient agar media, growth was observed to be possible at
45°C [
30].
Indol test:
The test tube containing the microorganism for inoculation received 5 drops of the Kovács reagent directly. Within seconds after introducing the reagent to the media, the reagent layer formed a pink to red colour (cherry-red ring), which was a sign of a positive indol test [
31].
Tolerance salinity test:
Its capacity to develop on nutrient agar while being responsive to 5% and
7 % NaCl was examined [
32].
Voges-Proskauer(VP) test:
For the test, Voges-Proskauer broth, a glucose-phosphate broth loaded with microorganisms, was added to alpha-naphthol and potassium hydroxide. A successful outcome was indicated by a cherry red tint, whereas an unfortunate outcome was indicated by a yellow-brown color [
33].
Casein hydrolysis test:
For testing the casein hydrolyzing activity of the test antibiotic, a single line streak of the given culture was made in the center of the skim milk agar plate under aseptic conditions and plate was incubated at
37°C in an incubator for
24-48 h [
34].
Saccharide fermentation tests:
Glucose fermentation test:
The fermentation reactions of glucose were investigated using glucose purple broth. Peptone and the
PH indicator bromcresol purple made up the purple broth. A
1% concentration of glucose was added. Isolated colonies from a 24-hour pure culture of microorganisms were added to the glucose purple broth as an inoculant. Parallel to the inoculation of the glucose-based medium, a control tube of purple broth base was used. The inoculated medium was incubated aerobically for
3 days at a temperature of
35–37 °C. The medium began to become yellow, which was a sign of a successful outcome. A poor carbohydrate fermentation response was indicated by the lack of yellow color development [
35].
Fructose fermentation test:
A pure culture’s inoculum was aseptically transferred to a sterile tube of phenol red fructose broth. The infected tube was incubated for
18–24 hours at
35–37 °C. A color shift from red to yellow, signifying an acidic PH alteration, was a sign of a favorable response [
36].
Maltose fermentation test:
A pure culture inoculum was aseptically transferred to a sterile tube containing phenol red maltose broth. The infected tube was incubated for
18–24 hours at
35–37 °C. A color shift from red to yellow, signifying an acidic PH alteration, was a sign of a favorable response [
37].
Sucrose fermentation test:
A pure culture’s inoculum was aseptically transferred to a sterile tube containing phenol red sucrose broth. For
24 hours, the infected tube was incubated at
35–37 0C. A colour shift from red to yellow, signifying an acidic
PH alteration, was a sign of a favourable response [
38].
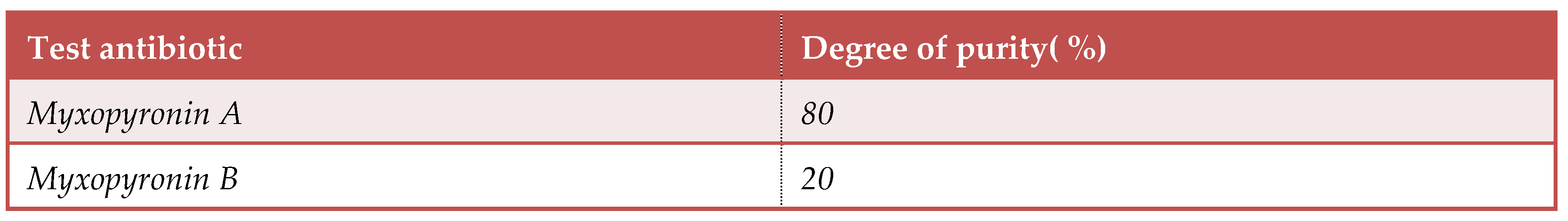
Purification of Myxopyronin A antibiotic:
This was achieved through reversed phase chromatography technique.
The aeration rate was
0.142 V/ V. min. The stirring rate was
500 rpm.
PO2 was about
90 % of saturation; but decreased to about
20 % after
18 hours). The fermentation was stopped after
40 hours via centrifugation at
500 rpm in a gyrator shaker. The supernatants were collected; then tested for antimicrobial sensitivity using broth dilution technique to detect MICs and agar paper diffusion discs technique. The test antibiotic was extracted from the
2 liters of culture broth with
2/ 10 volume ethyl acetate. The ethyl acetate was then removed under the reduced pressure at
40℃. Afterwards, the residue was dissolved in 398 ml of methanol-water(
90: 10) and chromatographed on reversed phase
HPLC. Methanol was the mobile phase. The eluent was
70 part
methanol:
16 part
water:
4 part
acetic acid with flow rate
300 ml/ min. Detection of the antibiotic components was achieved exploiting refractive index. The main peak with retention time
5 minutes contained the biological antibiotic activity which was determined via agar diffusion assay using paper discs and
Staphylococcus aureus as an indicator organism. On the other hand, the main peak was subjected to neutralization via
NaHCO3. Myxopyronin A was extracted using
10 % V/ V Ethylene chloride. After the evaporation of the solvent, about
85 % of the remaining antibiotic substance was pure. It was noticed that the retention rime of
Myxopyronin A was
11 minutes. Molecular formula of the purified
Myxopyronin A was detected through mass spectrometer( Quadrupole mass spectrometer, Advion, USA) [
39].
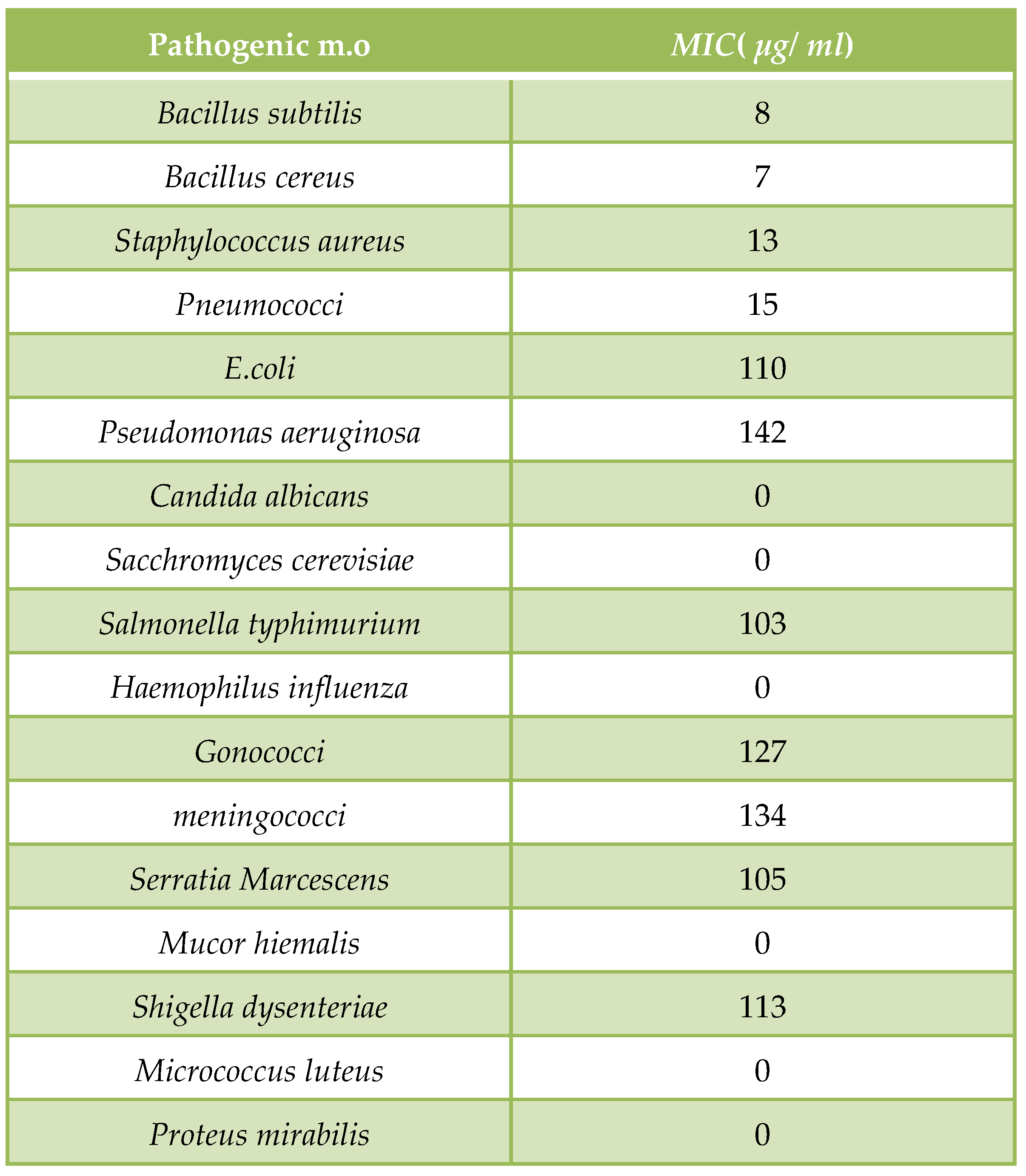
Procedure of Broth dilution assay for determination of MICs of Myxopyronin A:
During testing, multiple microtiter plates were filled with a certain broth, according to the needs of target bacteria. Varying concentrations of the antibiotics and the bacteria to be tested were then added to the plate. The plate was then placed into a non-
CO2 incubator and incubated at thirty-seven degrees Celsius for sixteen to twenty hours. Following the allotted time, the plate was removed and checked for bacterial growth. When the broth became cloudy, bacterial growth occurred. The results of the broth microdilution method were reported in Minimum Inhibitory Concentration(
MIC), or the lowest concentration of antibiotics that stopped bacterial expansion [
40].
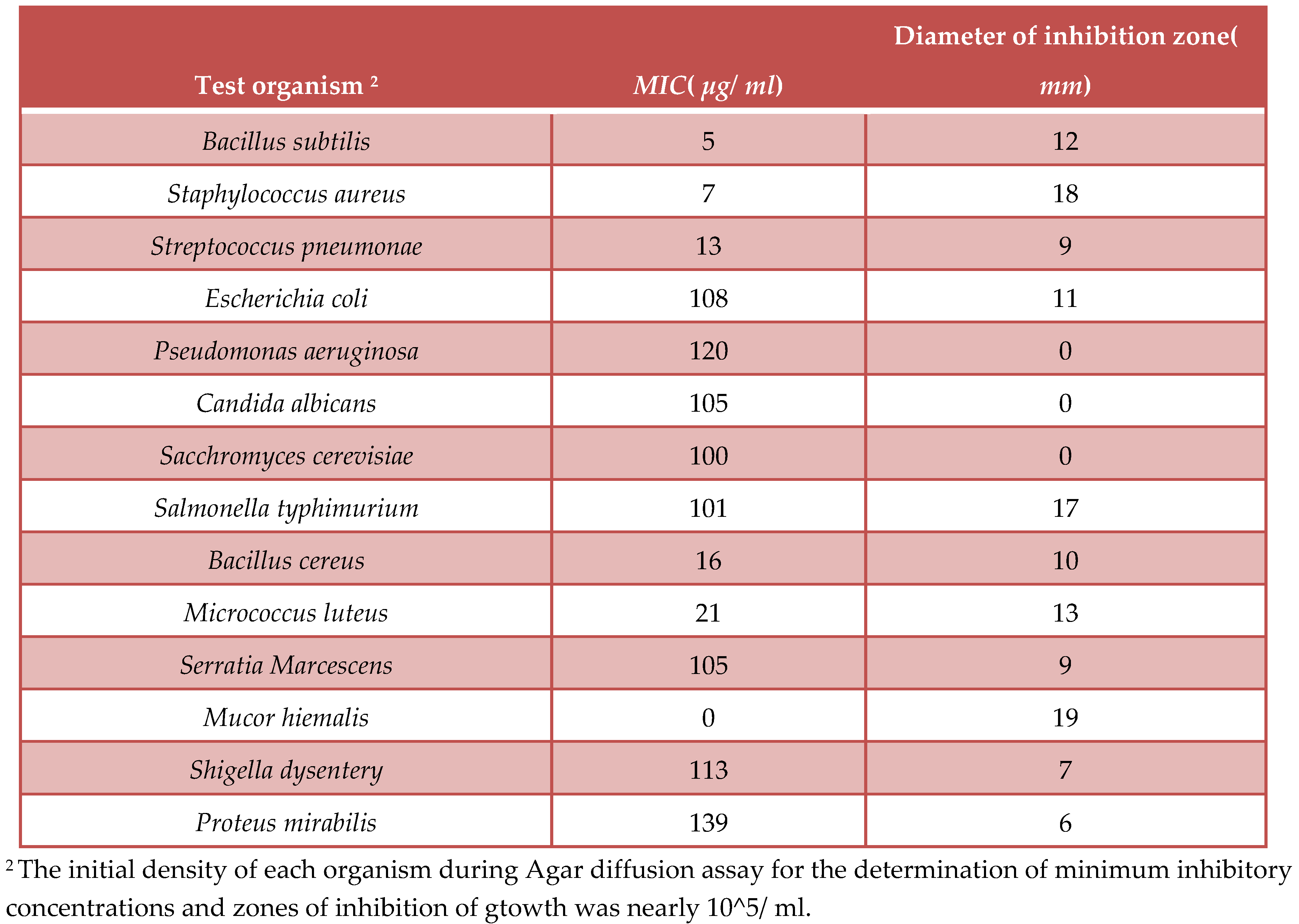
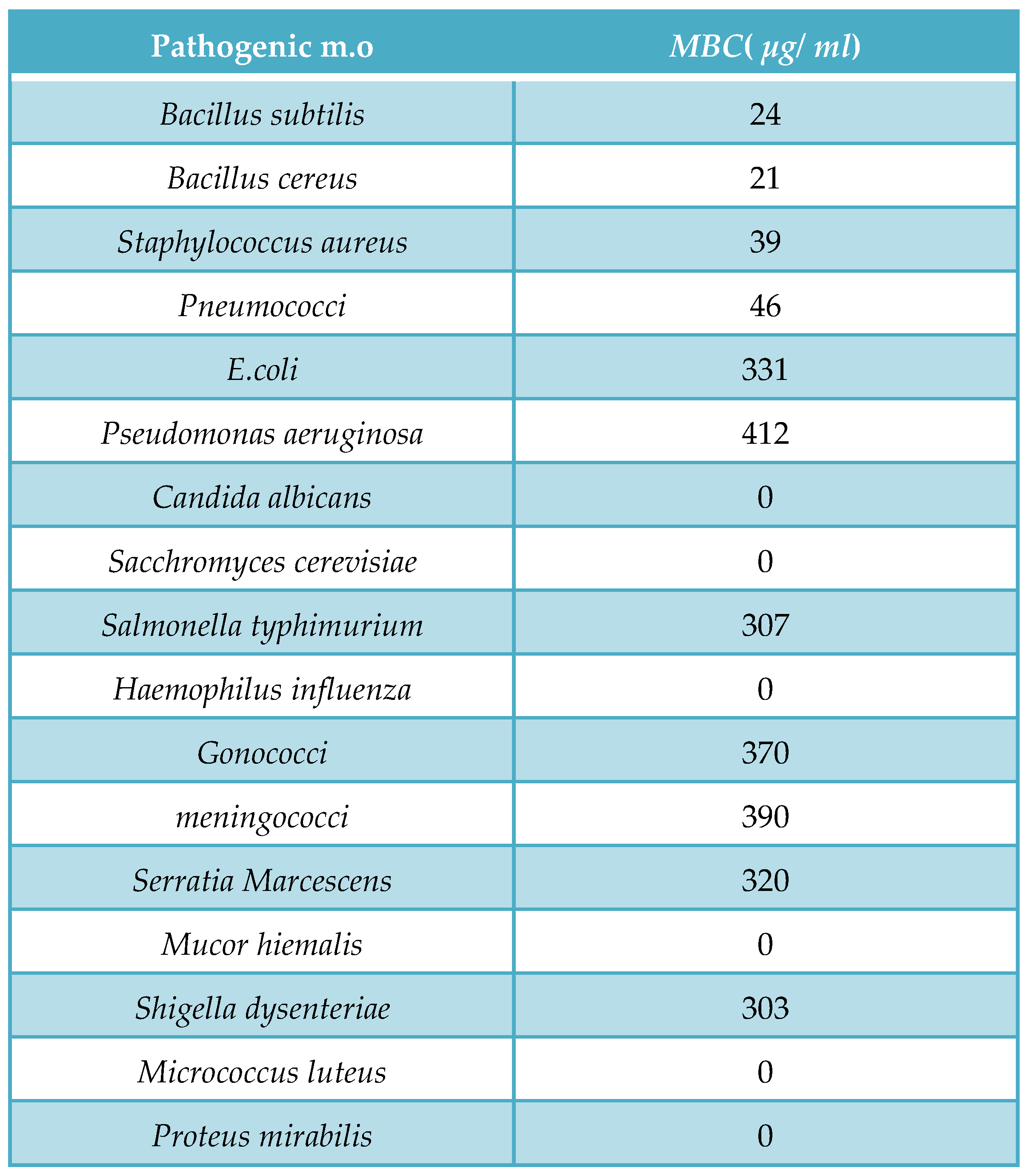
Agar diffusion assay with paper discs procedure for the determination of Myxopyronin A antimicrobial activity:
The disk diffusion method(
DDM) was classified as an agar diffusion method(
ADM) because the test antibiotic extract to be tested diffused from its reservoir through the agar medium seeded with the test microorganism. Generally, the reservoir was a filter paper disk, which was placed on top of an agar surface. When tested extracts compounds were microbiologically active, an inhibition zone developed around the filter paper disk after incubation. The diameter of the inhibition zone properly described the antimicrobial potency of test extract [
41]. The test microbes were isolated using either selective or enrich growth media or broth(
Table 2).
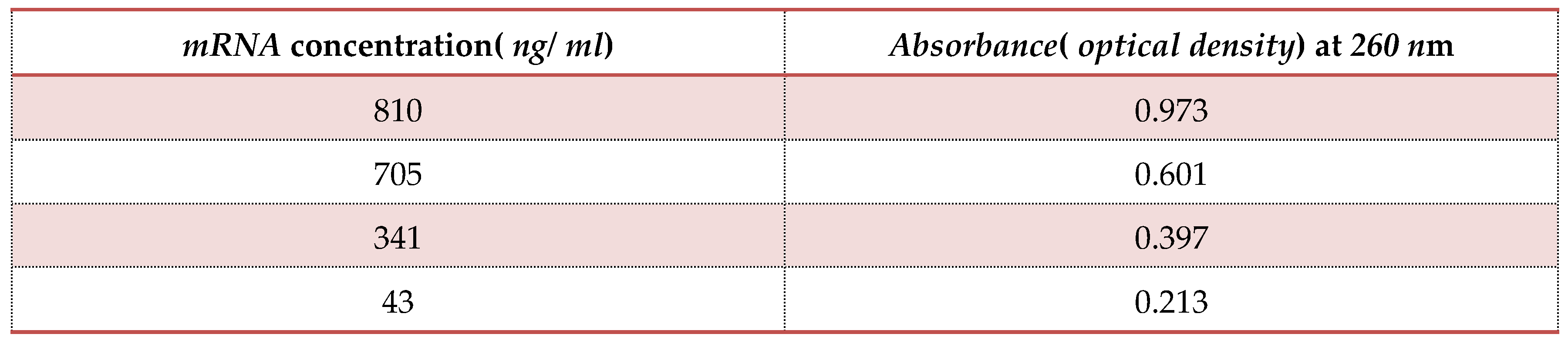
Estimation of Myxopyronin A effect on bacterial RNA synthesis:
The concentration of
RNA isolated with
RNeasy Kits( purchased from QIAGEN, USA) was determined by measuring the absorbance at
260 nm in a spectrophotometer. An absorbance of 1 unit at 260 nm corresponds to 40 µg of
RNA per ml(
A260 =
1 =
40 µg/ ml) [
42].
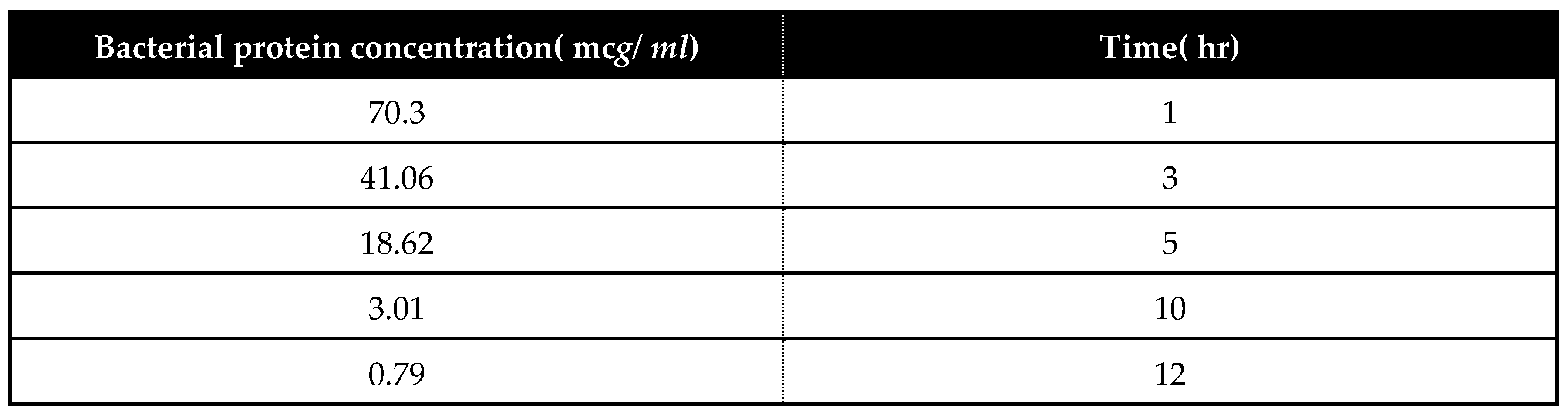
Estimation of Myxopyronin A effect on bacterial protein synthesis:
Absorbance was measured at
205 nm to calculate the protein concentration by comparison with a standard curve. A(
205) method could be used to quantify total protein in crude lysates and purified or partially purified protein. The
UV spectrophotometer was set to read at
205 nm allowing
15 min for the instrument to equilibrate. The absorbance reading was set to zero with a solution of the buffer and all components except the protein present. The protein solution was placed in the 1 mL cuvette and the absorbance was determined. The dilution and readings of samples were performed in duplicate.The matched cuvettes for samples and controls were utilized during the test procedure. The extinction coefficient of the protein was known, the following equation was employed.
Absorbance =
Extinction coefficient ×
concentration of protein ×
path length(
1 cm) to determine the concentration of the protein [
43].
Estimation of pharmacodynamic and pharmacokinetic effects of Myxopyronin A during experimental animal testing in preclinical clinical trials:
In the present study, the pharmacokinetics and the pharmacodynamics of
Myxopyronin A were evaluated after dosing in male rabbit animal models weighing about 2 kg. Furthermore, compound concentrations were determined in target compartments, such as lung, kidney and thigh tissue, using
LC-MS/ MS. Based on the pharmacokinetic results, the pharmacodynamic profile of
Myxopyronin A was assessed victimizing the standard neutropenic thigh and lung infection models [
44]
.
Estimation of pharmacodynamic and pharmacokinetic effects of Myxopyronin A in randomized human clinical trials phases 1/2:
This study was conducted in
150 human volunteer subjects to show the bioavailability, pharmacokinetics and the pharmacodynamics of the test antibiotic. The study was designed as randomized, single-dose,
2-treatment,
2-period crossover trial with a washout period of
1 week. Blood samples were collected at
0(
baseline),
10,
20, and
40 minutes and at
1, 1.5, 2, 3, 4, 6, 9, 12, and
24 hours postdose. Plasma concentrations of the 4 drugs were measured by using a rapid chromatography-tandem mass spectrometry method. Pharmacokinetic parameters were calculated by using noncompartmental methods. Bioequivalence was determined if the
90 % CIs of the log-transformed test/ reference ratios
AUC(
0-24),
AUC(
0-∞), and
Cmax were within the predetermined range of
80% to
125%. Tolerability was assessed by using clinical parameters and subject reports Pharmacodynamic effects were evaluated through the determination of
MICs via agar diffusion assay and broth dilution technique During randomized human clinical trials phases 1/2 all utilized infectious bacterial cell counts were estimated spectrophotometrically [
45].
Estimation of of phototoxicity, mutagenicity and carcinogenicity of the test antibiotic:
The phototoxicity was determined via
3T3 neutral red uptake phototoxicity technique [
46]. On the other hand, mutagenicity and carcinogenicity of the test antibiotic were assessed using Ames test [
47].
The determination of toxokinetics and toxodynamic effects:
Up and down method for acute toxicity detection of
Myxopyronin A was utilized for this purpose [
48].
The determination of maximum bactericidal activity of Myxopyronin A:
A pure culture of a specified microorganism was grown overnight, then diluted in growth-supporting broth( typically
Mueller Hinton Broth) to a concentration between
1 x 10^5 and
1 x 10^6 cfu/ ml. A stock dilution of the antimicrobial test substance was made at approximately
100 times the expected
MIC. Further
1:1 dilutions were made in test tubes. All dilutions of the test antibiotic were inoculated with equal volumes of the specified microorganism. A positive and negative control tube was included for every test microorganism to demonstrate adequate microbial growth over the course of the incubation period and media sterility, respectively. An aliquot of the positive control was plated and used to establish a baseline concentration of the microorganism used.The tubes were then incubated at the appropriate temperature and duration. Turbidity indicated growth of the microorganism and the
MIC was the lowest concentration where no growth was visually observed. To determine the
MBC, the dilution representing the
MIC and at least two of the more concentrated test product dilutions were plated and enumerated to determine viable
CFU/ ml. The
MBC was the lowest concentration that demonstrated a pre-determined reduction (such as 99.9%) in
CFU/ ml when compared to the
MIC dilution [
49].
Determination of plasma protein binding capacity of Myxopyronin A:
Victimizing of an ultrafiltration technique, the protein binding( PB) extent and changeability of the test antibiotic medicates were settled when given simultaneously to 30 patients inoculated with infectious pneumococci inside hospitals in Egypt. Clinical samples used were routinely received by microbiological laboratory inside the faculty of Pharmacy, Cairo University, Egypt. Plasma proteins were likewise plumbed. A protein-free medium was used to determine the nonspecific binding. Plasma samples from 30 patients were enclosed, of which plasma proteins were deliberated for 24 patients.
Determination of liver, kidney and heart function tests:
These functional tests were performed to assess the vitality of liver, kidney and heart during the randomized human clinical trials phases 1/2. On the other hand, Urine, stool analyses were achieved in addition to estimation of blood complete counts to all experimental subjects received graded doses of Myxopyronin A.
Figure 1.
It demonstrates the structure of Myxopyronin A extracted from bacterial isolates Myxococcus fulvus strain Mx f50 collected from different soil environments in Egypt. Molecular formula of the purified test antibiotic was noticed to be C23H31NO6 determined through mass spectrometer.
Figure 1.
It demonstrates the structure of Myxopyronin A extracted from bacterial isolates Myxococcus fulvus strain Mx f50 collected from different soil environments in Egypt. Molecular formula of the purified test antibiotic was noticed to be C23H31NO6 determined through mass spectrometer.
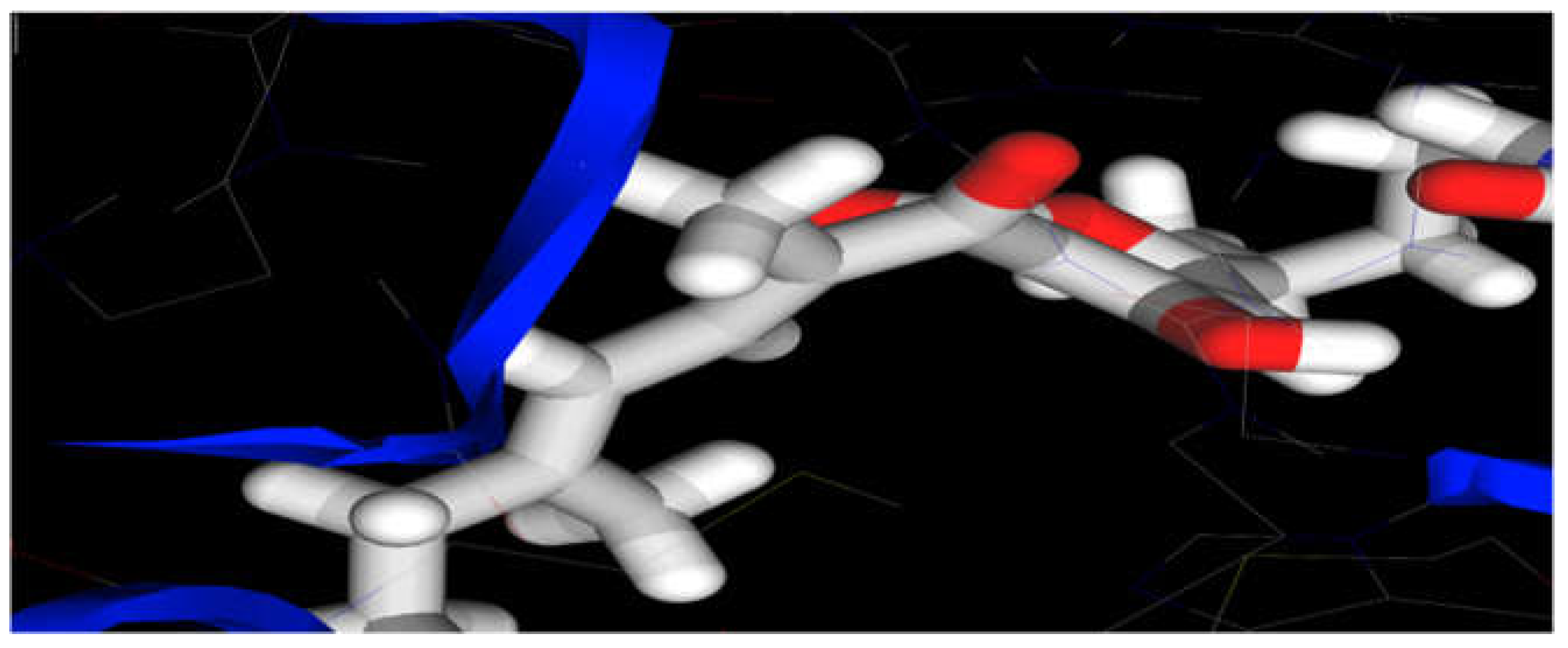
Figure 2.
It represents docking of Myxopyronin A ligand on Bacterial RNA polymerase. Myxopyronin showed high affinity and inhibitory effect towards the switch region.
Figure 2.
It represents docking of Myxopyronin A ligand on Bacterial RNA polymerase. Myxopyronin showed high affinity and inhibitory effect towards the switch region.
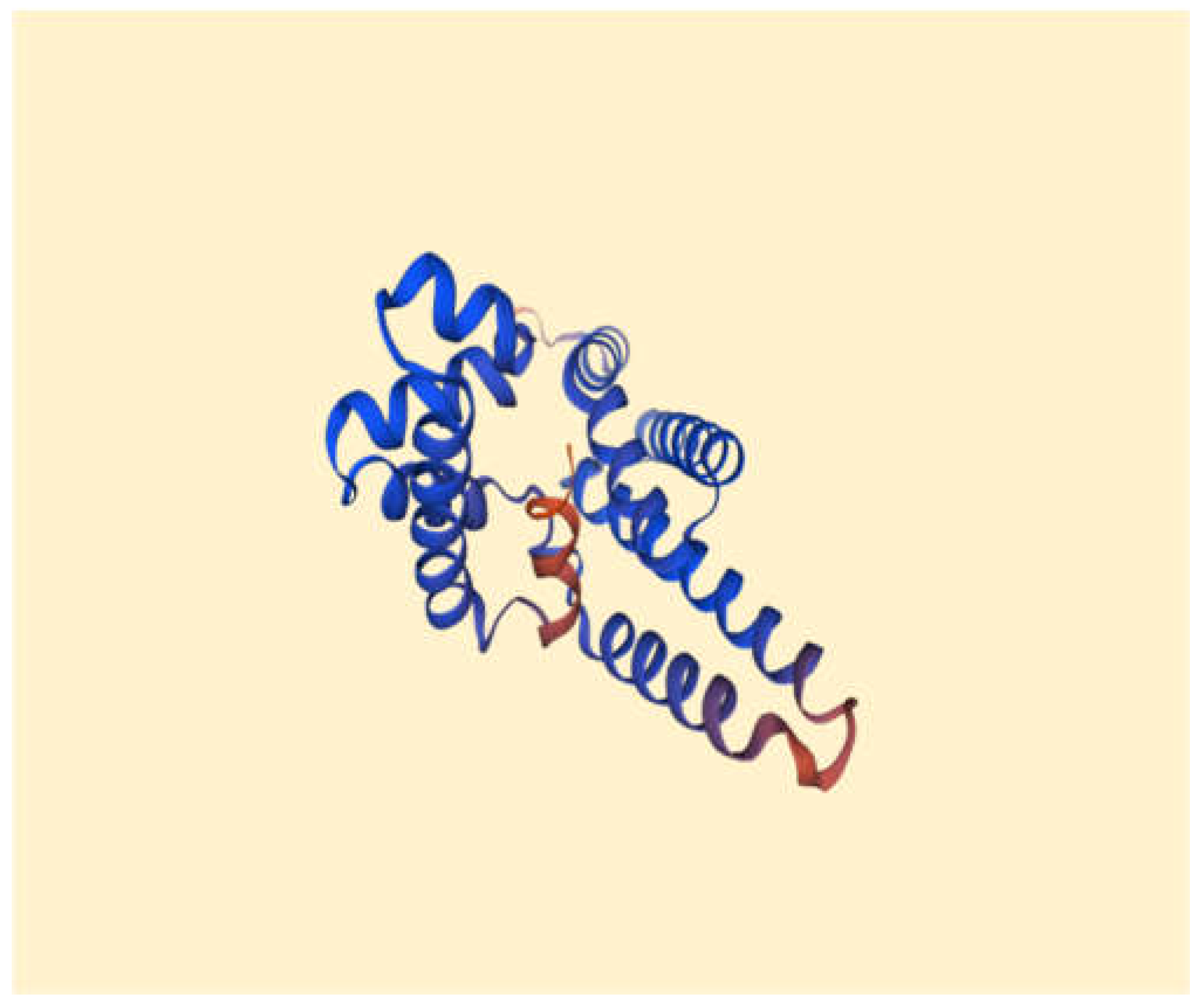
Figure 3.
It demonstrates 3D structure of bacterial prokaryotic RNA polymerase comprising the switch binding site to which Myxopyronin A Ligand strongly bound inhibiting bacterial RNA polymerase activity selectively leading to the inhibition of mRNA transcription and subsequently the mortality of the microbe. The secondary structure of RNA polymerase enzyme consisted of spiral alpha and beta sheets. Its molecular mass was approximately 198 amino-acids.
Figure 3.
It demonstrates 3D structure of bacterial prokaryotic RNA polymerase comprising the switch binding site to which Myxopyronin A Ligand strongly bound inhibiting bacterial RNA polymerase activity selectively leading to the inhibition of mRNA transcription and subsequently the mortality of the microbe. The secondary structure of RNA polymerase enzyme consisted of spiral alpha and beta sheets. Its molecular mass was approximately 198 amino-acids.
Figure 4.
It shows the impact of various concentrations of Soluble starch on the production of Myxopyronin A.
Figure 4.
It shows the impact of various concentrations of Soluble starch on the production of Myxopyronin A.
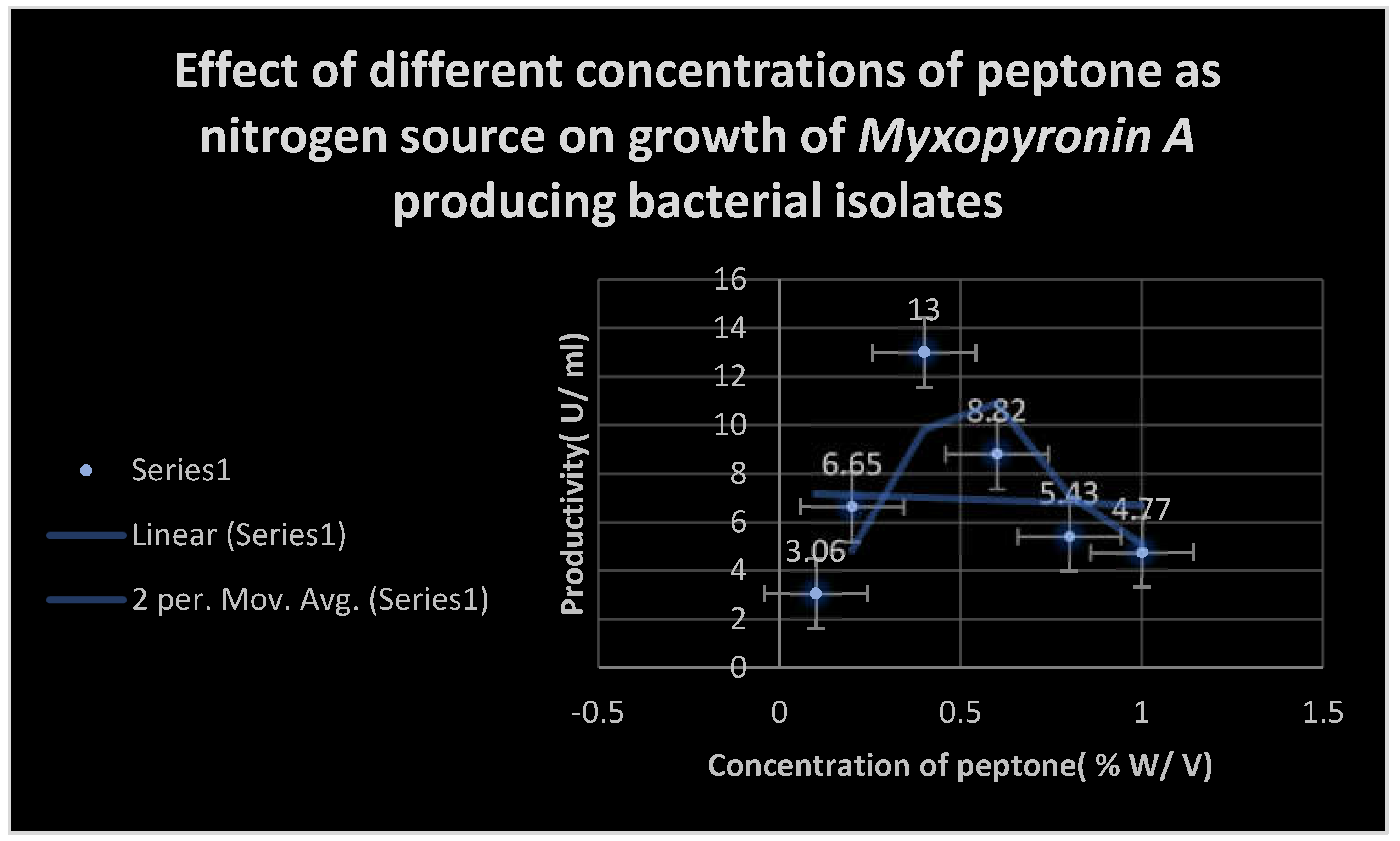
Figure 5.
It shows the effects0 of different Peptone concentrations as nitrogen growth factor on the productivity of Myxopyronin A.
Figure 5.
It shows the effects0 of different Peptone concentrations as nitrogen growth factor on the productivity of Myxopyronin A.
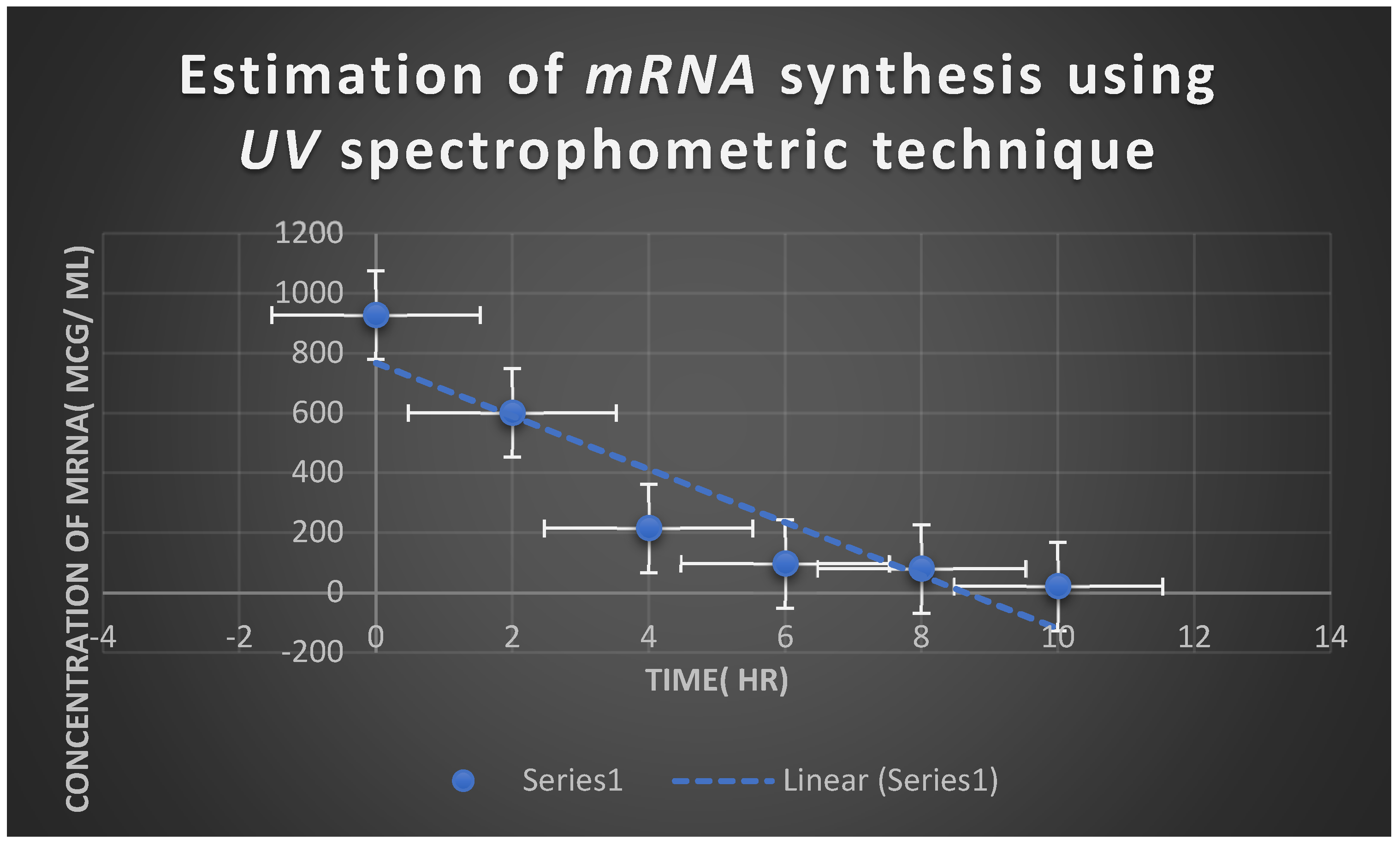
Figure 6.
It refers to the estimation of effect of Myxopyronin A on microbial mRNA productivity. mRNA synthesis was detected to be diminished proportionately up on employment of exploding doses of Myxopyronin A antibiotic.
Figure 6.
It refers to the estimation of effect of Myxopyronin A on microbial mRNA productivity. mRNA synthesis was detected to be diminished proportionately up on employment of exploding doses of Myxopyronin A antibiotic.
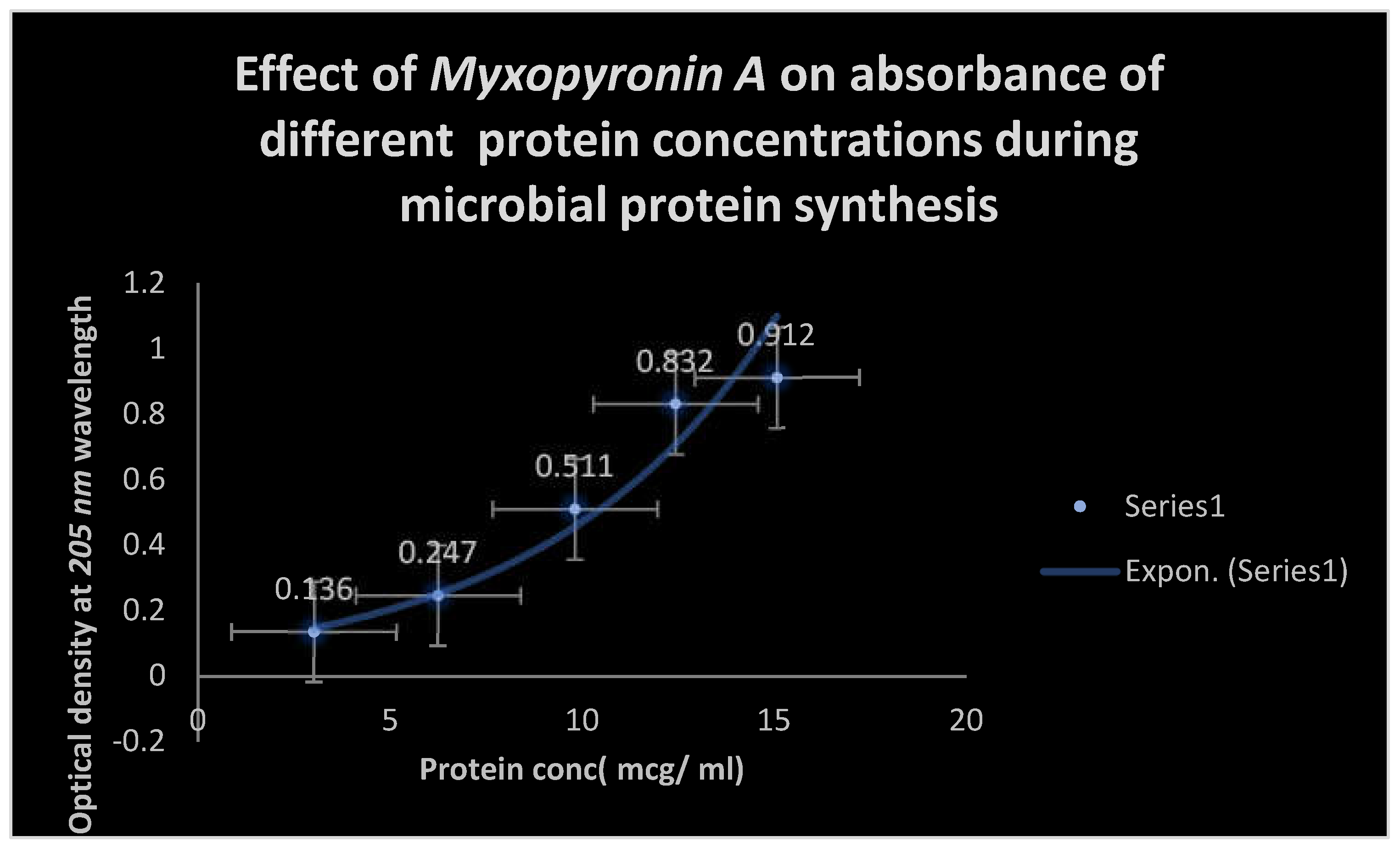
Figure 7.
It demonstrates the influence of Myxopyronin A on protein synthesis using UV spectrophotometer absorption at 205 nm. Protein synthesis was noticed to be decreased dramatically up on utilization of increasing doses of Myxopyronin A antibiotic.
Figure 7.
It demonstrates the influence of Myxopyronin A on protein synthesis using UV spectrophotometer absorption at 205 nm. Protein synthesis was noticed to be decreased dramatically up on utilization of increasing doses of Myxopyronin A antibiotic.
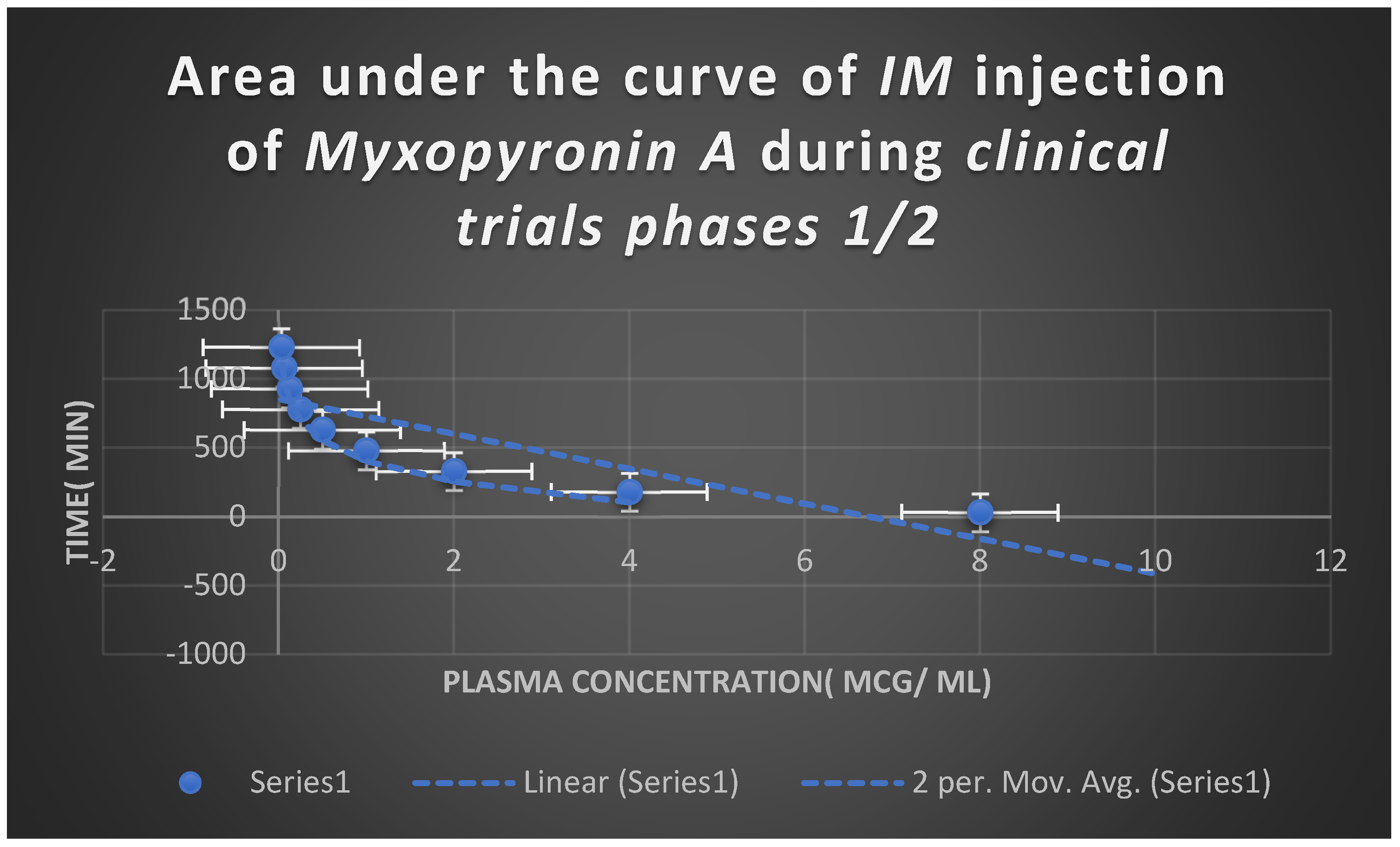
Figure 8.
It shows AUC of Myxopyronin A following IM administration in clinical trials stages 1/2. Efficacious dose ranged from 5-6 mg/ kg of body weight. Onset of action was observed following closely 15 minutes. It followed first order of elimination kinetics.
Figure 8.
It shows AUC of Myxopyronin A following IM administration in clinical trials stages 1/2. Efficacious dose ranged from 5-6 mg/ kg of body weight. Onset of action was observed following closely 15 minutes. It followed first order of elimination kinetics.
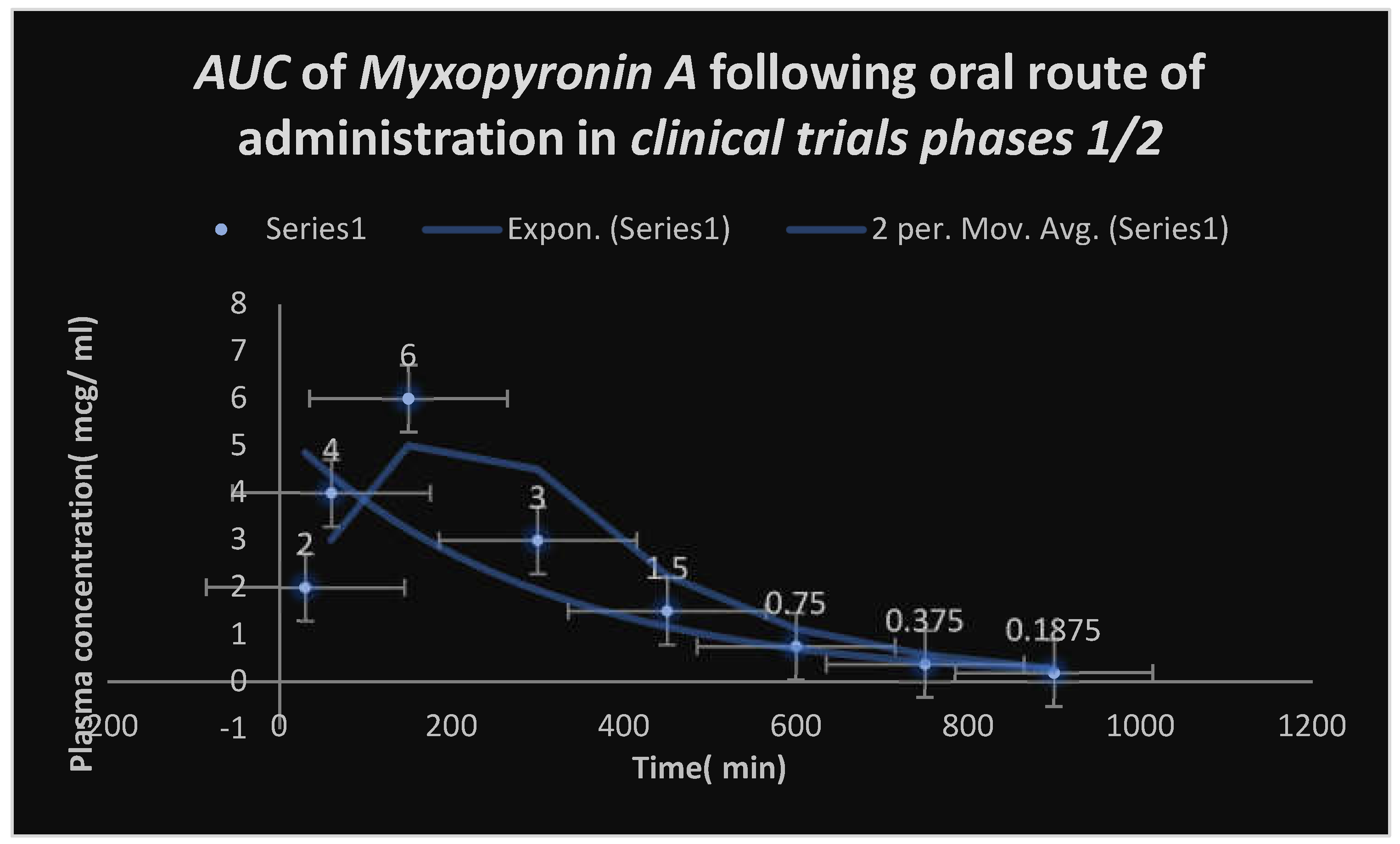
Figure 9.
Area under the curve( AUC) following oral administration of Myxopyronin A during clinical trials phases 1/2. Efficacious dose ranged from 7-8 mg/ kg of body weight. Onset of action was observed following nearly 30 minutes. It followed first order of elimination kinetics.
Figure 9.
Area under the curve( AUC) following oral administration of Myxopyronin A during clinical trials phases 1/2. Efficacious dose ranged from 7-8 mg/ kg of body weight. Onset of action was observed following nearly 30 minutes. It followed first order of elimination kinetics.
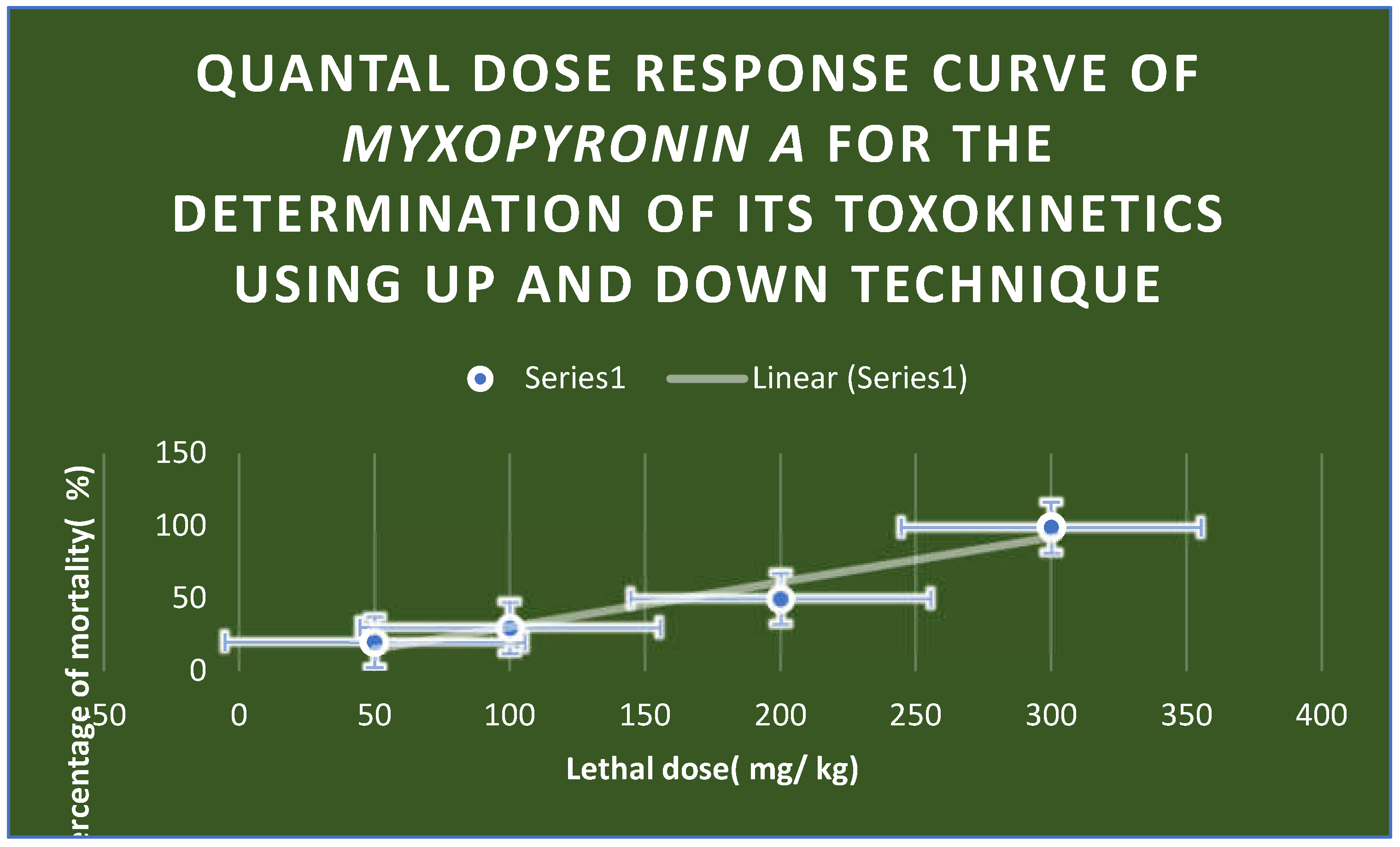
Figure 10.
Quantal dose response curve for the determination of toxokinetics of Myxopyronin A. LD50 % was found to be 200 mg / kg; while LD99 % was nearly 300 mg/ kg.
Figure 10.
Quantal dose response curve for the determination of toxokinetics of Myxopyronin A. LD50 % was found to be 200 mg / kg; while LD99 % was nearly 300 mg/ kg.