1. Introduction
Exosomes, ranging in size from 50 to 150 nm, represent a subtype of extracellular vesicles (EVs) found in several bodily fluids including serum, urine, seminal fluid, tears, breast milk, aqueous humor, saliva, and within diverse cell types, as well as
in vitro cultures [
1]. Exosomes work as carriers for biological material such as DNA, mRNA, microRNA, lipids, and proteins, thereby conveying information about the originating cells' state and potentially influencing the function of recipient cells [
2,
3,
4,
5,
6].
Adipose tissue (AT) is currently recognized as the primary source of exosomes, also known as adipocyte-derived extracellular vesicles (AdEVs), which function as a bridge between adipocytes and cells in the stromal fraction of the AT as well as with cells from other systems [
7]. AdEVs are filled with biological material that, in AT, plays a role in metabolic alterations such obesity, type 2 diabetes, and related illnesses [
8].
The significance of exosome content in developing AT dysfunction lies in their role as carriers of proteins that recruit macrophages to the AT and liver, mainly TNFα and IL-6 which contribute to the onset of insulin resistance (IR) [
2]. In cases of overweight and obesity, the accumulation of excess fat within adipocytes, as recognized by the World Health Organization (WHO), triggers continual cellular apoptosis. This process creates a hypoxic microenvironment characterized by chronic low-grade inflammation, leading to dysregulation in the secretion of cytokines, adipokines, and other factors essential for AT homeostasis [
9,
10].
To ensure precision and reproducibility, it is essential to compare new methods with the gold standard of exosome isolation, typically represented by differential centrifugation [
11]. Recently, techniques have been incorporated to reduce time, the use of specialized equipment, the amount of sample required, and supplies. Depending on the source and size of exosomes, as well as the research goals, these techniques include polymer precipitation, immunoaffinity capture, chromatography [
12], size exclusion [
13], and commercial kits [
14,
15].
Alternatively, to characterize exosomes, both optical and non-optical techniques are available, each chosen based on the specific information required for research purposes. Optical methods include dynamic light scattering, multi-angle light scattering, nanoparticle tracking analysis, flow cytometry, and surface plasmon resonance [
16,
17]. Non-optical approaches encompass scanning electron microscopy, transmission electron microscopy, cryogenic electron microscopy, atomic force microscopy, Fourier transform infrared spectroscopy, and labeling of exosomal membrane proteins such as CD9, CD63, CD81, and CD82 [
16,
18,
19,
20,
21].
In this study, to obtain whole exosomes in a concentration suitable for further protein and molecular biology assays, we compared three isolation techniques, differential centrifugation (DC), size exclusion chromatography (SEC), and a commercial kit (Invitrogen®), characterized by cryo-TEM, TEM, and western blot.
2. Results
2.1. The Successful Differentiation of 3T3-L1 Cells Results in Acquiring a Mature Adipocyte Phenotype
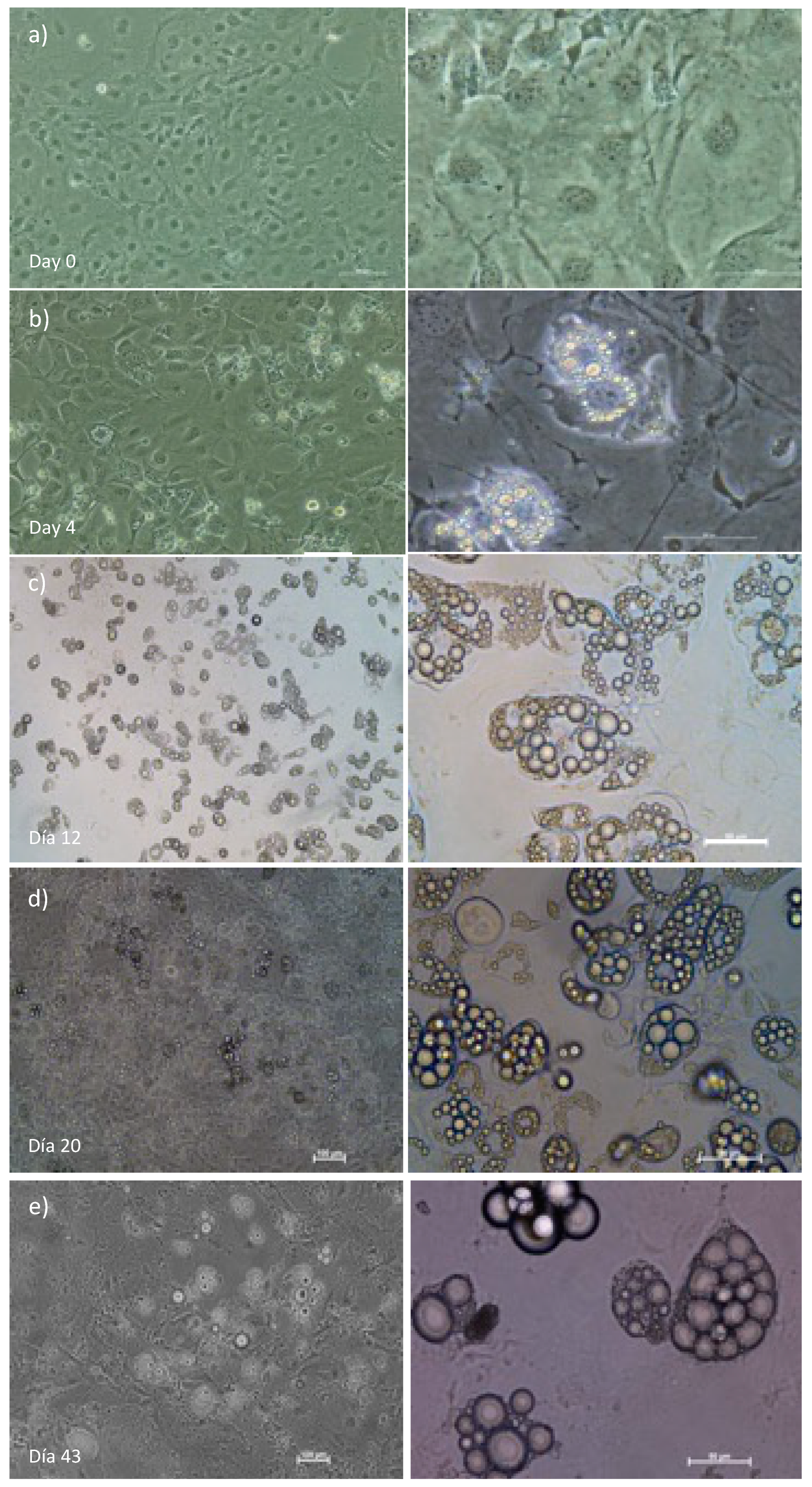
In the present study, we started with preadipocytes, which were stimulated to differentiate into cells with adipocyte phenotype. During the first three days, cell proliferation stops, and the differentiation of a few cells begins, as shown in
Figure 1a. By Day 0, cells of approximately 20 µm without lipid droplets pass to ∽50 µm with small lipid droplets on Day 4 (
Figure 1b). As the days pass, cell size increases due to the triglyceride’s accumulation in the lipid droplets, measuring up to 100 µm, as seen in
Figure 1c–e.
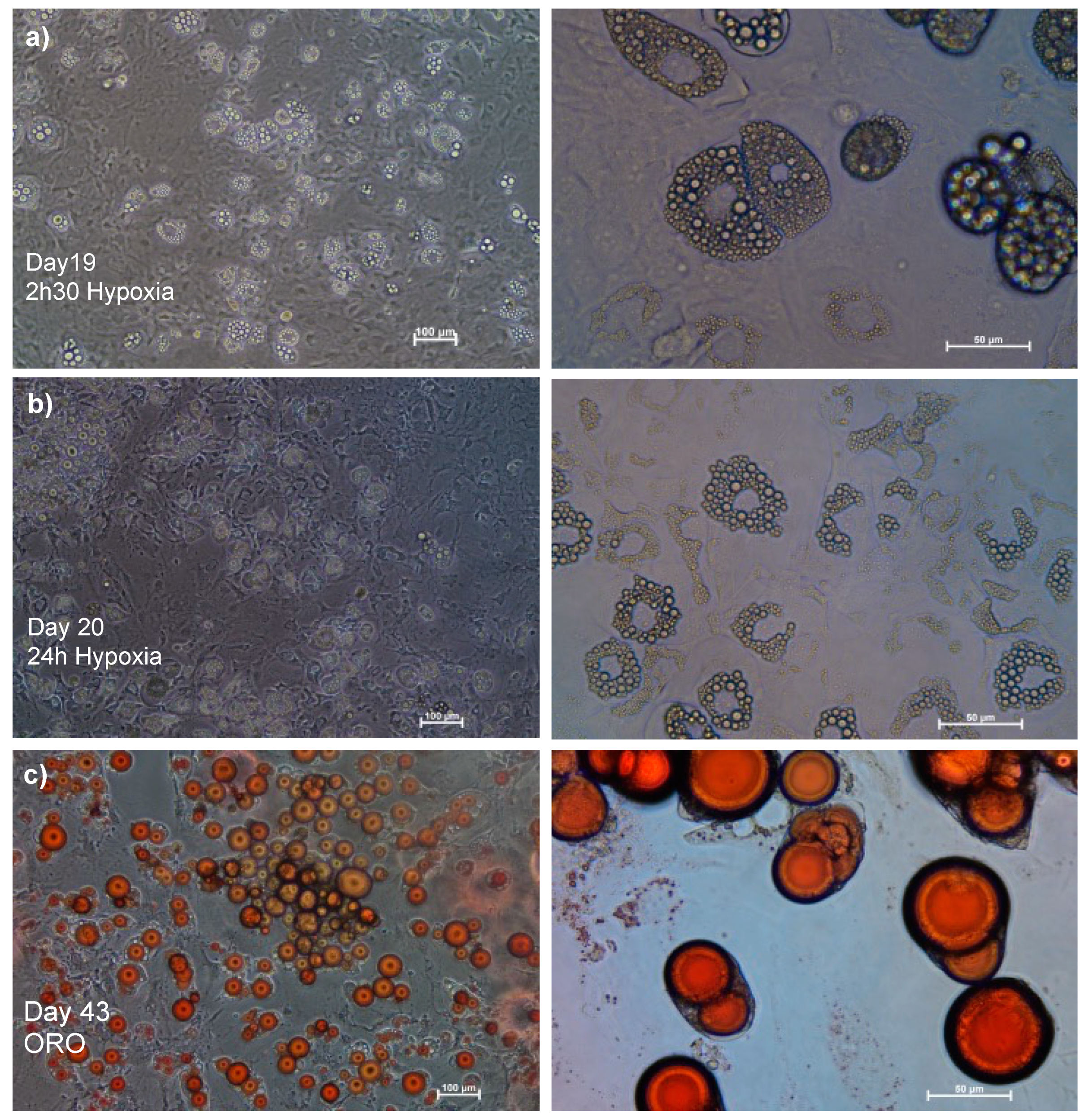
As seen in
Figure 2a, there was no morphological change on Day 19, when the cells were exposed to 2.5 hours of hypoxia. However, at 24 hours of hypoxia, we observed cell disintegration, which may indicate cell death, as well as a reversal of the size of the lipid droplets (
Figure 2b).
To verify that prolonged hypoxia, rather than just continuous intake was the cause of this morphological shift, the cells were maintained with MDII for 43 days and stained with oil-red O (ORO) to identify intracellular triglycerides. The form and size of the lipid droplets, which eventually filled almost the whole cytoplasm, changed, as seen in
Figure 2c.
2.2. Different Exosome Isolation Methods Yield Similar Amount of Total Protein of Intact Exosomes
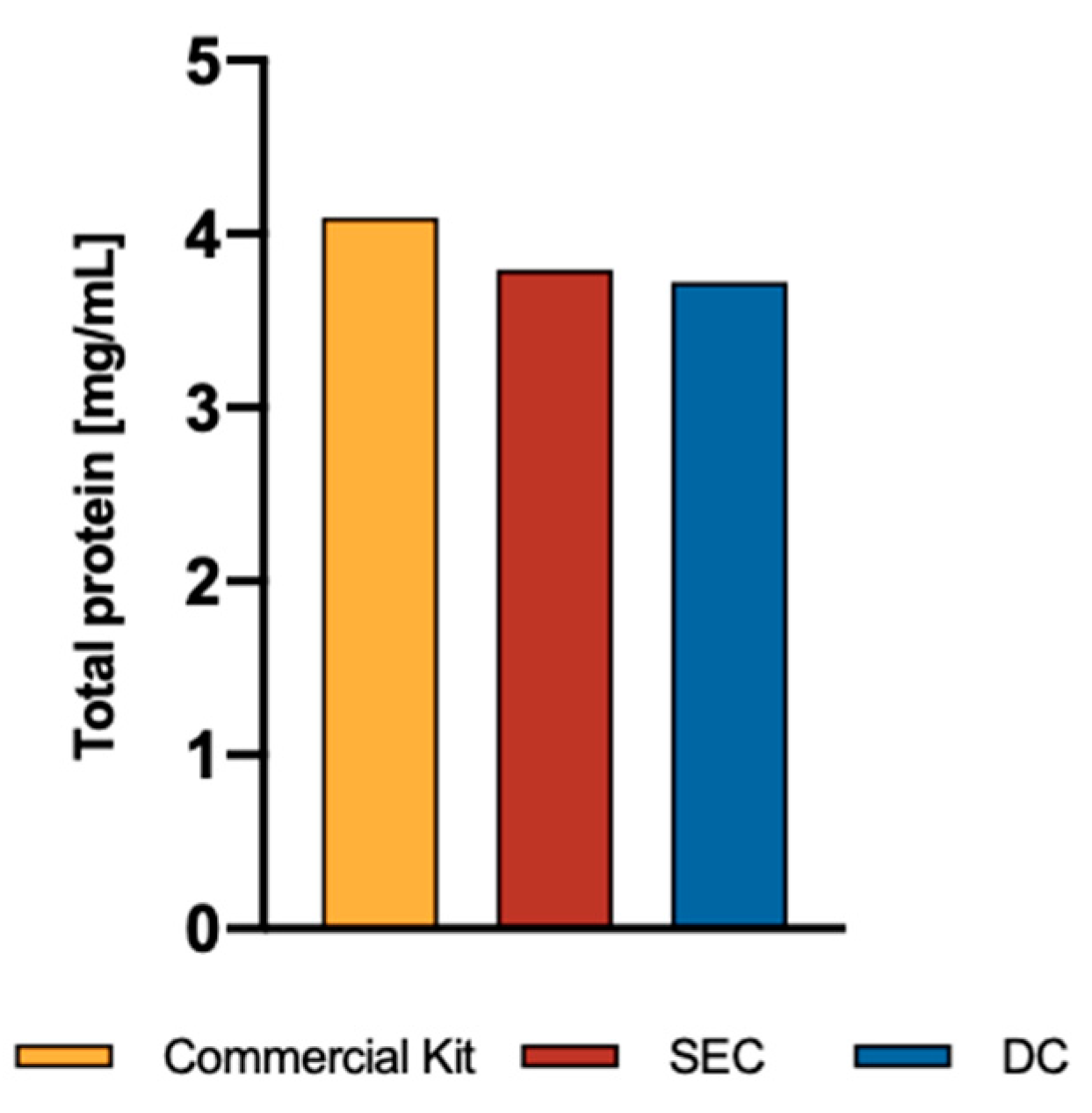
Exosomes were isolated according to the manufacturers’ recommended instructions for the commercial kit, whereas differential centrifugation and size exclusion chromatography were done by for classical ultracentrifugation and column fraction separation. Exosome yield was determined by total protein estimation from intact exosomes using spectrophotometer (NanoDrop UV/Vis 1000). Even though there was no significance, we observed that the precipitation-based Total Exosomes Isolation kit (Invitrogen®) had the maximum yield followed by SEC, and DC (
Figure 3).
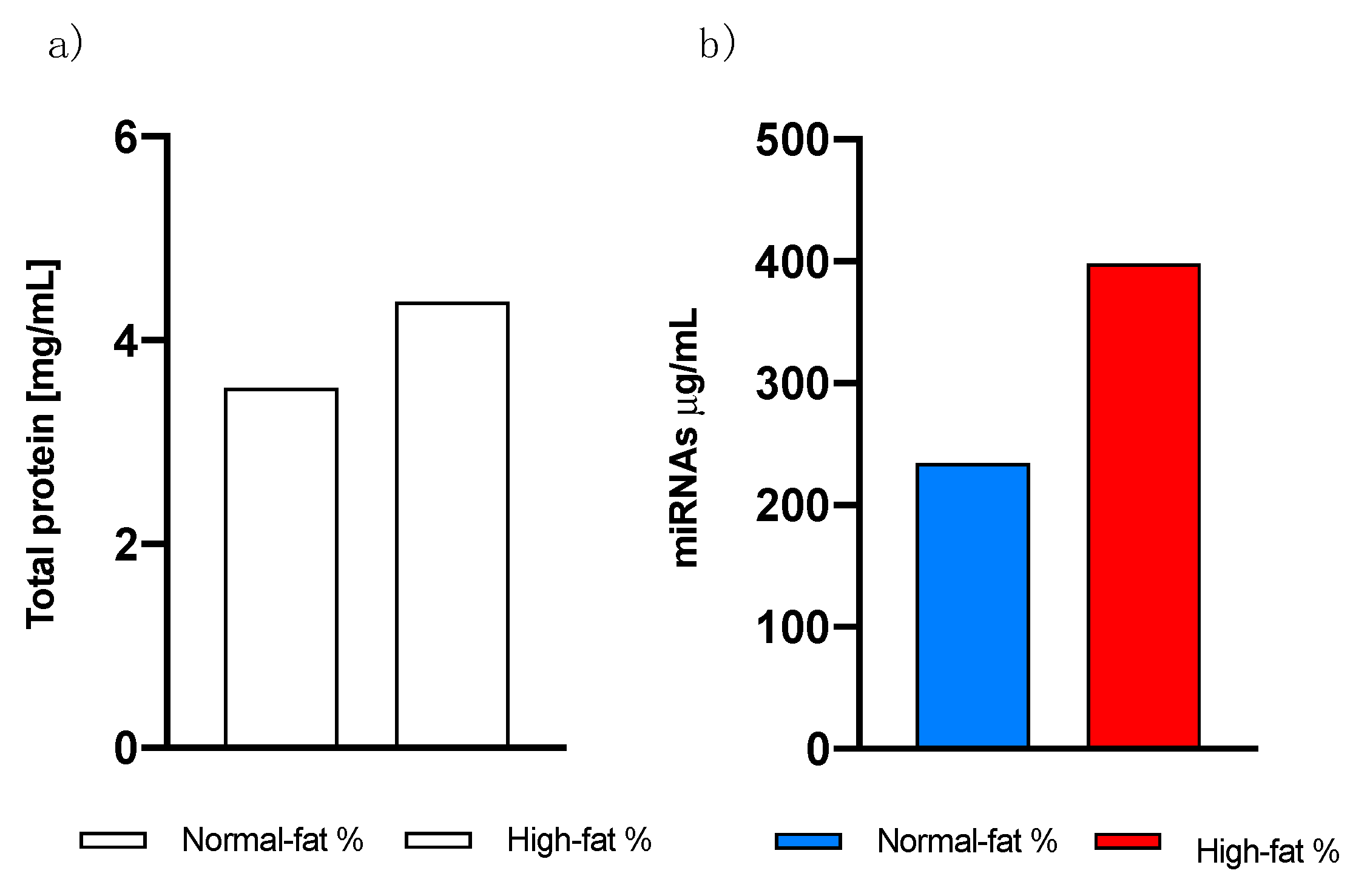
On the other hand, no difference in total exosome proteins was found between individuals with normal and high-fat percentages. As shown in
Figure 4, we saw an increasing pattern in high-fat individuals, suggesting that it is not the protein concentration that differs between these individuals but most likely the content in the exosomes.
2.3. No Differences in Exomes Morphology, Quality and Quantity Were Observed in Exosome Isolation Methods
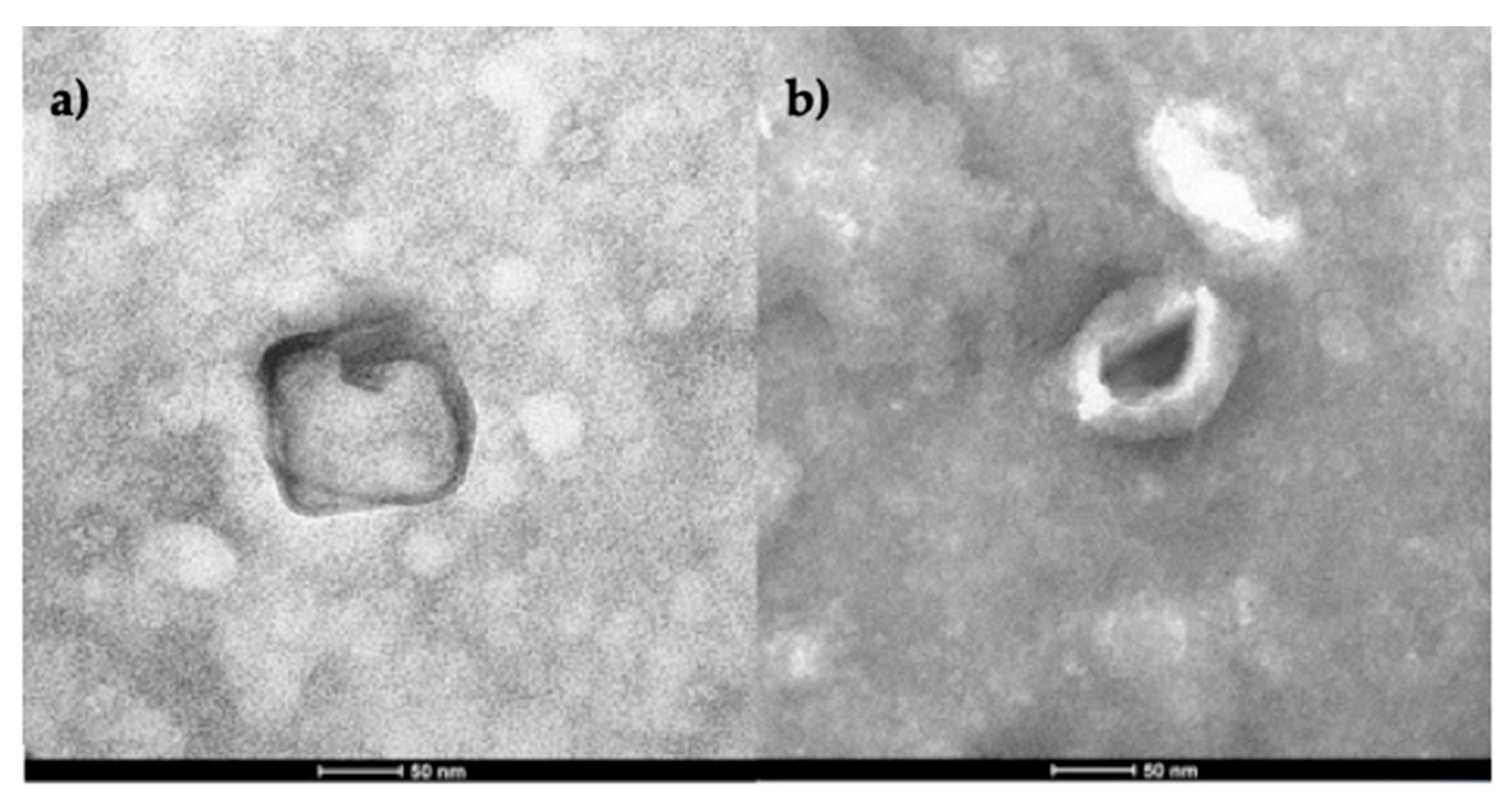
TEM and cryo-TEM were used to characterize exosomes isolated using the abovementioned techniques. In the next photomicrography, round and slightly elongated bilayer vesicles are compatible in morphology and size with exosomes. (
Figure 5).
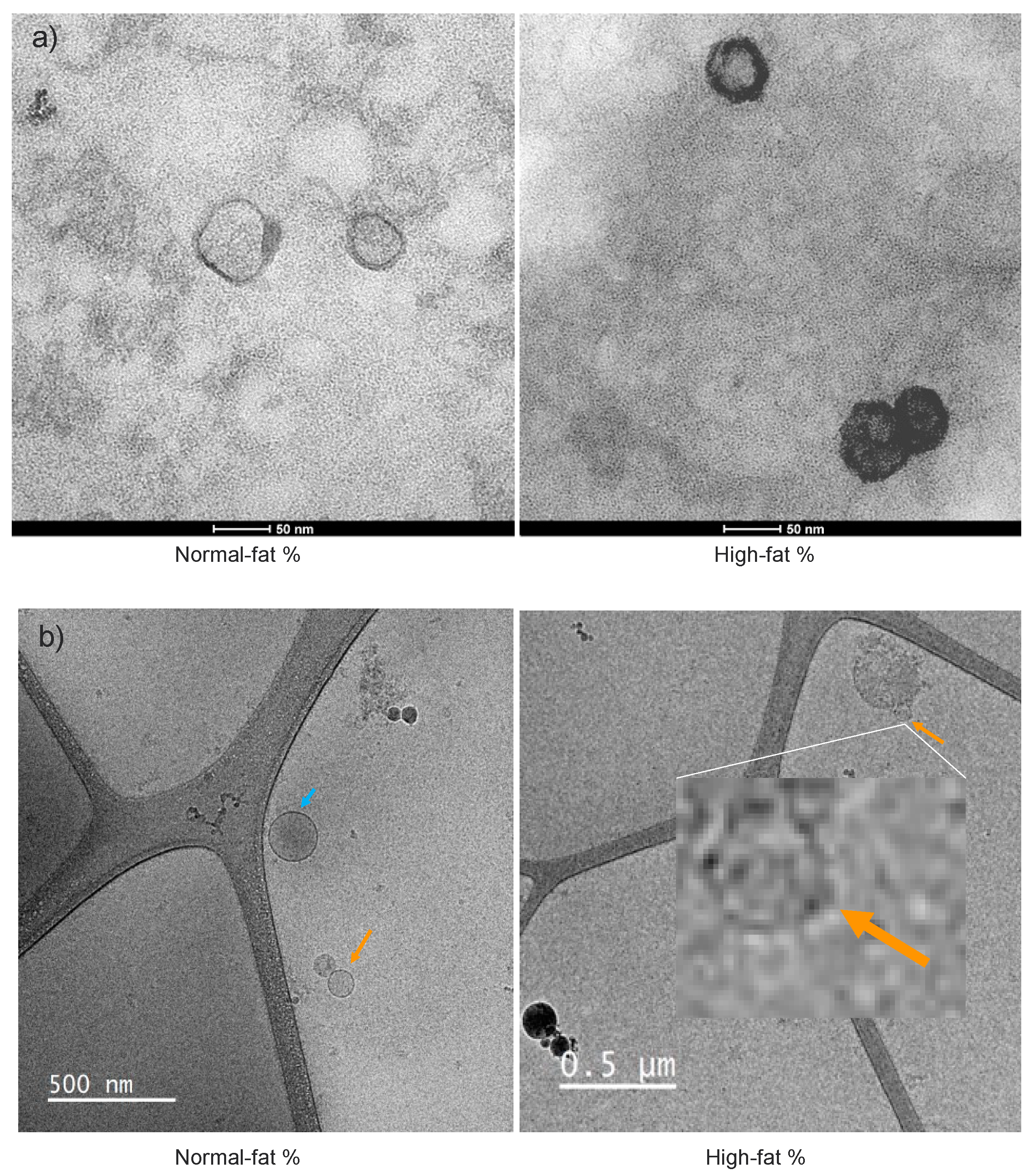
Additionally, exosomes were of similar quality, quantity, and morphology; at the same time, no differences were observed between individuals categorized by proportion of body fat (
Figure 6a). As seen in the same
Figure 6b, the SEC also entrains microvesicles larger than exosomes (∽230nm), and tiny contaminants.
2.4. CD9 and CD81 Differs in SEC Fractions between Normal and High-Fat Percentage Individuals
Since integrity, quality, and quantity in microscopy images and total protein did not differ, we chose an exosome isolation technique that was more cost-effective, time-efficient, and equipment-efficient for identifying CD9 and CD81 in plasma exosomes from individuals who had normal and high-fat percentages.
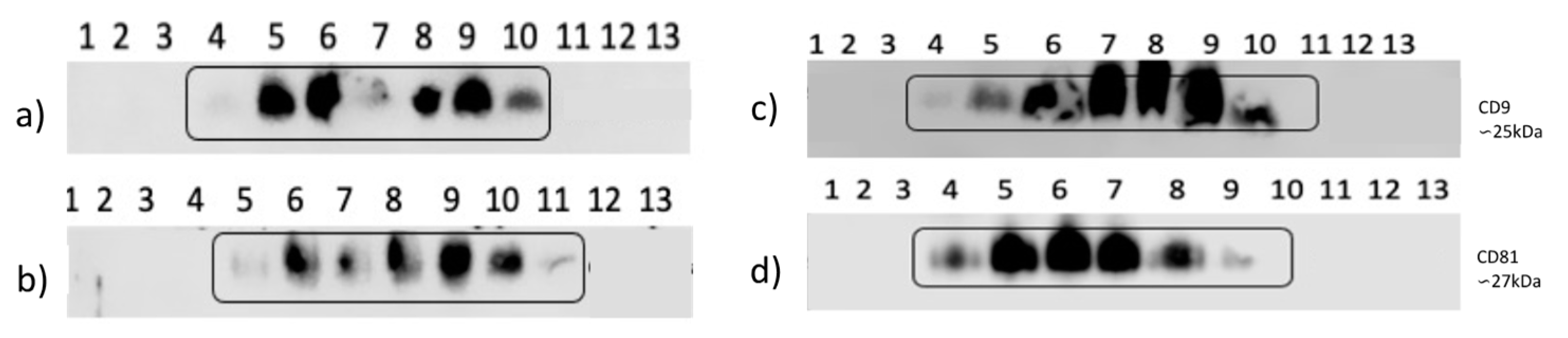
In normal-fat percentage individuals exists a displacement of the markers, CD9 is present in rails 4 to 10 (
Figure 7a) and for CD81, 5 to 11 (
Figure 7b), whereas in high- fat percentage individuals, there is only one rial more with presence of CD9 (rial 10) (
Figure 7c,d).
Under our experience we can resume some criteria for these three-exosome isolation technique in the
Table 1 below,
3. Discussion
The differentiation to cells with adipocyte phenotype allows simulating the hyper-trophy in the AT by stimulating with glucose uptake and storing in the of triglycerides [
22], suggesting that the excess and continuous intake plus the lack of energy expenditure is a delicate balance between the glucose concentration and the differentiation time as reported by Jackson et al. [
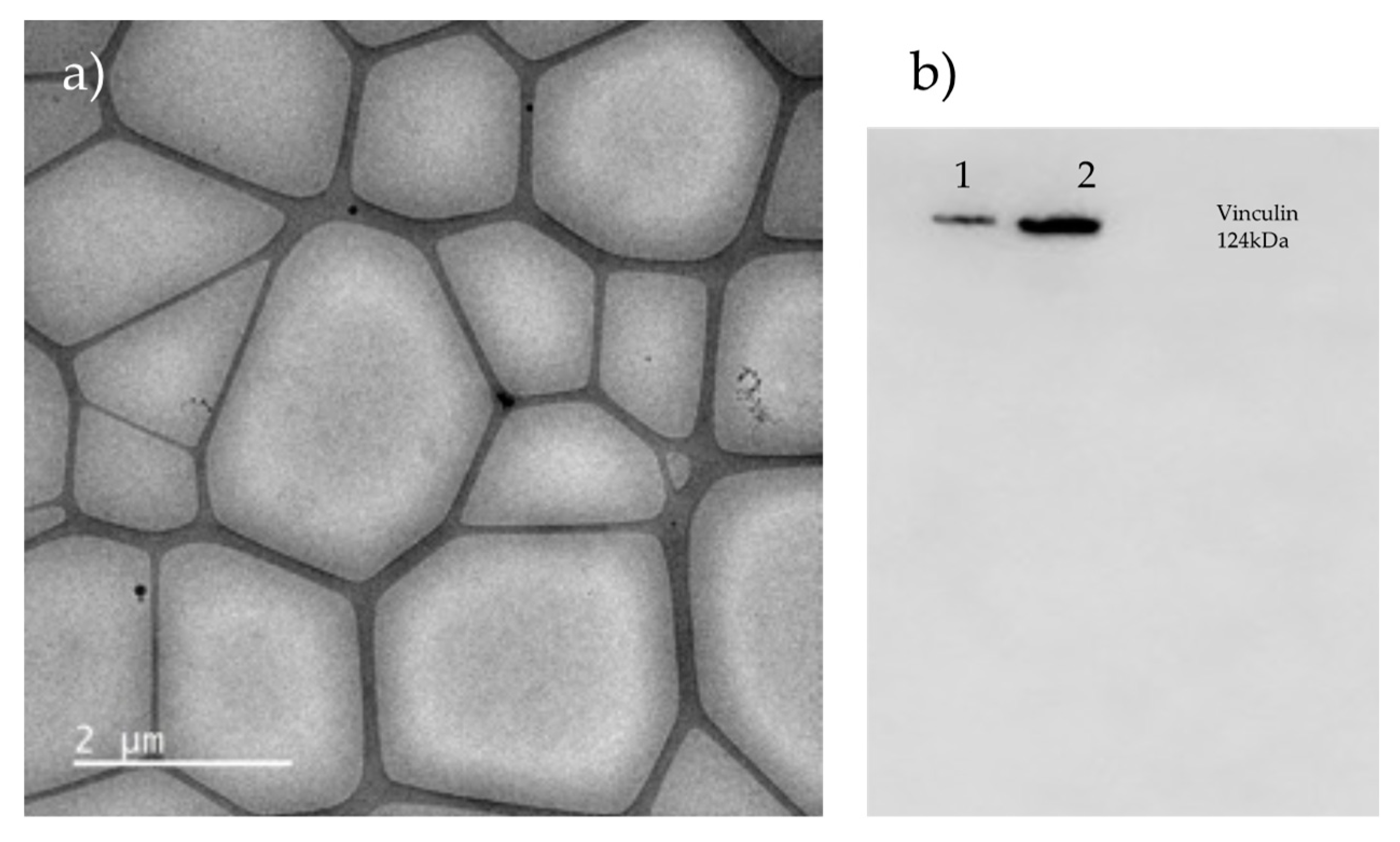
23], favoring fat accumulation in adipose tissue. Additionally, we corroborated that the morphology of cells subjected to 24 hours of hypoxia changes, as mentioned by Vogler M.
et al, who demonstrated the increase in vinculin positive focal cell contacts in hypoxia by microscopy imaging [
24], and like Synowiec
et al., did not detect a difference by western blot for vinculin [
25], as we did (
Figure A1.b).
Numerous methods have been employed for exosome isolation of 3T3-L1, each one with its advantage and purpose, as noted by Sidhom
et al. [
26], concluded that SEC is a good alternative in this instance, however, they advise an optimized combination method. We probe concentration particles (Centriprep YM-10, 10 kDa NMWL. Millipore) prior to SEC and DC respectively, and in conjunction with DC previously to SEC, as reported by Akbar
et al. [
27],. Regretfully, we were unable to obtained isolated exosomes, as confirmed by cryo-TEM and western blot (CD9 and CD81). Therefore, we propose that the supernatant’s initial volume and the additions of ultracentrifugation time, may be crucial, as the protocol of Nakatani et al. [
28].
After comparing the total protein content of the exosomes extracted using the three methods, we decided to assess the normal and high-fat percentage and total microRNAs using the simple, low-labor-time, and large-quantity samples. A commercial kit is a good alternative to nucleic acid subsequent analysis, even if we could not find a difference between them, as determined by Mohammad A. et al. when comparing one commercial kit with ultracentrifugation [
29]. Considering Wang Y. et al.'s evaluation of the exosome proteome in obese and non-obese individuals with T2DM [
30], we can propose that exosome concentration is linked to metabolic-associated disorders rather than fat content [
31,
32]
Many techniques have been used for exosome characterization [
33], depending on the information that the research looks for. In our case, we chose two that can be used for quantity (total exomes count) and quality (protein marker, structure, and size) analyses, electron microscopy (TEM and cryo-TEM) and western blot (CD9 and CD81 markers). Electron microscopy allows imaging of the single exosome, visualizing its size and morphology, counting per visual camp, and detecting the presence of contaminants. In our study, exosomes isolated by commercial kit and DC, demonstrated fewer contaminants, while SEC, presented tiny contamination and different microvesicles, as mentioned by Davidson S.
et al in their review [
34]., while DC and SEC allow isolated integral exosomes. To verify that we isolated exosomes we chose two tetraspanins present in the exome surface (CD9 and CD81), that were isolated by SEC. We analyzed each fraction of normal and high-fat percentage individuals. In both normal and high-fat percentage individuals, there is a shift of markers in SEC fractions, not because of the fat condition, but of the markers per se, the same pattern showed in carcinoma exosomes [
35], and circulating exosomes [
36], both isolated by SEC. We suggest that the length of the column, the volume fraction, and the biological sample may influence the presence of the marker in each fraction.
4. Materials and Methods
4.1. Samples
4.1.1. Plasma Sample
Blood collection was approved by the Comisión de Investigación y Ética del Antiguo Hospital Civil de Guadalajara “Fray Antonio Alcalde” O.P.D. HCG/CEI-0835/22, Nº. 130/22, and participants provided written, informed consent. Ten milliliters of blood were obtained from 120 individuals, 25-45 years old (women and men), in EDTA-coated tubes and allowed to sit at room temperature for 30 minutes. Whole blood was then centrifuged at 3000 g for 15 minutes at 4°C to separate plasma. The individuals were classified by fat percentage, excess was considered more than 25% in men and 35% in women.
4.1.2. Cell Culture
3T3-L1 cells were cultured on a polystyrene tissue culture flask t75 cm
2. Cell growth started with basal media, DMEM (Dulbecco’s modified Eagles medium. Merck. Cat D6429) with 10% FBS (Merck. F2442-500ML), for one week. This time point was referred to as day 0, to begin 3T3-L1 differentiation with MDI (DMEM Merck. Cat D6429 with 10% FBS (Merck. F2442-500ML) + 0.5 mM de IBMX (3-Isobutyl-1-methylxanthine) (SERVA. 26445.02. 500 mg), 1 μM dexamethasone (Dexa. MCE. Cat. HY-14648. 500 mg) + 10 μg/mL insulin (NeoBiotech. Cat. NB-58-0012. 5mg/ml)) for 4 days at 37 ºC and 5% CO
2, finally at day 5 we used MDII (DMEM Merck. Cat D6429 with 10% FBS (Deplete of exosomes by overnight ultracentrifugation at 100000 g 4ºC.
Figure A2.d–f) + 10 μg/mL insulin) for 43 days at 37 ºC and 5% CO
2, on this day cells were stained with oil-red O (ORO) ((Sigma-Aldrich Cat. O0625)). At day 19 one subculture was subjected to hypoxia by anaerobiosis (BD GasPak
TM EZ Lot. 581972) inside of a hermetic chamber (BD GasPak
TM EZ. Anaerobe Container System. Ref 260678) for 2.5 hours and 24 hours [
37].
4.2. Exosome Isolation and Characterization
4.2.1. Isolation, Differential Centrifugation
We used 2 mL of blood plasma and 100 mL of 3T3-L1 differentiated supernatant. All centrifugations were realized at 4ºC. We started by 300 g for 10 minutes (Heraeus Megafuge 2.0 R) discarded the pellet; 2000 g for 20 minutes (Heraeus Megafuge 2.0 R) discarded the pellet; 10000 g for 20 minutes (Beckman Coulter. Rotor JA-14. Jars NALGENE® 250 mL) discarded the pellet; 100000 g for 70 minutes (Beckman Couelter. Rotor T70i. Beckman Centrifuge Tubes 26.3 mL), discarded supernatant and resuspend the pellet with 500 μL of 1X PBS, 100000 g for 70 minutes (Thermo Sorvall Discovery M150 SE Floor Micro-Ultracentrifuge 150K RPM. Rotor S150-AT Eppendorf Tubes PP 1 mL) and resuspend in 200 μL of 1X PBS [
38].
4.2.2. Isolation, Size Exclusion Chromatography (SEC)
We used 1mL of blood plasma, and 1 mL of 3T3-L1 differentiated supernatant to pass through each column (Econo-Pac® Chromatography Columns, Pkg of 50 #7321010 with Sepharose™ CL-2B) with 1X PBS filtrated through a 0.22 μm filter (CORNING 45 mm diameter). We collected 200 μL of 15 fractions, and finally, 4 to 9 fractions and 12 to 15 fractions were mixed respectively [
13].
4.2.3. Isolation, Commercial KIT
Total Exosome Isolation (from plasma) (Invitrogen Cat. No. 4404450) was used. All centrifugations were realized at room temperature. Starting with 500 μL of blood plasma and 500 μL of 3T3-L1 differentiated supernatant, we centrifugated at 10000 g for 20 minutes, added 0.5 volumes of saline solution to the supernatant, mixed by vortex, added 0.2 volume of exosome precipitation reagent, pipetted up and down. This mix was incubated for 10 minutes at room temperature, then centrifugated at 10000 g for 5 minutes, the supernatant was discarded. Finally, the pellet was resuspended in 50 μL of saline solution.
4.2.4. Characterization, Cryo-TEM and TEM
For each electron microscopy, 3 μL of blood plasma and 3T3-L1 differentiated supernatant, was used. Transmission Electron Microscopy (FEI Technology, Model Tecnai Spirit BioTwin, software FEI TIA 4.15.) and cryo-TEM (Jeol JM-2011, Tokyo, Japan), were analyzed by technicians of the Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Ciudad de México-México and Serveis Científico-tècnics UAB, Barelona-España, respectively.
4.2.5. Characterization, Western Blot
Starting with exosome lysis by adding 20 μL to 150 μL of sample in ice-cold, then incubating in rotational agitation at 4ºC for 1 hour, then centrifugate at 15000 g for 15 minutes at 4ºC, the supernatant was measured in the spectrophotometer (NanoDrop 1000 UV Visible Spectrophotometer, Thermo Scientific). We charged 5 μg of total protein on 14% acrylamide gel. Exosomes markers were quantified using mouse monoclonal CD9 (1, 1000; antibodies.com. Anti-CD9 (MEM-61) (A86089)), CD81 (1, 1000; antibodies.com. Anti-CD81 (M38) (A86719), and Vinculin (1, 500 Invitrogen Ref. MA5-11690), we used anti-mouse HRP (1, 5000; abcam. NA931V ECL Anti-mouse IgG), revealed with ECL (BioRad Clarity Western ECL Substrate, 500 ml #1705061) in the spectrophotometer (Li-cor, Odyssey XF, LI-COR Acquisition Software).
4.3. Statistical Analyses
Mann-Whitney test was used to compare the differing of total protein and miRNAs concentration between normal and high-fat percentage. p value < 0.05 was considered significant. GraphPad Prism version 8.4.0 for macOS were used for data analysis and graphing.
5. Conclusions
According to our results, the concentration of the circulating exosomes is not different between normal and high-fat percentage individuals, but rather their composition. We encourage further analysis of the content of circulating exosomes as residents of adipose tissue that can show this difference.
We emphasize the importance of selecting the appropriate methodology for exosome isolation, which depends on the specific research objectives. When prioritizing exosome quantity and content, we suggest employing SEC or a commercial kit for plasma samples when seeking higher yields of intact exosomes. However, if purity is the priority, DC is the best method.
In this study, exosomes were not obtained from the supernatant of 3T3-L1 cells. Therefore, our results highlight the use of FBS depleted of microvesicles in 3T3-L1 differentiation. We recommend increasing the supernatant volume and/or the ultracentrifugation process or exploring a commercial exosome isolation kit designed specifically for cell culture samples.
Author Contributions
Conceptualization, J.N., I.Á. and Rosa Navarro; methodology, J.N. and J.L.; formal analysis, J.N., I.Á. and R.N.; investigation, J.N., J.L., J.C., A.F., I.Á. and R.N.; resources, J.C., A.F., I.Á. and R.N.; data curation, J.N., I.Á. and R.N., A; writing—original draft preparation, J.N., I.Á. and R.N.; writing—review and editing, J.N., J.L., J.C., A.F., P.M., I.Á. and R.N.; visualization, J.N., J.L., J.C., A.F., P.M., I.Á. and R.N.; supervision, I.Á. and R.N.; project administration, I.Á. and R.N.; funding acquisition, J.C., A.F., I.Á. and R.N.
Funding
This research was funded by Doctoral scholarship number (CVU), 1103690, by Universidad de Guadalajara.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of Comisión de Investigación y Ética del Antiguo Hospital Civil de Guadalajara “Fray Antonio Alcalde” O.P.D. HCG/CEI-0835/22, Nº. 130/22 at 23-05-2022” for studies involving humans.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Data are available on reasonable request to the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A
For the 3T3-L1 exosome, we combined two techniques, DC and SEC, and ultrafiltration (passed by filter 0,5 μm, filter 0,22 μm, 10 000 x g 30 minutes 4ºC, filter 0,15 μm) and SEC. Still, we didn’t identify exosomes by electron microscopy (
Figure A1.a) nor by western blot (
Figure A1.b). We suggest proving a commercial kit specific for cell culture and increasing the ultracentrifugation time; another option could be stimulating the cells for increase the exosome secretion.
Figure A1.
Exosome isolation from differentiated 3T3-L1 isolated by SEC. a) Cryo-TEM without exosomes eighter contaminants. b) Western blot for CD9, CD81 and Vinculin as constitutive protein. Rail 1= supernatant in normoxia. Rail 2= supernatant in hypoxia.
Figure A1.
Exosome isolation from differentiated 3T3-L1 isolated by SEC. a) Cryo-TEM without exosomes eighter contaminants. b) Western blot for CD9, CD81 and Vinculin as constitutive protein. Rail 1= supernatant in normoxia. Rail 2= supernatant in hypoxia.
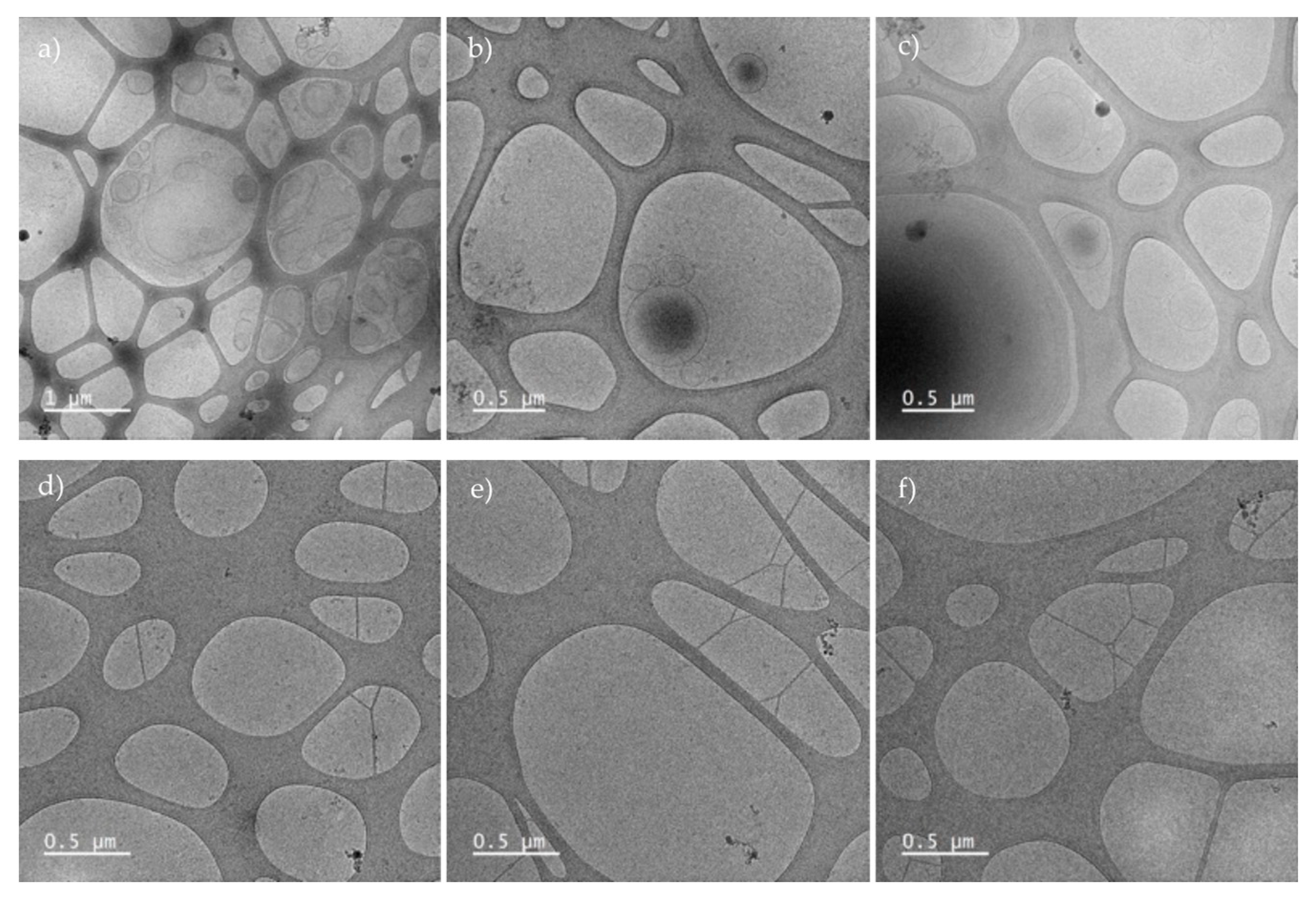
For cell culture, we obtained FBS depleted of exosomes, verify by cryo-TEM before and after overnight ultracentrifugation (
Figure A2). To ensure that the serum did not affect cell growth and division, the culture was maintained with this serum for one week prior to differentiation.
Figure A2.
Cryo-TEM of FBS microvesicles isolated by ultracentrifugation. a-c) microvesicles previous overnight ultracentrifugation d-f) FBS depleted of microvesicles ready for cell culture.
Figure A2.
Cryo-TEM of FBS microvesicles isolated by ultracentrifugation. a-c) microvesicles previous overnight ultracentrifugation d-f) FBS depleted of microvesicles ready for cell culture.
References
- Thery C, Amigorena S, Raposo G, Clayton A, Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol 2006, Chapter 3, Unit 3 22. [CrossRef]
- Lei LM, Lin X, Xu F, Shan SK, Guo B, Li FX, Zheng MH, Wang Y, Xu QS, Yuan LQ, Exosomes and Obesity-Related Insulin Resistance. Front Cell Dev Biol 2021, 9, 651996. [CrossRef]
- Dance A, The body’s tiny cargo carriers. In Knowable magazine. Edited by; 2019.
- Jiang X, You L, Zhang Z, Cui X, Zhong H, Sun X, Ji C, Chi X, Biological Properties of Milk-Derived Extracellular Vesicles and Their Physiological Functions in Infant. Frontiers in Cell and Developmental Biology 2021, 9. [CrossRef]
- Ragusa M, Barbagallo C, Statello L, Caltabiano R, Russo A, Puzzo L, Avitabile T, Longo A, Toro MD, Barbagallo D, et al. miRNA profiling in vitreous humor, vitreal exosomes and serum from uveal melanoma patients, Pathological and diagnostic implications. Cancer Biol Ther 2015, 16, 1387–1396. [CrossRef] [PubMed]
- Jan AT, Rahman S, Badierah R, Lee EJ, Mattar EH, Redwan EM, Choi I, Expedition into Exosome Biology, A Perspective of Progress from Discovery to Therapeutic Development. Cancers 2021, 13, 1157. [CrossRef]
- Shamsi F, Zhang H, Tseng YH, MicroRNA Regulation of Brown Adipogenesis and Thermogenic Energy Expenditure. Front Endocrinol 2017, 8, 205. [CrossRef] [PubMed]
- Mei R, Qin W, Zheng Y, Wan Z, Liu L, Role of Adipose Tissue Derived Exosomes in Metabolic Disease. Frontiers in Endocrinology 2022, 13. [CrossRef]
- Bond ST, Calkin AC, Drew BG, Adipose-Derived Extracellular Vesicles, Systemic Messengers and Metabolic Regulators in Health and Disease. Front Physiol 2022, 13, 837001. [CrossRef] [PubMed]
- Fasshauer M, Bluher M, Adipokines in health and disease. Trends Pharmacol Sci 2015, 36, 461–470. [CrossRef] [PubMed]
- Zhao R, Zhao T, He Z, Cai R, Pang W, Composition, isolation, identification and function of adipose tissue-derived exosomes. Adipocyte 2021, 10, 587–604. [CrossRef]
- Yang XX, Sun C, Wang L, Guo XL, New insight into isolation, identification techniques and medical applications of exosomes. Journal of Controlled Release 2019, 308, 119–129. [CrossRef]
- Muller L, Hong CS, Stolz DB, Watkins SC, Whiteside TL, Isolation of biologically-active exosomes from human plasma. J Immunol Methods 2014, 411, 55–65. [CrossRef] [PubMed]
- Cheng Y, Qu X, Dong Z, Zeng Q, Ma X, Jia Y, Li R, Jiang X, Williams C, Wang T, et al. Comparison of serum exosome isolation methods on co-precipitated free microRNAs. PeerJ 2020, 8, e9434. [CrossRef] [PubMed]
- Chen J, Li P, Zhang T, Xu Z, Huang X, Wang R, Du L, Review on Strategies and Technologies for Exosome Isolation and Purification. Front Bioeng Biotechnol 2021, 9, 811971. [CrossRef]
- Rech J, Getinger-Panek A, Gałka S, Bednarek I, Origin and Composition of Exosomes as Crucial Factors in Designing Drug Delivery Systems. Applied Sciences 2022, 12, 12259. [CrossRef]
- Koritzinsky EH, Street JM, Star RA, Yuen PS, Quantification of Exosomes. J Cell Physiol 2017, 232, 1587–1590. [CrossRef]
- Whitehead C, Luwor R, Morokoff A, Kaye A, Stylli S, Cancer exosomes in cerebrospinal fluid. Translational Cancer Research 2017, 6.
- Deng FY, Miller J, A review on protein markers of exosome from different bio-resources and the antibodies used for characterization. Journal of Histotechnology 2019, 42, 226–239. [CrossRef] [PubMed]
- Chernyshev VS, Rachamadugu R, Tseng YH, Belnap DM, Jia Y, Branch KJ, Butterfield AE, Pease LF, 3rd, Bernard PS, Skliar M, Size and shape characterization of hydrated and desiccated exosomes. Anal Bioanal Chem 2015, 407, 3285–3301. [CrossRef]
- Li D, Luo H, Ruan H, Chen Z, Chen S, Wang B, Xie Y, Isolation and identification of exosomes from feline plasma, urine and adipose-derived mesenchymal stem cells. BMC Vet Res 2021, 17, 272. [CrossRef]
- Lee MJ, Wu Y, Fried SK, A modified protocol to maximize differentiation of human preadipocytes and improve metabolic phenotypes. Obesity (Silver Spring) 2012, 20, 2334–2340. [CrossRef]
- Jackson HC, Pheiffer C, Jack B, Africander D, Time- and glucose-dependent differentiation of 3T3-L1 adipocytes mimics dysfunctional adiposity. Biochem Biophys Res Commun 2023, 671, 286–291. [CrossRef] [PubMed]
- Vogler M, Vogel S, Krull S, Farhat K, Leisering P, Lutz S, Wuertz CM, Katschinski DM, Zieseniss A, Hypoxia modulates fibroblastic architecture, adhesion and migration, a role for HIF-1alpha in cofilin regulation and cytoplasmic actin distribution. PLoS One 2013, 8, e69128. [CrossRef]
- Synowiec A, Brodaczewska K, Wcisło G, Majewska A, Borkowska A, Filipiak-Duliban A, Gawrylak A, Wilkus K, Piwocka K, Kominek A, et al. Hypoxia, but Not Normoxia, Reduces Effects of Resveratrol on Cisplatin Treatment in A2780 Ovarian Cancer Cells, A Challenge for Resveratrol Use in Anticancer Adjuvant Cisplatin Therapy. International Journal of Molecular Sciences 2023, 24, 5715. [CrossRef]
- Sidhom K, Obi PO, Saleem A, A Review of Exosomal Isolation Methods, Is Size Exclusion Chromatography the Best Option? International Journal of Molecular Sciences 2020, 21, 6466. [CrossRef] [PubMed]
- Akbar N, Pinnick KE, Paget D, Choudhury RP, Isolation and Characterization of Human Adipocyte-Derived Extracellular Vesicles using Filtration and Ultracentrifugation. J Vis Exp 2021. [CrossRef]
- Nakatani E, Naito Y, Ishibashi K, Ohkura N, Atsumi GI, Extracellular Vesicles Derived from 3T3-L1 Adipocytes Enhance Procoagulant Activity. Biol Pharm Bull 2022, 45, 178–183. [CrossRef] [PubMed]
- Aziz MA, Seo B, Hussaini HM, Hibma M, Rich AM, Comparing Two Methods for the Isolation of Exosomes. J Nucleic Acids 2022, 2022, 8648373. [CrossRef]
- Wang Y, Wu Y, Yang S, Chen Y, Comparison of Plasma Exosome Proteomes Between Obese and Non-Obese Patients with Type 2 Diabetes Mellitus. Diabetes Metab Syndr Obes 2023, 16, 629–642. [CrossRef]
- Kwan HY, Chen M, Xu K, Chen B, The impact of obesity on adipocyte-derived extracellular vesicles. Cell Mol Life Sci 2021. [CrossRef]
- Son T, Jeong I, Park J, Jun W, Kim A, Kim O-K, Adipose tissue-derived exosomes contribute to obesity-associated liver diseases in long-term high-fat diet-fed mice, but not in short-term. Frontiers in Nutrition 2023, 10. [CrossRef]
- Lai JJ, Chau ZL, Chen SY, Hill JJ, Korpany KV, Liang NW, Lin LH, Lin YH, Liu JK, Liu YC, et al. Exosome Processing and Characterization Approaches for Research and Technology Development. Adv Sci 2022, 9, e2103222. [CrossRef]
- Davidson SM, Boulanger CM, Aikawa E, Badimon L, Barile L, Binder CJ, Brisson A, Buzas E, Emanueli C, Jansen F, et al. Methods for the identification and characterization of extracellular vesicles in cardiovascular studies, from exosomes to microvesicles. Cardiovasc Res 2023, 119, 45–63. [CrossRef] [PubMed]
- Sakha S, Muramatsu T, Ueda K, Inazawa J, Exosomal microRNA miR-1246 induces cell motility and invasion through the regulation of DENND2D in oral squamous cell carcinoma. Sci Rep 2016, 6, 38750. [CrossRef]
- Yarana C, Siwaponanan P, Maneechote C, Khuanjing T, Ongnok B, Prathumsap N, Chattipakorn SC, Chattipakorn N, Pattanapanyasat K, Extracellular Vesicles Released after Doxorubicin Treatment in Rats Protect Cardiomyocytes from Oxidative Damage and Induce Pro-Inflammatory Gene Expression in Macrophages. Int J Mol Sci 2022, 23. [CrossRef]
- Etesami B, Ghaseminezhad S, Nowrouzi A, Rashidipour M, Yazdanparast R, Investigation of 3T3-L1 Cell Differentiation to Adipocyte, Affected by Aqueous Seed Extract of Phoenix Dactylifera L. Rep Biochem Mol Biol 2020, 9, 14–25. [CrossRef]
- Lobb RJ, Becker M, Wen SW, Wong CS, Wiegmans AP, Leimgruber A, Moller A, Optimized exosome isolation protocol for cell culture supernatant and human plasma. J Extracell Vesicles 2015, 4, 27031. [CrossRef]
Figure 1.
T3-L1 adipocyte differentiation. Images illustrate the transition of 3T3-L1 cells from induction (Day 0) to 40 days post-induction. a) Initially, cells exhibit a fibroblast-like phenotype. b-e) As differentiation progresses, their morphology changes, and they start accumulating lipid droplets internally. Phase-contrast images. Left side with 100 µm scale (10X), right 50 µm (40X).
Figure 1.
T3-L1 adipocyte differentiation. Images illustrate the transition of 3T3-L1 cells from induction (Day 0) to 40 days post-induction. a) Initially, cells exhibit a fibroblast-like phenotype. b-e) As differentiation progresses, their morphology changes, and they start accumulating lipid droplets internally. Phase-contrast images. Left side with 100 µm scale (10X), right 50 µm (40X).
Figure 2.
Differentiated 3T3-L1 subjected to hypoxia. a). Day 19, 2.5 of hypoxia without adipocyte morphological changes. b). Day 20, after 24 hours of hypoxia, morphological changes were observed (orange arrow). c). Staining with oil-red O (ORO) at day 43 in normoxia. The different patterns of lipid droplet accumulation can be seen in red. Phase-contrast images. Left side with 100 µm scale (10X), right 50 µm (40X).
Figure 2.
Differentiated 3T3-L1 subjected to hypoxia. a). Day 19, 2.5 of hypoxia without adipocyte morphological changes. b). Day 20, after 24 hours of hypoxia, morphological changes were observed (orange arrow). c). Staining with oil-red O (ORO) at day 43 in normoxia. The different patterns of lipid droplet accumulation can be seen in red. Phase-contrast images. Left side with 100 µm scale (10X), right 50 µm (40X).
Figure 3.
Exosomes total protein. Protein levels were estimated in intact exosomes using spectrophotometer (NanoDrop UV-Vis 1000).
Figure 3.
Exosomes total protein. Protein levels were estimated in intact exosomes using spectrophotometer (NanoDrop UV-Vis 1000).
Figure 4.
Analysis from exosomes isolated by commercial kit. a) Total protein classified by fat percentage. b) Total miRNAs concentration.
Figure 4.
Analysis from exosomes isolated by commercial kit. a) Total protein classified by fat percentage. b) Total miRNAs concentration.
Figure 5.
TEM plasma exosomes. Isolated by a) Commercial kit (Invitrogen Cat. No. 4404450), b). DC. In both cases, the morphology and size (∽50nm) correspond to exosomes. Scale 50 nm. Abbreviation DC= differential centrifugation.
Figure 5.
TEM plasma exosomes. Isolated by a) Commercial kit (Invitrogen Cat. No. 4404450), b). DC. In both cases, the morphology and size (∽50nm) correspond to exosomes. Scale 50 nm. Abbreviation DC= differential centrifugation.
Figure 6.
TEM and cryo-TEM exosomes photomicrography. a). TEM of exosomes of ∽50 nm isolated by commercial kit (Invitrogen Cat. No. 4404450). b). Cryo-TEM of exosomes of ∽125 nm (orange arrow) isolated by SEC, as well as microvesicles of highest dimension (blue arrow) ∽230 nm.
Figure 6.
TEM and cryo-TEM exosomes photomicrography. a). TEM of exosomes of ∽50 nm isolated by commercial kit (Invitrogen Cat. No. 4404450). b). Cryo-TEM of exosomes of ∽125 nm (orange arrow) isolated by SEC, as well as microvesicles of highest dimension (blue arrow) ∽230 nm.
Figure 7.
CD9 and CD81 markers of exosomes isolated from, normal-fat percentage individuals. a) CD9 western blot, rails 1-3 and 11-13 without exosomes in exclusion fractions, rails 4 -10 presence of exosomes in exclusion fractions. b). Western Blot of CD81, rails 1-4 and 12-13 without exosomes in the exclusion fractions, rails 5 -11 present exosomes in the exclusion fractions, isolated by SEC. High-fat percentage individuals. c). CD9 western blot, rails 1-3 and 11-13 without exosomes in exclusion fractions, rails 4 -10 present exosomes in exclusion fractions. d). Western Blot of CD81, rails 1-3 and 10-13 without exosomes in the exclusion fractions, rails 4 -10 present exosomes in the exclusion fractions, isolated by SEC.
Figure 7.
CD9 and CD81 markers of exosomes isolated from, normal-fat percentage individuals. a) CD9 western blot, rails 1-3 and 11-13 without exosomes in exclusion fractions, rails 4 -10 presence of exosomes in exclusion fractions. b). Western Blot of CD81, rails 1-4 and 12-13 without exosomes in the exclusion fractions, rails 5 -11 present exosomes in the exclusion fractions, isolated by SEC. High-fat percentage individuals. c). CD9 western blot, rails 1-3 and 11-13 without exosomes in exclusion fractions, rails 4 -10 present exosomes in exclusion fractions. d). Western Blot of CD81, rails 1-3 and 10-13 without exosomes in the exclusion fractions, rails 4 -10 present exosomes in the exclusion fractions, isolated by SEC.
Table 1.
Comparation between three methods for exosome isolation.
Table 1.
Comparation between three methods for exosome isolation.
| Isolation technique |
Advantage |
Disadvantage |
| Differential centrifugation (DC) |
Purity (exosome size)
Economic
Many samples at the same time (ultracentrifugation rotor tubes capacity)
High yield |
Time lab
Expensive equipment
Pressure damages the exosomes integrity |
| Size exclusion chromatography (SEC) |
Economical material
Non-destructive |
Time lab
Limit in the sample’s quantity to work at the same time.
Low yield |
| Commercial Kit |
Fast procedure
Many samples at the same time (centrifugation rotor tubes capacity)
No expensive either complicated equipment
Easy technique
High yield |
No economic
Kit stability
No high purity (for further proteomic analysis) |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).