1. Introduction
Breast cancer, constituting 31% of all female cancers, remains a significant health concern, necessitating precise diagnostic measures [
1]. Estimations by the American Cancer Society (ACS) project 297,790 newly diagnosed cases of invasive breast cancer among women in 2024 [
2]. Early detection and accurate diagnosis are crucial for effective treatment and better patient outcomes. Histopathology, considered the gold standard for breast cancer diagnosis, involves microscopic examination of tissue specimens to identify cancerous abnormalities. However, manual interpretation of histopathological images is time-consuming, prone to inter-observer discrepancies, and reliant on pathologist expertise [
3,
4,
5]. In response, computational methodologies, particularly deep learning-driven image analysis, offer promising avenues to enhance diagnostic precision and efficiency.
Several recent studies have investigated various methodologies to advance breast cancer histopathology image classification. Notably, investigations have been centered around ensemble techniques, direct utilization of specific Convolution Neural Network (CNN) architectures, and the integration of transfer learning with novel model designs. [
6] used ResNet [
7] models, emphasizing fine-tuning and down sampling strategies to address computational complexities, achieving commendable accuracies of 86% on the BACH (Grand Challenge on Breast Cancer Histology images) test set [
8] and 97.2% on the Bioimaging 2015 challenge dataset. [
9] employed ResNet-101 and DenseNet-161 [
10] networks, showcasing a notable accuracy of 87% on BACH, albeit with varying sensitivities across classes. Both studies elucidated the efficacy of ensemble approaches incorporating pre-trained CNNs. Similarly [
11] introduced a hybrid CNN architecture integrating local and global model branches, achieving competitive accuracies of 87.5% on the BACH Dataset and 85.2% on the Breast Cancer Histopathological Image Classification (BreakHis) Dataset [
12].
Furthermore, investigations underscored the efficacy of direct application of established CNN architectures. [
13] demonstrated the utility of VGG16 [
14] in breast cancer grading, achieving a commendable accuracy of 83% on the BACH Test set. Similarly, [
15] pioneered a hybrid convolutional and recurrent deep learning method, yielding notable accuracies of 82.1% and 91.3% at patch and image levels, respectively. Moreover, studies emphasized the integration of transfer learning with innovative model architectures. [
16] delineated pipeline for breast cancer histopathology image classification, leveraging a two-stage neural network approach and achieving particularly an image level accuracy of 97.5%.
For classification between benign and malignant, [
17] proposed DenTnet, a deep learning model that achieved accuracy of 99.28% on the BreakHis dataset. Comparably, [
32] introduced BreastNet, an end-to-end model that incorporates a convolutional block attention module (CBAM), dense block, residual block, and hypercolumn technique. Meanwhile, [
34] developed DenseNet121-AnoGAN, which consists of two components: an unsupervised anomaly detection segment using generative adversarial networks to screen mislabeled patches, and a densely connected CNN (DenseNet) for extracting multi-layered features from discriminative patches. Additionally, [
35,
36] utilized transfer learning on the BreakHis dataset employing ResNet-18 and Inception-V3, respectively. Further advancements were made by [
37], which proposed an enhanced autoencoder network utilizing a Siamese framework, and [
39], which designed DRDA-Net, a model featuring dense attention and dual-shuffle attention mechanisms to guide deep learning processes.
Concurrently, [
18] introduced a hierarchical structure utilizing ResNeXt50 models, achieving notable accuracies of 81% on the BACH Challenge test set.
As a summary, the collective endeavors in breast cancer histopathology image classification underscore the significance of ensemble strategies [
19,
20,
21,
22], direct application of CNN architectures [
23,
24,
25], and the fusion of transfer learning with innovative model designs [
26,
27,
28]. Inspired by these pioneering works, our study aims to build upon existing methodologies and enhance performance on the aforementioned datasets through innovative techniques and meticulous experimentation. This research addresses histopathology image analysis limitations through advanced deep learning algorithms, focusing on breast cancer classification using VGG16 and ResNet50 architectures. These CNN models were selected for their ability to extract distinctive features crucial for accurate classification. Employing an image patching technique, each image is divided into smaller patches for focused analysis, managing computational demands. Initial experiments achieved a promising 89.84% accuracy in patch-level classification. The study provides a comprehensive investigation into deep learning for breast cancer histopathology classification, detailing methodology, including image patching and CNN implementation, alongside ensemble learning techniques. Results, analysis, and discussions highlight the approach's efficacy in surpassing the performance of existing studies on the two specified datasets and potential implications for breast cancer diagnosis improvement.
2. Materials and Methods
2.1. Dataset and Implementation Details
The BACH dataset utilized in this study comprises Hematoxylin and eosin (H&E) stained breast histology microscopy images and whole-slide images. Hematoxylin and eosin (H&E) stained breast histology microscopy images are valuable in this study because they provide detailed visual information regarding the cellular structure, morphology, and spatial distribution of tissues within the breast, aiding in the interpretation of pathological features relevant to breast cancer research.
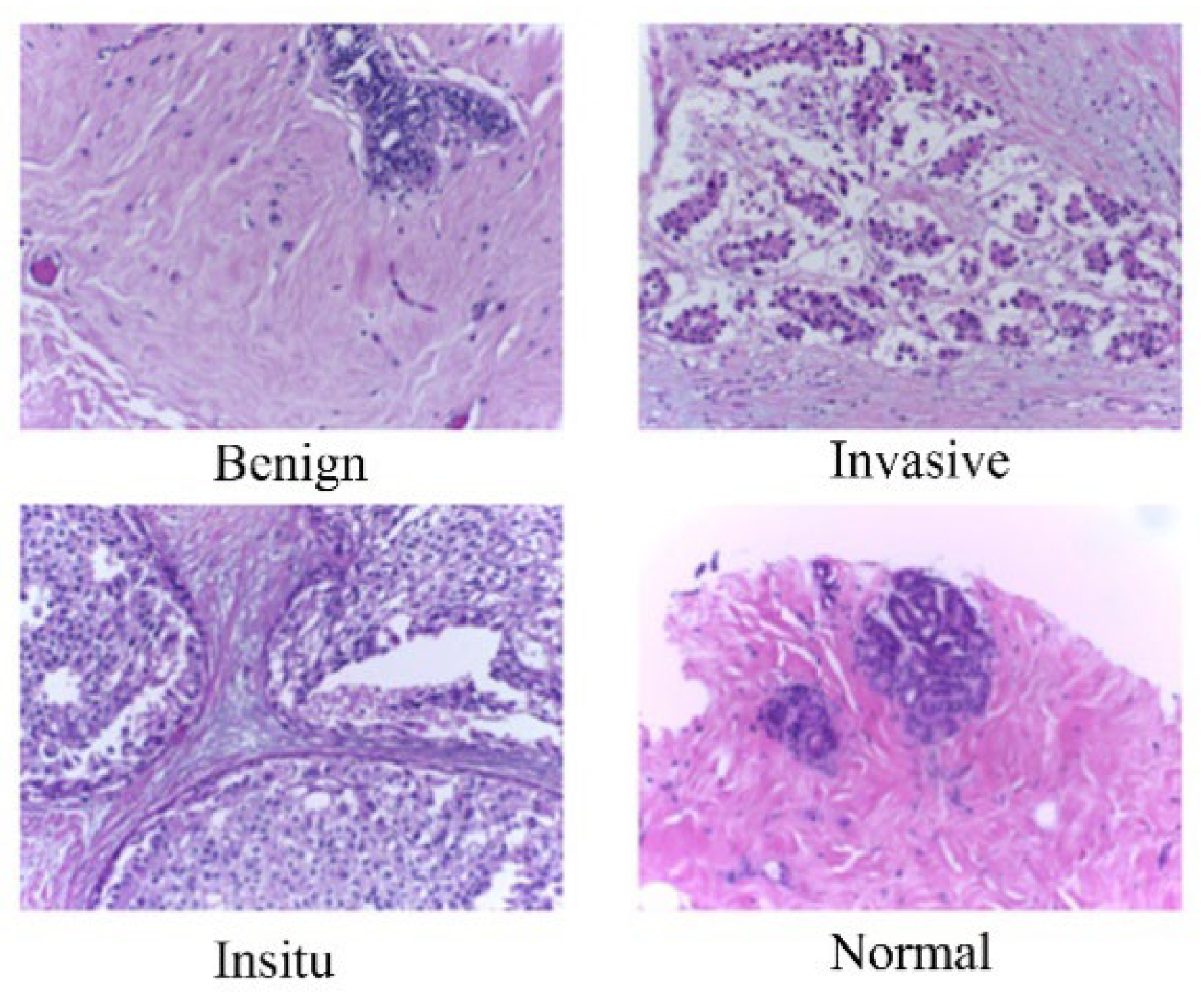
In BACH dataset, we focused on the microscopy images, which have been meticulously labeled by medical experts based on the predominant cancer type present in each image. Specifically, images are categorized into four classes: Normal, Benign, In Situ Carcinoma, and Invasive Carcinoma. The annotation process involved the assessment of each image by two medical experts, with any disagreements resulting in the exclusion of the respective image from the dataset. In total, the microscopy image dataset consists of 400 images, evenly distributed across the four classes, with each class containing 100 images. These images are stored in .tiff format and possess specific characteristics: a color model of Red-Green-Blue (RGB), dimensions of 2048 x 1536 pixels, and a pixel scale of 0.42 µm x 0.42 µm. Additionally, the memory space required for each image ranges from 10-20 MB approximately. Notably, the labeling of the dataset is image-wise, providing comprehensive information about the cancer type represented in each image. This meticulously curated dataset serves as a valuable resource for training and evaluating machine learning models aimed at breast cancer histopathology image analysis.
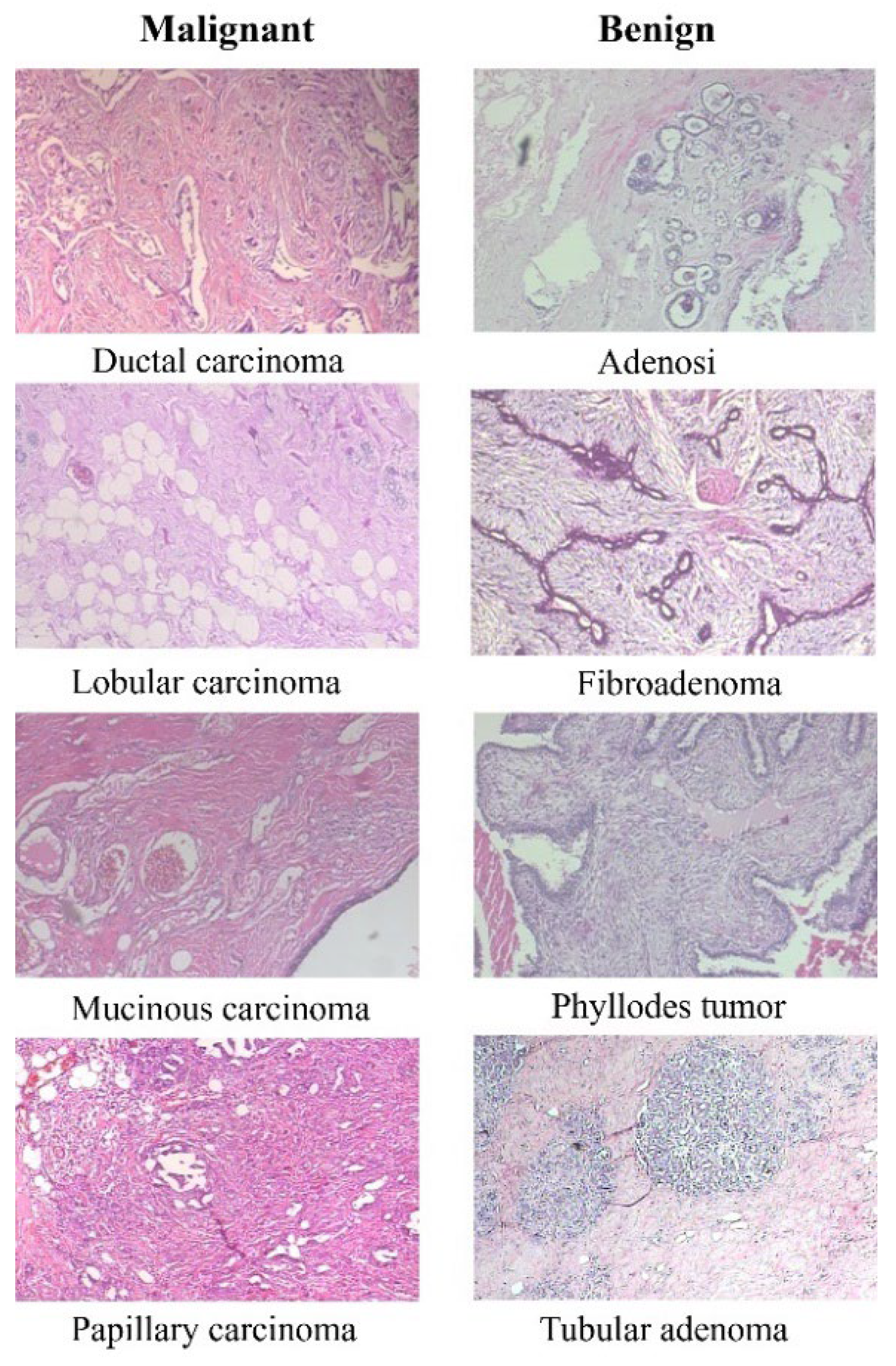
The BreakHis dataset consists of 9,109 microscopic images of breast tumor tissue. These images were collected from 82 patients and captured at various magnification factors (40X, 100X, 200X, and 400X). The dataset includes 2,480 benign and 5,429 malignant samples. Each sample is a 700X460 pixel, 3-channel RGB image with an 8-bit depth in each channel, stored in PNG format. The dataset categorizes tumors into benign and malignant classes, each with several subclasses. For benign tumors, these subclasses include Adenosis, Fibroadenoma, Tubular adenoma, and Phyllodes adenoma. For malignant tumors, the subclasses include Ductal carcinoma, Lobular carcinoma, Mucinous carcinoma, and Papillary carcinoma.
Figure 1 displays eight images, with the first column representing four types of malignant tissues and the second column representing four types of benign tissues.
2.2. Data Preprocessing
Breast cancer histopathology images, often characterized by large dimensions and high-resolution details, present challenges in computational analysis. To address this, we employed an image patching technique on BACH dataset to segment the images into smaller, more manageable patches. Each original image, with dimensions of 2048 x 1536 pixels and a pixel scale of 0.42 µm x 0.42 µm, underwent partitioning into six smaller images. This approach effectively reduced the computational burden and memory requirements, typically ranging from 10-20 MB per image. Moreover, employing image-wise labeling facilitated the categorization of patches into distinct classes, including Benign, InSitu, Invasive, and Normal. The patching algorithm involved dividing the image into two halves horizontally and further segmenting each half into three parts vertically. Subsequently, each patch was resized to a target size of 256 x 256 pixels and saved individually. This patching strategy not only facilitated efficient processing but also allowed for focused analysis on specific regions of interest within the histopathology images, thereby enhancing classification accuracy.
In the BreakHis dataset, no preprocessing has been applied. The data, which includes four scales (40X, 100X, 200X, and 400X), has been used for training a single model using multiscale magnification. The data is divided into five folds, with each fold containing all four scale magnifications. There are around 39,545 images of all magnifications, each with a size of 3x700x460, divided into five folds.
2.3. Implementation of VGG16 and ResNet50 Architectures
For our image classification tasks, we utilized two powerful convolutional neural network (CNN) models: VGG16 and ResNet50. These models have been proven effective in various computer vision tasks and provided a solid foundation for our classification efforts.
The VGG16 architecture is a well-known CNN model comprising multiple convolutional and max-pooling layers, followed by fully connected layers. To expedite our implementation, we employed transfer learning by utilizing pre-trained weights from the ImageNet dataset. We initialized the VGG16 model without its top classification layers and froze the weights of the convolutional layers to preserve the learned features. Custom fully connected layers were then added on top of the VGG16 architecture for classification purposes. During training, we fine-tuned these custom layers while keeping the convolutional layers frozen. This approach facilitated efficient training on our breast cancer histopathology dataset, resulting in accurate classification results.
In addition to VGG16, we incorporated the ResNet50 architecture, another prominent CNN model, into our classification framework. ResNet50 is characterized by its deep structure with skip connections, which helped alleviate the vanishing gradient problem and enabled the training of very deep neural networks. Similar to our approach with VGG16, we loaded the pre-trained ResNet50 model without its classification layers and froze the weights of the convolutional layers to retain learned features. Custom fully connected layers were then added for classification purposes. By leveraging ResNet50, we aimed to capture intricate features from the breast cancer histopathology images, potentially enhancing classification performance compared to shallower architectures.
2.4. Implementation of Ensemble Learning
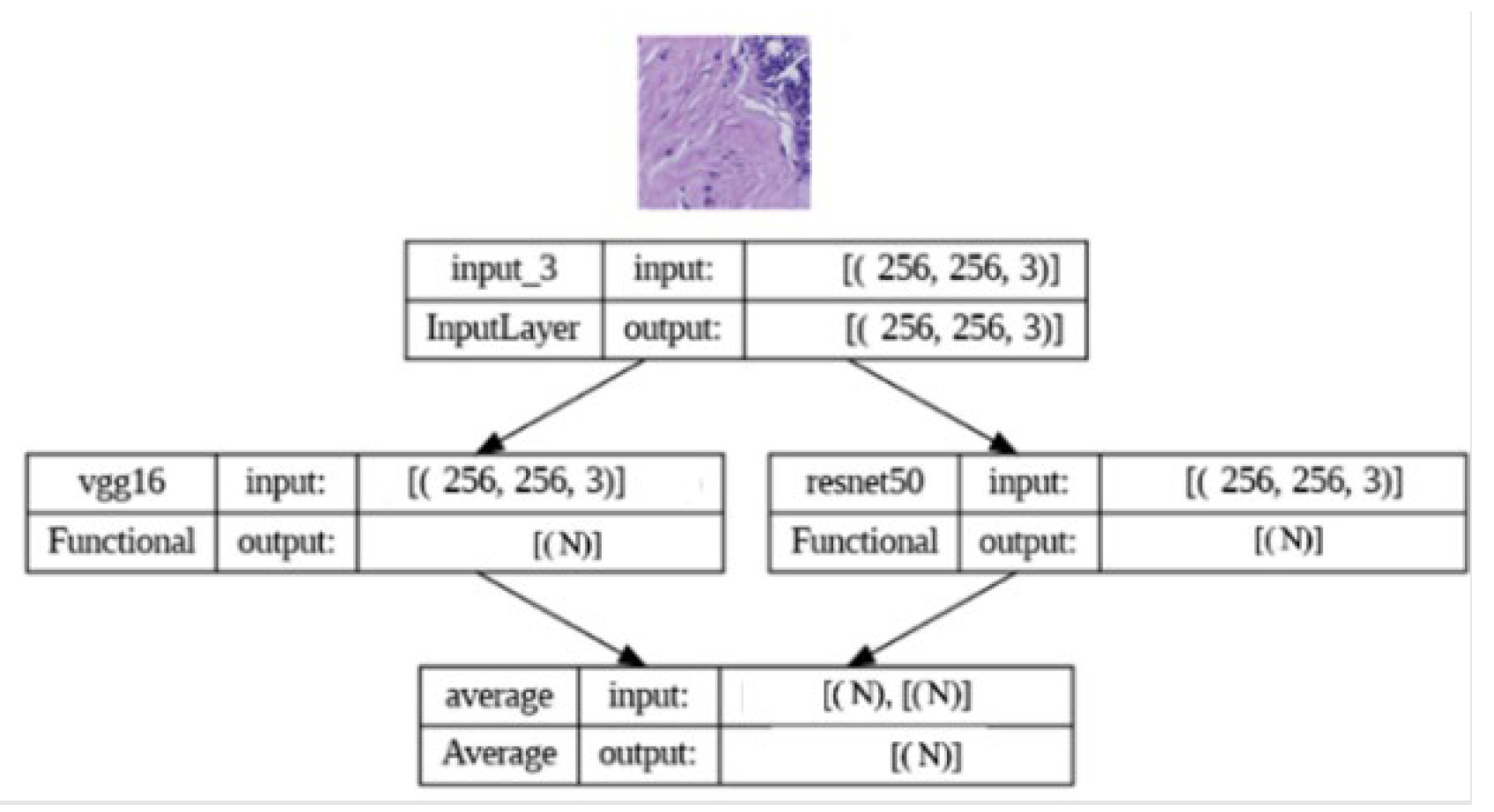
To enhance predictive performance, we adopted an ensemble learning approach by combining predictions from both the VGG16 and ResNet50 models. The ensemble model leverages the predicted probabilities generated by the individual models. The implementation involved loading the pre-trained VGG16 and ResNet50 models and creating a new model that combines their outputs using averaging. The ensemble model was then compiled with appropriate loss and optimization functions. By harnessing the complementary strengths of VGG16 and ResNet50, our ensemble model aimed to achieve enhanced classification accuracy and robustness, contributing to more reliable breast cancer histopathology image analysis.
Figure 3.
Ensemble model architectureTop of Form.
Figure 3.
Ensemble model architectureTop of Form.
Bottom of Form
3. Results
In this section, we present the classification outcomes obtained from our study on two distinct datasets. We evaluate the performance of machine learning algorithms, including VGG16, ResNet, and EfficientNet, in comparison with a novel model proposed in our research.
On the BACH dataset, we trained and evaluated three architectures: VGG16, ResNet, and an ensemble model. We assessed their performance using various metrics, including training and validation accuracies, obtained through both single-model and ensemble approaches.
Table 1 provides a comprehensive summary of the training and validation accuracies and losses achieved by each architecture on the BACH dataset.
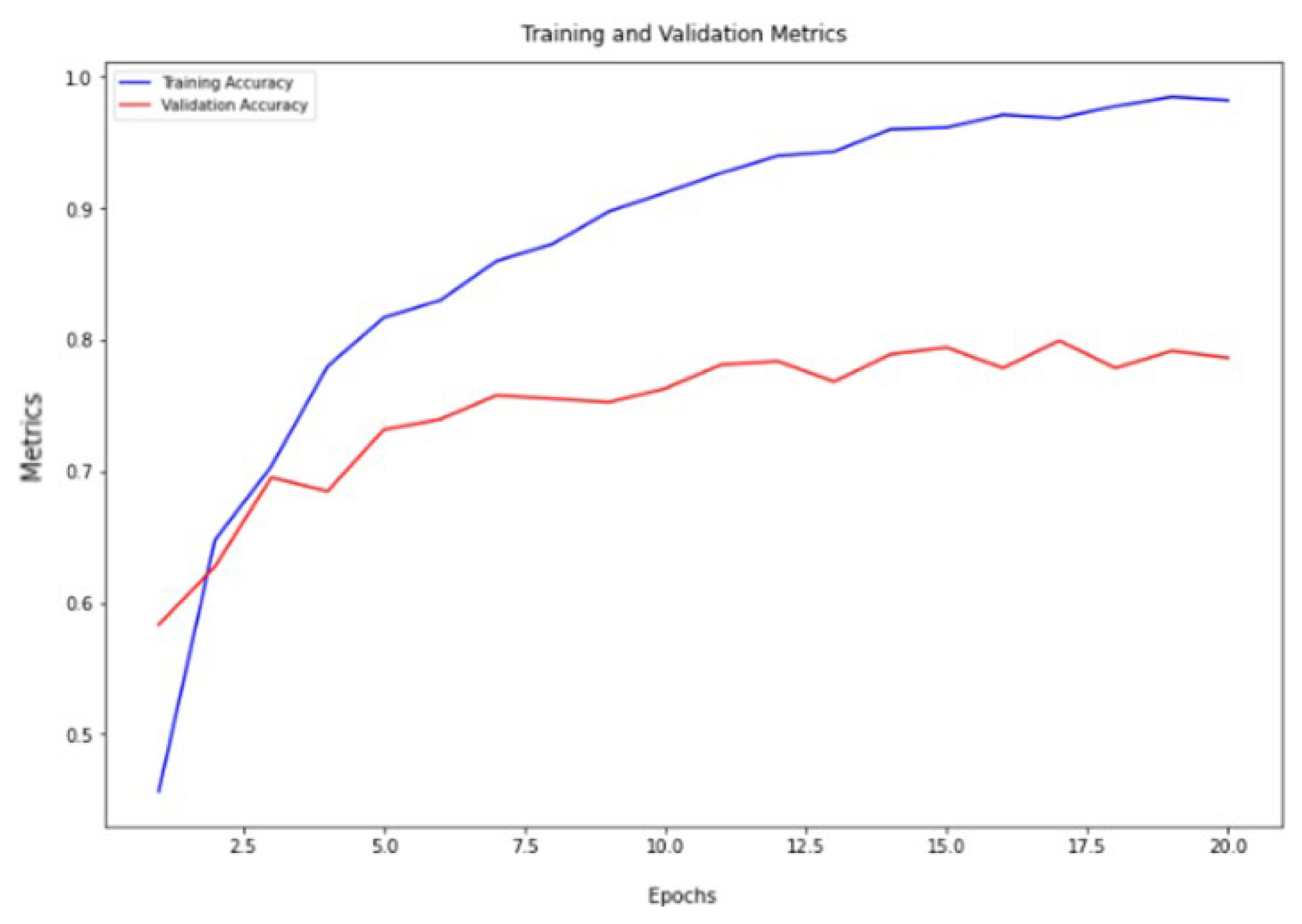
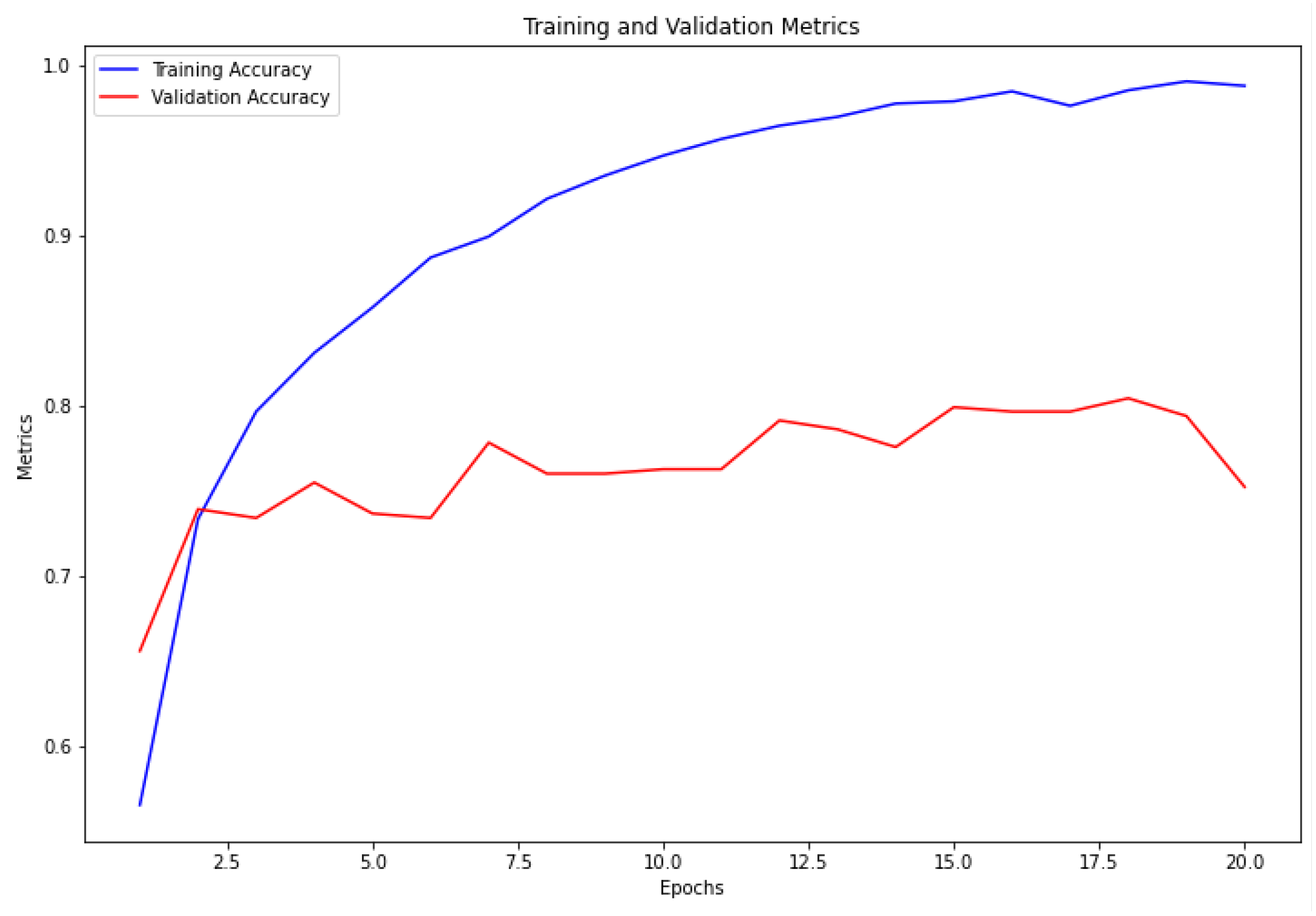
The VGG16 architecture demonstrated commendable results, achieving a training accuracy of 98.72% and a validation accuracy of 79.95%, as depicted in
Figure 4. Similarly, the ResNet architecture exhibited strong performance, with a training accuracy of 99.05% and a validation accuracy of 80.47%, as illustrated in
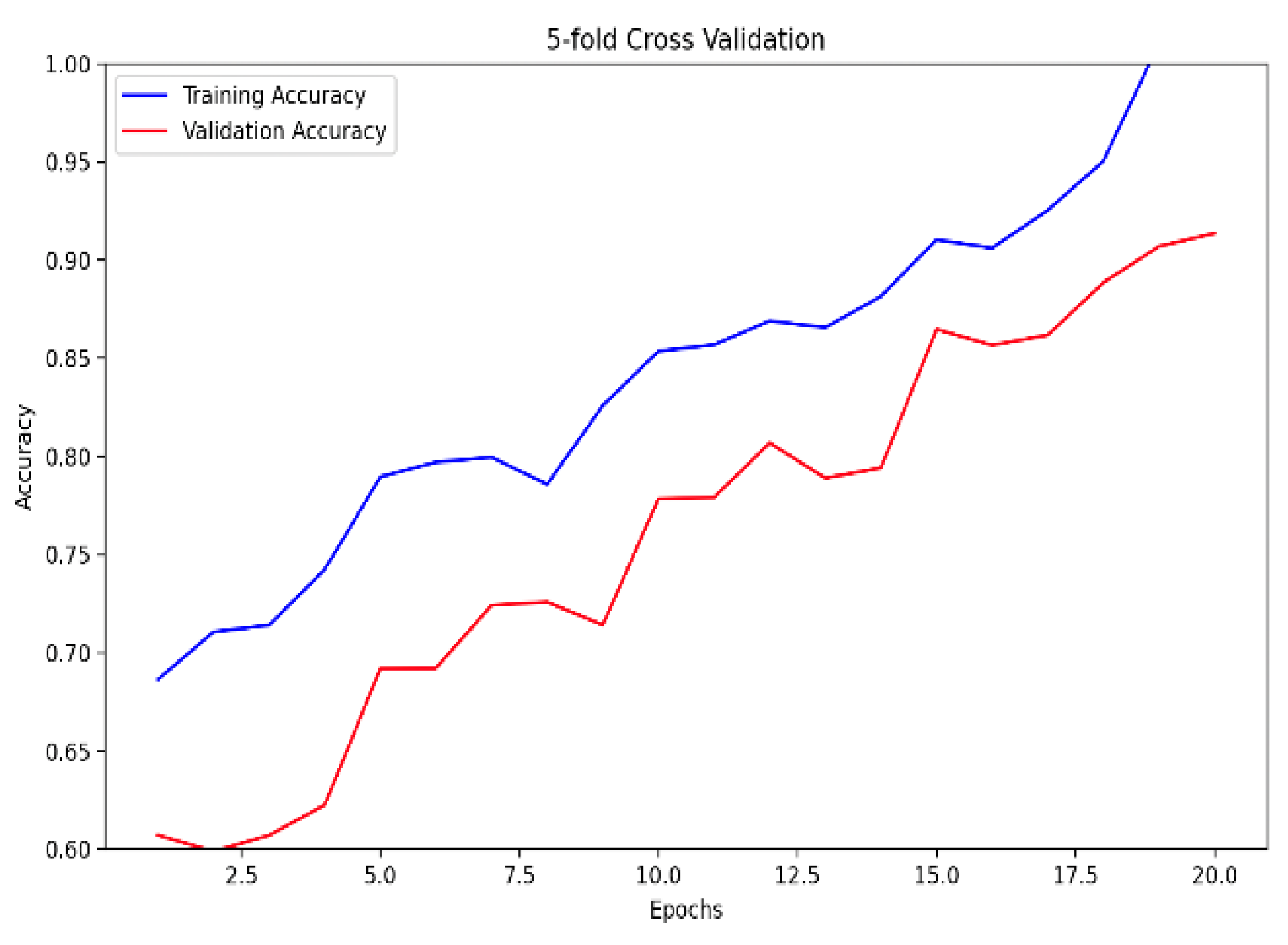
Figure 5. Notably, the ensemble model, trained with 5-fold cross-validation, showcased superior performance, boasting a training accuracy of 99.84% and a validation accuracy of 92.58%, as indicated in
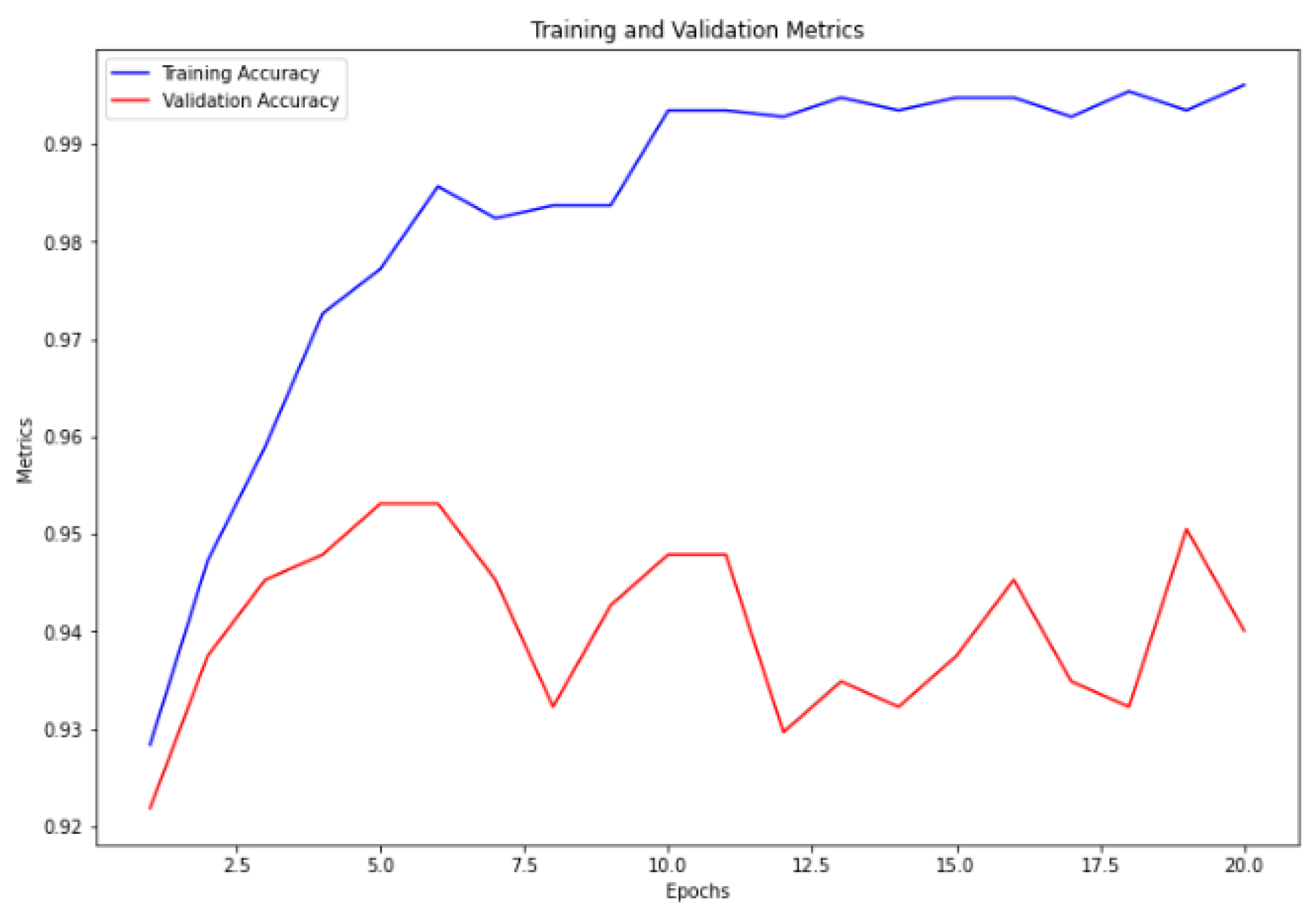
Figure 6. Furthermore, the ensemble model trained without cross-validation attained the highest accuracy, with a training accuracy of 99.78% and a validation accuracy of 95.31%, as illustrated in
Figure 7.
The notable improvement observed with the ensemble model compared to individually modified CNN architectures can be attributed to its ability to combine the strengths of individual architectures while mitigating their weaknesses. By leveraging the diversity among models generated through different initialization and training strategies, the ensemble model achieves robust predictions. Additionally, the incorporation of cross-validation helps generalize the model, reducing overfitting and enhancing its performance on unseen data. These results underscore the efficacy of ensemble learning in enhancing classification accuracy, particularly in the context of medical image analysis where subtle variations in image features can significantly impact diagnosis.
On the BreakHis dataset, the ensemble model demonstrated superior performance compared to state-of-the-art algorithms in terms of accuracy and Jaccard Index across fivefold validation.
Table 2 presents a comparative analysis of various machine learning models, including EfficientNet b0, VGGNet 16, ResNet 34, ResNet 50, and an Ensemble model. These models are evaluated based on their training and validation performance metrics such as accuracy, Jaccard Index, and loss. Remarkably, the Ensemble model outperforms all other models with the highest average validation accuracy of 98.43%, indicating its superior predictive capability on unseen data.
Table 3 presents an analysis of the Ensemble model designed for the classification of breast histopathological images from the BreakHis dataset into benign and malignant categories. This model achieved an exceptional average validation accuracy of 99.72%, significantly surpassing the performance of all prior models referenced in
Table 4. This can be seen with BreastNet [
32] which achieved an overall accuracy of 97.56%. Similarly, DenseNet121-AnoGAN [
34], an unsupervised anomaly detection model using generative adversarial networks recorded an accuracy of 91.44%. Transfer learning applications on the BreakHis dataset with ResNet-18 [
35] and Inception-V3 [
36] demonstrated accuracies of 92.15% and 98.97%, respectively. Additionally, [
37] proposed an enhanced autoencoder network utilizing a Siamese framework, achieving an accuracy of 96.97%. DRDA-Net [
39], a model featuring dual-shuffle attention mechanisms attained an accuracy of 96.10% and DenTnet obtained an accuracy of 99.28%.
The success of the ensemble model on the BreakHis dataset can be attributed to its capacity to capture intricate patterns and features present in histopathology images, which are crucial for accurate diagnosis. Furthermore, the ensemble approach helps alleviate the challenges posed by dataset heterogeneity and variability, leading to robust and reliable predictions across diverse tissue samples. These findings highlight the potential of ensemble learning in addressing the complexities inherent in medical image classification tasks, thereby facilitating more accurate and timely diagnose
Overall, our results demonstrate the effectiveness of deep learning techniques, particularly ensemble learning, in accurately classifying breast cancer histopathology images. These findings underscore the potential of artificial intelligence in enhancing breast cancer diagnosis and management, paving the way for improved patient outcomes and reduced healthcare burdens. The training and validation were conducted using an Nvidia Tesla V100 graphics card. The ensemble learning architecture was trained over 20 epochs, with each epoch comprising 48 steps. On average, each step of training took approximately 16 seconds. Additionally, our study highlights the importance of exploring novel approaches and ensemble strategies in medical image analysis, offering promising avenues for future research and clinical applications.
4. Discussion
The results obtained from our comprehensive study on breast cancer histopathology image classification underscore the pivotal role of advanced computational techniques, particularly deep learning algorithms, in improving diagnostic precision and efficiency. Our discussion delves into various aspects, including computational efficiency, algorithmic advancements, and future directions for leveraging these methodologies in clinical practice and beyond.
4.1. Computational Efficiency
One of the key challenges in histopathology image analysis lies in handling large-scale datasets with high-resolution images, which often pose computational burdens. To address this challenge, we employed innovative techniques such as image patching, downsampling, and ensemble learning. Our approach of segmenting high-resolution images into smaller patches enabled focused analysis of localized regions, reducing computational demands while preserving diagnostic accuracy. Additionally, leveraging pre-trained convolutional neural network (CNN) models like VGG16 and ResNet50 allowed us to harness learned features, thereby optimizing computational efficiency. Furthermore, the ensemble learning strategy, which combines predictions from multiple models, not only enhances classification accuracy but also mitigates computational costs by leveraging existing model architectures.
4.2. Algorithmic Advancements
Our study contributes to the ongoing advancements in algorithmic design for breast cancer histopathology image classification. By exploring diverse CNN architectures and ensemble learning techniques, we demonstrated the efficacy of leveraging deep learning for accurate classification across multiple datasets. The utilization of pre-trained models, fine-tuning strategies, and ensemble approaches enabled us to extract intricate features from histopathology images, leading to enhanced classification performance. Moreover, our investigation into multiscale analysis, particularly in the BreakHis dataset, highlights the importance of integrating information from different magnification levels to capture diverse pathological structures and improve diagnostic accuracy. This approach can enhance the performance of image classification by integrating information at different magnifications, thereby improving the recognition of various pathological structures that are more discernible at certain scales. It also increases the efficiency and accuracy of diagnosis by reducing operator variability. Moreover, multiscale analysis can assist in understanding disease progression and in developing targeted therapy strategies. Automated systems that employ this technique can reduce the time required for image analysis. Consequently, multiscale magnification offers a comprehensive view of histopathological images, facilitating more precise and efficient disease diagnosis and research. These algorithmic advancements not only facilitate more precise cancer diagnosis but also hold promise for accelerating the development of personalized treatment strategies.
4.3. Future Directions
Looking ahead, several avenues warrant exploration to further enhance the utility of deep learning in breast cancer diagnosis and management. Firstly, the development of user-friendly web applications incorporating our classification algorithms could empower healthcare professionals with accessible tools for rapid and accurate histopathology image analysis. Promising studies that validate efficacy of such AI-aided tools would be referred to create viable applied solutions to enhance healthcare workflows [
29,
30]. Such applications could streamline clinical workflows, facilitate remote consultations, and improve patient care outcomes. Additionally, extending our algorithmic framework to encompass other types of cancer, such as lung cancer or prostate cancer, holds promise for broadening the scope of AI-driven diagnostic solutions. Additional imaging modalities can also be leveraged to encompass end-to-end care paths aided by AI solutions as explained by authors in the following review study [
31]. By leveraging transfer learning and dataset adaptation techniques, our methodologies could be adapted to address diverse oncological challenges, thereby advancing cancer research and clinical practice. Furthermore, integrating multimodal data, such as genomic profiles or clinical metadata, into our classification models could enrich diagnostic insights and enable more comprehensive patient stratification.
5. Conclusions
In conclusion, our research represents a significant stride towards leveraging advanced computational techniques, particularly deep learning algorithms, to enhance breast cancer diagnosis and management through histopathology image analysis. By focusing on two prominent datasets, BACH and BreakHis, and employing innovative methodologies, we have contributed to the growing body of knowledge aimed at improving diagnostic precision and efficiency in breast cancer detection.
Through meticulous experimentation and implementation of deep learning models, including VGG16 and ResNet50, we demonstrated the efficacy of our approach in accurately classifying breast cancer histopathology images. Our ensemble learning strategy, which combines predictions from multiple models, showcased superior performance compared to state-of-the-art algorithms, underscoring the importance of leveraging complementary strengths of different CNN architectures.
Furthermore, our research addresses computational challenges inherent in histopathology image analysis by introducing techniques such as image patching and multiscale analysis. These strategies enhance computational efficiency and facilitate focused analysis on specific regions of interest within the histopathology images, improving classification accuracy.
Supplementary Materials
The following supporting information can be downloaded at: Prprints.org.
Funding
This research received no funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The public dataset used for modeling our deep learning algorithm can be found on the Kaggle website. Survey results presented in this study are available on request from the corresponding author.
Acknowledgments
In this section, you can acknowledge any support given which is not covered by the author's contribution or funding sections. This may include administrative and technical support, or donations in kind (e.g., materials used for experiments).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Siegel, R. L., Miller, K. D., Wagle, N. S., & Jemal, A. (2023). Cancer statistics, 2023. Ca Cancer J Clin, 73(1), 17-48.
- American Cancer Society. (2024). Cancer facts & figures 2024. American Cancer Society.
- Alyami, W., Kyme, A., & Bourne, R. (2022). Histological validation of MRI: A review of challenges in registration of imaging and whole-mount histopathology. Journal of Magnetic Resonance Imaging, 55(1), 11-22. [CrossRef]
- Çinar, U. (2023). Integrating hyperspectral imaging and microscopy for hepatocellular carcinoma detection from H&E stained histopathology images.
- Panayides, A. S., Amini, A., Filipovic, N. D., Sharma, A., Tsaftaris, S. A., Young, A., ... & Pattichis, C. S. (2020). AI in medical imaging informatics: Current challenges and future directions. IEEE journal of biomedical and health informatics, 24(7), 1837-1857. [CrossRef]
- Brancati, N., Frucci, M., & Riccio, D. (2018). Multi-classification of breast cancer histology images by using a fine-tuning strategy. In Image Analysis and Recognition: 15th International Conference, ICIAR 2018, Póvoa de Varzim, Portugal, June 27–29, 2018, Proceedings 15 (pp. 771-778). Springer International Publishing.
- He, K., Zhang, X., Ren, S., & Sun, J. (2016). Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 770-778).
- Aresta, G., Araújo, T., Kwok, S., Chennamsetty, S. S., Safwan, M., Alex, V., ... & Aguiar, P. (2019). BACH: Grand challenge on breast cancer histology images. Medical image analysis, 56, 122-139. [CrossRef]
- Chennamsetty, S. S., Safwan, M., & Alex, V. (2018). Classification of breast cancer histology image using ensemble of pre-trained neural networks. In Image Analysis and Recognition: 15th International Conference, ICIAR 2018, Póvoa de Varzim, Portugal, June 27–29, 2018, Proceedings 15 (pp. 804-811). Springer International Publishing.
- Huang, G., Liu, Z., Van Der Maaten, L., & Weinberger, K. Q. (2017). Densely connected convolutional networks. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 4700-4708).
- Zhu, C., Song, F., Wang, Y., Dong, H., Guo, Y., & Liu, J. (2019). Breast cancer histopathology image classification through assembling multiple compact CNNs. BMC medical informatics and decision making, 19, 1-17. [CrossRef]
- Spanhol, F. A., Oliveira, L. S., Petitjean, C., & Heutte, L. (2015). A dataset for breast cancer histopathological image classification. Ieee transactions on biomedical engineering, 63(7), 1455-1462. [CrossRef]
- Wang, Y., Sun, L., Ma, K., & Fang, J. (2018). Breast cancer microscope image classification based on CNN with image deformation. In Image Analysis and Recognition: 15th International Conference, ICIAR 2018, Póvoa de Varzim, Portugal, June 27–29, 2018, Proceedings 15 (pp. 845-852). Springer International Publishing. [CrossRef]
- Simonyan, K., & Zisserman, A. (2014). Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556.
- Yan, R., Ren, F., Wang, Z., Wang, L., Zhang, T., Liu, Y., ... & Zhang, F. (2020). Breast cancer histopathological image classification using a hybrid deep neural network. Methods, 173, 52-60. [CrossRef]
- Bagchi, A., Pramanik, P., & Sarkar, R. (2022). A multi-stage approach to breast cancer classification using histopathology images. Diagnostics, 13(1), 126. [CrossRef]
- Wakili, M. A., Shehu, H. A., Sharif, M. H., Sharif, M. H. U., Umar, A., Kusetogullari, H., ... & Uyaver, S. (2022). Classification of breast cancer histopathological images using DenseNet and transfer learning. Computational Intelligence and Neuroscience, 2022. [CrossRef]
- Koné, I., & Boulmane, L. (2018). Hierarchical ResNeXt models for breast cancer histology image classification. In Image Analysis and Recognition: 15th International Conference, ICIAR 2018, Póvoa de Varzim, Portugal, June 27–29, 2018, Proceedings 15 (pp. 796-803). Springer International Publishing.
- Ray, R. K., Linkon, A. A., Bhuiyan, M. S., Jewel, R. M., Anjum, N., Ghosh, B. P., ... & Shaima, M. (2024). Transforming Breast Cancer Identification: An In-Depth Examination of Advanced Machine Learning Models Applied to Histopathological Images. Journal of Computer Science and Technology Studies, 6(1), 155-161. [CrossRef]
- Addo, D., Zhou, S., Sarpong, K., Nartey, O. T., Abdullah, M. A., Ukwuoma, C. C., & Al-antari, M. A. (2024). A hybrid lightweight breast cancer classification framework using the histopathological images. Biocybernetics and Biomedical Engineering, 44(1), 31-54. [CrossRef]
- Sahran, S., Qasem, A., Omar, K., Albashih, D., Adam, A., Abdullah, S. N. H. S., ... & Abd Shukor, N. (2018). Machine learning methods for breast cancer diagnostic. Breast Cancer and Surgery, 57-76.
- Alirezazadeh, P., Dornaika, F., & Moujahid, A. (2023). Chasing a Better Decision Margin for Discriminative Histopathological Breast Cancer Image Classification. Electronics, 12(20), 4356. [CrossRef]
- Bayramoglu, N., Kannala, J., & Heikkilä, J. (2016, December). Deep learning for magnification independent breast cancer histopathology image classification. In 2016 23rd International conference on pattern recognition (ICPR) (pp. 2440-2445). IEEE.
- Gour, M., Jain, S., & Sunil Kumar, T. (2020). Residual learning based CNN for breast cancer histopathological image classification. International Journal of Imaging Systems and Technology, 30(3), 621-635. [CrossRef]
- Titoriya, A., & Sachdeva, S. (2019, November). Breast cancer histopathology image classification using AlexNet. In 2019 4th International conference on information systems and computer networks (ISCON) (pp. 708-712). IEEE.
- Deniz, E., Şengür, A., Kadiroğlu, Z., Guo, Y., Bajaj, V., & Budak, Ü. (2018). Transfer learning based histopathologic image classification for breast cancer detection. Health information science and systems, 6, 1-7. [CrossRef]
- Vesal, S., Ravikumar, N., Davari, A., Ellmann, S., & Maier, A. (2018). Classification of breast cancer histology images using transfer learning. In Image Analysis and Recognition: 15th International Conference, ICIAR 2018, Póvoa de Varzim, Portugal, June 27–29, 2018, Proceedings 15 (pp. 812-819). Springer International Publishing.
- Ahmad, N., Asghar, S., & Gillani, S. A. (2022). Transfer learning-assisted multi-resolution breast cancer histopathological images classification. The Visual Computer, 38(8), 2751-2770. [CrossRef]
- Singh, A., Randive, S., Breggia, A., Ahmad, B., Christman, R., & Amal, S. (2023). Enhancing Prostate Cancer Diagnosis with a Novel Artificial Intelligence-Based Web Application: Synergizing Deep Learning Models, Multimodal Data, and Insights from Usability Study with Pathologists. Cancers, 15(23), 5659. [CrossRef]
- Singh, A., Wan, M., Harrison, L., Breggia, A., Christman, R., Winslow, R. L., & Amal, S. (2023, March). Visualizing Decisions and Analytics of Artificial Intelligence based Cancer Diagnosis and Grading of Specimen Digitized Biopsy: Case Study for Prostate Cancer. In Companion Proceedings of the 28th International Conference on Intelligent User Interfaces (pp. 166-170).
- Milosevic, M., Jin, Q., Singh, A., & Amal, S. (2024). Applications of AI in multi-modal imaging for cardiovascular disease. Frontiers in Radiology, 3, 1294068. [CrossRef]
- Toğaçar, M.; Özkurt, K.B.; Ergen, B.; Cömert, Z. BreastNet: A Novel Convolutional Neural Network Model through Histopathological Images for the Diagnosis of Breast Cancer. Physica A: Statistical Mechanics and its Applications 2020, 545, 123592. [CrossRef]
- Parvin, F.; Mehedi Hasan, Md.A. A Comparative Study of Different Types of Convolutional Neural Networks for Breast Cancer Histopathological Image Classification. In Proceedings of the 2020 IEEE Region 10 Symposium (TENSYMP); June 2020; pp. 945–948.
- Man, R.; Yang, P.; Xu, B. Classification of Breast Cancer Histopathological Images Using Discriminative Patches Screened by Generative Adversarial Networks. IEEE Access 2020, 8, 155362–155377. [CrossRef]
- Boumaraf, S.; Liu, X.; Zheng, Z.; Ma, X.; Ferkous, C. A New Transfer Learning Based Approach to Magnification Dependent and Independent Classification of Breast Cancer in Histopathological Images. Biomedical Signal Processing and Control 2021, 63, 102192. [CrossRef]
- Soumik, Mohd.F.I.; Aziz, A.Z.B.; Hossain, Md.A. Improved Transfer Learning Based Deep Learning Model For Breast Cancer Histopathological Image Classification. In Proceedings of the 2021 International Conference on Automation, Control and Mechatronics for Industry 4.0 (ACMI); July 2021; pp. 1–4.
- Liu, M.; He, Y.; Wu, M.; Zeng, C. Breast Histopathological Image Classification Method Based on Autoencoder and Siamese Framework. Information 2022, 13, 107. [CrossRef]
- Zerouaoui, H.; Idri, A. Deep Hybrid Architectures for Binary Classification of Medical Breast Cancer Images. Biomedical Signal Processing and Control 2022, 71, 103226. [CrossRef]
- Chattopadhyay, S.; Dey, A.; Singh, P.K.; Sarkar, R. DRDA-Net: Dense Residual Dual-Shuffle Attention Network for Breast Cancer Classification Using Histopathological Images. Computers in Biology and Medicine 2022, 145, 105437. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).