Submitted:
09 May 2024
Posted:
10 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Cell Culture and CYP450 Enzyme Induction
2.3. In Vitro Cell Viability Assay
2.4. Lactate Dehydrogenase (LDH), Aspartate Aminotransferase (AST), and Aspartate Aminotransferase (AST) Assays for Detecting Liver Injury Markers
2.5. Albumin and Urea Secretion
2.6. Quantitative RT-PCR Analysis of CYP450 Enzyme mRNA
2.7. Statistical Analysis
3. Result
3.1. Cytotoxicities of ADR and AAP to HepG2 and HepaRG Cells Depending on Culture Type
3.2. Detection of Drug-Induced Hepatic Injury Markers
3.3. Analysis of Hepatocyte Specific Factors ALB and Urea Secretion
3.4. Compare Expression Levels CYP450 Enzymes after Drugs Treatment
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kaplowitz, N. Idiosyncratic drug hepatotoxicity. Nature reviews Drug discovery 2005, 4, 489–49. [Google Scholar] [CrossRef]
- Abboud, G.; Kaplowitz, N. Drug-induced liver injury. Drug safety 2007, 30, 277–294. [Google Scholar] [CrossRef]
- Olson, H.; Betton, G.; Robinson, D.; Thomas, K.; Monro, A.; Kolaja, G.; Lilly, P.; Sanders, J.; Sipes, G.; Bracken, W. Concordance of the toxicity of pharmaceuticals in humans and in animals. Regulatory toxicology and pharmacology 2000, 32, 56–67. [Google Scholar] [CrossRef]
- Lin, C.; Khetani, S.R. Advances in engineered liver models for investigating drug-induced liver injury. BioMed research international 2016, 2016. [Google Scholar] [CrossRef]
- Edmondson, R.; Broglie, J.J.; Adcock, A.F.; Yang, L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay and drug development technologies 2014, 12, 207–218. [Google Scholar] [CrossRef]
- Wrzesinski, K.; J Fey, S. From 2D to 3D-a new dimension for modelling the effect of natural products on human tissue. Current Pharmaceutical Design 2015, 21, 5605–5616. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, S.-T. High-throughput 3-D cell-based proliferation and cytotoxicity assays for drug screening and bioprocess development. Journal of biotechnology 2011, 151, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Fey, S.J.; Wrzesinski, K. Determination of drug toxicity using 3D spheroids constructed from an immortal human hepatocyte cell line. Toxicological sciences 2012, 127, 403–411. [Google Scholar] [CrossRef]
- Gunness, P.; Mueller, D.; Shevchenko, V.; Heinzle, E.; Ingelman-Sundberg, M.; Noor, F. 3D organotypic cultures of human HepaRG cells: a tool for in vitro toxicity studies. Toxicological sciences 2013, 133, 67–78. [Google Scholar] [CrossRef]
- Bell, C.C.; Hendriks, D.F.; Moro, S.M.; Ellis, E.; Walsh, J.; Renblom, A.; Fredriksson Puigvert, L.; Dankers, A.C.; Jacobs, F.; Snoeys, J. Characterization of primary human hepatocyte spheroids as a model system for drug-induced liver injury, liver function and disease. Scientific reports 2016, 6, 25187. [Google Scholar] [CrossRef]
- Hart, S.N.; Li, Y.; Nakamoto, K.; Subileau, E.-a.; Steen, D.; Zhong, X.-b. A comparison of whole genome gene expression profiles of HepaRG cells and HepG2 cells to primary human hepatocytes and human liver tissues. Drug metabolism and disposition 2010, 38, 988–994. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, P.J.; Edvardsson, A. Validation of a multiparametric, high-content-screening assay for predictive/investigative cytotoxicity: evidence from technology transfer studies and literature review. Chemical Research in Toxicology 2017, 30, 804–829. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, P.J.; Irwin, W.; Diaz, D.; Howard-Cofield, E.; Krejsa, C.M.; Slaughter, M.R.; Gao, B.; Kaludercic, N.; Angeline, A.; Bernardi, P. High concordance of drug-induced human hepatotoxicity with in vitro cytotoxicity measured in a novel cell-based model using high content screening. Archives of toxicology 2006, 80, 580–604. [Google Scholar] [CrossRef] [PubMed]
- McGinnity, D.F.; Zhang, G.; Kenny, J.R.; Hamilton, G.A.; Otmani, S.; Stams, K.R.; Haney, S.; Brassil, P.; Stresser, D.M.; Riley, R.J. Evaluation of multiple in vitro systems for assessment of CYP3A4 induction in drug discovery: human hepatocytes, pregnane X receptor reporter gene, and Fa2N-4 and HepaRG cells. Drug metabolism and disposition 2009, 37, 1259–1268. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Lechon, M.; Lahoz, A.; Gombau, L.; Castell, J.; Donato, M. In vitro evaluation of potential hepatotoxicity induced by drugs. Current pharmaceutical design 2010, 16, 1963–1977. [Google Scholar] [CrossRef] [PubMed]
- Tolosa, L.; Pinto, S.; Donato, M.T.; Lahoz, A.; Castell, J.V.; O’Connor, J.E.; Gómez-Lechón, M.J. Development of a multiparametric cell-based protocol to screen and classify the hepatotoxicity potential of drugs. Toxicological Sciences 2012, 127, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Tham, N.T.; Hwang, S.-R.; Bang, J.-H.; Yi, H.; Park, Y.-I.; Kang, S.-J.; Kang, H.-G.; Kim, Y.-S.; Ku, H.-O. High-content analysis of in vitro hepatocyte injury induced by various hepatotoxicants. Journal of Veterinary Science 2019, 20, 34. [Google Scholar] [CrossRef]
- Huang, S.M.; Temple, R.; Throckmorton, D.; Lesko, L. Drug interaction studies: study design, data analysis, and implications for dosing and labeling. Clinical Pharmacology & Therapeutics 2007, 81, 298–304. [Google Scholar]
- Kleijn, A.; Kloezeman, J.; Balvers, R.; Kaaij, M.v.d.; Dirven, C.; Leenstra, S.; Lamfers, M. A systematic comparison identifies an ATP-based viability assay as most suitable read-out for drug screening in glioma stem-like cells. Stem cells international 2016, 2016. [Google Scholar] [CrossRef]
- Crouch, S.; Kozlowski, R.; Slater, K.; Fletcher, J. The use of ATP bioluminescence as a measure of cell proliferation and cytotoxicity. Journal of immunological methods 1993, 160, 81–88. [Google Scholar] [CrossRef]
- LeCluyse, E.L.; Witek, R.P.; Andersen, M.E.; Powers, M.J. Organotypic liver culture models: meeting current challenges in toxicity testing. Critical reviews in toxicology 2012, 42, 501–548. [Google Scholar] [CrossRef]
- Gomez-Lechon, M.; Donato, M.; Lahoz, A.; Castell, J. Cell lines: a tool for in vitro drug metabolism studies. Current drug metabolism 2008, 9, 1–11. [Google Scholar] [CrossRef]
- Wrzesinski, K.; Magnone, M.C.; Hansen, L.V.; Kruse, M.E.; Bergauer, T.; Bobadilla, M.; Gubler, M.; Mizrahi, J.; Zhang, K.; Andreasen, C.M. HepG2/C3A 3D spheroids exhibit stable physiological functionality for at least 24 days after recovering from trypsinisation. Toxicology research 2013, 2, 163–172. [Google Scholar] [CrossRef]
- Bokhari, M.; Carnachan, R.J.; Cameron, N.R.; Przyborski, S.A. Culture of HepG2 liver cells on three dimensional polystyrene scaffolds enhances cell structure and function during toxicological challenge. Journal of anatomy 2007, 211, 567–576. [Google Scholar] [CrossRef]
- McGill, M.R. The past and present of serum aminotransferases and the future of liver injury biomarkers. EXCLI journal 2016, 15, 817. [Google Scholar]
- Spinella, R.; Sawhney, R.; Jalan, R. Albumin in chronic liver disease: structure, functions and therapeutic implications. Hepatology international 2016, 10, 124–132. [Google Scholar] [CrossRef]
- Nagamani, S.C.; Ali, S.; Izem, R.; Schady, D.; Masand, P.; Shneider, B.L.; Leung, D.H.; Burrage, L.C. Biomarkers for liver disease in urea cycle disorders. Molecular genetics and metabolism 2021, 133, 148–156. [Google Scholar] [CrossRef]
- Yan, M.; Huo, Y.; Yin, S.; Hu, H. Mechanisms of acetaminophen-induced liver injury and its implications for therapeutic interventions. Redox biology 2018, 17, 274–283. [Google Scholar] [CrossRef]
- Essrani, R.; Mehershahi, S.; Essrani, R.K.; Ravi, S.J.K.; Bhura, S.; Sudhakaran, A.; Hossain, M.; Mehmood, A. Amiodarone-induced acute liver injury. Case Reports in Gastroenterology 2020, 14, 87–90. [Google Scholar] [CrossRef]
- Behrends, V.; Giskeødegård, G.F.; Bravo-Santano, N.; Letek, M.; Keun, H.C. Acetaminophen cytotoxicity in HepG2 cells is associated with a decoupling of glycolysis from the TCA cycle, loss of NADPH production, and suppression of anabolism. Archives of toxicology 2019, 93, 341–353. [Google Scholar] [CrossRef]
- Zahno, A.; Brecht, K.; Morand, R.; Maseneni, S.; Török, M.; Lindinger, P.W.; Krähenbühl, S. The role of CYP3A4 in amiodarone-associated toxicity on HepG2 cells. Biochemical pharmacology 2011, 81, 432–441. [Google Scholar] [CrossRef] [PubMed]
- Allard, J.; Bucher, S.; Massart, J.; Ferron, P.-J.; Le Guillou, D.; Loyant, R.; Daniel, Y.; Launay, Y.; Buron, N.; Begriche, K. Drug-induced hepatic steatosis in absence of severe mitochondrial dysfunction in HepaRG cells: proof of multiple mechanism-based toxicity. Cell Biology and Toxicology 2021, 37, 151–175. [Google Scholar] [CrossRef]
- McGill, M.R.; Yan, H.M.; Ramachandran, A.; Murray, G.J.; Rollins, D.E.; Jaeschke, H. HepaRG cells: a human model to study mechanisms of acetaminophen hepatotoxicity. Hepatology 2011, 53, 974–982. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.-J.; Lee, H.-M.; Park, Y.-I.; Yi, H.; Lee, H.; So, B.; Song, J.-Y.; Kang, H.-G. Chemically induced hepatotoxicity in human stem cell-induced hepatocytes compared with primary hepatocytes and HepG2. Cell biology and toxicology 2016, 32, 403–417. [Google Scholar] [CrossRef]
- Tham, N.T.; Hwang, S.-R.; Bang, J.-H.; Yi, H.; Park, Y.-I.; Kang, S.-J.; Kang, H.-G.; Kim, Y.-S.; Ku, H.-O. High-content analysis of in vitro hepatocyte injury induced by various hepatotoxicants. Journal of Veterinary Science 2019, 20, 34. [Google Scholar] [CrossRef]
- Capdevila, J.; Harris, R.; Falck, J. Microsomal cytochrome P450 and eicosanoid metabolism. Cellular and Molecular Life Sciences 2002, 59, 780–789. [Google Scholar] [CrossRef]
- Nebert, D.W.; Dalton, T.P. The role of cytochrome P450 enzymes in endogenous signalling pathways and environmental carcinogenesis. Nature Reviews Cancer 2006, 6, 947–960. [Google Scholar] [CrossRef] [PubMed]
- Guengerich, F.P. Human cytochrome P450 enzymes. Cytochrome P450: structure, mechanism, and biochemistry. 2015, 523-785.
- Zhou, S.-F.; Wang, B.; Yang, L.-P.; Liu, J.-P. Structure, function, regulation and polymorphism and the clinical significance of human cytochrome P450 1A2. Drug metabolism reviews 2010, 42, 268–354. [Google Scholar] [CrossRef]
- Wang, B.; Wang, J.; Huang, S.-Q.; Su, H.-H.; Zhou, S.-F. Genetic polymorphism of the human cytochrome P450 2C9 gene and its clinical significance. Current drug metabolism 2009, 10, 781–834. [Google Scholar] [CrossRef]
- Guengerich, F.P. Cytochrome P-450 3A4: regulation and role in drug metabolism. Annual review of pharmacology and toxicology 1999, 39, 1–17. [Google Scholar] [CrossRef]
- Xie, C.; Pogribna, M.; Word, B.; Lyn-Cook Jr, L.; Lyn-Cook, B.D.; Hammons, G.J. In vitro analysis of factors influencing CYP 1A2 expression as potential determinants of interindividual variation. Pharmacology Research & Perspectives 2017, 5, e00299. [Google Scholar]
- Washio, I.; Maeda, M.; Sugiura, C.; Shiga, R.; Yoshida, M.; Nonen, S.; Fujio, Y.; Azuma, J. Cigarette smoke extract induces CYP2B6 through constitutive androstane receptor in hepatocytes. Drug metabolism and disposition 2011, 39, 1–3. [Google Scholar] [CrossRef]
- Horn, J.R.; Hansten, P.D. Get to know an enzyme: CYP1A2. Pharmacy Times 2007, 73, 76. [Google Scholar]
- Tolosa, L.; Gómez-Lechón, M.J.; Pérez-Cataldo, G.; Castell, J.V.; Donato, M.T. HepG2 cells simultaneously expressing five P450 enzymes for the screening of hepatotoxicity: identification of bioactivable drugs and the potential mechanism of toxicity involved. Archives of toxicology 2013, 87, 1115–1127. [Google Scholar] [CrossRef]
- Jover, R.; Bort, R.; Gómez-Lechón, M.J.; Castell, J.V. Re-expression of C/EBPα induces CYP2B6, CYP2C9 and CYP2D6 genes in HepG2 cells. FEBS letters 1998, 431, 227–230. [Google Scholar] [CrossRef]
- Sekretarska, J.; Szczepaniak, J.; Sosnowska, M.; Grodzik, M.; Kutwin, M.; Wierzbicki, M.; Jaworski, S.; Bałaban, J.; Daniluk, K.; Sawosz, E. Influence of selected carbon nanostructures on the CYP2C9 enzyme of the P450 cytochrome. Materials 2019, 12, 4149. [Google Scholar] [CrossRef]
- Negoro, R.; Tasaka, M.; Deguchi, S.; Takayama, K.; Fujita, T. Generation of HepG2 Cells with High Expression of Multiple Drug-Metabolizing Enzymes for Drug Discovery Research Using a PITCh System. Cells 2022, 11, 1677. [Google Scholar] [CrossRef]
- Gerets, H.; Tilmant, K.; Gerin, B.; Chanteux, H.; Depelchin, B.; Dhalluin, S.; Atienzar, F. Characterization of primary human hepatocytes, HepG2 cells, and HepaRG cells at the mRNA level and CYP activity in response to inducers and their predictivity for the detection of human hepatotoxins. Cell biology and toxicology 2012, 28, 69–87. [Google Scholar] [CrossRef]
- Kanebratt, K.P.; Andersson, T.B. HepaRG cells as an in vitro model for evaluation of cytochrome P450 induction in humans. Drug metabolism and disposition 2008, 36, 137–145. [Google Scholar] [CrossRef]
- Ueyama, T.; Tsuji, S.; Sugiyama, T.; Tada, M. Fluorometric evaluation of CYP3A4 expression using improved transgenic HepaRG cells carrying a dual-colour reporter for CYP3A4 and CYP3A7. Scientific Reports 2017, 7, 2874. [Google Scholar] [CrossRef]
- Mandon, M.; Huet, S.; Dubreil, E.; Fessard, V.; Le Hégarat, L. Three-dimensional HepaRG spheroids as a liver model to study human genotoxicity in vitro with the single cell gel electrophoresis assay. Scientific Reports 2019, 9, 10548. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, Y.; Kawai, K.; Kanaki, T.; Yamazaki, H.; Chesné, C.; Guguen-Guillouzo, C.; Suemizu, H. Functional polymer-dependent 3D culture accelerates the differentiation of HepaRG cells into mature hepatocytes. Hepatology Research 2016, 46, 1045–1057. [Google Scholar] [CrossRef] [PubMed]
- Ott, L.M.; Ramachandran, K.; Stehno-Bittel, L. An automated multiplexed hepatotoxicity and CYP induction assay using HepaRG cells in 2D and 3D. SLAS DISCOVERY: Advancing Life Sciences R&D 2017, 22, 614–625. [Google Scholar]
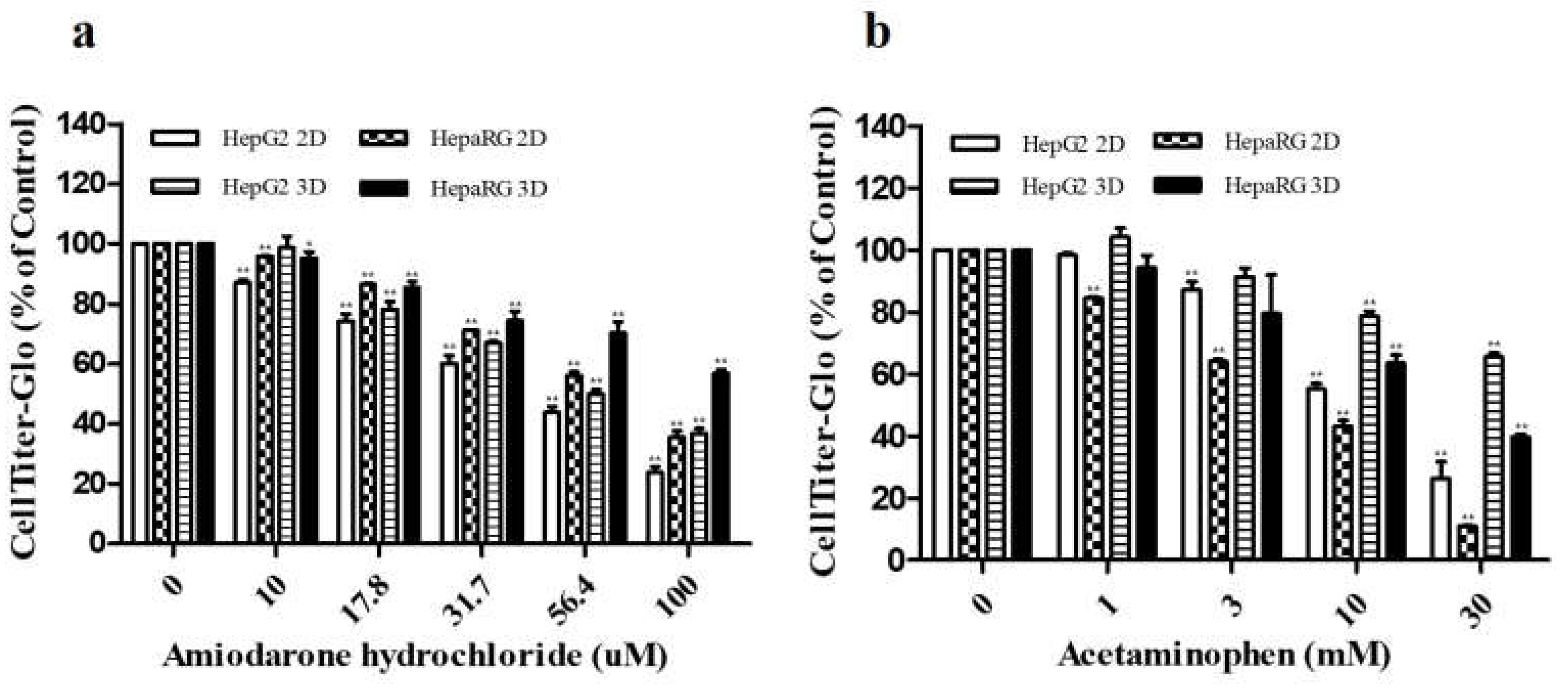
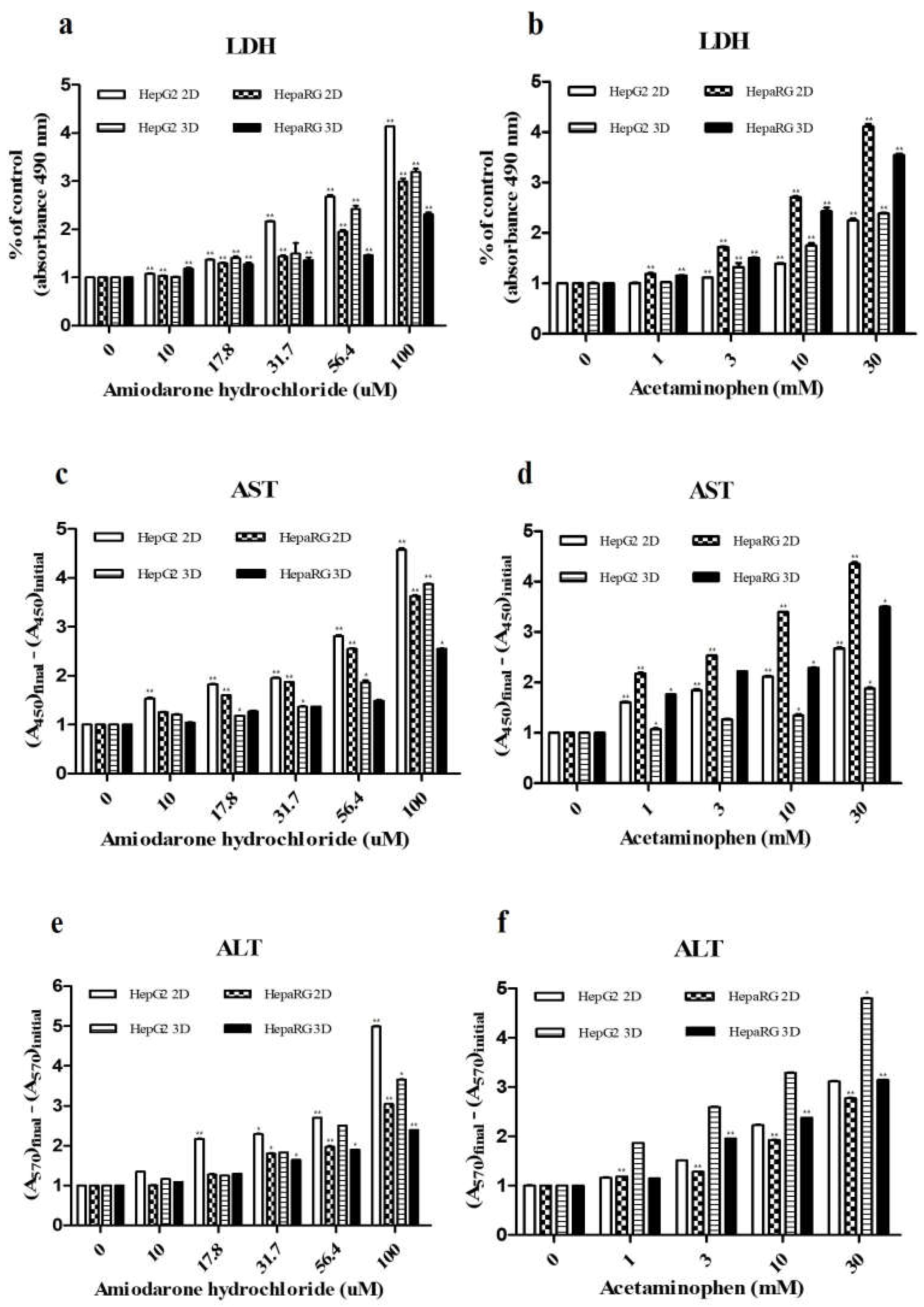
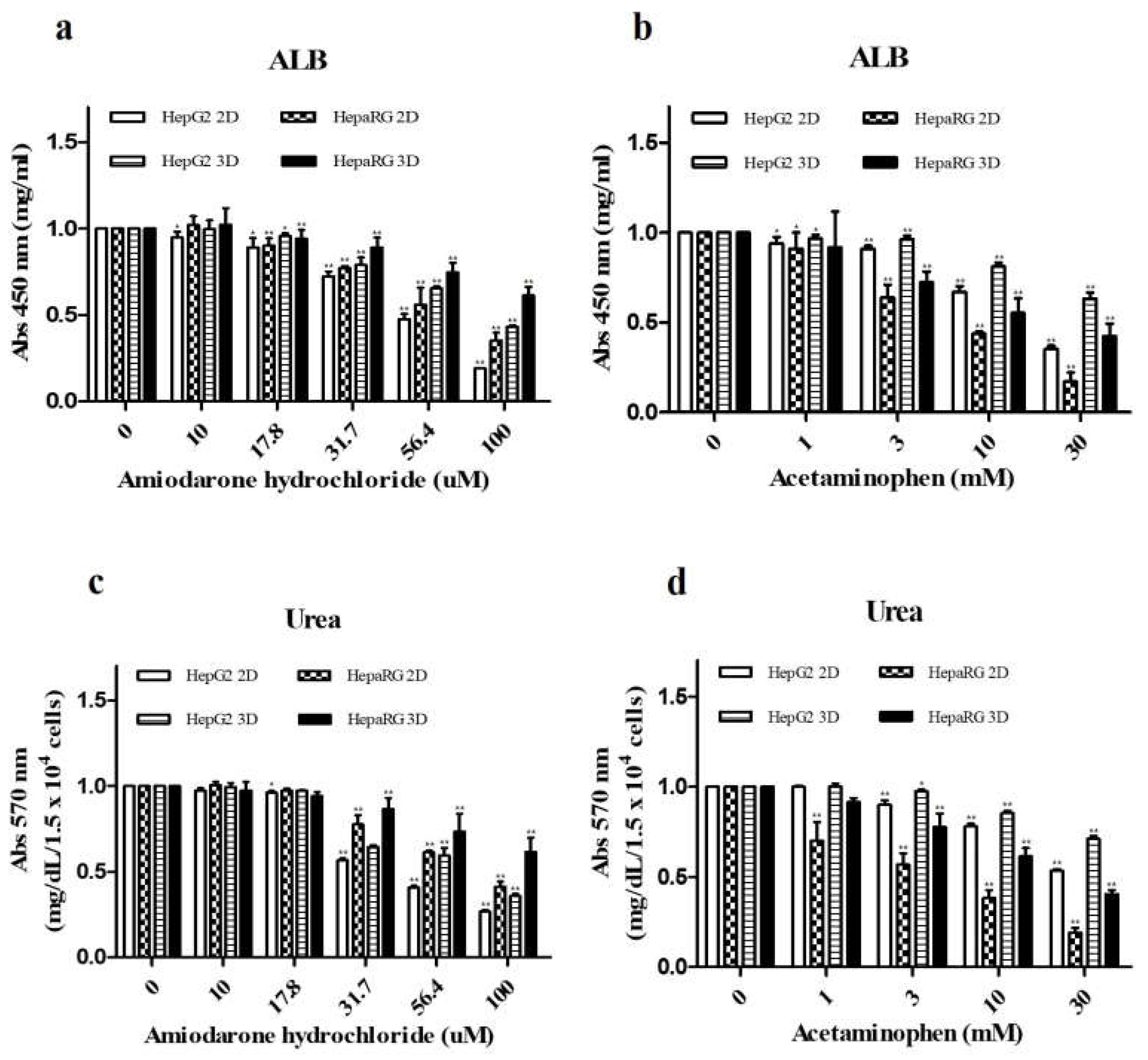
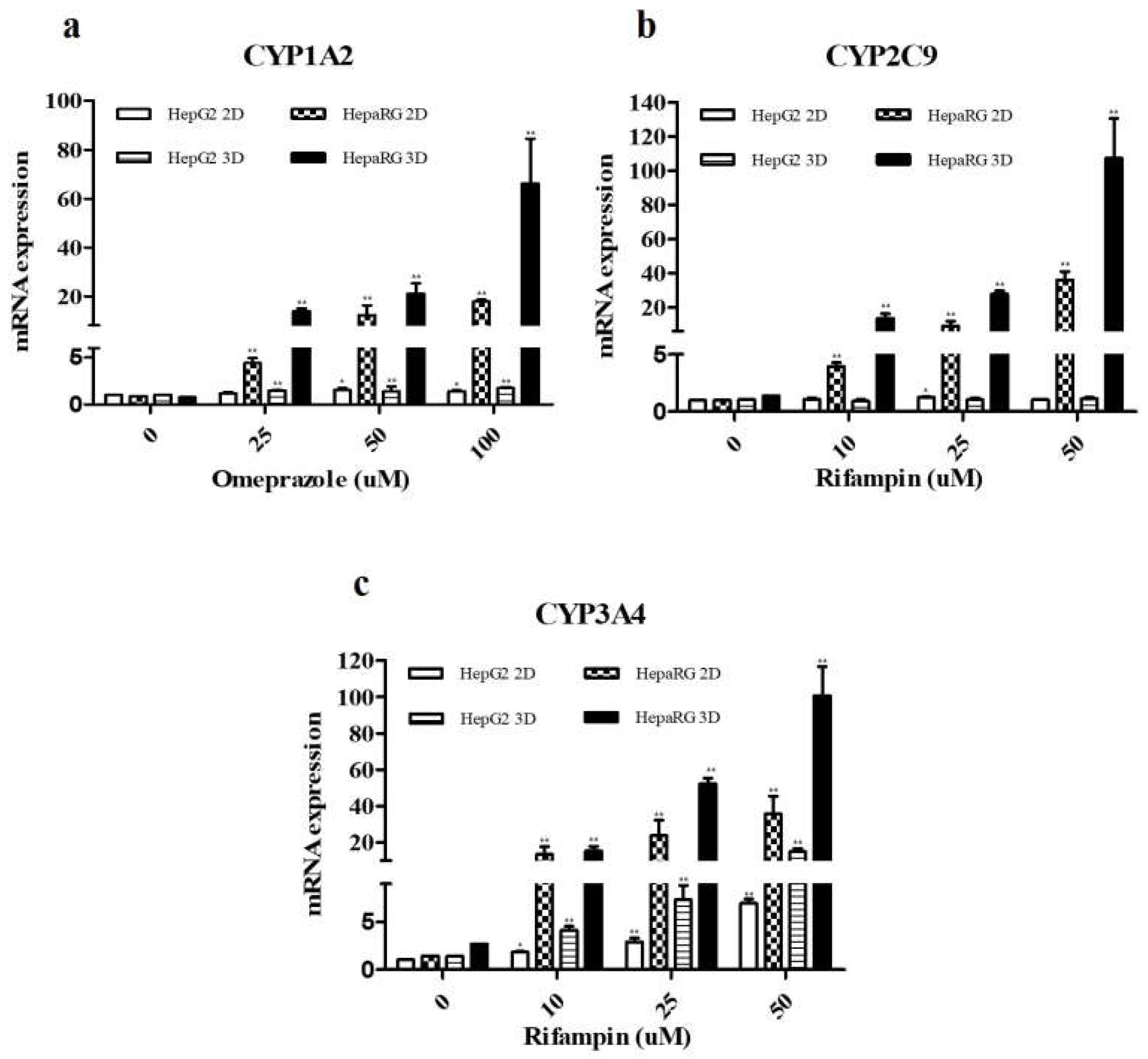
| CYP enzyme | Inducer | Final Concentration (μM) | Incubation time |
|---|---|---|---|
| CYP1A2 | Omeprazole | 0, 25, 50, 100 | |
| CYP2C9 | Refampin | 0, 10, 25, 50 | 48 hr |
| CYP3A4 | Refampin | 0, 10, 25, 50 |
| Primersets | Sequences | |
| CYP1A2 | Forward | 5‘-CACCATGGCATTGTCCCAGTCTG-3‘ |
| Reverse | 5‘-CCGCGGTCAGTTGATGGAGAAGCGC-3‘ | |
| CYP2C9 | Forward | 5‘-CAGATCTGCAATAATTTTTCTC-3‘ |
| Reverse | 5‘-CTTTCAATAGTAAATTCAGATG-3‘ | |
| CYP3A4 | Forward | 5‘-CTCTCATCCCAGACTTGGCCA-3‘ |
| Reverse | 5‘-ACAGGCTGTTGACCATCATAAAAG-3‘ | |
| β-actin | Forward | 5‘-CAAAGGCGAGGCTCTGTG-3‘ |
| Reverse | 5‘-CCGAAAGTTGCCTTTTATGG-3‘ | |
| Hepatotoxicants | ||
| Celltype | AmiodaroneHClIC50(μM) | AcetaminophenIC50(mM) |
| HepG2 2D | 44.85 | 14.95 |
| HepaRG 2D | 65.47 | 7.11 |
| HepG2 3D | 55.64 | 48.43 |
| HepaRG 3D | 109.38 | 26.19 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




