Submitted:
06 May 2024
Posted:
08 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
- Astrophysics Inspiration: BHO draws inspiration from astrophysics, specifically the fascinating concept of black holes. This unique property sets it apart from traditional statistical methods. By mimicking the gravitational interactions of black holes, BHO explores feature spaces in a novel and non-deterministic manner.
- Randomness-Based Exploration: BHO leverages randomness and domesticity techniques to explore wider areas within the search space. Unlike deterministic approaches, it embraces stochasticity, allowing it to escape the local optima problem, found in many FS methods, and avoid premature convergence. This adaptability is particularly valuable in complex optimization landscapes.
- Simplicity and Computational Efficiency: The mathematical model underlying BHO is not overly complex. Consequently, it requires less computational expense compared to more intricate approaches. This efficiency makes it practical for real-world applications.
- We propose a novel wrapper feature selection (FS) approach, that combines the black hole algorithm with inversion mutation, to select the most descriptive subset of features from datasets that cover different application domains.
- We modify a well-established multi-objective function that focuses on the interleaved classifier and the number of selected features in the decision-making process. Additionally, we enhance the decision-making process by considering the correlation among the selected subset of features and the correlation of each feature within that subset with the corresponding label.
- We assess our approach using fourteen benchmark datasets. We benchmark the performance of a wrapper FS approach called Binary Cuckoo Search (BCS). We also benchmark the performance of three filter-based FS, namely Mutual Information Maximisation (MIM), Joint Mutual Information (JMI) and minimum Redundancy Maximum Relevance (mRMR).
- We release the source codes of our framework for the research community. The implementation link is: https://github.com/Mohammed-Ryiad-Eiadeh/A-Modified-BHO-and-BCS-With-Mutation-for-FS-based-on-Modified-Objective-Function.
2. Related Work
2.1. Meta-Heuristic Algorithms for Feature Selection
2.2. Approximate Algorithms for Search
2.3. Filter-Based FS Approaches
2.4. Contributions of Our Work
3. Background on Mutual Information
4. Methodology
4.1. Optimization Problem
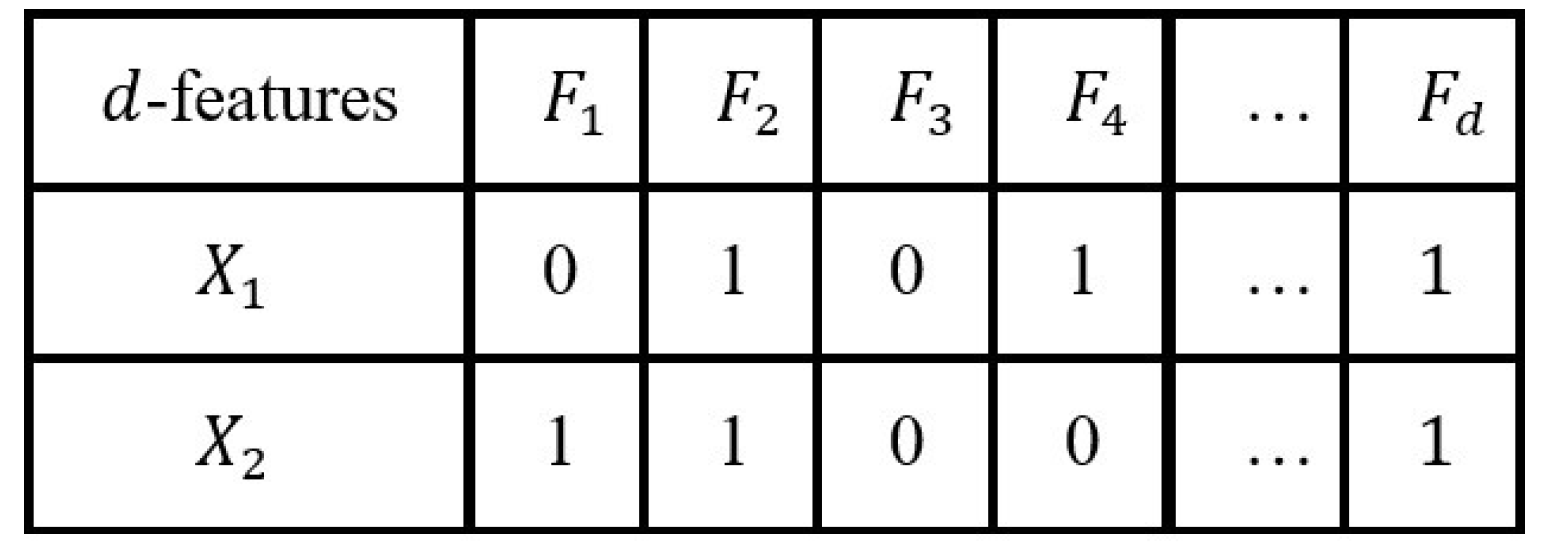
4.2. Population Representation
4.3. Evaluation Function
4.4. Modified Evaluation Function
4.4.1. The Dilemma Of the Weight Factor () in Wrapper Feature Selection (FS)
4.5. Correlation between Two Candidate Feature Vectors
4.5.1. Background about Correlation
4.5.2. Intuition of Adding Correlation Terms in Our Objective
4.5.3. Used Correlation Functions in This Study
4.6. Binary Improved Black Hole Optimizer (BHO)
4.6.1. Background about Motivation for BHO
4.6.2. Mathematical Modeling of Black Hole Optimization
4.6.3. Mitigating Main Issues in BHO Search Algorithm
| Algorithm 1: Pseudocode of MBHO |
 |
4.7. Time Complexity Analysis
5. Experimental Results
5.1. Used Datasets
5.2. Main Hyperparameters
5.3. Baselines
5.4. Evaluation Measurement
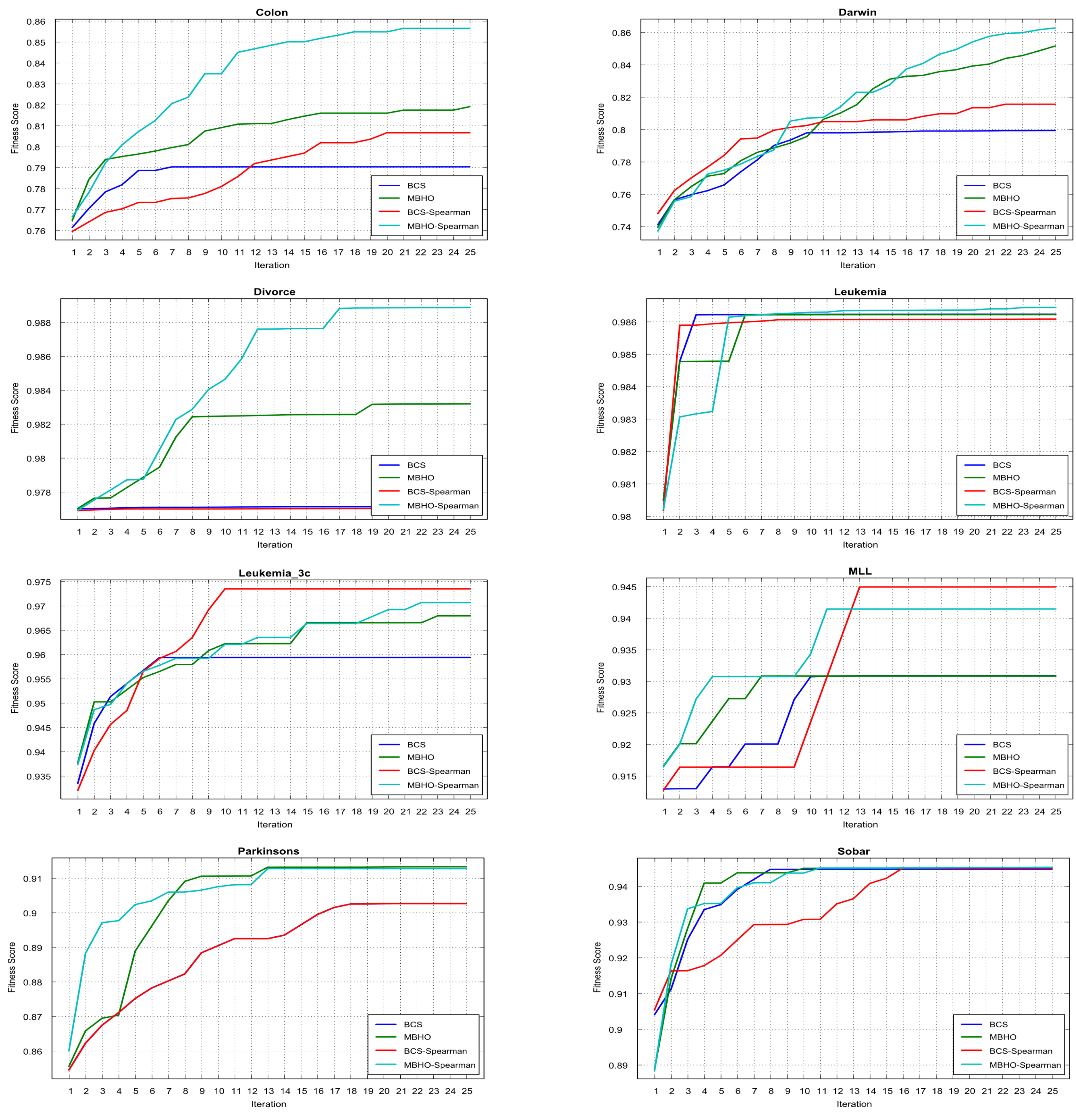
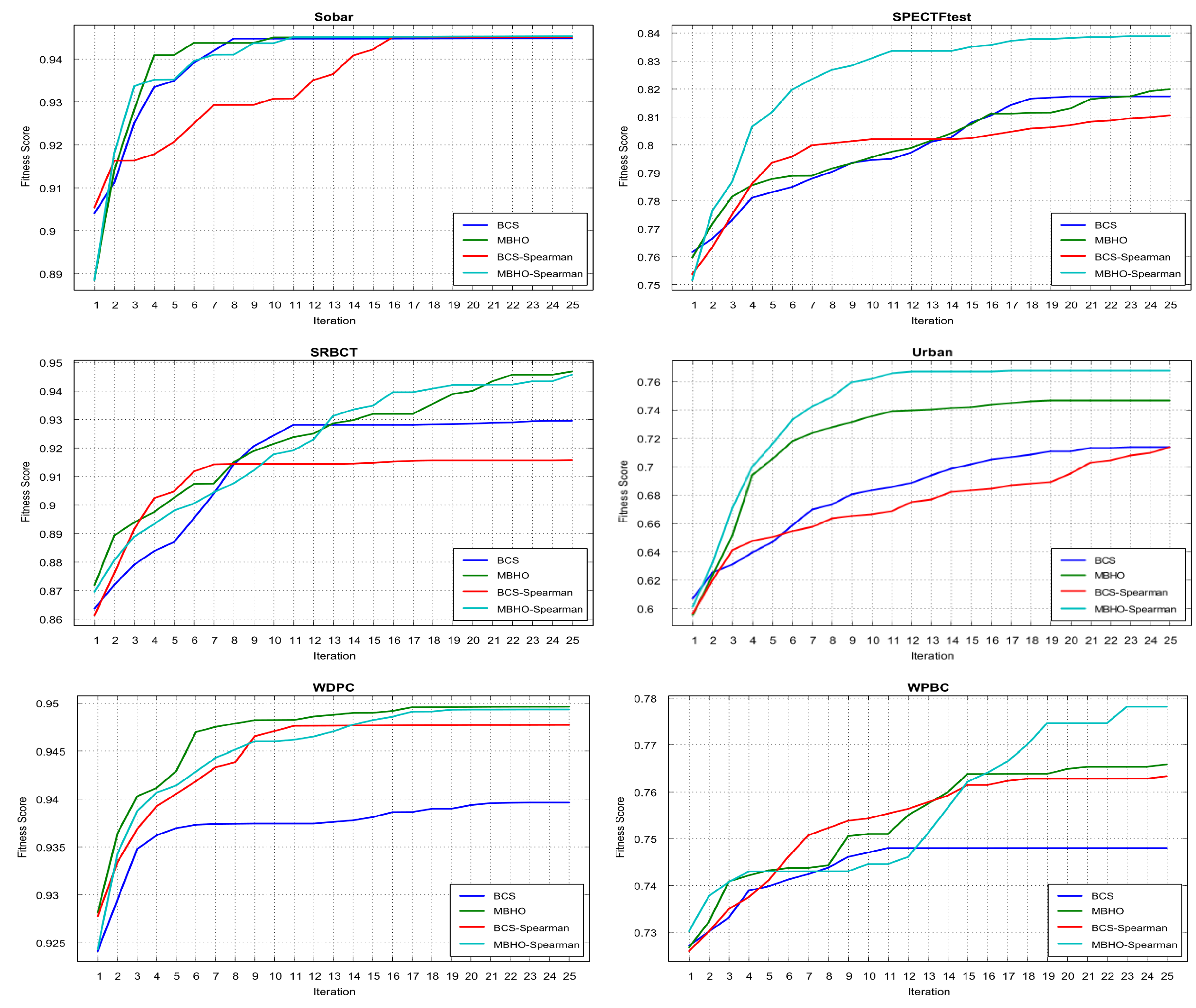
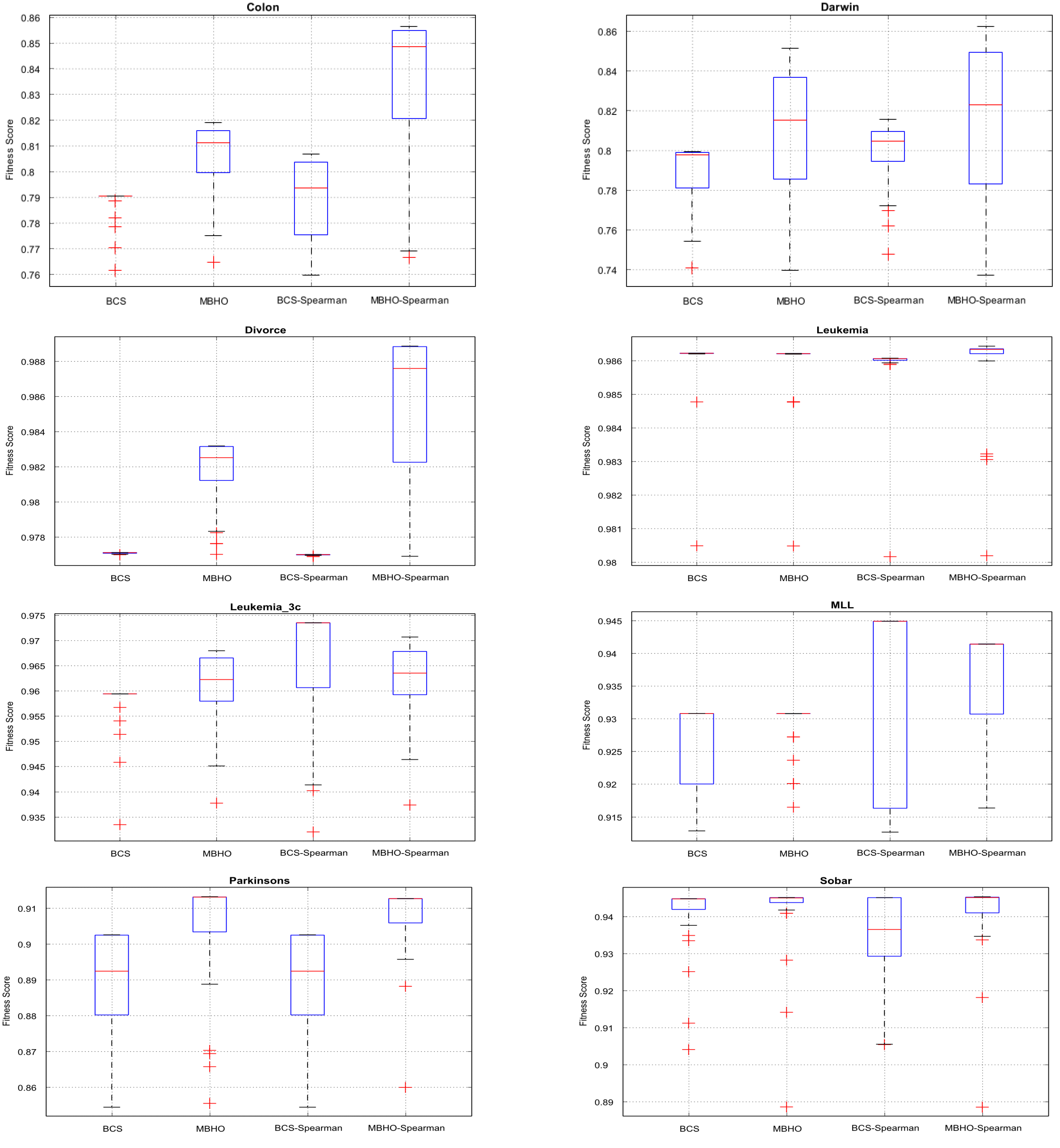
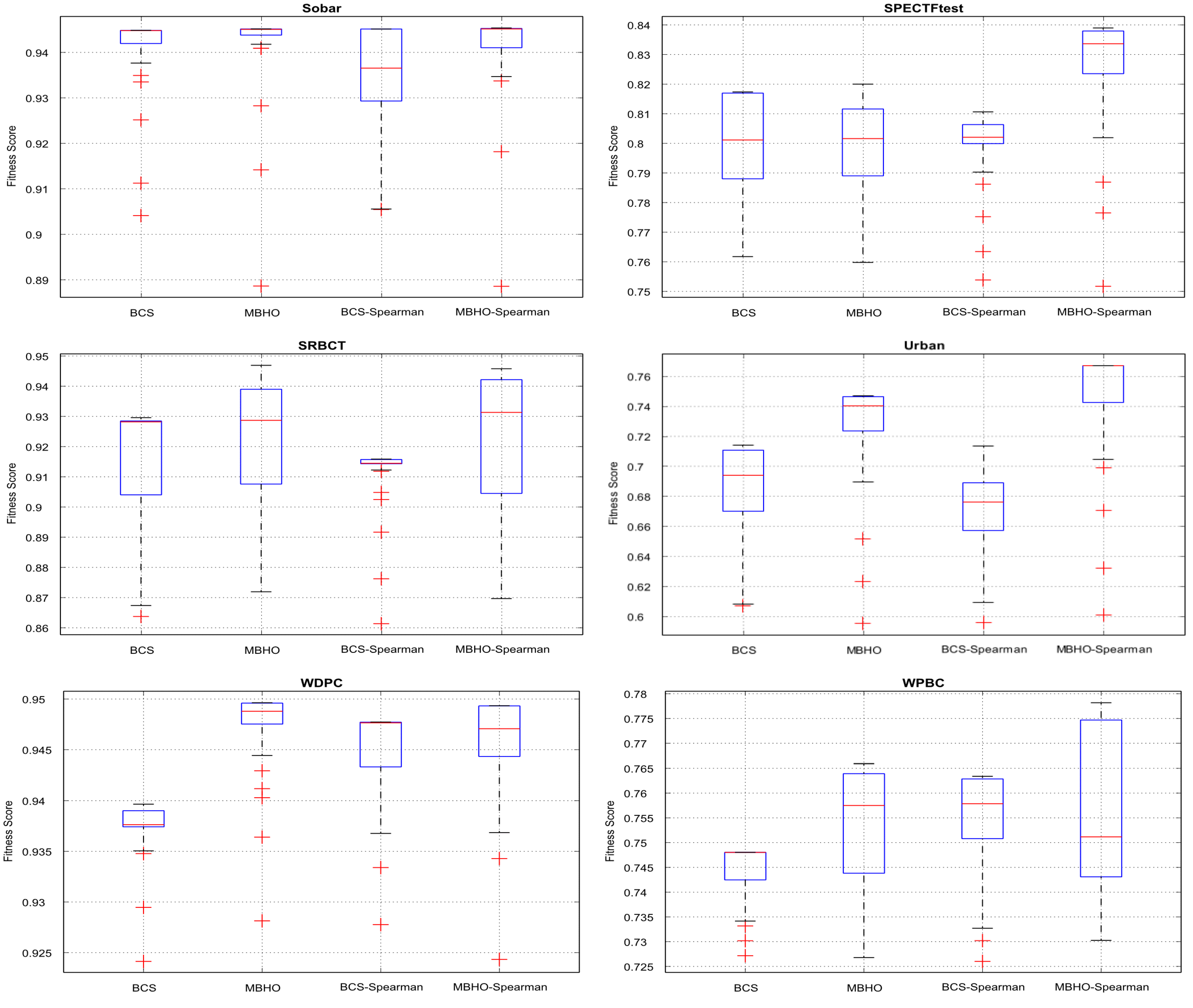
5.5. Evaluation Results
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
References
- García, S.; Luengo, J.; Herrera, F. Feature selection. Intelligent Systems Reference Library 2015, 72, 163–193. [Google Scholar] [CrossRef]
- Kursa, M.B.; Rudnicki, W.R. Feature selection with the boruta package. Journal of Statistical Software 2010, 36, 1–13. [Google Scholar] [CrossRef]
- Gao, Y.; Zhou, Y.; Luo, Q. An Efficient Binary Equilibrium Optimizer Algorithm for Feature Selection. IEEE Access 2020, 8, 140936–140963. [Google Scholar] [CrossRef]
- Xie, S.; Zhang, Y.; Lv, D.; Chen, X.; Lu, J.; Liu, J. A new improved maximal relevance and minimal redundancy method based on feature subset. Journal of Supercomputing 2023, 79, 3157–3180. [Google Scholar] [CrossRef]
- Vergara, J.R.; Estévez, P.A. A review of feature selection methods based on mutual information. Neural Computing and Applications 2014, 24, 175–186. [Google Scholar] [CrossRef]
- Lillywhite, K.; Lee, D.J.; Tippetts, B.; Archibald, J. A feature construction method for general object recognition. Pattern Recognition 2013, 46, 3300–3314. [Google Scholar] [CrossRef]
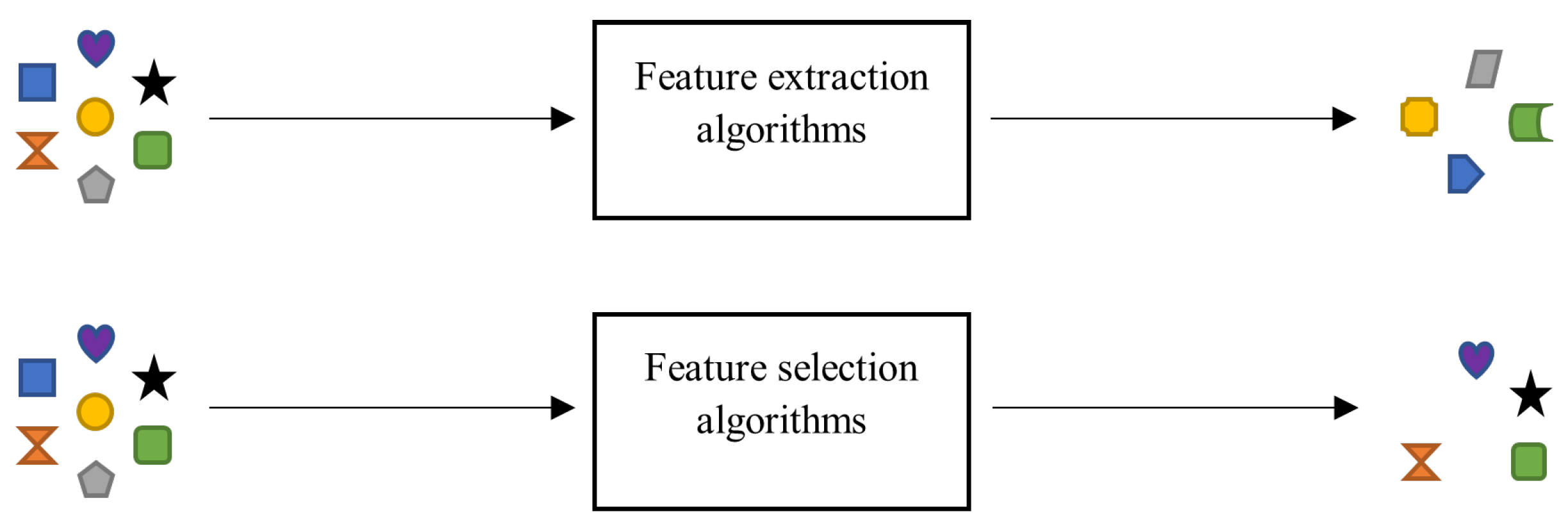
- Motoda, H.; Liu, H. Feature selection, extraction and construction. Communication of IICM 2002, 5, 67–72. [Google Scholar]
- Khalid, S.; Khalil, T.; Nasreen, S. A survey of feature selection and feature extraction techniques in machine learning. Proceedings of 2014 Science and Information Conference, SAI 2014 2014, pp. 372–378. [CrossRef]
- Remeseiro, B.; Bolon-Canedo, V. A review of feature selection methods in medical applications. Computers in Biology and Medicine 2019, 112. [Google Scholar] [CrossRef]
- Support vector machines in water quality management. Analytica Chimica Acta 2011, 703, 152–162. [CrossRef] [PubMed]
- Bolón-Canedo, V.; Remeseiro, B. Feature selection in image analysis: a survey. Artificial Intelligence Review 2020, 53, 2905–2931. [Google Scholar] [CrossRef]
- Mladenić, D., Feature Selection in Text Mining. In Encyclopedia of Machine Learning; Sammut, C.; Webb, G.I., Eds.; Springer US: Boston, MA, 2010; pp. 406–410. [CrossRef]
- Deng, Z.; Han, T.; Liu, R.; Zhi, F. A fault diagnosis method in industrial processes with integrated feature space and optimized random forest. In Proceedings of the 2022 IEEE 31st International Symposium on Industrial Electronics (ISIE); 2022; pp. 1170–1173. [Google Scholar] [CrossRef]
- Qaddoura, R.; Biltawi, M.M.; Faris, H. A Metaheuristic Approach for Life Expectancy Prediction based on Automatically Fine-tuned Models with Feature Selection. In Proceedings of the 2023 IEEE International Conference on Artificial Intelligence, Blockchain, and Internet of Things (AIBThings). IEEE, 2023, pp. 1–7.
- Biltawi, M.M.; Qaddoura, R. The impact of feature selection on the regression task for life expectancy prediction. In Proceedings of the 2022 International Conference on Emerging Trends in Computing and Engineering Applications (ETCEA). IEEE, 2022, pp. 1–5.
- Jović, A.; Brkić, K.; Bogunović, N. A review of feature selection methods with applications. 2015 38th International Convention on Information and Communication Technology, Electronics and Microelectronics, MIPRO 2015 - Proceedings 2015, pp. 1200–1205. [CrossRef]
- Brezočnik, L.; Fister, I.; Podgorelec, V. Swarm intelligence algorithms for feature selection: A review. Applied Sciences (Switzerland) 2018, 8. [Google Scholar] [CrossRef]
- Rais, H.M.; Mehmood, T. Dynamic Ant Colony System with Three Level Update Feature Selection for Intrusion Detection. International Journal of Network Security 2018, 20, 184–192. [Google Scholar] [CrossRef]
- Amierh, Z.; Hammad, L.; Qaddoura, R.; Al-Omari, H.; Faris, H. A Multiclass Classification Approach for IoT Intrusion Detection Based on Feature Selection and Oversampling. In Cyber Malware: Offensive and Defensive Systems; Springer, 2023; pp. 197–233.
- Biltawi, M.M.; Qaddoura, R.; Faris, H. Optimizing Feature Selection and Oversampling Using Metaheuristic Algorithms for Binary Fraud Detection Classification. In Proceedings of the IFIP International Conference on Artificial Intelligence Applications and Innovations. Springer; 2023; pp. 452–462. [Google Scholar]
- Kumar, V. Feature Selection: A literature Review. The Smart Computing Review 2014, 4. [Google Scholar] [CrossRef]
- Burger, S. The wrapper approach 2011. 97.
- Spolaôr, N.; Cherman, E.A.; Monard, M.C.; Lee, H.D. Filter approach feature selection methods to support multi-label learning based on relieff and information gain. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) 2012, 7589, 72–81. [Google Scholar] [CrossRef]
- Wang, P.; Xue, B.; Liang, J.; Zhang, M. Differential Evolution With Duplication Analysis for Feature Selection in Classification. IEEE Transactions on Cybernetics 2022, 46, 1–14. [Google Scholar] [CrossRef]
- Xue, B.; Zhang, M.; Browne, W.N.; Yao, X. A Survey on Evolutionary Computation Approaches to Feature Selection. IEEE Transactions on Evolutionary Computation 2016, 20, 606–626. [Google Scholar] [CrossRef]
- Suppers, A.; van Gool, A.J.; Wessels, H.J. Integrated chemometrics and statistics to drive successful proteomics biomarker discovery. Proteomes 2018, 6. [Google Scholar] [CrossRef]
- Pocock, A.C. Feature Selection Via Joint Likelihood. Thesis 2012, p. total 173.
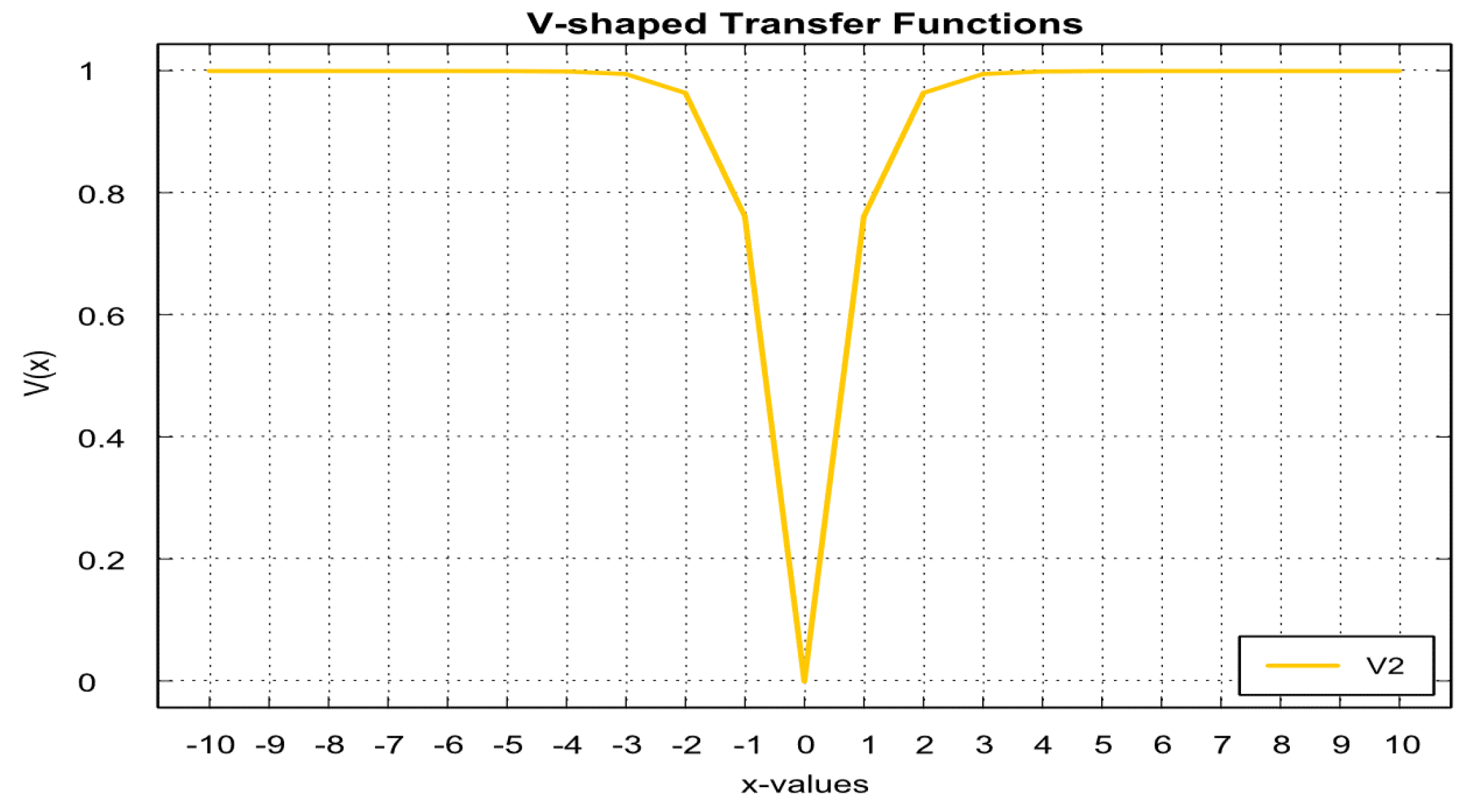
- Mafarja, M.; Eleyan, D.; Abdullah, S.; Mirjalili, S. S-shaped vs. V-shaped transfer functions for ant lion optimization algorithm in feature selection problem. ACM International Conference Proceeding Series 2017, Part F1305. [CrossRef]
- Liu, H.; Dougherty, E.R.; Dy, J.G.; Torkkola, K.; Tuv, E.; Peng, H.; Ding, C.; Long, F.; Berens, M.; Parsons, L.; et al. Evolving feature selection. IEEE Intelligent systems 2005, 20, 64–76. [Google Scholar] [CrossRef]
- Fakhraei, S.; Soltanian-Zadeh, H.; Fotouhi, F. Bias and stability of single variable classifiers for feature ranking and selection. Expert Systems with Applications 2014, 41, 6945–6958. [Google Scholar] [CrossRef] [PubMed]
- Ververidis, D.; Kotropoulos, C. Sequential forward feature selection with low computational cost. In Proceedings of the 2005 13th European Signal Processing Conference. IEEE; 2005; pp. 1–4. [Google Scholar]
- Abe, S. Modified backward feature selection by cross validation. In Proceedings of the ESANN; 2005; pp. 163–168. [Google Scholar]
- Sabzekar, M.; Aydin, Z. A noise-aware feature selection approach for classification. Soft Computing 2021, 25, 6391–6400. [Google Scholar] [CrossRef]
- Ramos, C.C.; Rodrigues, D.; De Souza, A.N.; Papa, J.P. On the study of commercial losses in Brazil: A binary black hole algorithm for theft characterization. IEEE Transactions on Smart Grid 2018, 9, 676–683. [Google Scholar] [CrossRef]
- Pashaei, E.; Aydin, N. Binary black hole algorithm for feature selection and classification on biological data. Applied Soft Computing 2017, 56, 94–106. [Google Scholar] [CrossRef]
- Qasim, O.S.; Al-Thanoon, N.A.; Algamal, Z.Y. Feature selection based on chaotic binary black hole algorithm for data classification. Chemometrics and Intelligent Laboratory Systems 2020, 204, 104104. [Google Scholar] [CrossRef]
- Winter, J.D.; Gosling, S.D. Supplemental Material for Comparing the Pearson and Spearman Correlation Coefficients Across Distributions and Sample Sizes: A Tutorial Using Simulations and Empirical Data. Psychological Methods 2016. [Google Scholar] [CrossRef]
- Rodrigues, D.; Pereira, L.A.M.; Almeida, T.N.S.; Papa, J.P.; Souza, A.N.; Ramos, C.C.O. ; Xin-She Yang. In BCS: A Binary Cuckoo Search algorithm for feature selection. In Proceedings of the 2013 IEEE International Symposium on Circuits and Systems (ISCAS2013). IEEE, may 2013; pp. 465–468. [Google Scholar] [CrossRef]
- Gu, X.; Guo, J.; Xiao, L.; Li, C. Conditional mutual information-based feature selection algorithm for maximal relevance minimal redundancy. Applied Intelligence 2022, 52, 1436–1447. [Google Scholar] [CrossRef]
- Can high-order dependencies improve mutual information based feature selection? Pattern Recognition 2016, 53, 46–58. [CrossRef]
- Angulo, A.P.; Shin, K. Mrmr+ and Cfs+ feature selection algorithms for high-dimensional data. Applied Intelligence 2019, 49, 1954–1967. [Google Scholar] [CrossRef]
- Abdel-Basset, M.; Abdel-Fatah, L.; Sangaiah, A.K. Metaheuristic algorithms: A comprehensive review. Computational intelligence for multimedia big data on the cloud with engineering applications 2018, pp. 185–231.
- Wu, S.; Hu, Y.; Wang, W.; Feng, X.; Shu, W. Application of global optimization methods for feature selection and machine learning. Mathematical Problems in Engineering 2013, 2013. [Google Scholar] [CrossRef]
- Wang, Y.; Li, T. Local feature selection based on artificial immune system for classification. Applied Soft Computing Journal 2020, 87, 105989. [Google Scholar] [CrossRef]
- Huang, Z.; Yang, C.; Zhou, X.; Huang, T. A Hybrid Feature Selection Method Based on Binary State Transition Algorithm and ReliefF. IEEE Journal of Biomedical and Health Informatics 2019, 23, 1888–1898. [Google Scholar] [CrossRef]
- Lee, C.Y.; Le, T.A. Optimised approach of feature selection based on genetic and binary state transition algorithm in the classification of bearing fault in bldc motor. IET Electric Power Applications 2020, 14, 2598–2608. [Google Scholar] [CrossRef]
- Kennedy, J.; Eberhart, R. Particle swarm optimization PAPER - IGNORE FROM REFS. ICNN’95-international conference on neural networks 1995, pp. 1942–1948.
- Sharkawy, R.; Ibrahim, K.; Salama, M.; Bartnikas, R. Particle swarm optimization feature selection for the classification of conducting particles in transformer oil. IEEE Transactions on Dielectrics and Electrical Insulation 2011, 18, 1897–1907. [Google Scholar] [CrossRef]
- Zhang, Y.; Gong, D.W.; Cheng, J. Multi-objective particle swarm optimization approach for cost-based feature selection in classification. IEEE/ACM Transactions on Computational Biology and Bioinformatics 2017, 14, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Sakri, S.B.; Abdul Rashid, N.B.; Muhammad Zain, Z. Particle Swarm Optimization Feature Selection for Breast Cancer Recurrence Prediction. IEEE Access 2018, 6, 29637–29647. [Google Scholar] [CrossRef]
- Mirjalili, S.; Lewis, A. The Whale Optimization Algorithm. Advances in Engineering Software 2016, 95, 51–67. [Google Scholar] [CrossRef]
- Zamani, H.; Nadimi-Shahraki, M.H. Feature Selection Based on Whale Optimization Algorithm for Diseases Diagnosis. International Journal of Computer Science and Information Security 2016, 14, 1243–1247. [Google Scholar]
- Tubishat, M.; Abushariah, M.A.; Idris, N.; Aljarah, I. Improved whale optimization algorithm for feature selection in Arabic sentiment analysis. Applied Intelligence 2019, 49, 1688–1707. [Google Scholar] [CrossRef]
- Guha, R.; Ghosh, M.; Mutsuddi, S.; Sarkar, R.; Mirjalili, S. Embedded chaotic whale survival algorithm for filter–wrapper feature selection. Soft Computing 2020, 24, 12821–12843. [Google Scholar] [CrossRef]
- Forrest, S.; Mitchell, M. What Makes a Problem Hard for a Genetic Algorithm? Some Anomalous Results and Their Explanation. Machine Learning 1993, 13, 285–319. [Google Scholar] [CrossRef]
- Babatunde, O.; Armstrong, L.; Leng, J.; Diepeveen, D. A Genetic Algorithm-Based Feature Selection. International Journal of Electronics Communication and Computer Engineering 2014, 5, 899–905. [Google Scholar]
- Desale, K.S.; Ade, R. Genetic algorithm based feature selection approach for effective intrusion detection system. 2015 International Conference on Computer Communication and Informatics, ICCCI 2015. [CrossRef]
- Khammassi, C.; Krichen, S. A GA-LR wrapper approach for feature selection in network intrusion detection. Computers and Security 2017, 70, 255–277. [Google Scholar] [CrossRef]
- Liu, X.Y.; Liang, Y.; Wang, S.; Yang, Z.Y.; Ye, H.S. A Hybrid Genetic Algorithm with Wrapper-Embedded Approaches for Feature Selection. IEEE Access 2018, 6, 22863–22874. [Google Scholar] [CrossRef]
- Bardamova, M.; Konev, A.; Hodashinsky, I.; Shelupanov, A. A fuzzy classifier with feature selection based on the gravitational search algorithm. Symmetry 2018, 10. [Google Scholar] [CrossRef]
- Taradeh, M.; Mafarja, M.; Heidari, A.A.; Faris, H.; Aljarah, I.; Mirjalili, S.; Fujita, H. An evolutionary gravitational search-based feature selection. Information Sciences 2019, 497, 219–239. [Google Scholar] [CrossRef]
- Faramarzi, A.; Heidarinejad, M.; Stephens, B.; Mirjalili, S. Equilibrium optimizer: A novel optimization algorithm. Knowledge-Based Systems 2020, 191. [Google Scholar] [CrossRef]
- Aarts, E.; Korst, J. Chapter 2 Simulated annealing 2.1 Introduction of the algorithm. Simulated Annealing: Theory and Application 1987, p. 7.
- Ghosh, K.K.; Guha, R.; Bera, S.K.; Sarkar, R.; Mirjalili, S. BEO: Binary Equilibrium Optimizer Combined with Simulated Annealing for Feature Selection 2020. [CrossRef]
- Too, J.; Mirjalili, S. General Learning Equilibrium Optimizer: A New Feature Selection Method for Biological Data Classification. Applied Artificial Intelligence 2021, 35, 247–263. [Google Scholar] [CrossRef]
- Sayed, G.I.; Khoriba, G.; Haggag, M.H. A novel Chaotic Equilibrium Optimizer Algorithm with S-shaped and V-shaped transfer functions for feature selection. Journal of Ambient Intelligence and Humanized Computing 2021. [Google Scholar] [CrossRef]
- Vazirani, V.V. Approximation algorithms; Vol. 1, Springer, 2001.
- Aziz, M.A.E.; Hassanien, A.E. Modified cuckoo search algorithm with rough sets for feature selection. Neural Computing and Applications 2018, 29, 925–934. [Google Scholar] [CrossRef]
- Wang, L.; Gao, Y.; Li, J.; Wang, X. A Feature Selection Method by using Chaotic Cuckoo Search Optimization Algorithm with Elitist Preservation and Uniform Mutation for Data Classification. Discrete Dynamics in Nature and Society 2021, 2021, 1–19. [Google Scholar] [CrossRef]
- Zhang, Z. Speech feature selection and emotion recognition based on weighted binary cuckoo search. Alexandria Engineering Journal 2021, 60, 1499–1507. [Google Scholar] [CrossRef]
- Askarzadeh, A. A novel metaheuristic method for solving constrained engineering optimization problems: Crow search algorithm. Computers and Structures 2016, 169, 1–12. [Google Scholar] [CrossRef]
- Jain, M.; Rani, A.; Singh, V. An improved Crow Search Algorithm for high-dimensional problems. Journal of Intelligent and Fuzzy Systems 2017, 33, 3597–3614. [Google Scholar] [CrossRef]
- De Souza, R.C.T.; Coelho, L.D.S.; De MacEdo, C.A.; Pierezan, J. A V-Shaped Binary Crow Search Algorithm for Feature Selection. 2018 IEEE Congress on Evolutionary Computation, CEC 2018 - Proceedings 2018, pp. 1–8. [CrossRef]
- Sayed, G.I.; Hassanien, A.E.; Azar, A.T. Feature selection via a novel chaotic crow search algorithm. Neural Computing and Applications 2019, 31, 171–188. [Google Scholar] [CrossRef]
- Yang, X.S.; Deb, S. Cuckoo Search via Levy Flights 2010. [1003.1594].
- Nakamura, R.Y.; Pereira, L.A.; Costa, K.A.; Rodrigues, D.; Papa, J.P.; Yang, X.S. BBA: A binary bat algorithm for feature selection. Brazilian Symposium of Computer Graphic and Image Processing 2012, pp. 291–297. [CrossRef]
- Liu, F.; Yan, X.; Lu, Y. Feature Selection for Image Steganalysis Using Binary Bat Algorithm. IEEE Access 2020, 8, 4244–4249. [Google Scholar] [CrossRef]
- Chu, S.c.; Tsai, P.w.; Pan, J.s. 2006-Cat_Swarm_Optimization.pdf 2006. pp. 854–858.
- Siqueira, H.; Santana, C.; MacEdo, M.; Figueiredo, E.; Gokhale, A.; Bastos-Filho, C. Simplified binary cat swarm optimization. Integrated Computer-Aided Engineering 2021, 28, 35–50. [Google Scholar] [CrossRef]
- Mirjalili, S.; Mirjalili, S.M.; Lewis, A. Grey Wolf Optimizer. Advances in Engineering Software 2014, 69, 46–61. [Google Scholar] [CrossRef]
- Pathak, Y.; Arya, K.V.; Tiwari, S. Feature selection for image steganalysis using levy flight-based grey wolf optimization. Multimedia Tools and Applications 2019, 78, 1473–1494. [Google Scholar] [CrossRef]
- Saabia, A.A.B.R.; El-Hafeez, T.A.; Zaki, A.M. Face Recognition Based on Grey Wolf Optimization for Feature Selection; Vol. 845, Springer International Publishing, 2019; pp. 273–283. [CrossRef]
- Al-Tashi, Q.; Rais, H.M.; Abdulkadir, S.J.; Mirjalili, S. Feature Selection Based on Grey Wolf Optimizer for Oil Gas Reservoir Classification. 2020 International Conference on Computational Intelligence, ICCI 2020 2020, pp. 211–216. [CrossRef]
- Venkata Rao, R. Jaya: A simple and new optimization algorithm for solving constrained and unconstrained optimization problems. International Journal of Industrial Engineering Computations 2016, 7, 19–34. [Google Scholar] [CrossRef]
- Awadallah, M.A.; Al-Betar, M.A.; Hammouri, A.I.; Alomari, O.A. Binary JAYA Algorithm with Adaptive Mutation for Feature Selection. Arabian Journal for Science and Engineering 2020, 45, 10875–10890. [Google Scholar] [CrossRef]
- Chaudhuri, A.; Sahu, T.P. Binary Jaya algorithm based on binary similarity measure for feature selection. Journal of Ambient Intelligence and Humanized Computing 2021. [Google Scholar] [CrossRef]
- Alijla, B.O.; Peng, L.C.; Khader, A.T.; Al-Betar, M.A. Intelligent water drops algorithm for rough set feature selection. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) 2013, 7803 LNAI, 356–365. [CrossRef]
- Duan, H.; Qiao, P. Pigeon-inspired optimization: A new swarm intelligence optimizer for air robot path planning. International Journal of Intelligent Computing and Cybernetics 2014, 7, 24–37. [Google Scholar] [CrossRef]
- Alazzam, H.; Sharieh, A.; Sabri, K.E. A feature selection algorithm for intrusion detection system based on Pigeon Inspired Optimizer. Expert Systems with Applications 2020, 148. [Google Scholar] [CrossRef]
- Buvana, M.; Muthumayil, K.; Jayasankar, T. Content-based image retrieval based on hybrid feature extraction and feature selection technique pigeon inspired based optimization. Annals of the Romanian Society for Cell Biology 2021, 25, 424–443. [Google Scholar]
- Pan, J.s.; Tian, A.q.; Chu, S.c. Improved binary pigeon-inspired optimization and its application for feature selection 2021.
- Luo, J.; Zhou, D.; Jiang, L.; Ma, H. A particle swarm optimization based multiobjective memetic algorithm for high-dimensional feature selection. Memetic Computing 2022, 14, 77–93. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, J.; Tao, D. Neurodynamics-driven supervised feature selection. Pattern Recognition 2023, 136, 109254. [Google Scholar] [CrossRef]
- Spencer, R.; Thabtah, F.; Abdelhamid, N.; Thompson, M. Exploring feature selection and classification methods for predicting heart disease. Digital Health 2020, 6, 1–10. [Google Scholar] [CrossRef]
- Jadhav, S.; He, H.; Jenkins, K. Information gain directed genetic algorithm wrapper feature selection for credit rating. Applied Soft Computing Journal 2018, 69, 541–553. [Google Scholar] [CrossRef]
- Barzegar, Z.; Jamzad, M. Fully automated glioma tumour segmentation using anatomical symmetry plane detection in multimodal brain MRI. IET Computer Vision 2021. [Google Scholar] [CrossRef]
- Bakhshandeh, S.; Azmi, R.; Teshnehlab, M. Symmetric uncertainty class-feature association map for feature selection in microarray dataset. International Journal of Machine Learning and Cybernetics 2020, 11, 15–32. [Google Scholar] [CrossRef]
- Aswani, C.; Amir, K. Integrated Intrusion Detection Model Using Chi-Square Feature Selection and Ensemble of Classifiers. Arabian Journal for Science and Engineering 2019, 44, 3357–3368. [Google Scholar] [CrossRef]
- Bachri, O.S.; Kusnadi. ; Hatta, M.; Nurhayati, O.D. Feature selection based on CHI square in artificial neural network to predict the accuracy of student study period. International Journal of Civil Engineering and Technology 2017, 8, 731–739. [Google Scholar]
- Senliol, B.; Gulgezen, G.; Yu, L.; Cataltepe, Z. Fast Correlation Based Filter (FCBF) with a different search strategy. 2008 23rd International Symposium on Computer and Information Sciences, ISCIS 2008 2008. [CrossRef]
- Dash, M.; Liu, H.; Motoda, H. Consistency based feature selection. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) 2000, 1805, 98–109. [Google Scholar] [CrossRef]
- Dash, M.; Liu, H. Consistency-based search in feature selection. Artificial Intelligence 2003, 151, 155–176. [Google Scholar] [CrossRef]
- Palma-Mendoza, R.J.; De-Marcos, L.; Rodriguez, D.; Alonso-Betanzos, A. Distributed correlation-based feature selection in spark. Information Sciences 2019, 496, 287–299. [Google Scholar] [CrossRef]
- De Tre, G.; Hallez, A.; Bronselaer, A. Performance optimization of object comparison. International Journal of intelligent Systems 2014, 29, 495–524. [Google Scholar] [CrossRef]
- Bugata, P.; Drotar, P. On some aspects of minimum redundancy maximum relevance feature selection. Science China Information Sciences 2020, 63, 1–15. [Google Scholar] [CrossRef]
- Gulgezen, G.; Cataltepe, Z.; Yu, L. Stable and accurate feature selection. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). [CrossRef]
- Yaramakala, S.; Margaritis, D. Speculative Markov blanket discovery for optimal feature selection. Proceedings - IEEE International Conference on Data Mining, ICDM. [CrossRef]
- Sumaiya Thaseen, I.; Aswani Kumar, C. Intrusion detection model using fusion of chi-square feature selection and multi class SVM. Journal of King Saud University - Computer and Information Sciences 2017, 29, 462–472. [Google Scholar] [CrossRef]
- Yijun, S.; Jian, L. Iterative RELIEF for feature weighting. ACM International Conference Proceeding Series 2006, 148, 913–920. [Google Scholar] [CrossRef]
- Abdulrazaq, M.B.; Mahmood, M.R.; Zeebaree, S.R.; Abdulwahab, M.H.; Zebari, R.R.; Sallow, A.B. An Analytical Appraisal for Supervised Classifiers’ Performance on Facial Expression Recognition Based on Relief-F Feature Selection. Journal of Physics: Conference Series 2021, 1804. [Google Scholar] [CrossRef]
- Peker, M.; Ballı, S.; Sağbaş, E.A. Predicting Human Actions Using a Hybrid of ReliefF Feature Selection and Kernel-Based Extreme Learning Machine. Cognitive Analytics. [CrossRef]
- Praveena, H.D.; Subhas, C.; Naidu, K.R. Automatic epileptic seizure recognition using reliefF feature selection and long short term memory classifier. Journal of Ambient Intelligence and Humanized Computing 2021, 12, 6151–6167. [Google Scholar] [CrossRef]
- Pang, Z.; Zhu, D.; Chen, D.; Li, L.; Shao, Y. A computer-aided diagnosis system for dynamic contrast-enhanced MR images based on level set segmentation and Relieff feature selection. Computational and Mathematical Methods in Medicine 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Liu, Y.L.; Feng, C.S.; Zhu, G.Q. Applying the fisher score to identify Alzheimer’s disease-related genes. Genetics and Molecular Research 2016, 15, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Gu, Q.; Li, Z.; Han, J. Generalized fisher score for feature selection. Proceedings of the 27th Conference on Uncertainty in Artificial Intelligence, UAI 2011.
- Song, Q.J.; Jiang, H.Y.; Liu, J. Feature selection based on FDA and F-score for multi-class classification. Expert Systems with Applications 2017, 81, 22–27. [Google Scholar] [CrossRef]
- Core, C. Entropy and mutual information 1.1; 2018.
- Sun, X.; Liu, Y.; Li, J.; Zhu, J.; Chen, H.; Liu, X. Feature evaluation and selection with cooperative game theory. Pattern Recognition 2012, 45, 2992–3002. [Google Scholar] [CrossRef]
- García, J.; Crawford, B.; Soto, R.; Astorga, G. A clustering algorithm applied to the binarization of Swarm intelligence continuous metaheuristics. Swarm and Evolutionary Computation 2019, 44, 646–664. [Google Scholar] [CrossRef]
- Chang, D.; Rao, C.; Xiao, X.; Hu, F.; Goh, M. Multiple strategies based Grey Wolf Optimizer for feature selection in performance evaluation of open-ended funds. Swarm and Evolutionary Computation 2024, 86, 101518. [Google Scholar] [CrossRef]
- Qu, L.; He, W.; Li, J.; Zhang, H.; Yang, C.; Xie, B. Explicit and size-adaptive PSO-based feature selection for classification. Swarm and Evolutionary Computation 2023, 77, 101249. [Google Scholar] [CrossRef]
- Tizhoosh, H.R. Opposition-based learning: A new scheme for machine intelligence. Proceedings - International Conference on Computational Intelligence for Modelling, Control and Automation, CIMCA 2005 and International Conference on Intelligent Agents, Web Technologies and Internet 2005, 1, 695–701. [Google Scholar] [CrossRef]
- Al-Batah, M.S.; Al-Eiadeh, M.R. An improved discreet Jaya optimisation algorithm with mutation operator and opposition-based learning to solve the 0-1 knapsack problem. International Journal of Mathematics in Operational Research 2023, 26, 143–169. [Google Scholar] [CrossRef]
- Deng, Z.; Zhu, X.; Cheng, D.; Zong, M.; Zhang, S. Efficient kNN classification algorithm for big data. Neurocomputing 2016, 195, 143–148. [Google Scholar] [CrossRef]
- Zhang, M.L.; Zhou, Z.H. ML-KNN: A lazy learning approach to multi-label learning. Pattern Recognition 2007, 40, 2038–2048. [Google Scholar] [CrossRef]
- Xiong, L.; Chitti, S.; Liu, L. Mining multiple private databases using a kNN classifier. Proceedings of the ACM Symposium on Applied Computing 2007, pp.435–440. [CrossRef]
- Abu Alfeilat, H.A.; Hassanat, A.B.; Lasassmeh, O.; Tarawneh, A.S.; Alhasanat, M.B.; Eyal Salman, H.S.; Prasath, V.B. Effects of Distance Measure Choice on K-Nearest Neighbor Classifier Performance: A Review. Big Data 2019, 7, 221–248. [Google Scholar] [CrossRef] [PubMed]
- Machine learning in DNA microarray analysis for cancer classification 2003. pp. 189–198.
- Chormunge, S.; Jena, S. Correlation based feature selection with clustering for high dimensional data. Journal of Electrical Systems and Information Technology 2018, 5, 542–549. [Google Scholar] [CrossRef]
- Cerda, P.; Varoquaux, G.; Kégl, B. Similarity encoding for learning with dirty categorical variables. Machine Learning 2018, 107, 1477–1494. [Google Scholar] [CrossRef]
- Hauke, J.; Kossowski, T. Comparison of values of pearson’s and spearman’s correlation coefficients on the same sets of data. Quaestiones Geographicae 2011, 30, 87–93. [Google Scholar] [CrossRef]
- Introducing the black hole. Physics Today 1971, 24, 30–41. [CrossRef]
- The little robot, black holes, and spaghettification. Physics Education 2022, 57, 2203–04759. [CrossRef]
- Black hole: A new heuristic optimization approach for data clustering. Information Sciences 2013, 222, 175–184. [CrossRef]
- Nitasha, S.; KUMAR, T. Study of various mutation operators in genetic algorithms. International Journal of Computer Science and Information Technologies 2014, 5, 4519–4521. [Google Scholar]
- Pandey, H.M.; Chaudhary, A.; Mehrotra, D. A comparative review of approaches to prevent premature convergence in GA. Applied Soft Computing 2014, 24, 1047–1077. [Google Scholar] [CrossRef]
- Andre, J.; Siarry, P.; Dognon, T. An improvement of the standard genetic algorithm fighting premature convergence in continuous optimization. Advances in engineering software 2001, 32, 49–60. [Google Scholar] [CrossRef]
- Leung, Y.; Gao, Y.; Xu, Z.B. Degree of population diversity-a perspective on premature convergence in genetic algorithms and its markov chain analysis. IEEE Transactions on Neural Networks 1997, 8, 1165–1176. [Google Scholar] [CrossRef] [PubMed]
- Paquete, L.; Chiarandini, M.; Stützle, T. Pareto local optimum sets in the biobjective traveling salesman problem: An experimental study. In Metaheuristics for multiobjective optimisation; Springer, 2004; pp. 177–199.
- Gharehchopogh, F.S. An improved tunicate swarm algorithm with best-random mutation strategy for global optimization problems. Journal of Bionic Engineering 2022, 19, 1177–1202. [Google Scholar] [CrossRef]
- Jafari-Asl, J.; Azizyan, G.; Monfared, S.A.H.; Rashki, M.; Andrade-Campos, A.G. An enhanced binary dragonfly algorithm based on a V-shaped transfer function for optimization of pump scheduling program in water supply systems (case study of Iran). Engineering Failure Analysis 2021, 123, 105323. [Google Scholar] [CrossRef]
- Dua, D.; Graff, C. UCI Machine Learning Repository, 2017.
- Pocock, A. Tribuo:Machine Learning with Provenance in Java. 2021; arXiv:cs.LG/2110.03022]. [Google Scholar]
- Wilcoxon, F.; Katti, S.; Wilcox, R.A.; et al. Critical values and probability levels for the Wilcoxon rank sum test and the Wilcoxon signed rank test. Selected tables in mathematical statistics 1970, 1, 171–259. [Google Scholar]
- Sheldon, M.R.; Fillyaw, M.J.; Thompson, W.D. The use and interpretation of the Friedman test in the analysis of ordinal-scale data in repeated measures designs. Physiotherapy Research International 1996, 1, 221–228. [Google Scholar] [CrossRef]
| Name | No. of Attributes | No. of Instances | No. of Classes |
|---|---|---|---|
| Colon | 2000 | 62 | 2 |
| Darwin | 450 | 174 | 2 |
| Leukemia | 3571 | 72 | 2 |
| Leukemia-3c | 7129 | 72 | 2 |
| MLL | 12582 | 72 | 3 |
| WDBC | 30 | 569 | 2 |
| SRBCT | 2308 | 83 | 4 |
| Sobar | 19 | 72 | 2 |
| Parkinsons | 22 | 197 | 2 |
| Sonar | 60 | 208 | 2 |
| Divorce | 54 | 170 | 2 |
| SpectTF | 44 | 267 | 2 |
| Urban | 146 | 675 | 4 |
| WPBC | 31 | 198 | 2 |
| Name | MBHO-no correlation | BCS-no correlation [38] | ||||||
|---|---|---|---|---|---|---|---|---|
| Acc | F1 | Time | Features | Acc | F1 | Time | Features | |
| Colon | 0.82 | 0.82 | 6:21:686 | 997 | 0.79 | 0.79 | 10:32:061 | 972 |
| Darwin | 0.86 | 0.86 | 3:06:425 | 260 | 0.80 | 0.79 | 1:59:366 | 257 |
| Leukemia | 0.99 | 0.94 | 9:18:925 | 1702 | 0.99 | 0.94 | 6:29:548 | 1706 |
| Leukemia-3c | 0.97 | 0.87 | 25:21:440 | 3619 | 0.96 | 0.87 | 18:22:045 | 3517 |
| MLL | 0.93 | 0.85 | 49:20:481 | 6223 | 0.93 | 0.85 | 33:45:589 | 6188 |
| WDBC | 0.95 | 0.94 | 48:443 | 10 | 0.95 | 0.94 | 34:002 | 11 |
| SRBCT | 0.95 | 0.91 | 7:58:261 | 1250 | 0.93 | 0.86 | 4:27:249 | 1127 |
| Sobar | 0.94 | 0.85 | 31:860 | 8 | 0.94 | 0.85 | 19:685 | 14 |
| Parkinsons | 0.91 | 0.87 | 45:745 | 7 | 0.90 | 0.85 | 29:326 | 4 |
| Sonar | 0.90 | 0.90 | 47:031 | 26 | 0.88 | 0.87 | 32:396 | 36 |
| Divorce | 0.98 | 0.98 | 52:827 | 8 | 0.98 | 0.97 | 21:664 | 18 |
| SpectTF | 0.82 | 0.74 | 48:776 | 25 | 0.82 | 0.71 | 36:733 | 22 |
| Urban | 0.75 | 0.68 | 2:06:484 | 89 | 0.71 | 0.65 | 56:283 | 102 |
| WPBC | 0.77 | 0.65 | 37:776 | 13 | 0.75 | 0.65 | 27:797 | 15 |
| Name | MBHO-correlation | BCS-correlation [38] | ||||||
|---|---|---|---|---|---|---|---|---|
| Acc | F1 | Time | Features | Acc | F1 | Time | Features | |
| Colon | 0.86 | 0.83 | 19:16:256 | 1064 | 0.81 | 0.77 | 9:56:465 | 1069 |
| Darwin | 0.87 | 0.86 | 4:43:962 | 258 | 0.82 | 0.80 | 3:18:72 | 262 |
| Leukemia | 0.99 | 0.94 | 54:23:664 | 1732 | 0.99 | 0.94 | 31:49:948 | 1730 |
| Leukemia-3c | 0.97 | 0.87 | 13:26:19:141 | 3508 | 0.97 | 0.87 | 5:52:38:346 | 3508 |
| MLL | 0.94 | 0.87 | 14:32:17:233 | 6500 | 0.94 | 0.87 | 11:37:18:083 | 6223 |
| WDBC | 0.95 | 0.94 | 3:03:488 | 14 | 0.95 | 0.94 | 1:39:856 | 11 |
| SRBCT | 0.95 | 0.90 | 30:13:617 | 1201 | 0.92 | 0.87 | 15:04:977 | 1141 |
| Sobar | 0.94 | 0.85 | 26:661 | 6 | 0.94 | 0.85 | 18:519 | 8 |
| Parkinsons | 0.91 | 0.87 | 44:680 | 10 | 0.90 | 0.85 | 29:326 | 4 |
| Sonar | 0.92 | 0.91 | 1:13:362 | 29 | 0.88 | 0.87 | 44:256 | 41 |
| Divorce | 0.99 | 0.99 | 1:00:577 | 14 | 0.98 | 0.98 | 35:622 | 18 |
| SpectTF | 0.84 | 0.84 | 1:30:154 | 26 | 0.82 | 0.72 | 49:101 | 24 |
| Urban | 0.77 | 0.72 | 8:24:944 | 80 | 0.72 | 0.66 | 4:07:742 | 99 |
| WPBC | 0.78 | 0.65 | 1:02:621 | 13 | 0.77 | 0.63 | 35:621 | 16 |
| Name | MBHO-no correlation | MBHO-correlation | ||||||
|---|---|---|---|---|---|---|---|---|
| Acc | F1 | Time | Features | Acc | F1 | Time | Features | |
| Colon | 0.82 | 0.82 | 6:21:686 | 997 | 0.86 | 0.83 | 19:16:256 | 1064 |
| Darwin | 0.86 | 0.86 | 3:06:425 | 260 | 0.87 | 0.86 | 4:43:962 | 258 |
| Leukemia | 0.99 | 0.94 | 9:18:925 | 1702 | 0.99 | 0.94 | 54:23:664 | 1732 |
| Leukemia-3c | 0.97 | 0.87 | 25:21:440 | 3619 | 0.97 | 0.87 | 13:26:19:141 | 3508 |
| MLL | 0.93 | 0.85 | 49:20:481 | 6223 | 0.94 | 0.87 | 14:32:17:233 | 6500 |
| WDBC | 0.95 | 0.94 | 48:443 | 10 | 0.95 | 0.94 | 3:03:488 | 14 |
| SRBCT | 0.95 | 0.91 | 7:58:261 | 1250 | 0.95 | 0.90 | 30:13:617 | 1201 |
| Sobar | 0.94 | 0.85 | 31:860 | 8 | 0.94 | 0.85 | 26:661 | 6 |
| Parkinsons | 0.91 | 0.87 | 45:745 | 7 | 0.91 | 0.87 | 44:680 | 10 |
| Sonar | 0.90 | 0.90 | 47:031 | 26 | 0.92 | 0.91 | 1:13:362 | 29 |
| Divorce | 0.98 | 0.98 | 52:827 | 8 | 0.99 | 0.99 | 1:00:577 | 14 |
| SpectTF | 0.82 | 0.74 | 48:776 | 25 | 0.84 | 0.84 | 1:30:154 | 26 |
| Urban | 0.75 | 0.68 | 2:06:484 | 89 | 0.77 | 0.72 | 8:24:944 | 80 |
| WPBC | 0.77 | 0.65 | 37:776 | 13 | 0.78 | 0.65 | 1:02:621 | 13 |
| Name | Features | MBHO-correlation | MIM [39] | JMI [40] | mRMR [41] | ||||
|---|---|---|---|---|---|---|---|---|---|
| Acc | F1 | Acc | F1 | Acc | F1 | Acc | F1 | ||
| Colon | 1064 | 0.86 | 0.83 | 0.79 | 0.76 | 0.79 | 0.76 | 0.76 | 0.73 |
| Darwin | 258 | 0.87 | 0.86 | 0.76 | 0.74 | 0.75 | 0.72 | 0.75 | 0.72 |
| Leukemia | 1732 | 0.99 | 0.94 | 0.99 | 0.94 | 0.99 | 0.94 | 0.99 | 0.94 |
| Leukemia-3c | 3508 | 0.97 | 0.87 | 0.93 | 0.85 | 0.93 | 0.85 | 0.93 | 0.85 |
| MLL | 6500 | 0.94 | 0.87 | 0.94 | 0.87 | 0.93 | 0.85 | 0.93 | 0.85 |
| WDBC | 14 | 0.95 | 0.94 | 0.93 | 0.93 | 0.93 | 0.93 | 0.94 | 0.93 |
| SRBCT | 1201 | 0.95 | 0.90 | 0.95 | 0.89 | 0.90 | 0.85 | 0.95 | 0.89 |
| Sobar | 6 | 0.94 | 0.85 | 0.89 | 0.77 | 0.86 | 0.74 | 0.92 | 0.79 |
| Parkinsons | 10 | 0.91 | 0.87 | 0.86 | 0.79 | 0.87 | 0.88 | 0.85 | 0.77 |
| Sonar | 29 | 0.92 | 0.91 | 0.87 | 0.86 | 0.83 | 0.83 | 0.86 | 0.86 |
| Divorce | 14 | 0.99 | 0.99 | 0.98 | 0.97 | 0.98 | 0.97 | 0.98 | 0.97 |
| SpectTF | 26 | 0.84 | 0.84 | 0.76 | 0.63 | 0.76 | 0.65 | 0.72 | 0.58 |
| Urban | 80 | 0.77 | 0.72 | 0.64 | 0.60 | 0.70 | 0.66 | 0.69 | 0.62 |
| WPBC | 13 | 0.78 | 0.65 | 0.70 | 0.53 | 0.68 | 0.56 | 0.70 | 0.54 |
| Rank First | 14 | 3 | 1 | 2 | |||||
| Sum of Ranks | 53.06 | 33.04 | 26.04 | 28 | |||||
| Mean of Ranks | 3.79 | 2.36 | 1.86 | 2 | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





