1. Introduction
The immune response of teleost fish represents a crucial evolutionary stage for the development of defences against pathogens in biological systems [
1,
2]. Indeed, the study of the cellular and molecular components involved in such responses using fish models has provided important insights into the evolution of the immune system in higher vertebrates [
3,
4,
5]. One of these molecular components is the tripartite motif-containing (TRIM) protein family, a group with over 70 members in mammals that is associated with several biological processes, including apoptosis, oncogenesis, cellular proliferation, differentiation, development, and innate immunity [
6,
7]. TRIM proteins consist of an N-terminal RING finger domain, followed by one or two B-box zinc finger domain and a coiled-coil region, which is collectively known as the RBCC motif [
8]. The RBCC motif is fairly conserved among the members of the TRIM superfamily, with more evident differences and specific functions linked to their less conserved C-terminal domain [
9].
Accumulating evidence suggests that TRIM proteins have an important role in the innate immune response [
10,
11], with a crucial impact in the modulation of particular pro-inflammatory components [
12], such as regulation of antiviral restriction [
13], cellular autophagy [
14], and inflammasome activation [
15]. For this reason, TRIM proteins could also be considered as intracellular cytokines, and deciphering further information about their function and evolution in the immune system may provide important insights into the molecular mechanisms that govern host defence.
Over the last decade, greatest advancements have been made in the discovery and characterisation of TRIM proteins involved in innate immunity in fish models [
16,
17]. Indeed, a number of TRIM members found in mammals have already been confirmed to be expressed as orthologues in different fish, including for example TRIM32 and TRIM47 in carp (
Cyprinus carpio) [
18,
19], and TRIM69 in zebrafish (
Danio rerio) [
20]. On the other hand, a large new subset of TRIM genes has been specifically identified in rainbow trout (
Oncorhynchus mykiss) and zebrafish as virus-induced transcripts [
21]. These TRIM-like proteins exclusively identified in the fish were called fish novel TRIMs (finTRIMs). Nevertheless, although increasing data support the immunomodulatory potential of fish TRIMs against microbial infections [
17,
22], the molecular determinants associated with their antimicrobial mechanisms are still being investigated in cellular and animal models.
Therefore, the purpose of the present study was to explore and characterise the presence of novel TRIM proteins involved in innate immunity in rainbow trout using in vitro and in vivo approaches. In particular, we identified and sequenced novel TRIM transcripts in the rainbow trout gill cell line RTgill-W1. In addition, their expression patterns were examined both in RTgill-W1 cells and rainbow trout primary gill cell cultures following a stimulation with lipopolysaccharide (LPS), a cell wall component of Gram negative bacteria, and polyinosinic:polycytidylic acid (poly(I:C)), a molecule structurally similar to viral double-stranded RNA (dsRNA).
Furthermore, we examined the time-dependent expression of these TRIM transcripts in rainbow trout gills after the challenge with the salmonid pathogen Flavobacterium psychrophilum. We correlated the expression of these TRIM transcripts with the level of pro-inflammatory cytokines TNF-α2 and IL-1β, as well as the immunomodulatory protein type I IFN in these cellular and animal models. Finally, we determined that one of the TRIM-like proteins found in our study is critically necessary to trigger the expression of LPS-induced pro-inflammatory cytokines, as revealed in a post-transcriptional knock down assay. These data provide fundamental insights into the implications of TRIM proteins in the antimicrobial defence of the fish, and shed light on the potential conserved mechanisms that these intracellular cytokines may modulate the innate immune response in higher vertebrates.
2. Results
2.1. Novel TRIM-like Sequences Identified in RTgill-W1 Cells
Considering the specific finTRIMs published by van der Aa et al (2009), we were able to isolate the sequences corresponding to finTRIM1 and finTRIM2. However, we did not detect the expression of finTRIM3 under the PCR conditions utilised in RTgill-W1 cells.
Using the non-specific PCR amplification method, we isolated and sequenced two TRIM-like transcripts corresponding to human orthologues, named OmTRIM25 and OmTRIM16. Finally, the EST contig sequence search allowed us to identify other two TRIM orthologues, named OmTRIM8 and OmTRIM62. All the novel transcripts found (not including the finTRIMs already published) were submitted and registered in the Nucleotide NCBI database as RTgill-W1 novel TRIM-like mRNA sequences (
Table 1).
A multiple alignment of the predicted amino acidic sequences shows high conservation in the N-terminus regions (
Figure 1) along all OmTRIMs and finTRIMs found. In addition, a search in the Conserved Domains Database (NCBI) revealed that all the sequences display the presence of the main domains composing the RBCC motif –the structural signature of the TRIM superfamily–, including the RING, B-box and coiled-coil.
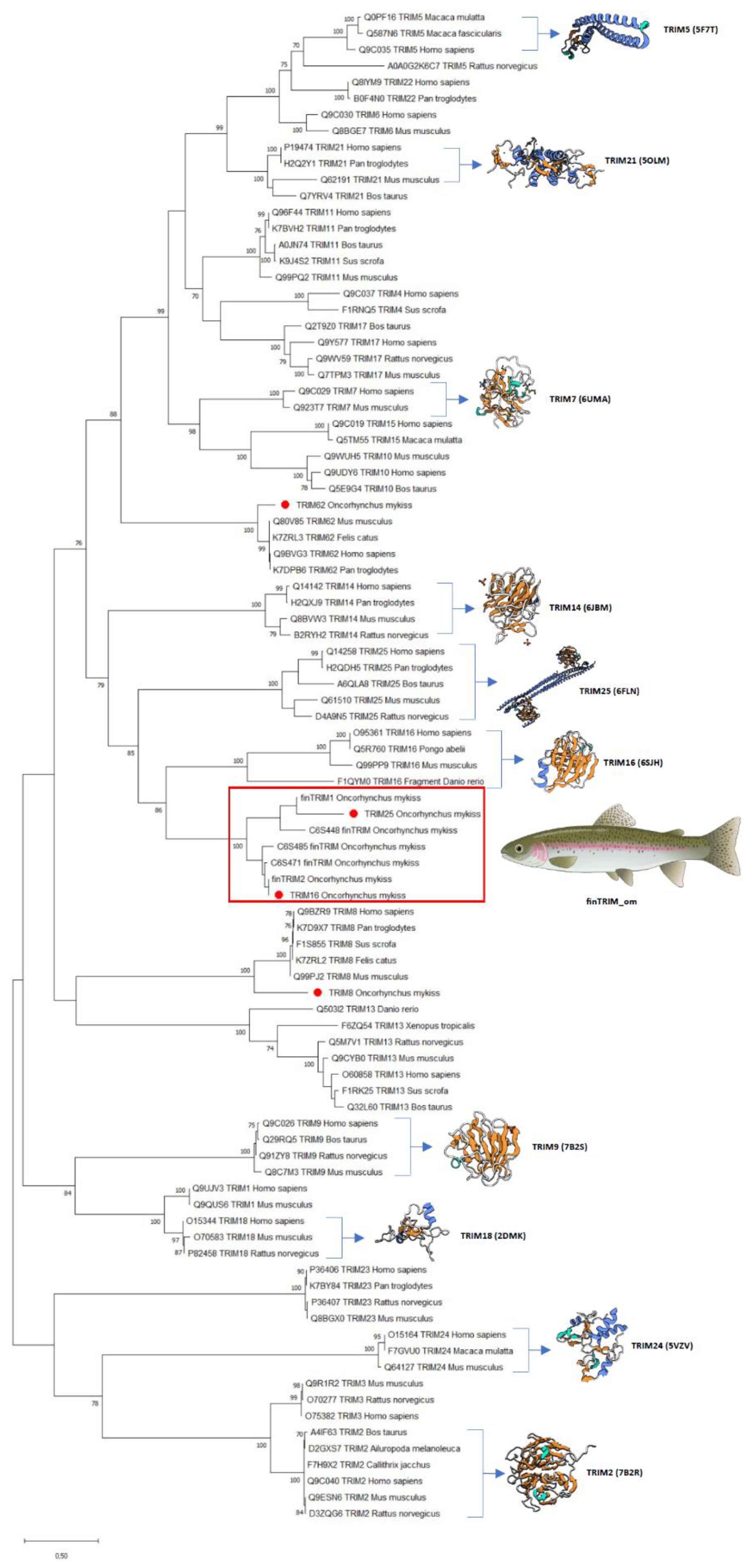
After a phylogenetic analysis, data showed that OmTRIM8 and OmTRIM62 are grouped closely to other homologue members, while OmTRIM16 and OmTRIM25 fit better within the finTRIM family (
Figure 2; see red box).
2.2. Differential Expression of Novel TRIM-like Genes upon Stimulation by MAMPs
To evaluate the capacity of TRIM genes to respond in an immune-related context, RTgill-W1 cells and primary gill cultures were stimulated with the microbe-associated molecular patterns (MAMPs) LPS or poly(I:C).
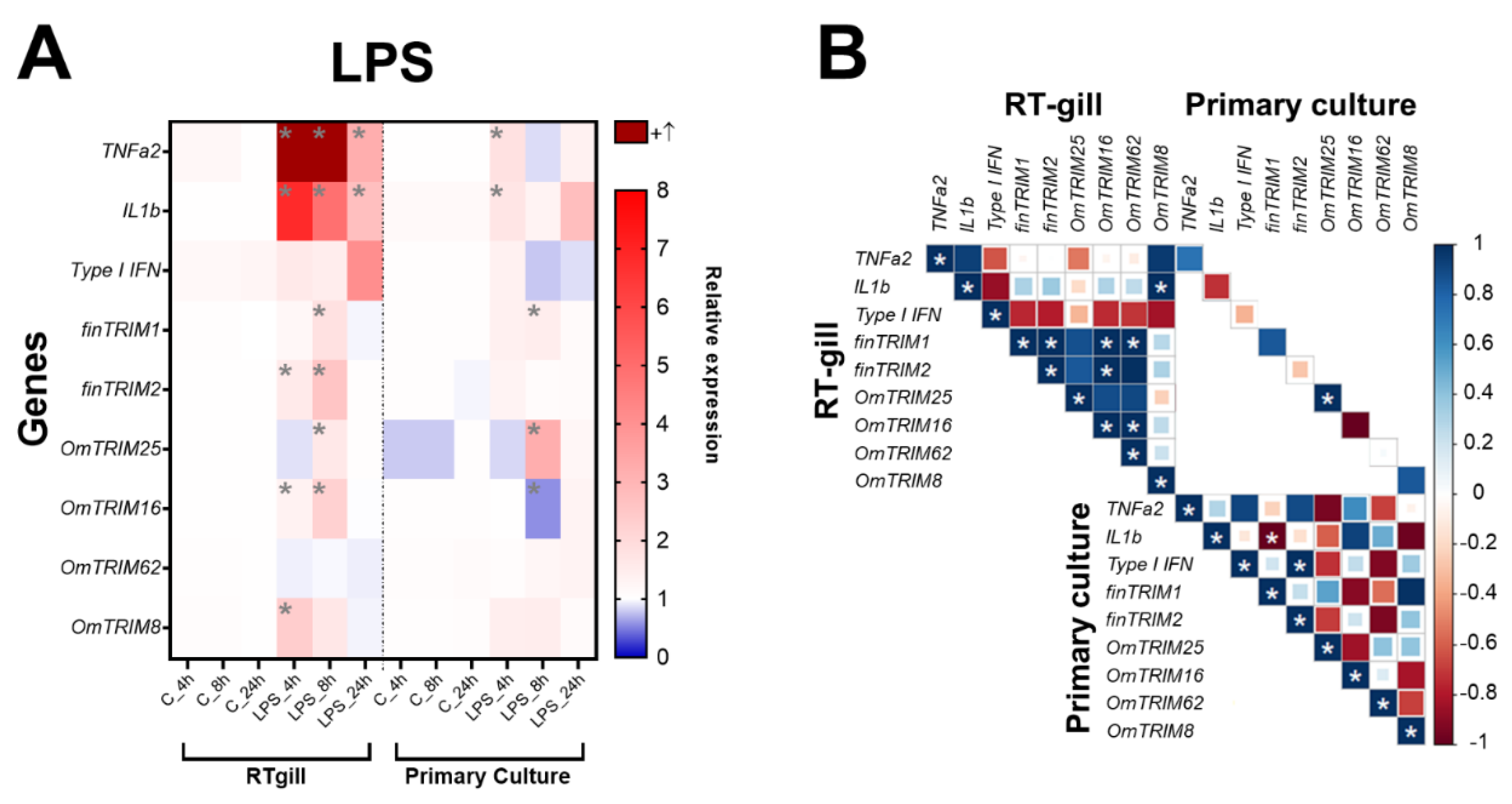
LPS was able to significantly induce the expression of TNF-α2 and IL-1β at 4; 8 and 24 hours post-stimulation (
Figure 3A). Most of TRIM-like genes were overexpressed at 8 hours of LPS treatment with the exception of OmTRIM8 and OmTRIM62. Moreover, OmTRIM8 was specifically induced at 4 hours only. Interestingly, the expression of some TRIM transcripts positively correlated between them, i.e. finTRIM 1 correlated with finTRIM 2, OmTRIM16 and OmTRIM62. In addition, finTRIM 2 significantly positive correlated with OmTRIM16, and OmTRIM16 with OmTRIM62 (
Figure 3B). Nevertheless, OmTRIM25 was the only one to correlate its own expression between RTgill-W1 cells and primary gill cultures.
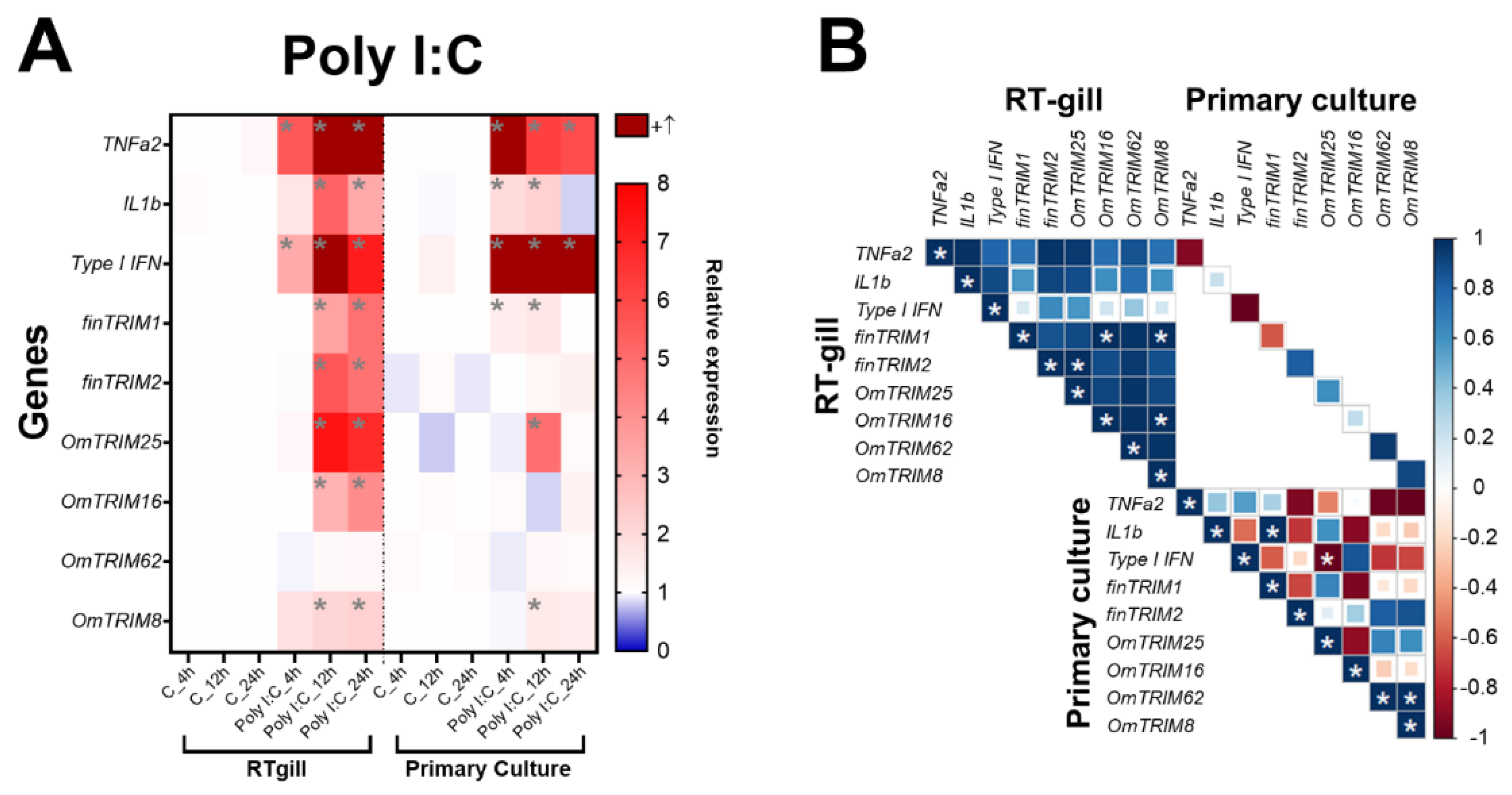
In contrast, poly(I:C) caused a significant modulation in the gene expression of TNF-α2 and type I IFN at all time points, while IL-1β was induced at 12 and 24 hours only (
Figure 4A). In comparison, the stimulus with poly(I:C) caused a higher expression of TRIM-like transcripts than LPS, but in the later time points 12 and 24 hours. Notably, OmTRIM16 and OmTRIM62 were not induced at any time point of stimulation with either LPS or poly(I:C). Analysis of correlation indicate that almost all the molecules induced by poly(I:C) are positively correlated, but only few comparisons reached significance, including finTRIM 1 with OmTRIM16 and OmTRIM8, and OmTRIM16 with OmTRIM8 (
Figure 4B).
In the case of primary cultures, the poly(I:C) stimulation was significantly more prone to induce a immune response than LPS. Indeed, poly(I:C) produced a significant overexpression of type I IFN and TNF-α2 in every time point examined, while LPS only induce IL-1β after 24 hours of stimulation (
Figure 3A). In terms of TRIM expression, both LPS and poly(I:C) induced the expression of OmTRIM25 only at 8 and 12 hours of stimulation respectively (
Figure 3A and 4A). Only few correlations were detected in primary cultures. IL-1β negatively correlated with finTRIM 1 under LPS stimulation, while poly(I:C) treatment produced the opposite effect, a positive correlation between IL-1β and finTRIM1 (
Figure 3B and 4B).
2.3. Rainbow Trout Infected with a Bacterial Pathogen Showed a Time-Depending Expression of TRIMs in Gill Tissue
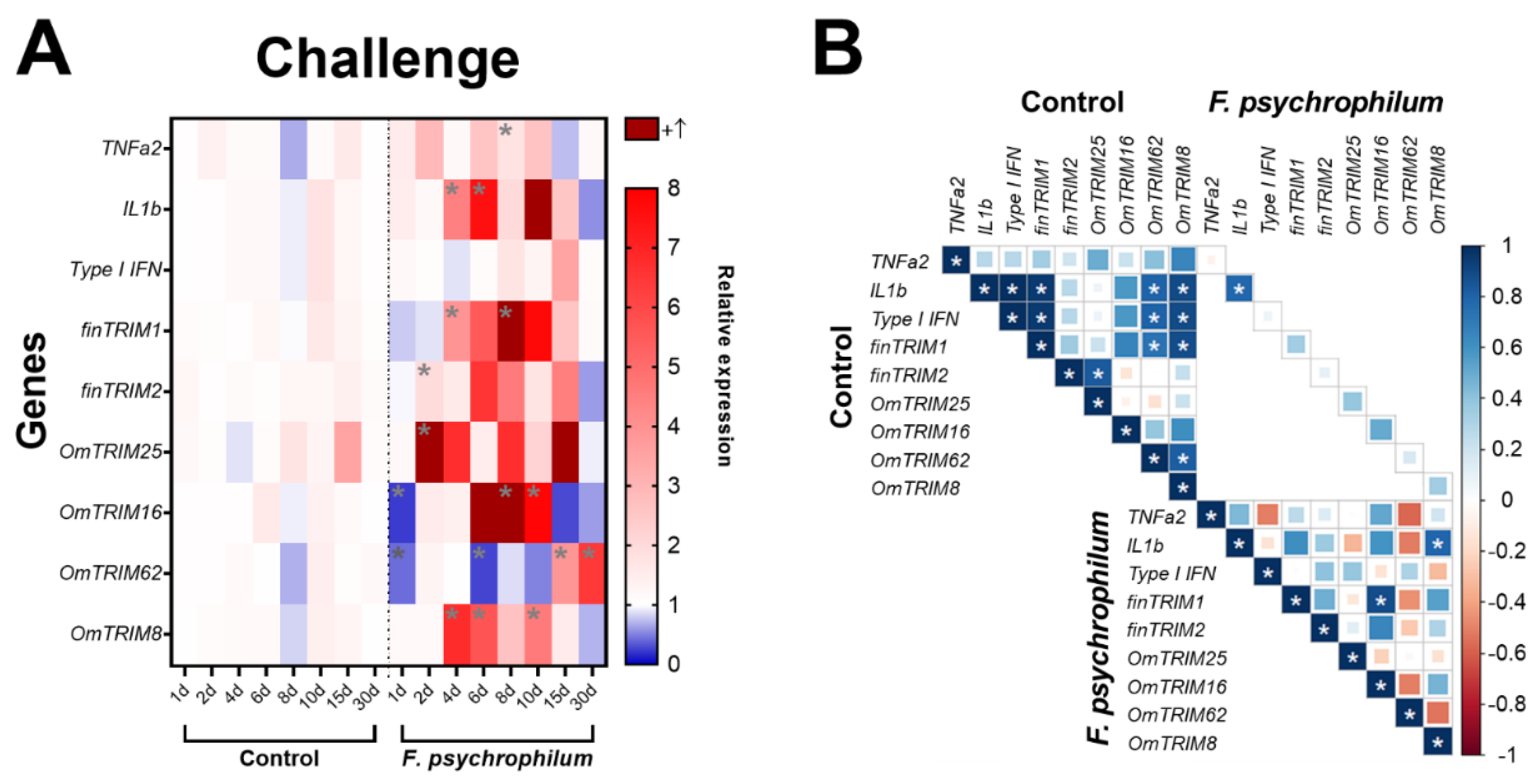
To explore the expression patterns of TRIM proteins at in vivo level, we measured the levels of TRIM transcripts in gills of rainbow trouts infected with the fish freshwater pathogen F. psychrophilum at different time points during 30 days of infection. The analysis of cytokines expression revealed that this bacterium significantly induced the expression of TNF-α2 at day 8, while IL-1β was at days 4 and 6 of infection. However, the bacterial challenge did not alter the basal levels of type I IFN (
Figure 5A). Interestingly, TRIM transcripts showed different patterns of expression. For instance, OmTRIM25 and finTRIM2 were significantly up-regulated earlier (after 2 days of infection), and then finTRIM 1 and OmTRIM8 showed peaks of expression from the 4th day post infection. Moreover, OmTRIM16 significantly increased its expression after 8 days, and this increase was maintained later as well at day 10.
Finally, OmTRIM62 presented their expression peaks in the last days of analysis at 15 and 30 days of infection (
Figure 5A). Although basal levels of gene expression are positively correlated between different molecules, we detected only a significant correlation between IL-1β and OmTRIM8, and between finTRIM 1 and OmTRIM16 (
Figure 5B).
2.4. OmTRIM25 Is Required to Trigger the Expression of TNF-α2 and IL-1β in RTgill-W1 Cells during LPS Stimulation
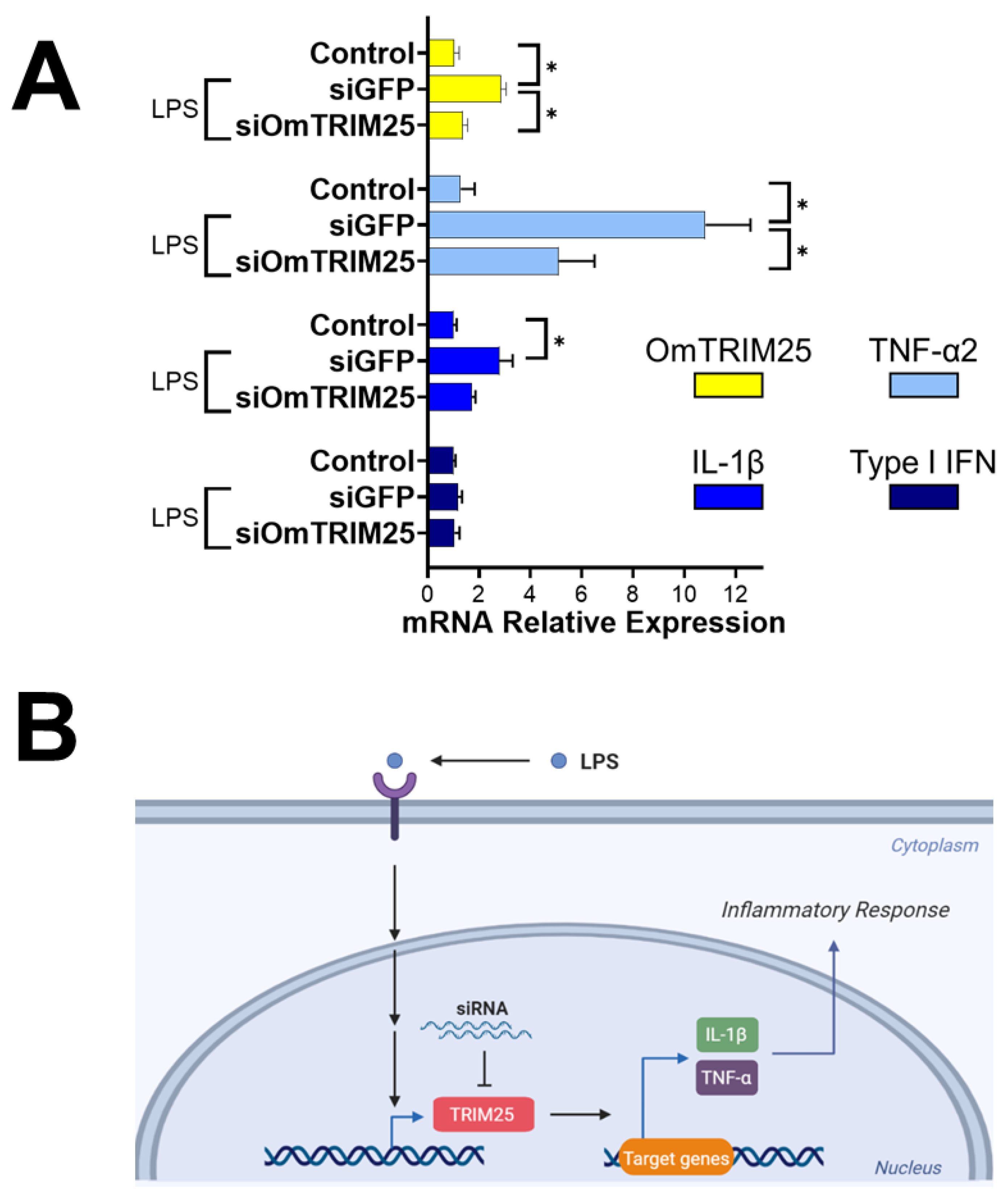
To investigate the potential role of OmTRIM25 in modulating the immune response of RTgill-W1 cells during LPS stimulation, we blocked its transcription using siRNAs. After 8-hour treatment with LPS, cells pre-incubated with siGFP displayed a significant increase of OmTRIM25, TNF-α2 and IL-1β mRNA levels, but not for type I IFN (
Figure 6) accordingly to our previous results (
Figure 3A). Cells pre-incubated with siOmTRIM25 showed adown-regulation in the expression of OmTRIM25; and moreover, the expression of TNF-α2 and IL-1β was significantly attenuated.
3. Discussion
There has been increasing attention given to the fish immunology to understand the molecular and cellular evolution of host defence systems in higher vertebrates [
2,
4]. In the present study, we characterised novel transcripts of TRIM proteins in rainbow trout involved in immune response of the gills using
in vitro and
in vivo models. Fish gills are a specialized tissue with respiratory function, and for this reason it is an organ in continuous contact with the external environment and potential pathogens. Thus, gills represent an important focus of attention for the study of the immune response since it also has mucosa associated lymphoid tissues [
23]. Indeed, we isolated and identified novel human TRIM orthologues and already described finTRIMs (OmTRIM8, OmTRIM16, OmTRIM25, OmTRIM62, finTRIM 1, and finTRIM 2), as well as their expression patterns in the gill cell line RTgill-W1, rainbow trout primary gill cultures and in gills from infected rainbow trout. In addition, we demonstrated that OmTRIM25 has a crucial role in the regulation of the molecular immunology in the fish by modulating the expression of TNF-α2 and IL-1β. Thus, our findings suggest the existence of novel TRIM orthologues in rainbow trout with immunomodulatory capacity and highlight the potential of fish models to elucidate the molecular signalling underpinning the conserved defence systems across lower and higher vertebrates.
Notably, this is the first study where it has been described at transcriptional level the presence of those TRIM orthologues in rainbow trout, and the first one to demonstrate that finTRIM 1 and finTRIM 2 can be expressed in gills apart from immune-specialized tissue [
21]. It is remarkable that some of the TRIM orthologues found in this study have been previously linked to the immune system in other organisms, including TRIM8 [
24,
25]; TRIM25 [
26,
27]; and TRIM62 [
28]. Indeed, we detected that all TRIM and finTRIMs, but TRIM62, are overexpressed after the challenge with either LPS or poly(I:C) in RTgill-W1 cells and primary gill cultures along with classical cytokines, such as TNF-α2, IL-1β and type I IFN, which suggests that these TRIM members may be involved in the local immune response of the gill. Nevertheless, the expression levels of these TRIMs were significantly higher in cells treated with poly(I:C) than LPS, indicating a greater sensitivity for viral infection. Accordingly, accumulating evidence suggest that several TRIM members have been proven to possess anti-viral mechanisms, including inhibition of distinct steps in the viral life cycle [
13], modulation of the retinoic acid-inducible gene-I (RIG-I) [
29], and regulation of the signalling cascade after TLR3 activation [
30]. Although we found that some TRIM proteins correlate between them at gene expression levels, it remains unclear whether this relationship is mutually dependent, synergic or independent in terms of signalling and mechanism. Future studies are warranted to address potential interactions between TRIM members in immune contexts.
We further investigated the immune reactivity of these TRIMs by exploring their gene expression in gills from infected rainbow trout with
F. psychrophilum, a Gram-negative bacterial rod responsible of rainbow trout fry syndrome and bacterial cold water disease and rainbow trout fry syndrome [
31]. Indeed, we detected overexpressed TRIMs at different time points after the challenge. A plethora of pro- and anti-inflammatory molecules are elevated during an infection, reaching peaks of expression at distinct stages depending of their function [
32,
33], and TRIM proteins with immunological activity may follow this assumption. In this experiment, OmTRIM25 and finTRIM2 were significantly elevated after only two days of infection, while other classic cytokines like IL-1β and TNF-α2 reached their peaks of expression at four- and six-days post-infection, respectively. This result shed light into a potential involvement of OmTRIM25 and finTRIM2 in the regulation of bacterial pattern recognition with direct effects on triggering the expression of IL-1β and TNF-α2, and even for other TRIMs which we found expressed at later time points. This hypothesis can be supported by previous literature pointing out the role of TRIM proteins in modulating immune signalling pathways [
34]. On the other hand, we detected expression peaks at the very end of the experiment for OmTRIM62 (15- and 30-day post infection), suggesting potential implications for adaptive and long-term immunity. Accordingly, the role of TRIM proteins in adaptive immunity has been noted to be crucial for T cell differentiation and production of immunomodulatory cytokines [
35,
36]; however, the capacity of TRIM62 to regulate adaptive immunity in higher vertebrates has not been confirmed yet.
Considering the high correlation of the induction of OmTRIM25 found in RTgill-W1 cells and primary gill cultures after the challenge with LPS, and the early expression of this TRIM in gills from infected rainbow trout with
F. psychrophilum, we decided to examine more profoundly into its potential interactions with immune signalling linked to bacterial defense. Thus, we silenced the expression of OmTRIM25 at transcriptional level by using siRNA in RTgill-W1 cells stimulated with LPS. We found a significant decrease in the mRNA levels of OmTRIM25 confirming the efficacy of the gene
knock down. Moreover, we detected that blocking OmTRIM25 caused a significant reduction in the expression of TNF-α2 and IL-1β as well, implying a modulatory function of OmTRIM25 over the expression of these cytokines after an LPS stimulation and potential bacterial infection. The precise mechanism of how this could happen was not investigated in this study, but open interesting molecular options for future studies. TRIM proteins are known to possess E3 ubiquitin ligase activity [
37], and such activity has been demonstrated for TRIM25 [
29,
38]. For instance, it was reported that TRIM7 is able to promote the activation of TLR4, a specific receptor for LPS recognition, through its E3 ubiquitin ligase activity [
39]. Other proteins can regulate IκB levels and modulate the activation of the nuclear factor NF-κB using the same enzymatic activity [
40,
41]. Additional experiments are necessary to prove whether OmTRIM25 utilises some of the aforementioned mechanisms to modulate the expression of TNF-α2 and IL-1β
in vitro.
In conclusion, our present work confirmed that gill tissue expresses novel TRIM protein transcripts not previously described in rainbow trout by using different molecular biology techniques and validated through bioinformatic tools. We further demonstrated that these TRIM transcripts can be induced by specific viral and bacterial pathogen-associated molecular pattern in gill using in vitro models, and that they present specific time-dependent expression peaks in gills from infected rainbow trout over a period of 30 days. In addition, our findings revealed that OmTRIM25 is needed to trigger LPS-induced expression of pro-inflammatory cytokines in RTgill-W1 cells, suggesting a potential immunomodulatory function in the gill. Together, these data provide novel insights into the role of TRIM proteins in rainbow trout immunology and potential molecular mechanisms underpinning their function.
4. Materials and Methods
4.1. Chemical and Reagents
L-15 medium was obtained from Life Technologies. Foetal bovine serum (FBS) was obtained from (Biological Industries). Penicillin/streptomycin was obtained from Gibco. Poly(I:C) was purchased in Sigma. LPS from Pseudomonas aeruginosa was provided by Dr. Alejandro Dinamarca (Universidad de Valparaíso, Chile). FuGENE® was obtained from Promega, and TRIzol® was obtained from Invitrogen.
4.2. Culturing of RTgill-W1 Cells and Treatment
The gill epithelial cell line RTgill-W1 from rainbow trout (ATCC® CRL-2523™) was gently provided by Dr. Brian Dixon (University of Waterloo, Canada), and cultured as previously described by Álvarez et al. 2017 [
42]. Briefly, RTgill-W1 cells were maintained in L-15 medium with 4 mM glutamine and supplemented with 5% FBS, 200 U mL
-1 penicillin and 200 µg mL
-1 streptomycin at 20 °C in 6-well plates at a density of 6 × 10
5 cells per well. RTgill-W1 cells were treated with either 10 µg mL
-1 LPS or 30 µg mL
-1 poly(I:C) in FuGENE® for 4, 8, 12 and 24 hours. Times and doses of LPS and poly(I:C) were chosen according to the molecular inflammatory pattern observed in RTgill-W1 cells [
42].
4.3. Primary Culturing of Rainbow Trout Gill Cells
Adult rainbow trouts (40–50 g; obtained from Río Blanco, Los Andes, Chile) were sacrificed using an overdose of benzocaine. The gills were dissected out and incubated in a PBS-antibiotic mix buffer solution (200 U mL
-1 penicillin, 200 μg mL
-1 streptomycin, and 400 μg mL
-1 gentamicin) three times for 15 min at 17 °C. The tissue was enzymatically dissociated with 1 mg mL
-1 collagenase (type I) in fresh PBS-antibiotic mix buffer. The cell suspension was passed through a 100 μm
2 cell strainer and centrifuged at 1200 × g for 5 min at 17 °C. The pellet was resuspended in L-15 media supplemented with 5% FBS, 200 U mL
-1 penicillin and 200 µg mL
-1 streptomycin. The suspended cells were maintained at 20 °C in 12-well plates at 2.5 × 105 cells per well. The following day, primary gill cells were treated with either 10 µg mL
-1 LPS or 30 µg mL
-1 poly(I:C) in FuGENE® for 4, 8, 12 and 24 hours. Times and doses of LPS and poly(I:C) were chosen according to the molecular inflammatory pattern observed in RTgill-W1 cells [
42].
4.4. Fish Handling and F. psychrophilum Growth Conditions
Juvenile and healthy rainbow trout (weighing 6 ± 2 g, n = 108) were procured from the Río Blanco fish farm, a hatchery with no disease history and certifications for being patho-gen-free as per Chilean Lists 1, 2, and 3 [
43]. These fish were kept in two separate 200 L plastic tanks containing aerated dechlorinated water at a temper-ature of 15 ± 1 °C and were given a week to acclimate before undergoing the bacterial challenge. The fish were fed daily ad libitum with a commercial feed (Skretting, Chile), amounting to 1.5% of body weight, and were exposed to a 12:12 h light:dark photoperiod, with the tank water being refreshed every alternate day to eliminate faecal and ni-trogenous waste.
The study utilized Flavobacterium psychrophilum CC5, an isolate recovered from rainbow trout in 2014 and authenticated as F. psychrophilum through conventional phenotyping and 16S rDNA-based PCR [
44,
45]. The isolate's growth medium was TYES agar plates (comprising 0.4% tryptone, 0.05% yeast extract, 0.02% anhydrous calcium chloride, 0.05% magnesium sulphate heptahydrate, and 1% bacteriological agar, with a pH of 7.2), and the isolate was aerobically incubated at 15 °C for a duration of 3–5 days [
46] . The medium was initially prepared in a solid state for the first growth, ensuring not more than two sub-cultured growths from stock cultures, and subsequently in a liquid state (omitting the bacteriological agar) with agitation set at 100 rpm, for generating the inoculum that was administered to the fish. The stock cultures were preserved at -80 °C within Criobilles tubes (AES Laboratory).
4.5. Experimental Design and Fish Sampling
To investigate and detail the existence of new TRIM proteins engaged in the innate immunity of rainbow trout through in vivo methodologies, infectivity tests utilizing the CC5 isolate were executed, anticipating a 10% mortality rate in fish. The methodology adhered to is congruent with the one delineated in Muñoz et al. (2019) [
47]. All experimental workflows and animal handlings were orchestrated in compliance with the ethical guidelines for live animals stipulated by the Chilean National Commission of Scientific and Technological Research (CONICYT) and the Ethics Committee for Animal Experiments at the Universidad Andrés Bello.
Two distinct challenges were performed in duplicate (Experiments 1 and 2), and preceding experiments 1 and 2, all fish were anesthetized using a 15 °C benzocaine solution (30 mg L-1). In Experiment 1, 54 rainbow trout were divided into two groups of 27 and housed in 6-L plastic tanks with aerated dechlorinated water at 15 ± 1 °C. These were inoculated via intramuscular injection with 0.1 mL of 5.6 ± 0.41×105 CFU fish-1. For Experiment 2, a separate group of 54 fish were injected with only 0.1 mL of TYES broth per trout (TYES group). The duration of all tests was maintained up to 30 days, with bi-daily water changes in each tank to eliminate faecal and nitrogenous waste, and daily monitoring for disease symptoms. Deceased fish, which were excluded from analysis samples, were removed daily from each tank, and analysed by direct streaking of various samples (skin lesion, kidney, liver, and spleen) onto TYES agar plates, incubated at 15 °C for 5 days. Biochemical and PCR analyses were conducted on isolates to validate whether the injected bacterium was the mortality cause. Additionally, to preserve tank density, an equivalent number of fish were randomly removed from the non-infected tank whenever a fish inoculated with the CC5 isolate was removed.
Fish samples were swiftly netted from all groups at multiple post-infection days (1, 2, 4, 6, 8, 10, 15, and 30). At each sampling interval, three individuals were sampled (n = 6, 3 fish per tank). Before sampling, all fish were first anesthetized using a benzocaine solution (30 mg L-1) and subsequently euthanized with an overdose of the same solution (100 mg L-1). A section of the second gill tissue was extracted, instantly frozen in liquid nitrogen, and stored at -80 °C for subsequent processing.
4.6. Molecular Cloning and Sequencing
PCR products were gel purified and sequenced as previously described [
42]. Briefly, amplicons were isolated with an E.Z.N.A.® Gel Extraction Kit (Omega Biotek) and cloned using the TOPO TA with the pCR 2.1 TOPO® vector (Thermo Scientific). Plasmids were transformed into DH5α Escherichia coli cells and purified using an E.Z.N.A.® Plasmid Mini Kit I (Omega Biotek). Plasmid constructs were verified by PCR and the sequences were obtained by pyrosequencing under the directions and services provided by Macrogen Inc., Seoul, Korea.
4.7. Identification of TRIM CDS in RTgill-W1
To obtain the TRIM coding DNA sequences (CDS) presented in this work, we followed three different PCR strategies: 1) Specific PCR primers (
Table 2) were designed to amplify the entire CDS region of three virus-induced finTRIMs previously described in spleen of O. mykiss [
21]. These sequences were arbitrary named in this study as finTRIM1 (AF483536), finTRIM2 (AM887838), and finTRIM3 (AM887799); 2) Degenerate PCR primers were produced to non-specifically amplify any TRIM belonging to the C-IV family, currently the group of TRIM proteins with the highest number of members associated with immunological activity in higher vertebrates [
48]. Specifically, two forward primers were designed in the RING/B-box N-terminal domain, and two reverse primers were designed in the PRYSPRY (B30.2) C-terminal domain (
Table 3); 3) Additional TRIM CDS were obtained through an EST contig sequence search in O. mykiss using the GenBank database.
4.8. RNAi Gene-Silencing Assay
To verify the involvement of OmTRIM3 in the immune response of rainbow trout against LPS in vitro, we established a RNA-mediated interference (RNAi) platform to silence its gene expression as previously demonstrated [
49,
50]. Small interfering RNA (siRNA) were designed using the Custom Dicer-Substrate siRNA Tool provided by Integrated DNA Technologies, Inc. (
https://www.idtdna.com), and ordered to the same company. Briefly, RTgill-W1 cells were pre-incubated with either 50 nM of two sets of OmTRIM25-siRNA or GFP-siRNA (technique control) using FuGENE® for 14 hours (
Table 4). The L15-media was replaced with fresh media containing 10 µg mL
-1 LPS and maintained 8 hours. The dose and duration of treatment of siRNA were chosen based on previous publications using RNAi platforms in different fish cell lines [
51,
52].
4.9. Total RNA Extraction, cDNA Synthesis and Reverse Transcription Quantitative Real-Time PCR (RT-qPCR)
Total RNA was isolated using the TRIzol® reagent and the RNA E.Z.N.A.® Total RNA Kit I (Omega Bio-Tek). Briefly, cells/tissue were lysated using TRIzol®, and transferred into the kit filter tubes to proceed with the manufacturer directions. RNA concentration was assessed using a ND-1000 spectrophotometer. Subsequently, isolated RNA was reverse transcribed into cDNA using the AffinityScript cDNA Synthesis kit (Agilent). The GOTaq polymerase (Promega) and an ESCO® Aerins™ thermal cycler were used for agarose gel electrophoresis PCR. For RT-qPCR, cDNA amplification was performed using the Brilliant II SYBR® Green QPCR Master Mix (Agilent) and the mRNA expression levels were measured using a Mx3000P qPCR System (Agilent). Each sample was analysed in duplicate for both target gene and reference gene, and the expression rations were calculated using the 2
-ΔΔCt method [
53]. Specific qPCR primers were design to detect the expression of the TRIM transcripts found in this study (
Table 5).
4.10. Bioinformatics
A bioinformatic approach was performed to determine the identity at protein level of each sequence using the protein database UniProt. To confirm the presence of the main domains of TRIM proteins, the Conserved Domain Database (NCBI) was used along with a prediction of the secondary structure using the server JPred4 [
54]. Comparison between sequences were performed by generating multiple sequence alignments with Clustal Omega, and analysed with Jalview [
55]. Finally, phylogenetic trees were produced by using the Maximum-Likelihood (ML) method provided by the MEGA (X) and were bootstrapped 1000 times. Three-dimensional structures were obtained from the Protein Data Bank database. For correlation analysis of gene expression, the R package “corrplot” (
https://github.com/taiyun/corrplot) was used in Rstudio to analyse and generate the correlation matrices.
4.11. Statistical Analysis
Statistical analysis and heatmaps were performed using GraphPad Prism 8.01 software. Data from cytokines and TRIM gene expression were analysed by multiple t-test against the experimental control group (within each time point). In addition, for the knock-down experiment, results were analysed using one-way ANOVA and Tukey´s multiple comparison test. A p-value < 0.05 was considered statistically significant.
Author Contributions
Conceptualization, L.M., C.A.A., and R.A.; Methodology, C.A.A, R.D.R, and P.S.; Software, N.S-P and B.M-L.; Validation, F.D. and F.R.; Formal analysis, F.D., F.R., N.S-P, and B.M-L.; Investigation, F.D., F.R., C.A.A. and L.M.; Data curation, N.S-P and B.M-L.; Writing – Original draft preparation, F.D. and L.M.; Writing – review and editing, C.A.A., P.S. B.M-L and R.A.; Visualization, F.D., N.S-P, B.M-L., and L.M.; Supervision, C.A.A. and L.M.; Project administration, L.M.; Funding acquisition, C.A.A., P.S., and L.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by FONDECYT 1140797 and 1231206 (L.M.), FONDECYT 1230712 (C.A.) and FONDECYT 1231088 (P.S.), from ANID Chile. BM-L was funded by Foods of Norway (Centre for Research-based Innovation: 237841/030). R.A-H. Acknowledges the Fondo de Financiamiento de Centros de Investigación en Áreas Prioritarias, Centro Interdisciplinario de Investigación en Acuicultura Sustentable (Project Number 1522A0004). R.D.R. was founded by CNPq (grant number 312047/2021-5).
Institutional Review Board Statement
The study was conducted according to National Research Council guidelines for the care and use of laboratory animals (National Research Council (US) Committee, 2010). In addition, all protocols were approved by the Committee on the Ethics of Animal Experiments of the Pontificia Universidad Católica de Valparaíso, Chile (Fondecyt No 1140797) and by the Universidad Andrés Bello (Chile) under approval act No 016/2010.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
We thank Dr. Brian Dixon for providing the cell line RTgill-W1 and suggesting the culture conditions. We thank Dr. Alejandro Dinamarca for providing Pseudomonas aeruginosa LPS.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Malmstrøm, M.; Matschiner, M.; Tørresen, O.K.; Star, B.; Snipen, L.G.; Hansen, T.F.; Baalsrud, H.T.; Nederbragt, A.J.; Hanel, R.; Salzburger, W.; et al. Evolution of the Immune System Influences Speciation Rates in Teleost Fishes. Nat. Genet. 2016, 48, 1204–1210. [Google Scholar] [CrossRef] [PubMed]
- Yatim, K.M.; Lakkis, F.G. A Brief Journey through the Immune System. Clin. J. Am. Soc. Nephrol. CJASN 2015, 10, 1274–1281. [Google Scholar] [CrossRef]
- Plouffe, D.A.; Hanington, P.C.; Walsh, J.G.; Wilson, E.C.; Belosevic, M. Comparison of Select Innate Immune Mechanisms of Fish and Mammals. Xenotransplantation 2005, 12, 266–277. [Google Scholar] [CrossRef]
- Xu, Y.; Zhu, S.-W.; Li, Q.-W. Lamprey: A Model for Vertebrate Evolutionary Research. Zool. Res. 2016, 37, 263–269. [Google Scholar] [CrossRef]
- Zhang, X.-J.; Zhang, X.-Y.; Wang, P.; Zhang, Y.-A. Identification of Another Primordial CD80/86 Molecule in Rainbow Trout: Insights into the Origin and Evolution of CD80 and CD86 in Vertebrates. Dev. Comp. Immunol. 2018, 89, 73–82. [Google Scholar] [CrossRef]
- Hatakeyama, S. TRIM Family Proteins: Roles in Autophagy, Immunity, and Carcinogenesis. Trends Biochem. Sci. 2017, 42, 297–311. [Google Scholar] [CrossRef] [PubMed]
- Vunjak, M.; Versteeg, G.A. TRIM Proteins. Curr. Biol. CB 2019, 29, R42–R44. [Google Scholar] [CrossRef]
- Esposito, D.; Koliopoulos, M.G.; Rittinger, K. Structural Determinants of TRIM Protein Function. Biochem. Soc. Trans. 2017, 45, 183–191. [Google Scholar] [CrossRef]
- Stevens, R.V.; Esposito, D.; Rittinger, K. Characterisation of Class VI TRIM RING Domains: Linking RING Activity to C-Terminal Domain Identity. Life Sci. Alliance 2019, 2. [Google Scholar] [CrossRef]
- Ozato, K.; Shin, D.-M.; Chang, T.-H.; Morse, H.C. TRIM Family Proteins and Their Emerging Roles in Innate Immunity. Nat. Rev. Immunol. 2008, 8, 849–860. [Google Scholar] [CrossRef]
- Versteeg, G.A.; Benke, S.; García-Sastre, A.; Rajsbaum, R. InTRIMsic Immunity: Positive and Negative Regulation of Immune Signaling by Tripartite Motif Proteins. Cytokine Growth Factor Rev. 2014, 25, 563–576. [Google Scholar] [CrossRef]
- Rajsbaum, R.; García-Sastre, A.; Versteeg, G.A. TRIMmunity: The Roles of the TRIM E3-Ubiquitin Ligase Family in Innate Antiviral Immunity. J. Mol. Biol. 2014, 426, 1265–1284. [Google Scholar] [CrossRef]
- van Gent, M.; Sparrer, K.M.J.; Gack, M.U. TRIM Proteins and Their Roles in Antiviral Host Defenses. Annu. Rev. Virol. 2018, 5, 385–405. [Google Scholar] [CrossRef]
- Kimura, T.; Jain, A.; Choi, S.W.; Mandell, M.A.; Johansen, T.; Deretic, V. TRIM-Directed Selective Autophagy Regulates Immune Activation. Autophagy 2017, 13, 989–990. [Google Scholar] [CrossRef]
- Weng, L.; Mitoma, H.; Trichot, C.; Tricot, C.; Bao, M.; Liu, Y.; Zhang, Z.; Liu, Y.-J. The E3 Ubiquitin Ligase Tripartite Motif 33 Is Essential for Cytosolic RNA-Induced NLRP3 Inflammasome Activation. J. Immunol. Baltim. Md 1950 2014, 193, 3676–3682. [Google Scholar] [CrossRef]
- Du Pasquier, L. Fish “n” TRIMs. J. Biol. 2009, 8, 50. [Google Scholar] [CrossRef]
- Langevin, C.; Levraud, J.-P.; Boudinot, P. Fish Antiviral Tripartite Motif (TRIM) Proteins. Fish Shellfish Immunol. 2019, 86, 724–733. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Z.; Lu, Y.; Hu, G.; Lin, L.; Zeng, L.; Zhou, Y.; Liu, X. Molecular Characterization, Tissue Distribution and Expression, and Potential Antiviral Effects of TRIM32 in the Common Carp (Cyprinus Carpio). Int. J. Mol. Sci. 2016, 17. [Google Scholar] [CrossRef]
- Wang, Y.; Kuang, M.; Lu, Y.; Lin, L.; Liu, X. Characterization and Biological Function Analysis of the TRIM47 Gene from Common Carp (Cyprinus Carpio). Gene 2017, 627, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Han, R.; Wang, R.; Zhao, Q.; Han, Y.; Zong, S.; Miao, S.; Song, W.; Wang, L. Trim69 Regulates Zebrafish Brain Development by Ap-1 Pathway. Sci. Rep. 2016, 6, 24034. [Google Scholar] [CrossRef] [PubMed]
- van der Aa, L.M.; Levraud, J.-P.; Yahmi, M.; Lauret, E.; Briolat, V.; Herbomel, P.; Benmansour, A.; Boudinot, P. A Large New Subset of TRIM Genes Highly Diversified by Duplication and Positive Selection in Teleost Fish. BMC Biol. 2009, 7, 7. [Google Scholar] [CrossRef]
- Kim, J.; Kim, J.-W.; Kim, D.-G.; Nam, B.-H.; Kim, Y.-O.; Park, J.Y.; Kong, H.J. Molecular Characterization of Rhodeus Uyekii Tripartite Motif Protein 1 (TRIM1) Involved in IFN-γ/LPS-Induced NF-ΚB Signaling. Fish Shellfish Immunol. 2018, 79, 42–51. [Google Scholar] [CrossRef]
- Koppang, E.O.; Kvellestad, A.; Fischer, U. 5 - Fish Mucosal Immunity: Gill. In Mucosal Health in Aquaculture; Beck, B.H., Peatman, E., Eds.; Academic Press: San Diego, 2015; pp. 93–133. ISBN 978-0-12-417186-2. [Google Scholar]
- Huang, Y.; Yu, Y.; Yang, Y.; Yang, M.; Zhou, L.; Huang, X.; Qin, Q. Fish TRIM8 Exerts Antiviral Roles through Regulation of the Proinflammatory Factors and Interferon Signaling. Fish Shellfish Immunol. 2016, 54, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Maarifi, G.; Smith, N.; Maillet, S.; Moncorgé, O.; Chamontin, C.; Edouard, J.; Sohm, F.; Blanchet, F.P.; Herbeuval, J.-P.; Lutfalla, G.; et al. TRIM8 Is Required for Virus-Induced IFN Response in Human Plasmacytoid Dendritic Cells. Sci. Adv. 2019, 5, eaax3511. [Google Scholar] [CrossRef]
- Koliopoulos, M.G.; Lethier, M.; van der Veen, A.G.; Haubrich, K.; Hennig, J.; Kowalinski, E.; Stevens, R.V.; Martin, S.R.; Reis e Sousa, C.; Cusack, S.; et al. Molecular Mechanism of Influenza A NS1-Mediated TRIM25 Recognition and Inhibition. Nat. Commun. 2018, 9, 1820. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, K.; Huang, Y.; Sun, M.; Tian, Q.; Zhang, S.; Qin, Y. TRIM25 Promotes TNF-α-Induced NF-ΚB Activation through Potentiating the K63-Linked Ubiquitination of TRAF2. J. Immunol. Baltim. Md 1950 2020, 204, 1499–1507. [Google Scholar] [CrossRef]
- Cao, Z.; Conway, K.L.; Heath, R.J.; Rush, J.S.; Leshchiner, E.S.; Ramirez-Ortiz, Z.G.; Nedelsky, N.B.; Huang, H.; Ng, A.; Gardet, A.; et al. Ubiquitin Ligase TRIM62 Regulates CARD9-Mediated Anti-Fungal Immunity and Intestinal Inflammation. Immunity 2015, 43, 715–726. [Google Scholar] [CrossRef]
- Gack, M.U.; Shin, Y.C.; Joo, C.-H.; Urano, T.; Liang, C.; Sun, L.; Takeuchi, O.; Akira, S.; Chen, Z.; Inoue, S.; et al. TRIM25 RING-Finger E3 Ubiquitin Ligase Is Essential for RIG-I-Mediated Antiviral Activity. Nature 2007, 446, 916–920. [Google Scholar] [CrossRef]
- Shen, Y.; Li, N.L.; Wang, J.; Liu, B.; Lester, S.; Li, K. TRIM56 Is an Essential Component of the TLR3 Antiviral Signaling Pathway. J. Biol. Chem. 2012, 287, 36404–36413. [Google Scholar] [CrossRef] [PubMed]
- Nematollahi, A.; Decostere, A.; Pasmans, F.; Haesebrouck, F. Flavobacterium Psychrophilum Infections in Salmonid Fish. J. Fish Dis. 2003, 26, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Schulte, W.; Bernhagen, J.; Bucala, R. Cytokines in Sepsis: Potent Immunoregulators and Potential Therapeutic Targets—An Updated View. Mediators Inflamm. 2013, 2013, e165974. [Google Scholar] [CrossRef]
- Cellular and Molecular Immunology - 9th Edition. Available online: https://www.elsevier.com/books/cellular-and-molecular-immunology/abbas/978-0-323-47978-3 (accessed on 24 June 2021).
- Kawai, T.; Akira, S. Regulation of Innate Immune Signalling Pathways by the Tripartite Motif (TRIM) Family Proteins. EMBO Mol. Med. 2011, 3, 513–527. [Google Scholar] [CrossRef]
- Chikuma, S.; Suita, N.; Okazaki, I.-M.; Shibayama, S.; Honjo, T. TRIM28 Prevents Autoinflammatory T Cell Development in Vivo. Nat. Immunol. 2012, 13, 596–603. [Google Scholar] [CrossRef]
- Ahn, Y.; Hwang, J.-H.; Zheng, Z.; Bang, D.; Kim, D.-Y. Enhancement of Th1/Th17 Inflammation by TRIM21 in Behçet’s Disease. Sci. Rep. 2017, 7, 3018. [Google Scholar] [CrossRef] [PubMed]
- Venuto, S.; Merla, G. E3 Ubiquitin Ligase TRIM Proteins, Cell Cycle and Mitosis. Cells 2019, 8, E510. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.M.; Choi, S.S.; Lee, Y.H.; Khim, K.W.; Yoon, S.; Kim, B.-G.; Nam, D.; Suh, P.-G.; Myung, K.; Choi, J.H. The E3 Ubiquitin Ligase TRIM25 Regulates Adipocyte Differentiation via Proteasome-Mediated Degradation of PPARγ. Exp. Mol. Med. 2018, 50, 1–11. [Google Scholar] [CrossRef]
- Lu, M.; Zhu, X.; Yang, Z.; Zhang, W.; Sun, Z.; Ji, Q.; Chen, X.; Zhu, J.; Wang, C.; Nie, S. E3 Ubiquitin Ligase Tripartite Motif 7 Positively Regulates the TLR4-Mediated Immune Response via Its E3 Ligase Domain in Macrophages. Mol. Immunol. 2019, 109, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.-J.; Liu, T.-T.; Ran, Y.; Li, Y.; Zhang, X.-D.; Shu, H.-B.; Wang, Y.-Y. The E3 Ubiquitin Ligase MIB1 Negatively Regulates Basal IκBα Level and Modulates NF-ΚB Activation. Cell Res. 2012, 22, 603–606. [Google Scholar] [CrossRef] [PubMed]
- Zemirli, N.; Pourcelot, M.; Dogan, N.; Vazquez, A.; Arnoult, D. The E3 Ubiquitin Ligase RNF121 Is a Positive Regulator of NF-ΚB Activation. Cell Commun. Signal. 2014, 12, 72. [Google Scholar] [CrossRef]
- Álvarez, C.A.; Ramírez-Cepeda, F.; Santana, P.; Torres, E.; Cortés, J.; Guzmán, F.; Schmitt, P.; Mercado, L. Insights into the Diversity of NOD-like Receptors: Identification and Expression Analysis of NLRC3, NLRC5 and NLRX1 in Rainbow Trout. Mol. Immunol. 2017, 87, 102–113. [Google Scholar] [CrossRef]
- 43. Enfermedades infecciosas del cultivo de salmónidos en Chile y el Mundo | ISBN 978-956-8861-01-8 - Libro.
- Urdaci, M.C.; Chakroun, C.; Faure, D.; Bernardet, J.F. Development of a Polymerase Chain Reaction Assay for Identification and Detection of the Fish Pathogen Flavobacterium Psychrophilum. Res. Microbiol. 1998, 149, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Bernardet, J.-F.; Nakagawa, Y.; Holmes, B. ; Subcommittee On The Taxonomy Of Flavobacterium And Cytophaga-Like Bacteria Of The International Committee On Systematics Of Prokaryotes, null Proposed Minimal Standards for Describing New Taxa of the Family Flavobacteriaceae and Emended Description of the Family. Int. J. Syst. Evol. Microbiol. 2002, 52, 1049–1070. [Google Scholar] [CrossRef] [PubMed]
- Valdebenito, S.; Avendaño-Herrera, R. Phenotypic, Serological and Genetic Characterization of Flavobacterium Psychrophilum Strains Isolated from Salmonids in Chile. J. Fish Dis. 2009, 32, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, J.L.P.; Ocampos, D.; Poblete-Morales, M.; Oyarzún, R.; Morera, F.J.; Tapia-Cammas, D.; Avendaño-Herrera, R.; Vargas-Chacoff, L. Effect of Flavobacterium Psychrophilum on the Neuroendocrine Response of Rainbow Trout (Oncorhynchus Mykiss) in a Time Course Experiment. Comp. Biochem. Physiol. A. Mol. Integr. Physiol. 2019, 236, 110525. [Google Scholar] [CrossRef]
- Watanabe, M.; Hatakeyama, S. TRIM Proteins and Diseases. J. Biochem. (Tokyo) 2017, 161, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and Specific Genetic Interference by Double-Stranded RNA in Caenorhabditis Elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Kalil, S.P.; Rosa, R.D. da; Capelli-Peixoto, J.; Pohl, P.C.; Oliveira, P.L. de; Fogaça, A.C.; Daffre, S. Immune-Related Redox Metabolism of Embryonic Cells of the Tick Rhipicephalus Microplus (BME26) in Response to Infection with Anaplasma Marginale. Parasit. Vectors 2017, 10, 613. [Google Scholar] [CrossRef] [PubMed]
- Bohle, H.; Lorenzen, N.; Schyth, B.D. Species Specific Inhibition of Viral Replication Using Dicer Substrate SiRNAs (DsiRNAs) Targeting the Viral Nucleoprotein of the Fish Pathogenic Rhabdovirus Viral Hemorrhagic Septicemia Virus (VHSV). Antiviral Res. 2011, 90, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Schyth, B.D.; Lorenzen, N.; Pedersen, F.S. Antiviral Activity of Small Interfering RNAs: Specificity Testing Using Heterologous Virus Reveals Interferon-Related Effects Overlooked by Conventional Mismatch Controls. Virology 2006, 349, 134–141. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods San Diego Calif 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Drozdetskiy, A.; Cole, C.; Procter, J.; Barton, G.J. JPred4: A Protein Secondary Structure Prediction Server. Nucleic Acids Res. 2015, 43, W389–W394. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2--a Multiple Sequence Alignment Editor and Analysis Workbench. Bioinforma. Oxf. Engl. 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
Figure 1.
Amino acid alignment between the TRIM-like proteins found in RTgill-W1. A multiple alignment was performed using the software Clustal Omega, for the predicted amino acidic sequences of the TRIM-like proteins identified in RTgill-W1 cell line. The main domains of the RBCC motif are highlighted in colours: RING = orange; B-box 1 = red; B-box 2= purple; coiled-coil = blue.
Figure 1.
Amino acid alignment between the TRIM-like proteins found in RTgill-W1. A multiple alignment was performed using the software Clustal Omega, for the predicted amino acidic sequences of the TRIM-like proteins identified in RTgill-W1 cell line. The main domains of the RBCC motif are highlighted in colours: RING = orange; B-box 1 = red; B-box 2= purple; coiled-coil = blue.
Figure 2.
Phylogenetic tree grouping the TRIM-like proteins identified in RTgill-W1 cells. An unrooted phylogenetic tree of TRIM proteins from fish and other species was produced using an amino acid multiple alignment and the ML method within the software MEGAX. The percentage of replicate trees in which the associated taxa clustered together in a bootstrap test (1000 replicates) is shown next to the branches. Novel TRIMs are highlighted with red dots.
Figure 2.
Phylogenetic tree grouping the TRIM-like proteins identified in RTgill-W1 cells. An unrooted phylogenetic tree of TRIM proteins from fish and other species was produced using an amino acid multiple alignment and the ML method within the software MEGAX. The percentage of replicate trees in which the associated taxa clustered together in a bootstrap test (1000 replicates) is shown next to the branches. Novel TRIMs are highlighted with red dots.
Figure 3.
LPS induced the gene expression of cytokines and OmTRIM-like proteins in RTgill-W1 cells and primary gill cultures in a time dependent manner. RTgill-W1 cells and primary gill cultures were stimulated with 10 µg mL-1 LPS for up to 24 hours. A: The gene expression of the cytokines TNF-α2, IL-1β and type I IFN, and OmTRIM-like proteins was quantitatively measured using RT-qPCR at 4; 8; and 24 hours post LPS treatment. Results are expressed as the mean ± SEM of six experimental replicates (*: p < 0.05 vs Control groups). B: Correlation between LPS-induced transcripts in RTgill-W1 cells and primary gill cultures (*: p < 0.05).
Figure 3.
LPS induced the gene expression of cytokines and OmTRIM-like proteins in RTgill-W1 cells and primary gill cultures in a time dependent manner. RTgill-W1 cells and primary gill cultures were stimulated with 10 µg mL-1 LPS for up to 24 hours. A: The gene expression of the cytokines TNF-α2, IL-1β and type I IFN, and OmTRIM-like proteins was quantitatively measured using RT-qPCR at 4; 8; and 24 hours post LPS treatment. Results are expressed as the mean ± SEM of six experimental replicates (*: p < 0.05 vs Control groups). B: Correlation between LPS-induced transcripts in RTgill-W1 cells and primary gill cultures (*: p < 0.05).
Figure 4.
Poly(I:C) induced the gene expression of cytokines and OmTRIM-like proteins in RTgill-W1 cells and primary gill cultures in a time dependent manner. RTgill-W1 cells were stimulated with 30 µg mL-1 Poly(I:C) for up to 24 hours. A: gene expression of the cytokines TNF-α2, IL-1β and type I IFN, and OmTRIM-like proteins was quantitatively measured using RT-qPCR at 4; 8; and 24 hours post poly(I:C) treatment. Results are expressed as the mean ± SEM of six experimental replicates (*: p < 0.05 vs Control groups). B: Correlation between Poly(I:C)-induced transcripts in RTgill-W1 cells and primary gill cultures (*: p < 0.05).
Figure 4.
Poly(I:C) induced the gene expression of cytokines and OmTRIM-like proteins in RTgill-W1 cells and primary gill cultures in a time dependent manner. RTgill-W1 cells were stimulated with 30 µg mL-1 Poly(I:C) for up to 24 hours. A: gene expression of the cytokines TNF-α2, IL-1β and type I IFN, and OmTRIM-like proteins was quantitatively measured using RT-qPCR at 4; 8; and 24 hours post poly(I:C) treatment. Results are expressed as the mean ± SEM of six experimental replicates (*: p < 0.05 vs Control groups). B: Correlation between Poly(I:C)-induced transcripts in RTgill-W1 cells and primary gill cultures (*: p < 0.05).
Figure 5.
Infection with F. psychrophilum CC5 induced the gene expression of OmTRIM-like proteins in rainbow trout. Animals were infected intramuscularly with a LD50 of F. psychrophilum and gill tissue samples were analysed in a period of 30 days. A: gene expression of the cytokines TNF-α2, IL-1β and type I IFN, and OmTRIM-like proteins was quantitatively measured using RT-qPCR at different time points. Results are expressed as the mean ± SEM, n = 3 (*: p < 0.05 vs Control groups). B: Correlation between F. psychrophilum-induced transcripts in rainbow trout gills (*: p < 0.05).
Figure 5.
Infection with F. psychrophilum CC5 induced the gene expression of OmTRIM-like proteins in rainbow trout. Animals were infected intramuscularly with a LD50 of F. psychrophilum and gill tissue samples were analysed in a period of 30 days. A: gene expression of the cytokines TNF-α2, IL-1β and type I IFN, and OmTRIM-like proteins was quantitatively measured using RT-qPCR at different time points. Results are expressed as the mean ± SEM, n = 3 (*: p < 0.05 vs Control groups). B: Correlation between F. psychrophilum-induced transcripts in rainbow trout gills (*: p < 0.05).
Figure 6.
LPS-induced inflammatory response in RTgill-W1 cells mediated by OmTRIM25. A: RTgill-W1 cells were incubated with 50 nM siOmTRIM25 for 14 hours, and then stimulated with 10 µg mL-1 LPS for 8 hours. The gene expression of OmTRIM25 and the cytokines TNF-α2, IL-1β and type I IFN was quantitatively measured using RT-qPCR. Results are expressed as the mean ± SEM of six experimental replicates (*: p < 0.05). B: Schematic pathway representing the proposed mechanism of action of OmTRIM25.
Figure 6.
LPS-induced inflammatory response in RTgill-W1 cells mediated by OmTRIM25. A: RTgill-W1 cells were incubated with 50 nM siOmTRIM25 for 14 hours, and then stimulated with 10 µg mL-1 LPS for 8 hours. The gene expression of OmTRIM25 and the cytokines TNF-α2, IL-1β and type I IFN was quantitatively measured using RT-qPCR. Results are expressed as the mean ± SEM of six experimental replicates (*: p < 0.05). B: Schematic pathway representing the proposed mechanism of action of OmTRIM25.
Table 1.
Novel TRIM-like sequences identified in the RTgill-W1 cell line.
Table 1.
Novel TRIM-like sequences identified in the RTgill-W1 cell line.
| Identified TRIMs in RTgill-W1 |
GenBank accession |
| OmTRIM25 |
KY073243 |
| OmTRIM16 |
KY073245 |
| OmTRIM62 |
KY073247 |
| OmTRIM8 |
KY073248 |
Table 2.
Specific PCR primers to amplify O. mykiss finTRIM CDS. F: forward, R: reverse.
Table 2.
Specific PCR primers to amplify O. mykiss finTRIM CDS. F: forward, R: reverse.
| Name |
GenBank |
Primers |
PCR product size |
| finTRIM1 |
AF483536 |
F: ATGGCTCAACAGGGAGTTCT
R: TCACTGCCTCTGTTTCTCAGTC |
606 bp |
| finTRIM2 |
AM887838 |
F: ATGGCTCAACAGGGAGTTCT
R: TCAATGACTCTTTCTGTTCCCTT |
1227 bp |
| finTRIM3 |
AM887799 |
F: ATGGCTCAGCAGGGAGTTT
R: CTACAGTTTAACCAGCTCAGCAGTAC |
1656 bp |
Table 3.
Degenerate primers to amplify potential C-IV TRIM members.
Table 3.
Degenerate primers to amplify potential C-IV TRIM members.
| Primer |
Forward (RING) |
Reverse (B30.2) |
| Set1 |
TGTGGACACASTTACTGYA |
AGTYCAGACCACATTCACTSA |
| Set2 |
GGMTGCTGGGAYCAGGA |
CAGACCACATTCACT |
Table 4.
Designed siRNAs to silence OmTRIM25 gene expression.
Table 4.
Designed siRNAs to silence OmTRIM25 gene expression.
| siRNA |
Sense sequence |
Antisense sequence |
| siGFP |
rArCrArArCrArGrCrCrArCrArArCrGrUrGrUrArCrArUrCAT
|
rArUrGrArUrGrUrArCrArCrGrUrUrGrUrGrGrCrUrGrUrUrGrUrArG |
| siTRIM25-1 |
rGrCrArGrCrArGrArGrArGrGrArCrUrGrArGrArArArCrAGA |
rUrCrUrGrUrUrUrCrUrCrArGrUrCrCrUrCrUrCrUrGrCrUrGrCrArG |
| siTRIM25-2 |
rGrGrArGrGrArCrArGrUrGrArUrCrArGrArUrCrUrUrUrACT |
rArGrUrArArArGrArUrCrUrGrArUrCrArCrUrGrUrCrCrUrCrCrArC |
Table 5.
Specific primers designed for this study for quantitative RT-qPCR.
Table 5.
Specific primers designed for this study for quantitative RT-qPCR.
| Gene |
Forward |
Reverse |
| finTRIM 1 |
CTACTGAAGGAGCCGGTGG |
CAGTGCAGACATCACACGC |
| finTRIM 2 |
CTGGACCTGGAAATGTGACA |
TGCAGGAAATTCATAGTGAGGTTT |
| OmTRIM25 |
TCACCAACTGGTACCAGTTACA |
AGAGCACTGGAAACTCCAGGACTT |
| OmTRIM16 |
AAAGGTGACCTGTACACACCTCT |
TCTCTGTTCTGCTGATGTCTTTA |
| OmTRIM62 |
GATTTCCCGACCTCCAAGTACA |
GCAGGTTACCATAGGCTACGAT |
| OmTRIM8 |
GGAAGTGGAAGTGGCTCTCTAA |
TCCATGGTACACACCAGGATCT |
| EF1α |
TGGAGACTGGCACCCTGAAG |
CCAACATTGTCACCAGGCATGG |
| GAPDH |
CCTGCAGAAGGGAATCAAAGTCGT |
TCTCATGGGGCTTCATACACTGGA |
| TNF-α2 |
GTGTGGCGTTCTCTTAATAGCAGC |
ATTCCGTCCTGCATCGTTGC |
| IL-1β |
GTCACATTGCCAACCTCATCATCG |
GTTGAGCAGGTCCTTGTCCTTGA |
| Type I IFN |
GATGCTGAGTTTGAGGACAAAGTC |
GTTTCATGGCAGGTGATACACAGGA |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).