1. Introduction
Mpox virus (MPXV) is a double-stranded enveloped DNA virus, belonging to the
Poxvirus family, genus
Orthopoxvirus, and is related to the variola virus, the causative agent of smallpox. In May 2022, a Mpox epidemic outside of Africa gained international attention. On July 23, 2022, the World Health Organization declared the Mpox epidemic a public health emergency of international concern and on July 29, 2022, the Brazilian Ministry of Health established the Center of National Public Health Emergency Operations for Mpox (COE-MPOX) to coordinate the country’s public health responses to the disease [
1].
In Brazil, the first Mpox confirmed case was reported on June 9 in the state of São Paulo and on June 15 Brazil had five confirmed cases, according to the Ministry of Health [
2]. The first fatal case was reported, on July 28, in Belo Horizonte (state of Minas Gerais), Southeast Brazil [
3]. On August 18, the municipal health secretariat of Belo Horizonte reported 106 confirmed and 14 probable cases in its first report (First Monkeypox report, Prefeitura de Belo Horizonte, 2022) [
4]. From June 1, 2022, to February 28, 2023, Brazil recorded 10,656 confirmed and probable Mpox cases, according to the 20th Epidemiological Bulletin of Monkeypox by the COE — Ministério da Saúde [
1].In March 2023, when the Ministry of Health initiated Mpox vaccinations, prioritizing high- risk groups. At this time, Brazil had reported 50,803 suspected cases, of which 10,301 (20.3%) were confirmed [
5].
Sewage surveillance has been recognized as a valuable method for monitoring pathogen circulation at the community level. Its effectivness was demonstrated during the COVID-19 pandemic, which enabled the detection of SARS-CoV-2 by tracking the virus in sewage samples from numerous cities worldwide [6 8], including the city of Belo Horizonte [
9]. Previous studies have detected MPXV in sewage samples in the Netherlands and Rome [10, 11], as the virus is excreted into wastewater through skin lesions, nasal secretion, urine, and feces [
12]. In our research, we investigated the circulation of MPXV (and other viruses) in sewage samples collected in Belo Horizonte using whole genome sequencing. Belo Horizonte is a largest Brazilian city from the state of Minas Gerais, with a population around 2.7 million people and with a metropolitan area of 6 million people [
13]. This study demonstrated the potential of genomic sewage surveillance for the early detection of emerging and reemerging pathogens in the community for aiding in outbreak response.
2. Material and Methods
Sewage samples were collected from four locations in the metropolitan area of Belo Horizonte: two hospitals (Hospital A, the reference hospital for infectious disease treatment in the state of Minas Gerais and Hospital B, a general practice hospital offering assistance to pregnant women and high-risk prenatal care) and two municipal wastewater treatment plants (WWTP), which together process sewage from approximately 2.2 million people. The mean influent loads are1994 L.s-¹ for WWTP-A and 1646 L.s-¹ for WWTP-B.
Sewage samples were collected bi-monthly from July 23, 2022, to March 15, 2023. In total, 60 samples were collected: 30 from the two hospitals (15 samples each), and 32 from the municipal WWTPs (16 samples from WWTP-A and 14 from WWTP-B).
Hospital sewage samples were collected as described previously [
14]. For the WWTPs, 24-hour composite samples were collected at the entrance and filtered through electronegative membranes (30 to 50 mL) [
9], with slight modifications from previous methods: MgCl₂ (2.5M) was not added and the pH was not adjusted to 3.5. Viral genetic material was extracted from the membranes using a commercial kit (AllPrep PowerViral DNA/RNA, Qiagen®, Hilden, Germany) according to the manufacturer's instructions. The extracted genetic material was resuspended in 100 µL of ultrapure water (free of RNases) and stored at -80 °C.
Samples were tested for MPXV using a quantitative PCR (qPCR) with five PLEX assays diagnostic kit from Bio-Manguinhos, following the manufacturer’sinstructions. The tests targeted the detection of Varicella Zoster Virus (VZV), Molluscum Contagiosum Virus (MOCV), MPXV, and other Orthopoxviruses (OPV) using the LightCycler® 480 System (Roche).
All samples underwent whole-genome sequencing through target enrichment using a hybrid capture method. The sequencing utilized the Illumina VSP panel, which characterizes 66 viruses, including DNA and RNA viruses such as Polyomavirus, HPV, Mpox, Poliovirus, Influenza, SARS-CoV-2, among others (
Figure S1).
Sequencing libraries were prepared by first synthesizing cDNA from concentrated wastewater samples. This process employed the Illumina RNA Prep with Enrichment Indexes Set A (for 96 samples) (Illumina) and the VSP (Illumina). Libraries were sequenced on the NextSeq™ 2000 System with a read length of 2 × 150 bp and a sequencing. FASTQ raw sequencing data were analyzed using the Illumina DRAGEN™ Microbial enrichment pipeline (available on the BaseSpace™ Sequence Hub) for viral detection using default parameters. Raw data were also analyzed using the Genome Detective software [
15].
3. Results and Discussions
MPXV DNA was detected via hybrid capture method for whole-genome sequencing (WGS) and confirmed using qPCR in 10/15 (66.7%) samples from Hospital A, spanning from July 23, 2022, to March 15, 2023 (
Table 1). In Hospital B, two out of 13 samples (15.4%) tested positive for MPXV. In municipal wastewaters, MPXV DNA was detected in four out of 30 samples (13.3%), using both methods of detection (WGS and qPCR,
Table 1).
The higher frequency of detection in the sewage samples from Hospital A (66.7%) can be attributed to its role as the reference hospital for infectious disease treatment in the state of Minas Gerais, which serves patients from many cities including Belo Horizonte. Notably, the first fatal case reported in Brazil, on July 28 2022, was a patient being treated at Hospital A, where this study was conducted (see
Table 1, sewage from Hospital A was positive on July 23rd). Furthermore, hospital sewage samples were collected directly from the hospital manhole before the sewage was discharged in the sewer network, thus avoiding any dilution.
Overall, MPXV DNA was detected in 16/60 (26.6%) of the total sewage samples combining both methods of detection (qPCR and WGS).
The low detection frequency of MPXV DNA in municipal sewage (13.3% in WWTP A and B, 4 out of 30 samples) could be attributed to the limited number of MPXV cases reported in the city: 106 confirmed cases on August 18, 2022, 324 on November 30, 2022, 339 on January 11, 2023 [
16] and 8 new cases (2 confirmed and 6 suspected) on February 24, 2023 [
17].
Additionally, the dilution factor should be considered, as each WWTP serves a population of approximately 1.1 million people. A previous study in the Netherlands [
11] reported the detection of MPXV DNA using qPCR in sewage samples from Amsterdam city districts and Schiphol airport when the national case count was 1,087, with most cases located in Amsterdam. The number of inhabitants connected to each sampling location varied from 16,000 to 121,000 and from 310,000 to 648,000 in WWTPs investigated [
10].
This is the first report of MPXV DNA in sewage in Brazil, from an area with relatively few cases. In Brazil, as reported by the Ministry of Health up to 31st of March 2023, the proportion of confirmed Mpox cases was 29.9% (10,378 out of 52,109 suspected cases) with an additional 330 cases (0.6%) classified as probable (according to the 21st Epidemiological Bulletin of Monkeypox by the COE — Ministério da Saúde) [
5]. These findings underscore the utility of wastewater surveillance as an invaluable epidemiological tool for monitoring emerging infectious diseases, highlighting the importance of employing both detection methods (WGS and qPCR) and extending surveillance to multiple sampling locations, including hospitals and municipal WWTPs.
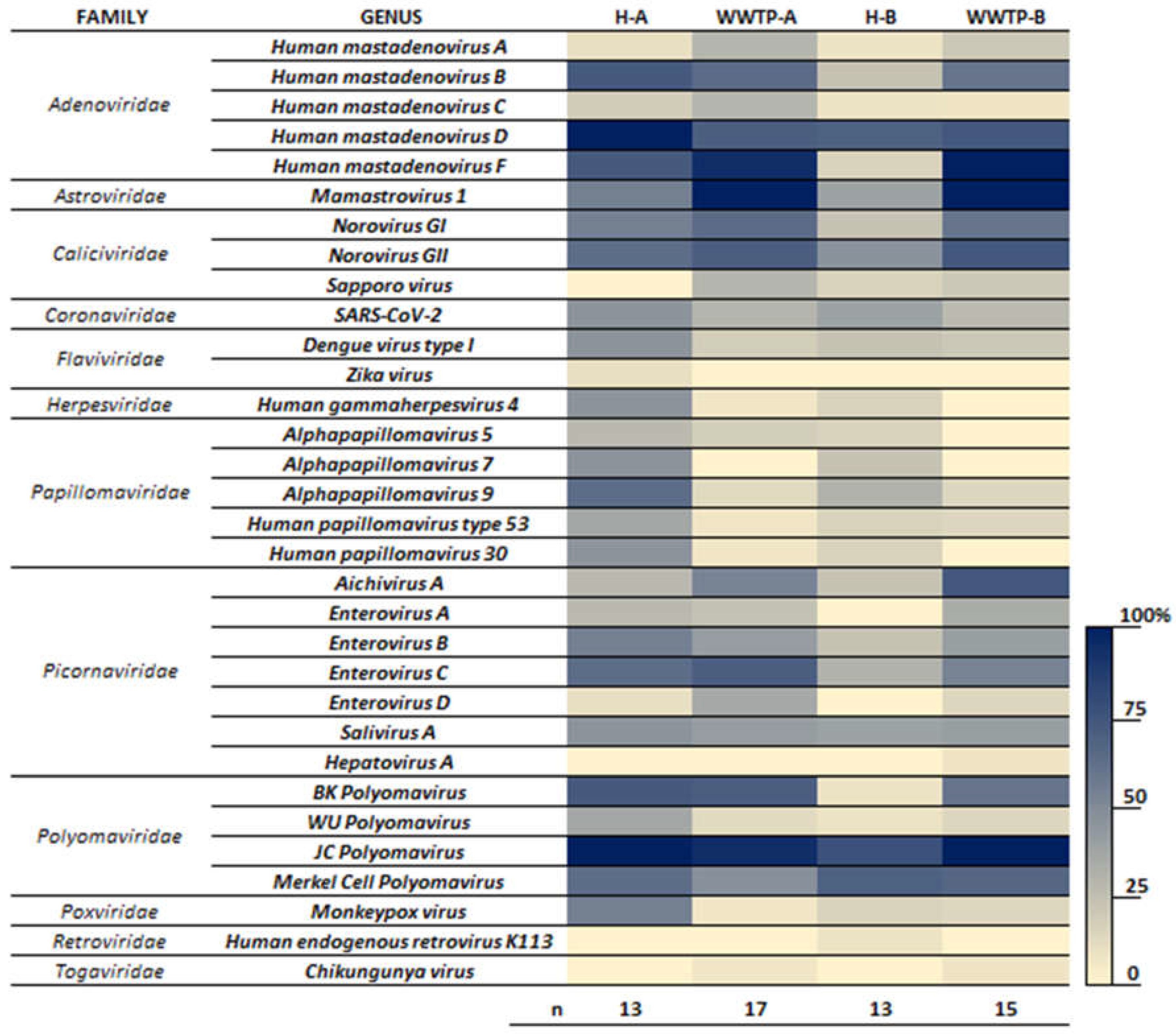
In addition to the Mpox virus, nine other virus families were identified in samples from Hospital A and municipal WWTPs (
Figure 1). These include Adenoviridade, Astroviridae, Caliciviridae, Picornaviridade, Polyomaviridae, Coronaviridae (Sars-CoV-2), Herspesviridae, Papillomaviridae and Flaviviridae (
Figure 1). Members of these families represent some of the most significant excreted human pathogens know to be shed in human waste and have previously been identified as part of the sewage virome [
18], with the exception of Mpox and Dengue viruses.
Human Adenovirus and JC Polyomavirus, detected in over 80% of samples from both hospitals and WWTPs (
Figure 1), are commonly used as fecal indicators due to their prevalence in sewage and their correlation with fecal pollution [
18].
Figure 1 also show, the more prevalent viruses in hospital wastewaters, including species from Adenoviridae (human mastadenovirus B, D and F), Astroviridae (Mamastrovirus 1), Caliciviridae (Norovirus GI and GII), Picornaviridade (Enterovirus B, C and Salivirus A) Herpesviridae (Human gammaherpesvirus 4) and Papillomaviridae (Alphapapillomavirus and Human papillomavirus).
Most viruses discussed in this study cause gastroenteritis in humans (19). Hospital A, which treats patients with HIV and other infectious disease, frequently encounters diarrhea among these patients. Polyomaviridae, such as BK Polyomavirus and JC Polyomavirus, are widely distributed across global populations [
20]. Typically, the primary infection of these viruses is asymptomatic and latent, becoming active due to immunosuppression. Previous studies have detected these viruses in urine samples from both HIV-positive patients and HIV-negative individuals who are immunocompromised [
21]. Human papillomavirus (HPV) detected includes high-risk HPV types, which are implicated in the development of cervical cancer [
22] and are the
most prevalent pathogens responsible for female cancers, particularly in developing countries [
23]
.
Enterovirus cause severe respiratory illness in children and association between EV-D68 and neurological disease such as Acute Flaccid Paralysis has been reported [
24]. Wu Polyomavirus is commonly found in the respiratory tract samples of immunocompromised patients [
25].
Concerning RNA viruses that cause respiratory infections, such as SARS-CoV-2 and Influenza A and B, the results are detailed in
Table S1. Notably, SARS-CoV-2 RNA was detected in about 53% of the sewage samples tested by RT-PCR and 68% by WGS. The number of COVID-19 confirmed cases reported in Belo Horizonte during the investigated period were: 4,000 (September/2022), 6,000 (November/2022), 14,000 (December/2022), 1,000 in February and 1,000 in March/2023 (according to Prefeitura de Belo Horizonte, 2023)[
26]. Influenza A RNA was detected only by RT-PCR (21.4% as shown in
Table S1) and not by hybrid capture and WGS, indicating a sensitivity difference between the two methods. There was no detection of Influenza B RNA by either method, suggesting that this virus was not circulating in the investigated period or the prevalence was too low. In fact, the prevalence of Influenza B in Brazil in the investigated period was: 0.2% of positive cases for respiratory viruses (August/2022), 1.2%(September/2022), 0.6% (October/2022), 0.3% (November/2022), 0.1% (December/2022), 0.7% (January/2023), 1.6% (February/2023) and 3.0% (March/2023) (according to the Respiratory diseases Information Bulletins of Fundação Oswaldo Cruz) [
27]. To our knowledge, this is the first study that has applied the hybrid capture and WGS method to the successful detection and genetic characterization of multiple viruses in wastewaters during the Mpox outbreak in Brazil. This could provide crucial insights for viral surveillance and support public health authorities in guiding control actions.
4. Conclusions
In conclusion, this study underscores the effectiveness of genomic sewage surveillance, utilizing the hybrid-capture method and whole genome sequencing (WGS), as a powerful tool for monitoring viruses in both community and hospital settings, including MPXV. Additionally, the study demonstrated that the qPCR diagnostic kit used for clinical samples (five PLEX assays diagnostic kit from Bio-Manguinhos), which is standard across Brazilian public health laboratories during the Mpox outbreak, also performed exceptionally well for monitoring wastewater samples, exhibiting a higher frequency of detection compared to WGS. This suggests that wastewater surveillance for Mpox and other viruses could be effectively conducted by central public health laboratories and should encompass both hospital and municipal wastewater systems. This approach could serve as a complement to clinical case studies, providing critical insights that help guide public health responses effectively.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, Figure S1: Figure showing the viruses included in the enrichment workflow of the Viral Surveillance Panel – VSP (Illumina); Table S1: WGS and RT-PCR results for SARS-CoV-2, Influenza A, Influenza B detection using BioManguinhos Kit.
Author Contributions
Conceptualization, J.C.A,LCJA methodology, J.C.A., C.D.L., AP.A.C, M.N.,MA, FMI; software,V.F., M.G, MB.; validation, F.M.I., T.E.A.R.., N.C.; formal analysis, C.D.L., A.P.A.C, J.C.A., T.E.A.R., V.F.; investigation, APAC, N.C,MB,MN, CDL,MA; resources, A.G., A.S.; F.M.I., L.C.J.A., J.C.A.; writing- original draft preparation, J.C.A., A.A.C., FMI, MG,VF,LCJA, TEAR; writing- review and editing JCA,FMI, MG,VF,LCJA; supervision, J.C.A., F.C.M.I, A.G., A.S., LCJA; funding acquisition, J.C.A., L.C.J.A., F.S., M.C.M.C., F.M.I, LVF. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by CNPq (424004/2021-6) and in part by the United World Arbovirus Research Network (UWARN), FAPESP (2021/11944-6). MG is funded by PON “Ricerca e Innovazione” 2014-2020. JCA was also supported by CNPq (research grant 306899/2022-1).
Acknowledgments
We gratefully acknowledge the support of Minas Gerais Sanitation Company (COPASA), for sewage samples collection from the two WWTPs. We also would like to thank FAPEMIG, CAPES and CNPq for the student scholarships. Authors would like to acknowledge the Global Consortium to Identify and Control Epidemics – CLIMADE, (L.C.J.A., M.G., J.C.A.: Principal Investigators from Latin America) (
https://climade.health/).
Conflicts of Interest
The authors declare no conflicts of interest.
Biographical Sketch
Dr. Calabria de Araújo is an associate professor at the Federal University of Minas Gerais. Her research interests and work include the molecular detection and sequencing of different pathogens, viruses, resistant bacteria and antibiotic resistance genes in water, wastewaters, sludge samples and wastewater treatment plants. She coordinated the first wastewater surveillance project to monitor SARS-CoV-2 in Brazil (Belo Horizonte).
References
- “Ministério da Saúde, 2023- Secretaria de Vigilância em Saúde e Ambiente. Boletim epidemiológico especial Mpox. Boletim Epidemiológico de Monkeypox n. 20 do Centro de Operações de Emergências (COE) 1/2/2023 a 28/2/2023”. Available online: www.gov.br/saúde.
- “Ministério da Saúde, 2022-Nota a imprensa 2022, Atualização sobre os casos e Monkeypox no Brasil, 15/06/2022”. Available online: www.gov.br/saúde.
- Guimarães, N.R.; Tomé, L.M.R.; Lamounier, L.O.; Silva, M.V.F.; Lima, M.T.; da Costa, A.V.B.; Luiz, K.C.M.; de Jesus, R.; Trindade, G.S.; Oliveira, D.B.; da Fonseca, F.G.; Fernandes, A.P.S.M.; de Oliveira, J.S.; Moura, J.B.P.; Kroon, E.G.; Giovanetti, M.; Fonseca, V.; Alcantara, L.; Adelino, T.E.R.; de Melo Iani, F.C. Genomic Surveillance of Monkeypox Virus, Minas Gerais, Brazil, 2022. Emerg Infect Dis. 2023, 29, 1270–1273, Epub 2023 Apr 17. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- “Prefeitura de Belo Horizonte, 2022- Secretaria municipal de saúde Boletim epidemiológico edição 1/2022, 18/8/2022.” Available online: www.prefeitura.pbh.gov.br.
- “Ministério da Saúde, 2023- Secretaria de Vigilância em Saúde e Ambiente. Boletim epidemiológico especial Mpox. Boletim Epidemiológico de Monkeypox n. 21 do Centro de Operações de Emergências (COE) 1/3/2023 a 31/3/2023”. Available online: www.gov.br/saúde.
- Ahmed, W.; Angel, N.; Edson, J.; Bibby, K.; Bivins, A.; Brien, J.W.O.; et al. First confirmed detection of SARSCoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci Total Environ 2020, 728, 138764. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, S.; Orschler, L.; Lackner, S. Long-term monitoring of SARS-CoV-2 RNA in wastewater of the Frankfurt metropolitan area in Southern Germany. Scientific Reports 2021, 11, 537. [Google Scholar] [CrossRef]
- Gonzalez, R.; Curtis, K.; Bivins, A.; Bibby, K.; Weir, M.H.; Yetka, K.; et al. COVID-19 surveillance in Southeastern Virginia using wastewater-based epidemiology. Water Research 2020, 186, 116296. [Google Scholar] [CrossRef] [PubMed]
- Araujo, J.C.; Mota, V.T.; Teodoro, A.; Leal, C.; Leroy, D.; Madeira, C.; et al. Long-term monitoring of SARS-CoV-2 RNA in sewage samples from specific public places and STPs to track COVID-19 spread and identify potential hotspots. Sci Total Environ 2022, 838, 155959. [Google Scholar] [CrossRef] [PubMed]
- De Jonge, E.F.; Peterse, C.M.; Koelewijn, J.M.; van der Drift, A.R.; van der Beek, R.F.H.J.; Nagelkerke, E.; et al. The detection of monkeypox virus DNA in wastewater samples in the Netherlands. Sci Total Environ 2022, 852, 158265, Epub ahead of print. [Google Scholar] [CrossRef]
- La Rosa, G.; Mancini, P.; Veneri, C.; Ferraro, G.B.; Luncentini, L.; Iaconelli, M. Detection of Monkeypox virus DNA in airport wastewater, Rome, Italy. J. of Emerging Infectious Disease 2023, 29, N.1. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, A.; Adhikari, S.; Kaya, D.; Islam, A.; Malla, B.; Sherchan, S.P.; et al. Monkeypox outbreak: Wastewater and environmental surveillance perspective. Sci Total Environ 2023, 856, 159166. [Google Scholar] [CrossRef] [PubMed]
- IBGE. Panorama de Belo Horizonte. Available online: https://cidades.ibge.gov.br/brasil/mg/belo-horizonte/panorama (acessed on 19 march 2024).
- Araujo, J.C.; Madeira, C.L.; Bressani, T.; LeaL, C.; Leroy, D.; Machado, E.C.; et al. Quantification of SARS-CoV-2 in wastewater samples from hospitals treating COVID-19 patients during the first wave of the pandemic in Brazil. Sci Total Environ 2023, 856, 160498. [Google Scholar] [CrossRef] [PubMed]
- Vilsker, M.; Moosa, Y.; Nooij, S.; Fonseca, V.; Ghysens, Y.; Dumon, K.; et al. Genome Detective: an automated system for virus identification from high-throughput sequencing data. Bioinformatics 2019, 35, 871–873. [Google Scholar] [CrossRef] [PubMed]
- Prefeitura de Belo Horizonte, 2023a- Secretaria municipal de saúde Boletim epidemiológico edição 21/2023, 11/1/2023.” Available on line: www.prefeitura.pbh.gov.br.
- Prefeitura de Belo Horizonte, 2023b- Secretaria municipal de saúde Boletim epidemiológico edição 27/2023, 24/2/2023.” Available on line: www.prefeitura.pbh.gov.br.
- Mejías-Molina, C.; Pico-Tomàs, A.; Beltran-Rubinat, A.; Martinez-Puchol, S.; Corominas, L.; Rusiñol, M.; Bofil-Mas, S. Effectiveness of passive sampling for the detection and genetic characterization of human viroses in wastewater. Environ. Sci: Water Res. Technol 2023, 9, 1195. [Google Scholar]
- Desselberger, U.; Gray, J. Viral Gastroenteritis. Medicine 2009, 37, 594–598, Epub 2009 Oct 31. [Google Scholar] [CrossRef] [PubMed]
- Hussain, I.; Tasneem, F.; Gilani, U.S.; Arshad, M.I.; Ul Haque, M.F.; Abbas, Z.; et al. Human BK and JC polyomaviruses: Molecular insights and prevalence in Asia. Virus Research 2020, 278, 197860. [Google Scholar] [CrossRef] [PubMed]
- Behzad-Behbahani, A.; Klapper, P.E.; Valley, P.J.; Cleator, G.M.; Khoo, S.H. Detection of BK virus and JC virus DNA in urine samples from immunocompromised (HIV-infected) and immunocompetent (HIV-non-infected) patients using polymerase chain reaction and microplate hybridization. J Clin Virol. 2004, 29, 224–9. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Han, S.; Li, X.; Wang, X.; Wang, S.; Ma, L. Prevalence and distribution of human papillomavirus (HPV) in Luoyang city of Henan province during 2015-2021 and the genetic variability of HPV16 and 52. Virol J. 2022, 19, 37. [Google Scholar] [CrossRef] [PubMed]
- Kombe Kombe, A.J.; Li, B.; Zahid, A.; Mengist, H.M.; Bounda, G.A.; Zhou, Y.; et al. Epidemiology and Burden of Human Papillomavirus and Related Diseases, Molecular Pathogenesis, and Vaccine Evaluation. Front. Public Health 2021, 8, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Gonzales-Sanz, R.; Taravillo, I.; Reina, J.; Navascués, A.; Moreno-Docón, A.; Aranzamendi, M.; et al. Enterovirus D68-associated respiratory and neurological illness in Spain, 2014-2018. Emerg Microbes Infect. 2019, 8, 1438–1444. [Google Scholar] [CrossRef] [PubMed]
- Babakir-Mina, M.; Ciccozzi, M.; Perno, C.F.; Ciotti, M. The human polyomaviruses KI and WU: virological background and clinical implications. J. of Pathology, Microbiology and Immunology. 2013, 121, 746–54, Epub 2013 Jun 19. [Google Scholar] [CrossRef] [PubMed]
- Prefeitura de Belo Horizonte, 2023c- Boletim epidemiológico da COVID-19 N. 533/2022 to 575/2023. Available on line: www.prefeitura.pbh.gov.br.
- INFOGripe Oswaldo Cruz Foundation, Boletim Infogripe 2022 a 2023. Available online: https://portal.fiocruz.br/documento/boletim-infogripe and http://info.gripe.fiocruz.br (accessed on 5 May 2024).
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).