Submitted:
09 May 2024
Posted:
09 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
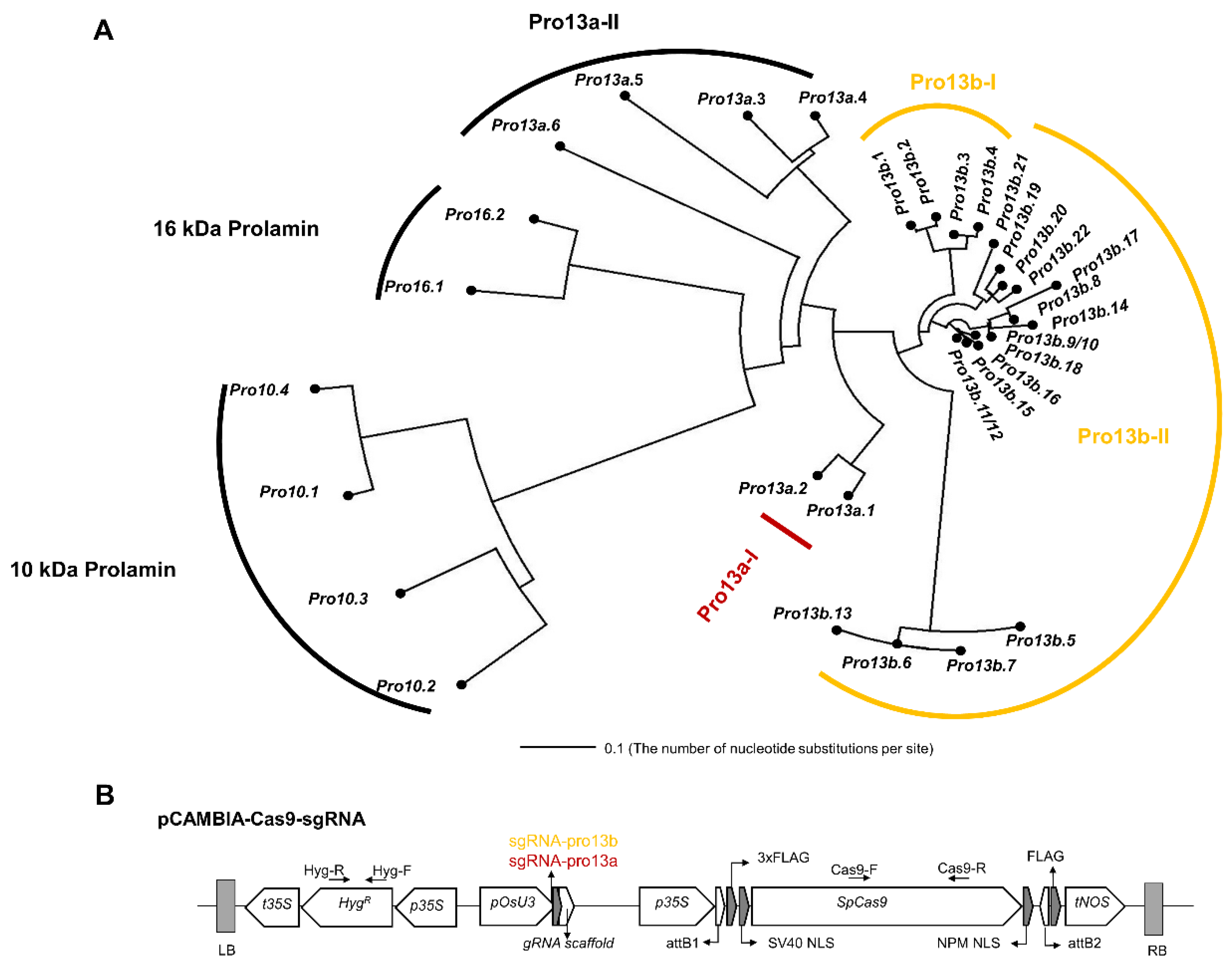
2.1. Design of sgRNA for Targeting 13 kDa Prolamin Genes Using CRISPR-Cas9 Technology
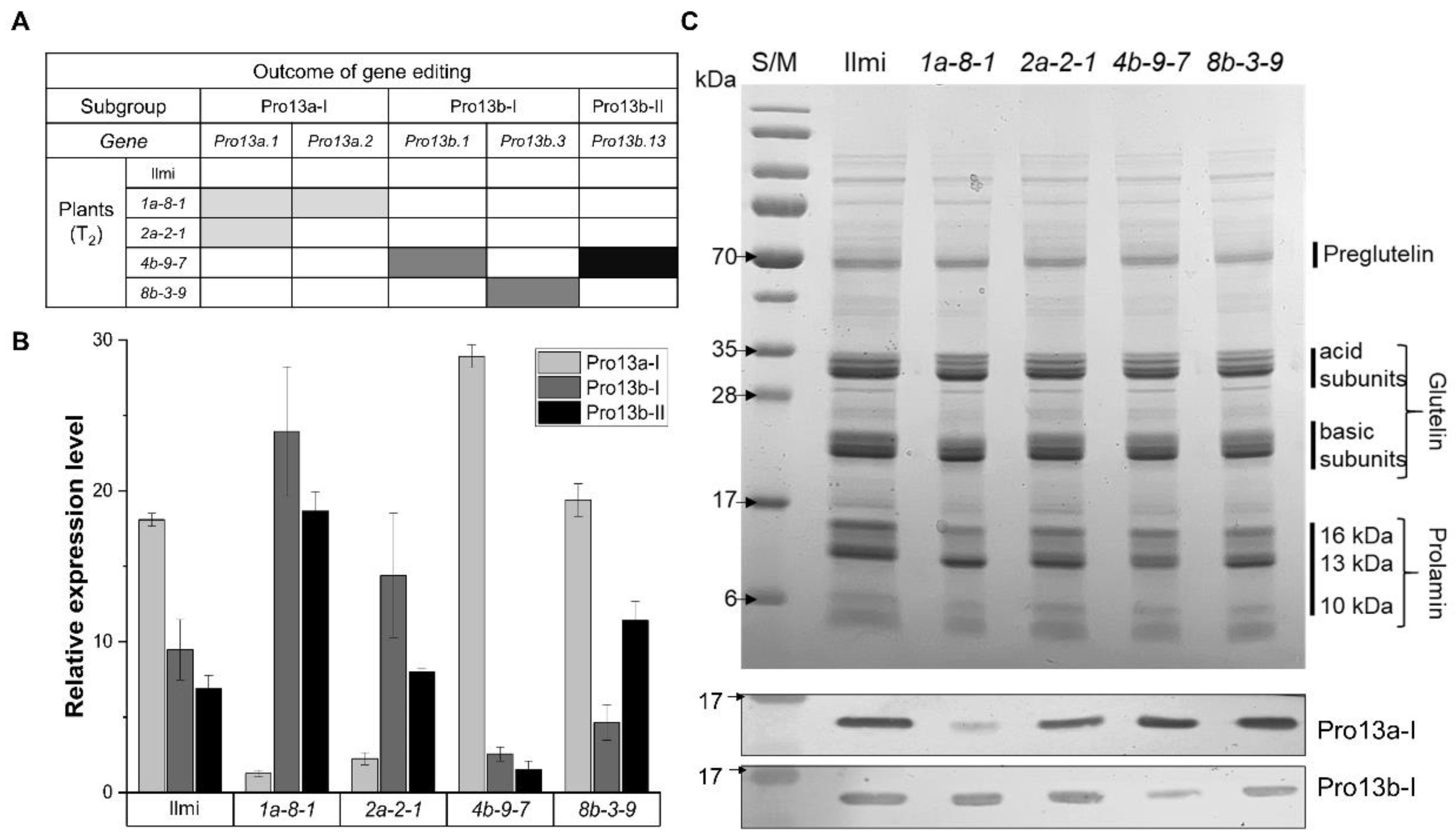
2.2. Generating and Characterizing 13 kDa Prolamin-Knockout Rice
2.3. Target Gene Expression at Transcriptional and Translational Levels
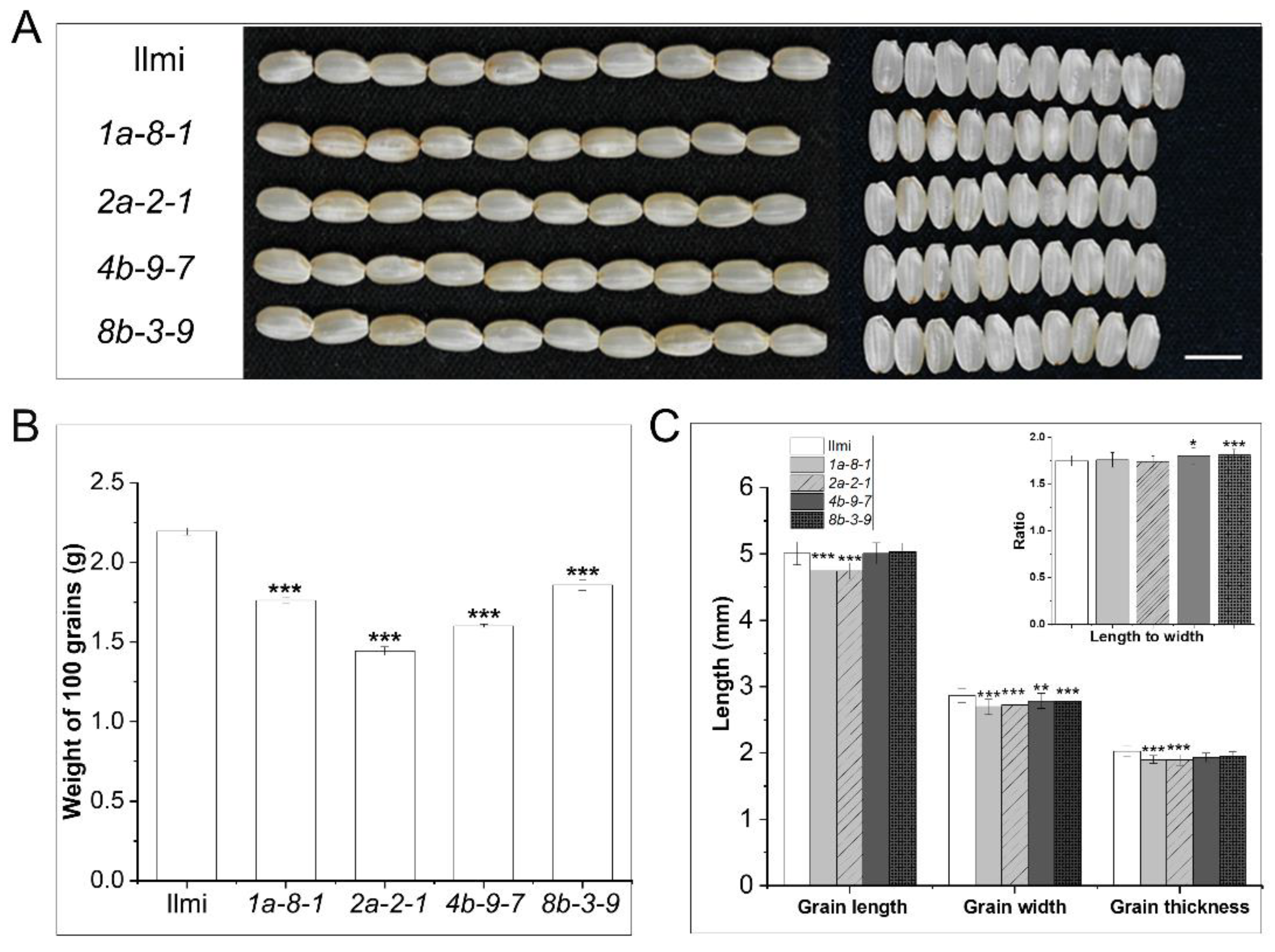
2.4. Morphological Features of 13 kDa Prolamin-Knockout Plants
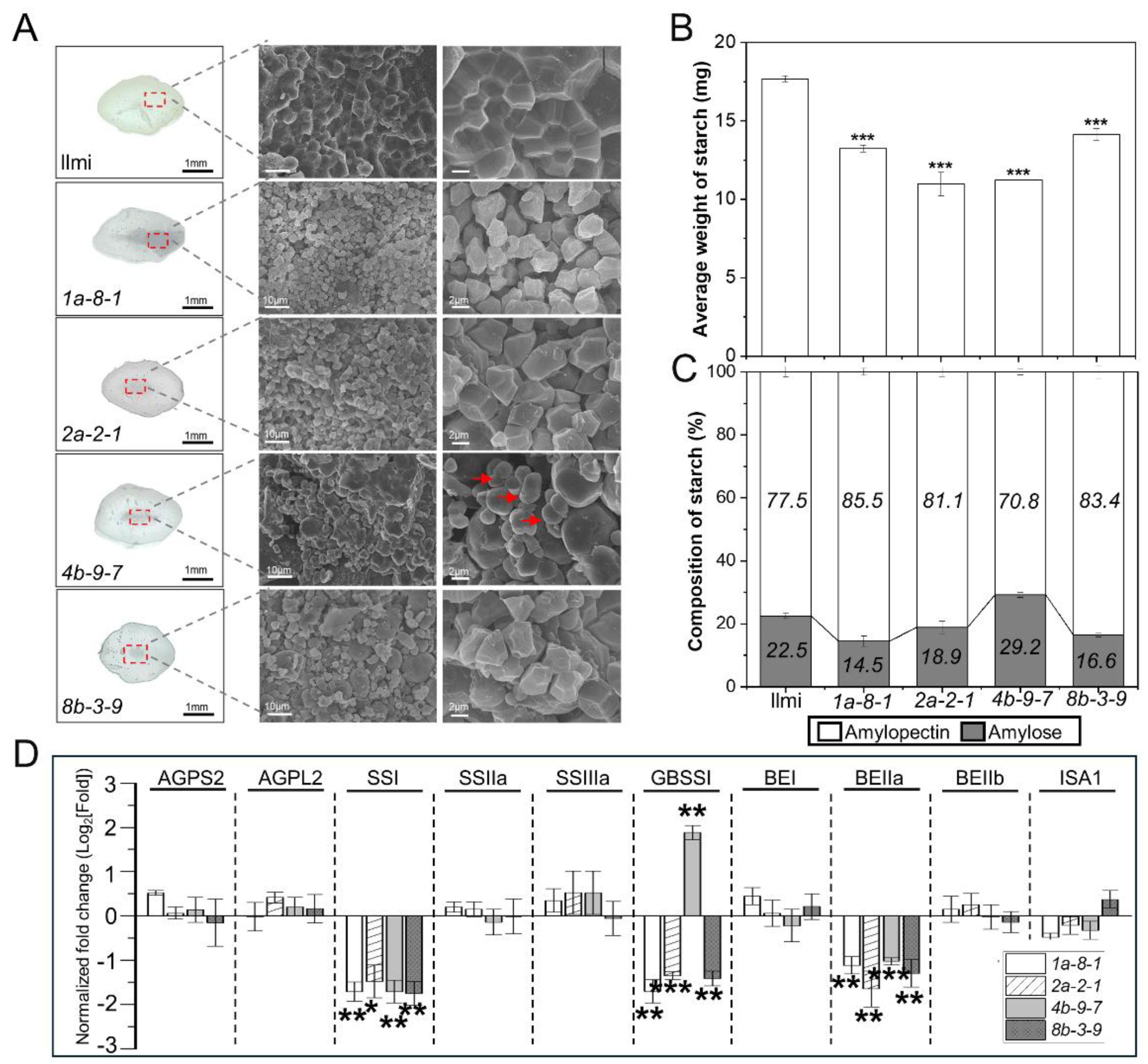
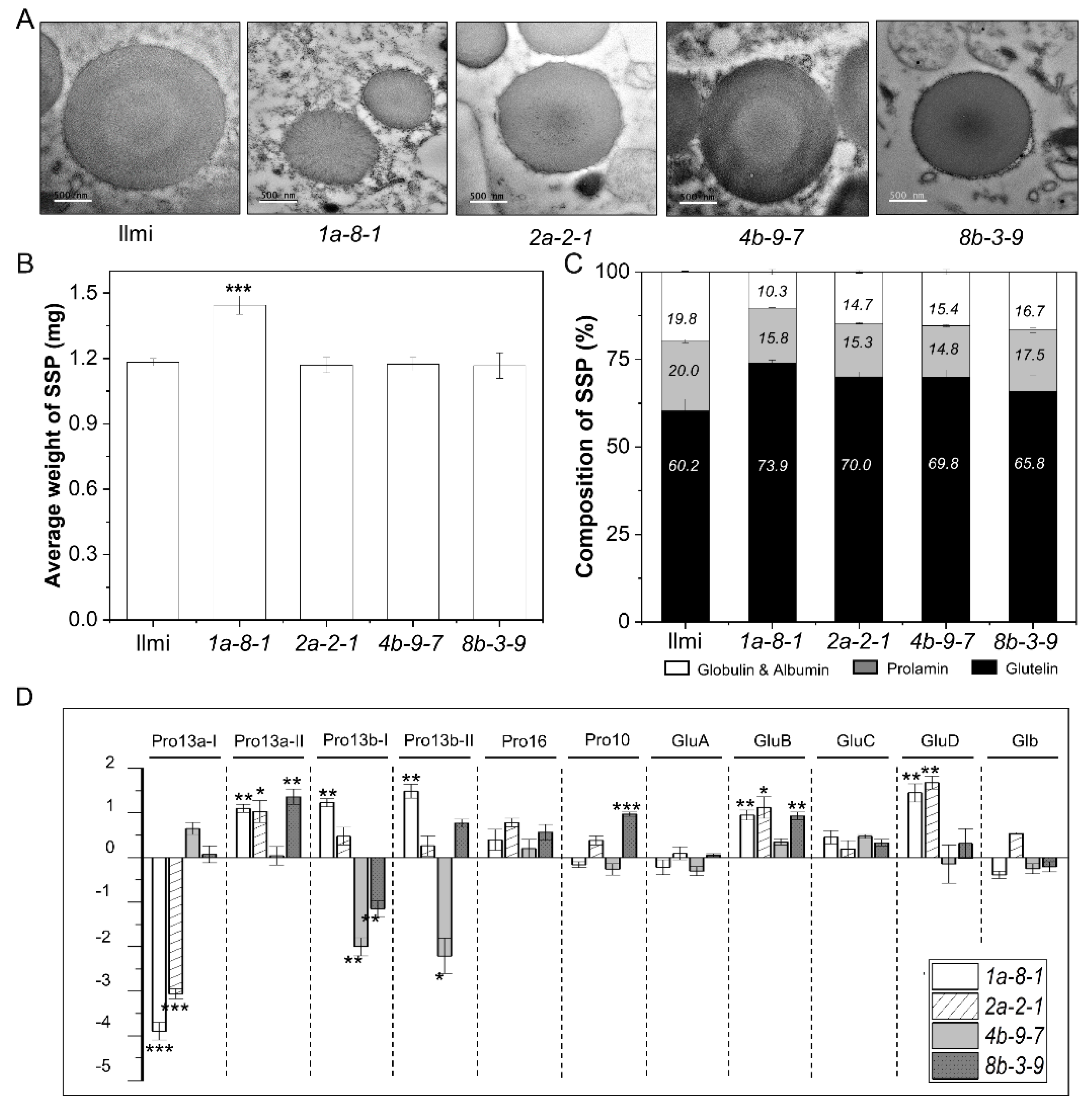
2.5. Protein Body Formation and SSP Composition in 13 kDa Prolamin-Knockout Lines
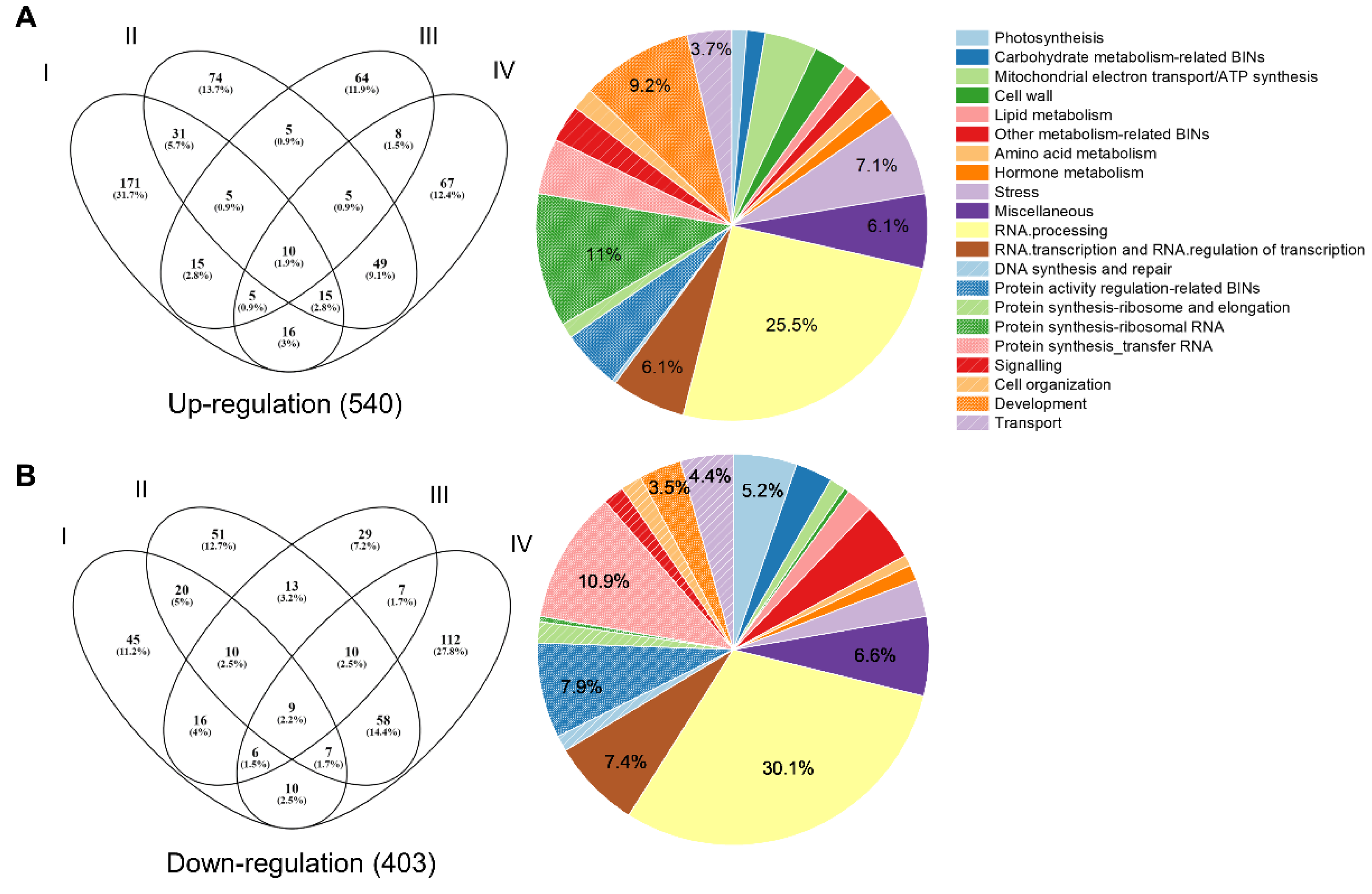
2.6. Identification of DEGs in Immature Seeds of 13 kDa Prolamin-Knockout Plants
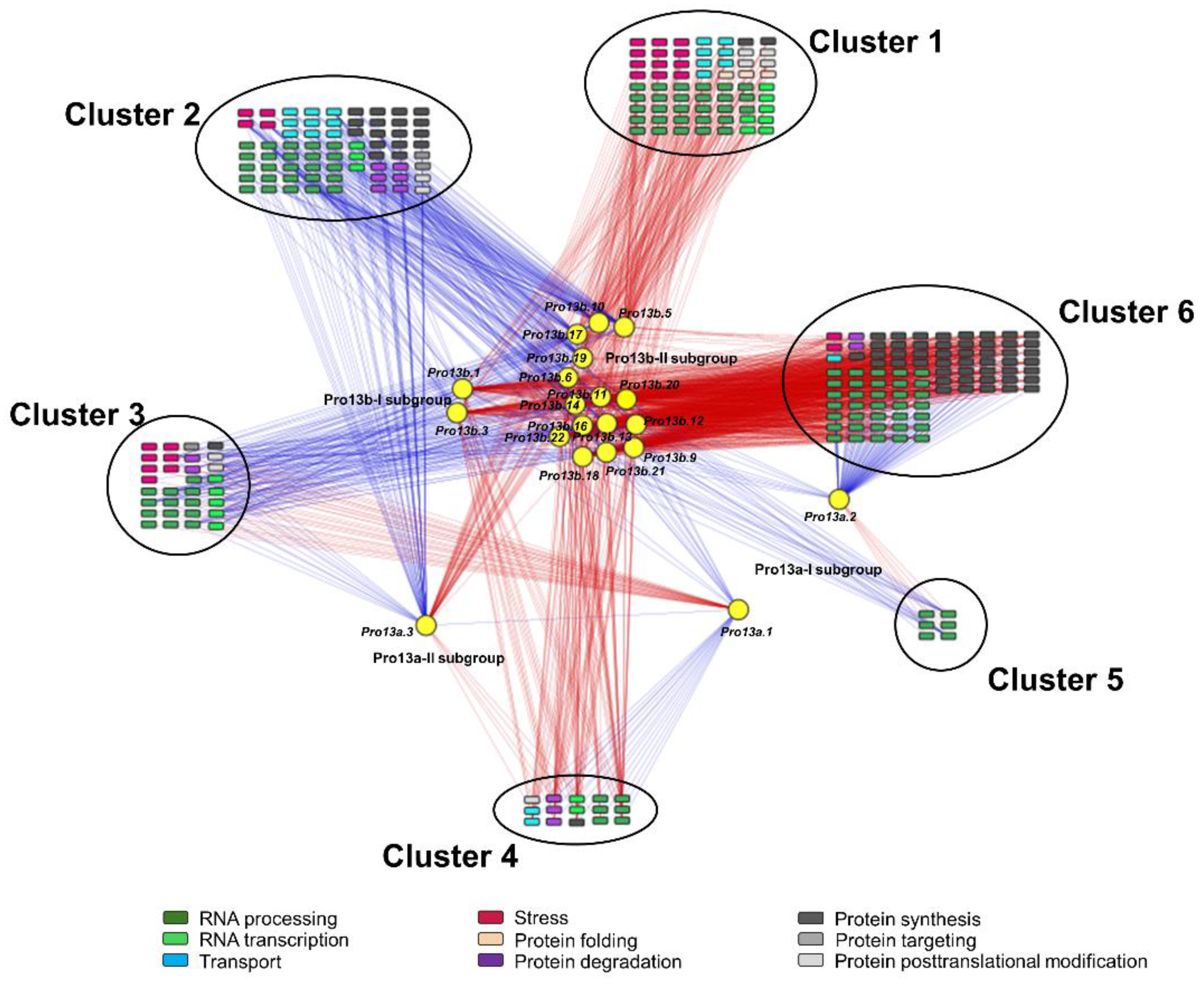
2.7. Correlation Network Involving DEGs and 13 kDa Prolamins
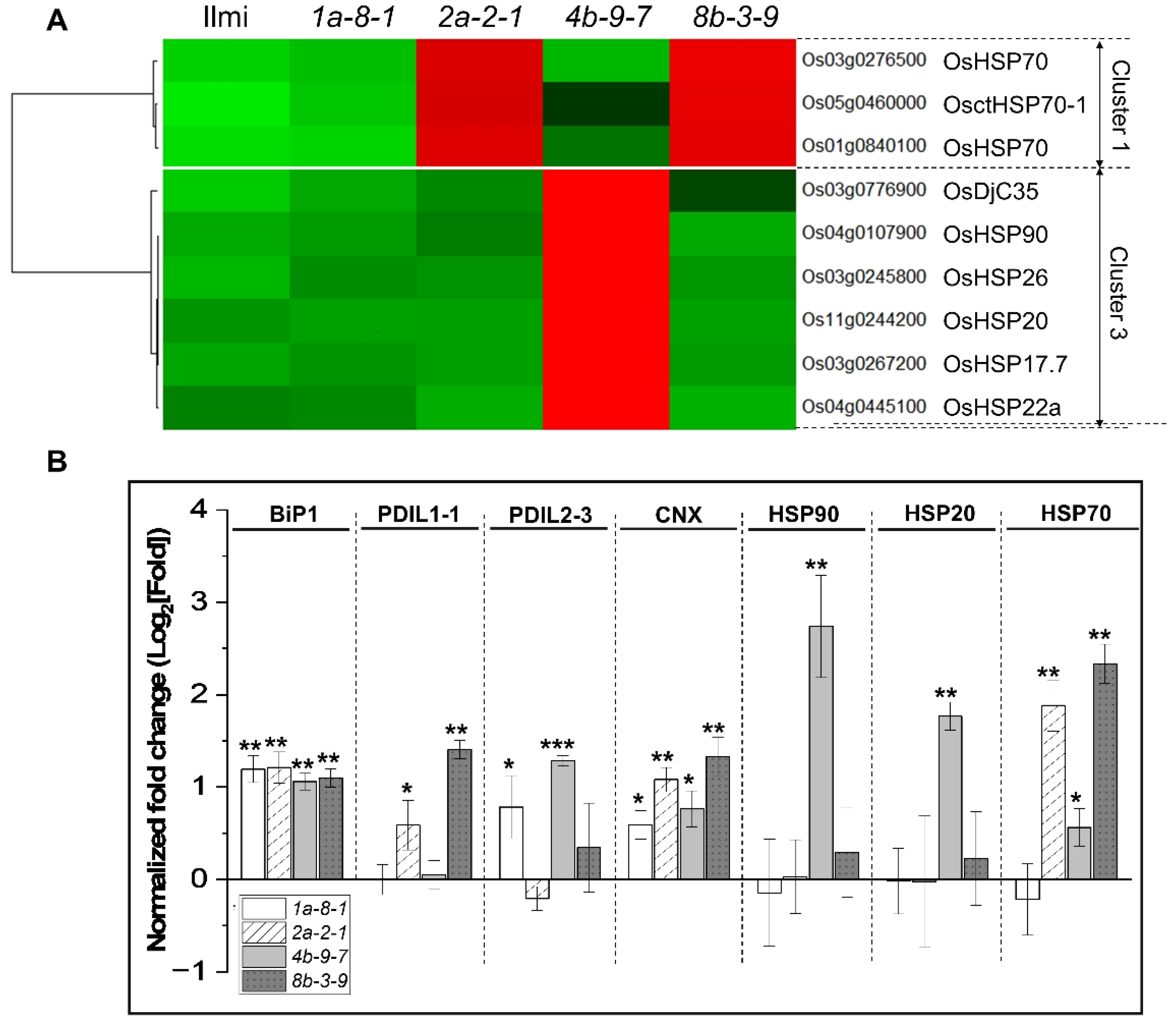
2.8. Analysis of Endoplasmic Reticulum Pathway Genes
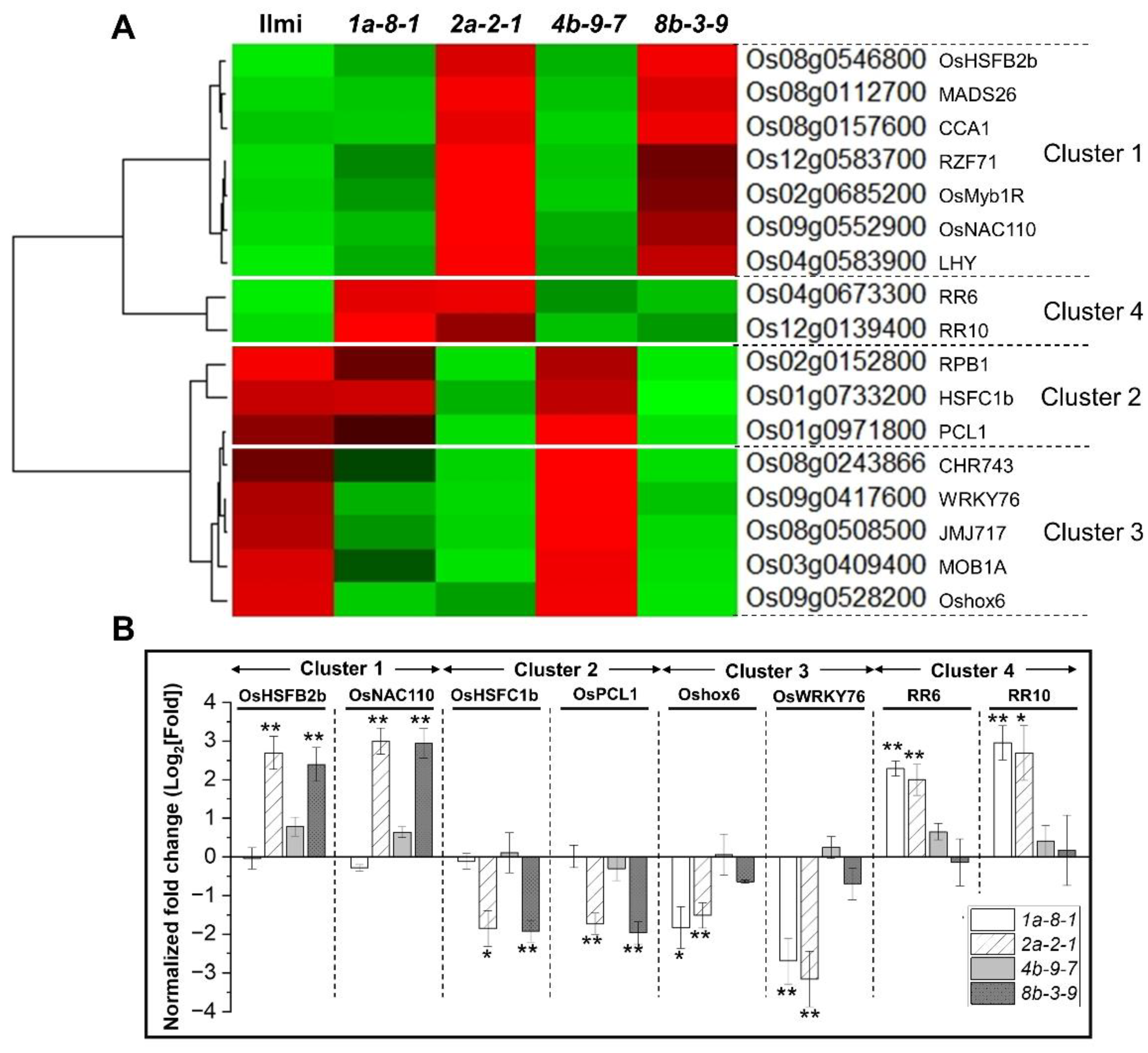
2.9. Role of Transcription Factors in Regulating 13 kDa Prolamin Correlation Network
3. Discussion
3.1. Morphological Differences between Starch Granules in WT and 13 kDa Prolamin-Knockout Rice Plants
3.2. Abnormal PB-1 Morphology in 13 kDa Prolamin-Knockout Plants
3.3. Functional Analysis of Genes Correlated with 13 kDa Prolamin Genes
4. Materials and Methods
4.1. Design of sgRNA
4.2. Cloning CRISPR-Cas9 Vector and Agrobacterium-Mediated Generation of 13 kDa Prolamin-Knockout Rice Plants
4.3. Screening Transgenic Rice Plants
4.4. RNA Isolation and qRT-PCR
4.5. Total Seed Storage Protein Extraction, SDS-PAGE, and Western Blot Analysis
4.6. SSP Fractionation
4.7. Measurement of Rice Agronomic Traits and Starch Content
4.8. Microscopic Analysis
4.9. RNA Sequencing
4.10. Transcriptome Analysis and Network Visualization
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bechtel DB, Juliano BO: Formation of Protein Bodies in the Starchy Endosperm of Rice (Oryza sativa L. ): A Re-investigation*. Annals of Botany 1980, 45, 503–509. [Google Scholar]
- Kim HJ, Lee JY, Yoon UH, Lim SH, Kim YM: Effects of reduced prolamin on seed storage protein composition and the nutritional quality of rice. Int J Mol Sci 2013, 14, 17073–17084. [CrossRef] [PubMed]
- Yamagata H, Tanaka K: The Site of Synthesis and Accumulation of Rice Storage Proteins. Plant and Cell Physiology 1986, 27, 135–145.
- Kawakatsu T, Hirose S, Yasuda H, Takaiwa F: Reducing rice seed storage protein accumulation leads to changes in nutrient quality and storage organelle formation. Plant Physiol 2010, 154, 1842–1854. [CrossRef] [PubMed]
- Krishnan HB, Okita TW: Structural Relationship among the Rice Glutelin Polypeptides. Plant Physiol 1986, 81, 748–753. [CrossRef]
- Xu JH, Messing J: Amplification of prolamin storage protein genes in different subfamilies of the Poaceae. Theor Appl Genet 2009, 119, 1397–1412. [CrossRef] [PubMed]
- Muench DG, Ogawa M, Okita TW: The Prolamins of Rice. In: Seed Proteins. Edited by Shewry PR, Casey R. Dordrecht: Springer Netherlands; 1999: 93-108.
- Saito Y, Shigemitsu T, Yamasaki R, Sasou A, Goto F, Kishida K, Kuroda M, Tanaka K, Morita S, Satoh S et al. : Formation mechanism of the internal structure of type I protein bodies in rice endosperm: Relationship between the localization of prolamin species and the expression of individual genes. Plant J 2012, 70, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Li X, Wu Y, Zhang DZ, Gillikin JW, Boston RS, Franceschi VR, Okita TW: Rice prolamine protein body biogenesis: A BiP-mediated process. Science 1993, 262, 1054–1056. [CrossRef] [PubMed]
- Tanaka K, Sugimoto T, Ogawa M, Kasai Z: Isolation and Characterization of Two Types of Protein Bodies in the Rice Endosperm. Agricultural and Biological Chemistry 1980, 44, 1633–1639.
- Sasou A, Shigemitsu T, Morita S, Masumura T: Accumulation of foreign polypeptides to rice seed protein body type I using prolamin portion sequences. Plant Cell Reports 2017, 36, 481–491. [CrossRef]
- Yamamoto MP, Onodera Y, Touno SM, Takaiwa F: Synergism between RPBF Dof and RISBZ1 bZIP Activators in the Regulation of Rice Seed Expression Genes. Plant Physiology 2006, 141, 1694–1707. [CrossRef] [PubMed]
- Kawakatsu T, Yamamoto MP, Touno SM, Yasuda H, Takaiwa F: Compensation and interaction between RISBZ1 and RPBF during grain filling in rice. Plant J 2009, 59, 908–920. [CrossRef] [PubMed]
- Wang J, Chen Z, Zhang Q, Meng S, Wei C: The NAC Transcription Factors OsNAC20 and OsNAC26 Regulate Starch and Storage Protein Synthesis. Plant Physiol 2020, 184, 1775–1791. [CrossRef]
- 15. Yang J, Zhou Y, Jiang Y: Amino Acids in Rice Grains and Their Regulation by Polyamines and Phytohormones. Plants (Basel).
- 16. Santos KF, Silveira RDD, Martin-Didonet CCG, Brondani C: Storage protein profile and amino acid content in wild rice Oryza glumaepatula. Pesquisa Agropecuária Brasileira.
- 17. Resurreccion AP, Li X, Okita TW, Juliano BO: Characterization of poorly digested protein of cooked rice protein bodies. Cereal chemistry (USA).
- 18. Wang Z, Li H, Liang M, Yang L: Glutelin and prolamin, different components of rice protein, exert differently in vitro antioxidant activities. Journal of Cereal Science.
- Cai J, Yang L, He H-J, Xu T, Liu H-B, Wu Q, Ma Y, Liu Q-H, Nie M-H: Antioxidant capacity responsible for a hypocholesterolemia is independent of dietary cholesterol in adult rats fed rice protein. Gene 2014, 533, 57–66. [CrossRef] [PubMed]
- Yang L, Chen J-H, Xu T, Zhou A-S, Yang H-K: Rice protein improves oxidative stress by regulating glutathione metabolism and attenuating oxidative damage to lipids and proteins in rats. Life Sciences 2012, 91, 389–394. [CrossRef]
- Kawakatsu T, Takaiwa F: Reduction of 13 kD prolamins increases recombinant protein yield and recovery rate in rice endosperm. Plant Signal Behav 2012, 7, 1402–1403. [CrossRef] [PubMed]
- Yang L, Hirose S, Takahashi H, Kawakatsu T, Takaiwa F: Recombinant protein yield in rice seed is enhanced by specific suppression of endogenous seed proteins at the same deposit site. Plant Biotechnology Journal 2012, 10, 1035–1045. [CrossRef]
- Cho K, Lee H-J, Jo Y-M, Lim S-H, Rakwal R, Lee J-Y, Kim Y-M: RNA Interference-Mediated Simultaneous Suppression of Seed Storage Proteins in Rice Grains. Frontiers in Plant Science 2016, 7.
- 24. Rowling PJ, Freedman RB: Folding, assembly, and posttranslational modification of proteins within the lumen of the endoplasmic reticulum. Subcell Biochem.
- Kaufman RJ: Stress signaling from the lumen of the endoplasmic reticulum: Coordination of gene transcriptional and translational controls. Genes Dev 1999, 13, 1211–1233. [CrossRef]
- Kleizen B, Braakman I: Protein folding and quality control in the endoplasmic reticulum. Curr Opin Cell Biol 2004, 16, 343–349. [CrossRef]
- Satoh-Cruz M, Crofts AJ, Takemoto-Kuno Y, Sugino A, Washida H, Crofts N, Okita TW, Ogawa M, Satoh H, Kumamaru T: Protein Disulfide Isomerase Like 1-1 Participates in the Maturation of Proglutelin Within the Endoplasmic Reticulum in Rice Endosperm. Plant and Cell Physiology 2010, 51, 1581–1593. [CrossRef] [PubMed]
- Muench DG, Wu Y, Zhang Y, Li X, Boston RS, Okita TW: Molecular cloning, expression and subcellular localization of a BiP homolog from rice endosperm tissue. Plant Cell Physiol 1997, 38, 404–412. [CrossRef] [PubMed]
- Wakasa Y, Yasuda H, Oono Y, Kawakatsu T, Hirose S, Takahashi H, Hayashi S, Yang L, Takaiwa F: Expression of ER quality control-related genes in response to changes in BiP1 levels in developing rice endosperm. Plant J 2011, 65, 675–689. [CrossRef] [PubMed]
- Tu BP, Weissman JS: Oxidative protein folding in eukaryotes: Mechanisms and consequences. J Cell Biol 2004, 164, 341–346. [CrossRef] [PubMed]
- Onda Y, Kawagoe Y: Oxidative protein folding: Selective pressure for prolamin evolution in rice. Plant Signal Behav 2011, 6, 1966–1972. [CrossRef] [PubMed]
- 32. Li J, Kong X, Ai Y: Modification of granular waxy, normal and high-amylose maize starches by maltogenic α-amylase to improve functionality. Carbohydrate Polymers.
- Lv X, Hong Y, Zhou Q, Jiang C: Structural Features and Digestibility of Corn Starch With Different Amylose Content. Frontiers in Nutrition 2021, 8.
- Nagamine A, Matsusaka H, Ushijima T, Kawagoe Y, Ogawa M, Okita TW, Kumamaru T: A role for the cysteine-rich 10 kDa prolamin in protein body I formation in rice. Plant Cell Physiol 2011, 52, 1003–1016. [CrossRef] [PubMed]
- Saito Y, Shigemitsu T, Yamasaki R, Sasou A, Goto F, Kishida K, Kuroda M, Tanaka K, Morita S, Satoh S et al. : Formation mechanism of the internal structure of type I protein bodies in rice endosperm: Relationship between the localization of prolamin species and the expression of individual genes. The Plant Journal 2012, 70, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- 36. Mitsukawa N, Konishi R, Kidzu K, Ohtsuki K, Masumura T, Tanaka K: Amino Acid Sequencing and cDNA Cloning of Rice Seed Storage Proteins, the 13kDa Prolamins, Extracted from Type I Protein Bodies. Plant Biotechnology.
- 37. Momma M, Saito M, Chikuni K, Saio K: Ultrastructure of prolamin accumulating protein bodies in endosperm of mutant rice for storage proteins (ultrastructure of protein bodies in mutant rice). Journal of The Japanese Society for Food Science and Technology-nippon Shokuhin Kagaku Kogaku Kaishi.
- Ogawa M, Kumamaru T, Satoh H, Iwata N, Omura T, Kasai Z, Tanaka K: Purification of Protein Body-I of Rice Seed and its Polypeptide Composition. Plant and Cell Physiology 1987, 28, 1517–1527.
- Qian D, Chen G, Tian L, Qu LQ: OsDER1 Is an ER-Associated Protein Degradation Factor That Responds to ER Stress. Plant Physiol 2018, 178, 402–412. [CrossRef]
- Kim Y-M, Lee J-Y, Lee T, Lee Y-H, Kim S-H, Kang S-H, Yoon U-H, Ha S-H, Lim S-H: The suppression of the glutelin storage protein gene in transgenic rice seeds results in a higher yield of recombinant protein. Plant Biotechnology Reports 2012, 6, 347–353. [CrossRef]
- He W, Wang L, Lin Q, Yu F: Rice seed storage proteins: Biosynthetic pathways and the effects of environmental factors. J Integr Plant Biol 2021, 63, 1999–2019. [CrossRef] [PubMed]
- Ohta M, Takaiwa F: OsHrd3 is necessary for maintaining the quality of endoplasmic reticulum-derived protein bodies in rice endosperm. J Exp Bot 2015, 66, 4585–4593. [CrossRef] [PubMed]
- Hu C, Yang J, Qi Z, Wu H, Wang B, Zou F, Mei H, Liu J, Wang W, Liu Q: Heat shock proteins: Biological functions, pathological roles, and therapeutic opportunities. MedComm (2020) 2022, 3, e161.
- Chaudhury S, Keegan BM, Blagg BSJ: The role and therapeutic potential of Hsp90, Hsp70, and smaller heat shock proteins in peripheral and central neuropathies. Med Res Rev 2021, 41, 202–222. [CrossRef] [PubMed]
- Chen S, Smith DF: Hop as an Adaptor in the Heat Shock Protein 70 (Hsp70) and Hsp90 Chaperone Machinery*. Journal of Biological Chemistry 1998, 273, 35194–35200. [CrossRef] [PubMed]
- Taipale M, Jarosz DF, Lindquist S: HSP90 at the hub of protein homeostasis: Emerging mechanistic insights. Nature Reviews Molecular Cell Biology 2010, 11, 515–528. [CrossRef] [PubMed]
- Pratt WB, Toft DO: Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp Biol Med (Maywood) 2003, 228, 111–133. [CrossRef] [PubMed]
- Park CJ, Seo YS: Heat Shock Proteins: A Review of the Molecular Chaperones for Plant Immunity. Plant Pathol J 2015, 31, 323–333. [CrossRef]
- Pobre KFR, Poet GJ, Hendershot LM: The endoplasmic reticulum (ER) chaperone BiP is a master regulator of ER functions: Getting by with a little help from ERdj friends. J Biol Chem 2019, 294, 2098–2108. [CrossRef]
- Mayer MP: Hsp70 chaperone dynamics and molecular mechanism. Trends Biochem Sci 2013, 38, 507–514. [CrossRef] [PubMed]
- Stolz A, Wolf DH: Endoplasmic reticulum associated protein degradation: A chaperone assisted journey to hell. Biochim Biophys Acta 2010, 1803, 694–705. [CrossRef] [PubMed]
- Chandra D, Cho K, Pham HA, Lee J-Y, Han O: Down-Regulation of Rice Glutelin by CRISPR-Cas9 Gene Editing Decreases Carbohydrate Content and Grain Weight and Modulates Synthesis of Seed Storage Proteins during Seed Maturation. International Journal of Molecular Sciences 2023, 24, 16941. [CrossRef] [PubMed]
- 53. Howell SH: Endoplasmic reticulum stress responses in plants. Annu Rev Plant Biol.
- Cao L, Liu P, Chen J, Deng L: Prediction of Transcription Factor Binding Sites Using a Combined Deep Learning Approach. Frontiers in Oncology 2022, 12.
- Li C, Li Y, Zhou Z, Huang Y, Tu Z, Zhuo X, Tian D, Liu Y, Di H, Lin Z et al. : Genome-wide identification and comprehensive analysis heat shock transcription factor (Hsf) members in asparagus (Asparagus officinalis) at the seeding stage under abiotic stresses. Scientific Reports 2023, 13, 18103. [Google Scholar] [CrossRef] [PubMed]
- Xiang J, Ran J, Zou J, Zhou X, Liu A, Zhang X, Peng Y, Tang N, Luo G, Chen X: Heat shock factor OsHsfB2b negatively regulates drought and salt tolerance in rice. Plant Cell Reports 2013, 32, 1795–1806. [CrossRef] [PubMed]
- Ma L, Wang C, Hu Y, Dai W, Liang Z, Zou C, Pan G, Lübberstedt T, Shen Y: GWAS and transcriptome analysis reveal MADS26 involved in seed germination ability in maize. Theoretical and Applied Genetics 2022, 135, 1717–1730. [CrossRef]
- Khong GN, Pati PK, Richaud F, Parizot B, Bidzinski P, Mai CD, Bès M, Bourrié I, Meynard D, Beeckman T et al. : OsMADS26 Negatively Regulates Resistance to Pathogens and Drought Tolerance in Rice. Plant Physiol 2015, 169, 2935–2949. [Google Scholar]
- Liang T, Yu S, Pan Y, Wang J, Kay SA: The interplay between the circadian clock and abiotic stress responses mediated by ABF3 and CCA1/LHY. Proc Natl Acad Sci U S A 2024, 121, e2316825121. [CrossRef]
- Xu DQ, Huang J, Guo SQ, Yang X, Bao YM, Tang HJ, Zhang HS: Overexpression of a TFIIIA-type zinc finger protein gene ZFP252 enhances drought and salt tolerance in rice (Oryza sativa L. ). FEBS Lett 2008, 582, 1037–1043. [Google Scholar]
- Xie L-n, Chen M, Min D-h, Feng L, Xu Z-s, Zhou Y-b, Xu D-b, Li L-c, Ma Y-z, Zhang X-h: The NAC-like transcription factor SiNAC110 in foxtail millet (Setaria italica L. ) confers tolerance to drought and high salt stress through an ABA independent signaling pathway. Journal of Integrative Agriculture 2017, 16, 559–571. [Google Scholar] [CrossRef]
- 62. Huang P, Ding Z, Duan M, Xiong Y, Li X, Yuan X, Huang J: OsLUX Confers Rice Cold Tolerance as a Positive Regulatory Factor. Int J Mol Sci.
- 63. Schmidt R, Schippers JH, Welker A, Mieulet D, Guiderdoni E, Mueller-Roeber B: Transcription factor OsHsfC1b regulates salt tolerance and development in Oryza sativa ssp. japonica. AoB Plants.
- Yokotani N, Sato Y, Tanabe S, Chujo T, Shimizu T, Okada K, Yamane H, Shimono M, Sugano S, Takatsuji H et al. : WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J Exp Bot 2013, 64, 5085–5097. [Google Scholar] [CrossRef] [PubMed]
- Maeda N, Matsuta F, Noguchi T, Fujii A, Ishida H, Kitagawa Y, Ishikawa A: The Homeodomain–Leucine Zipper Subfamily I Contributes to Leaf Age- and Time-Dependent Resistance to Pathogens in Arabidopsis thaliana. In: International Journal of Molecular Sciences. vol. 24; 2023.
- Hou Y, Wang L, Wang L, Liu L, Li L, Sun L, Rao Q, Zhang J, Huang S: JMJ704 positively regulates rice defense response against Xanthomonas oryzae pv. oryzae infection via reducing H3K4me2/3 associated with negative disease resistance regulators. BMC Plant Biology 2015, 15, 286. [Google Scholar]
- Wang W-C, Lin T-C, Kieber J, Tsai Y-C: Response Regulators 9 and 10 Negatively Regulate Salinity Tolerance in Rice. Plant and Cell Physiology 2019, 60, 2549–2563. [CrossRef]
- 68. Zhou J, Li D, Zheng C, Xu R, Zheng E, Yang Y, Chen Y, Yu C, Yan C, Chen J et al.: Targeted Transgene Expression in Rice Using a Callus Strong Promoter for Selectable Marker Gene Control. Frontiers in Plant Science.
- Hiei Y, Ohta S, Komari T, Kumashiro T: Efficient transformation of rice (Oryza sativa L. ) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. The Plant Journal 1994, 6, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Dellaporta SL, Wood J, Hicks JB: A plant DNA minipreparation: Version II. Plant Molecular Biology Reporter 1983, 1, 19–21. [CrossRef]
- Li Z, Trick HN: Rapid method for high-quality RNA isolation from seed endosperm containing high levels of starch. BioTechniques 2005, 38, 872–876. [CrossRef] [PubMed]
- Rao X, Huang X, Zhou Z, Lin X: An improvement of the 2ˆ(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat Bioinforma Biomath 2013, 3, 71–85.
- Cho K, Han Y, Woo JC, Baudisch B, Klösgen RB, Oh S, Han J, Han O: Cellular localization of dual positional specific maize lipoxygenase-1 in transgenic rice and calcium-mediated membrane association. Plant Science 2011, 181, 242–248. [CrossRef]
- Lee H-J, Jo Y-M, Lee J-Y, Lim S-H, Kim Y-M: Lack of Globulin Synthesis during Seed Development Alters Accumulation of Seed Storage Proteins in Rice. International Journal of Molecular Sciences 2015, 16, 14717–14736. [CrossRef]
- Bray NL, Pimentel H, Melsted P, Pachter L: Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 2016, 34, 525–527. [CrossRef] [PubMed]
- Li B, Dewey CN: RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011, 12, 323.
- McCarthy DJ, Chen Y, Smyth GK: Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 2012, 40, 4288–4297. [CrossRef] [PubMed]
- Love MI, Huber W, Anders S: Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 2014, 15, 550.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).




