Submitted:
11 May 2024
Posted:
13 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
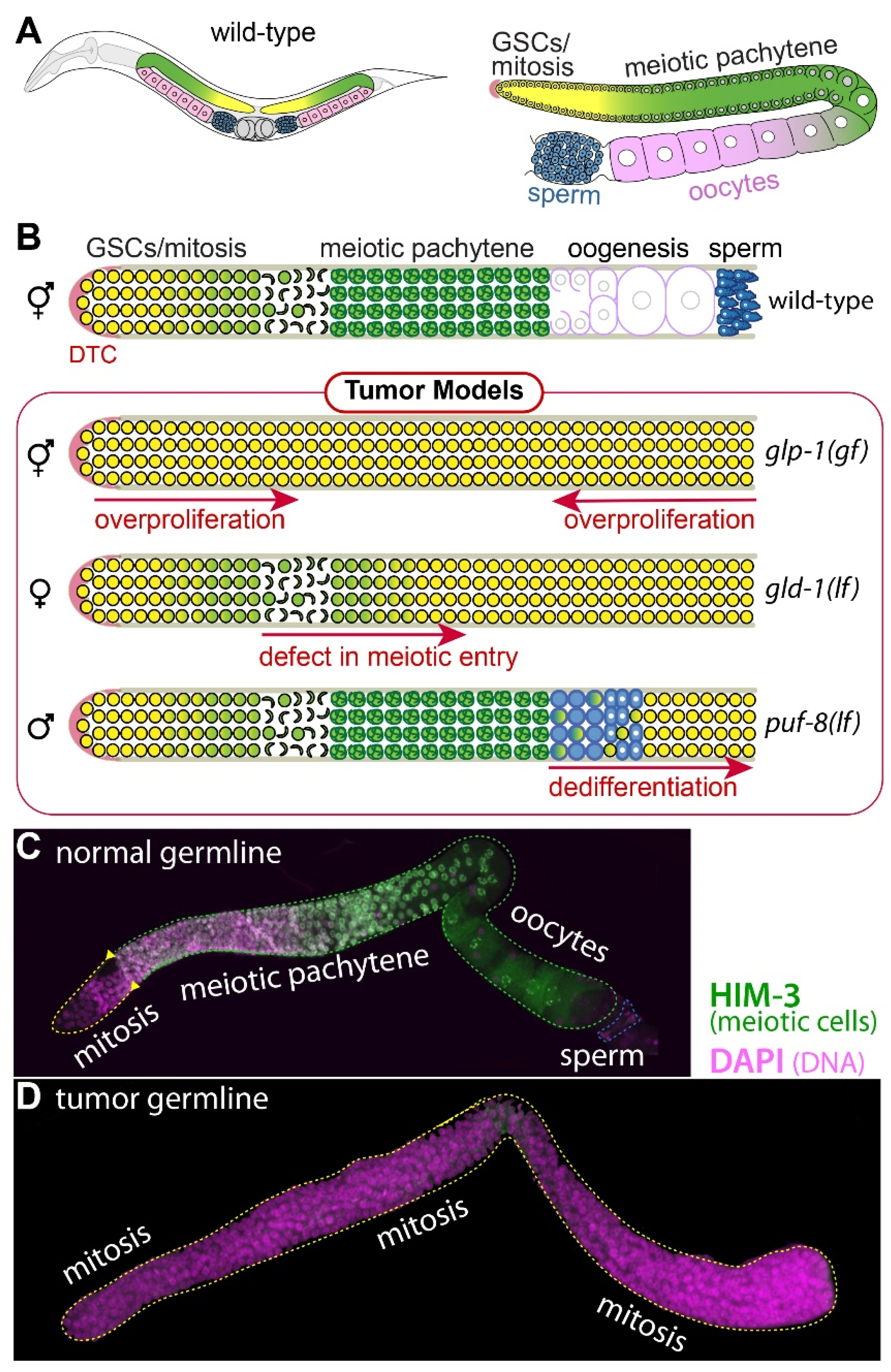
1.1. C. elegans Germline Development
1.2. Three Distinct Mechanisms of C. elegans Germline Tumorigenesis
2. GLP-1/Notch Activation-Mediated Tumorigenesis: Ectopic Proliferation
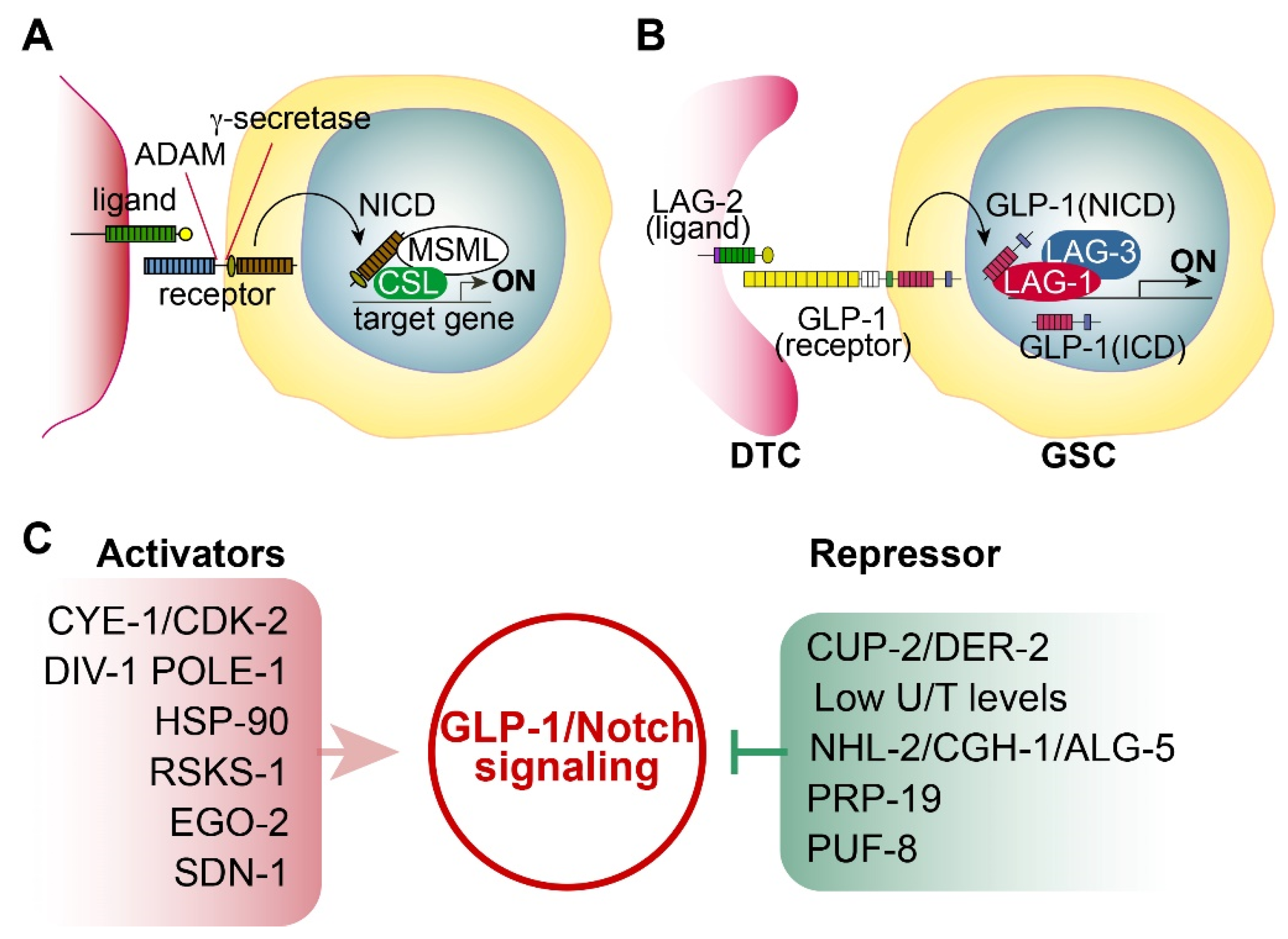
2.1. Notch Signaling
2.2. C. elegans Notch Signaling and Its Core Regulators
2.3. GLP-1 Mutant Alleles
2.4. Positive or Negative Regulators of GLP-1/Notch Signaling
- CYE-1 and CDK-2: Fox et al. found that germline CYE-1 and CDK-2 are required for GLP-1/Notch-mediated germ cell proliferation. RNAi-mediated depletion of CYE-1 or CDK-2 significantly suppressed germ cell proliferative fate in glp-1(bn18) mutants even at 20°C [15]. Additional genetic analysis suggests that CYE-1 and CDK-2 act independently of GLP-1/Notch signaling to promote proliferative fate [15].
- Subunits of the DNA polymerase alpha-primase complex: Yoon et al. found that DIV-1 (regulatory subunit) is indispensable for GLP-1/Notch-mediated germ cell proliferation during early larval development, whereas POLA-1 (catalytic subunit) and two primase subunits, PRI-1 and PRI-2, play a crucial role in GLP-1/Notch-mediated maintenance of proliferative cell fate during adulthood [16]. Robinson-Thiewes et al. also identified POLE-1 (the catalytic subunit of DNA polymerase e) as a regulator of germ cell proliferation [17].
- Chaperone HSP90: Lissemore et al. performed a genetic screening to identify genes that promote GLP-1/Notch signaling and found that HSP-90, a molecular chaperone, plays an essential role in stem cell maintenance [18]. It was a novel finding demonstrating the essential role of HSP90 in Notch signaling in development.
- Ribosomal Protein S6 kinase (S6K): Roy et al. identified RSKS-1/S6K as a positive regulator of GLP-1/Notch signaling-mediated germline proliferation [19]. Additional screening also found that Hedgehog-related ligands may share functional relationships with GLP-1/Notch and RSKS-1/S6K in maintaining GSCs [19].
- Bro1-domain protein: Liu and Maine identified the ego-2 (enhancer of glp-1) gene as a positive regulator of germline proliferation that interacts genetically with the GLP-1/Notch signaling pathway [20]. Notably, ego-2 also promotes LIN-12/Notch signaling in somatic tissues [20]. They found that the EGO-1 protein contains a Bro1 domain, which localizes to specific endosomal compartments in other systems. Thus, they suggest that EGO-2 may promote GLP-1/Notch signaling through endocytic process function [20].
- Derlin family proteins: Singh et al. demonstrated that reduced CUP-2 and DER-2 function suppresses GLP-1/Notch-mediated germline tumorigenesis [21]. CUP-2 and DER-2 are Derlin family proteins that function in endoplasmic reticulum-associated degradation (ERAD). Therefore, they suggest that Derlin activity may be associated with GLP-1/Notch-mediated tumorigenesis [21].
- U/T level: Chi et al. demonstrated that GLP-1 expression in the distal gonad region is repressed by low Uridine/Thymidine (U/T) levels [22]. C. elegans CDD-1/-2 Cytidine Deaminases are involved in uridine biosynthesis. Notably, worms lacking both CDD-1 and CDD-2 exhibited germline proliferation defects, whose phenotype was rescued by Uridine supplementation [22].
- TRIM-NHL protein: Brenner et al. identified nhl-2 as an inhibitor of glp-1(ar202)-mediated tumorigenesis [23]. NHL-2, a conserved TRIM-NHL protein family member, suppresses germ cell proliferation by inhibiting PUF-3 and PUF-11. They also found that CGH-1 RNA helicase and ALG-5 work with NHL-2 to inhibit glp-1(ar202)-mediated tumorigenesis.
- E3 Ubiquitin ligase: Gutnik et al. reported that the splicing factor PRP-19 (a candidate E3 ubiquitin ligase) inhibits the nuclear accumulation of the GLP-1/Notch intracellular domain [24].
- PUF RNA-binding protein: PUF-8 is a conserved PUF RNA-binding protein that inhibits the translation of target mRNAs. In C. elegans germline, PUF-8 is involved in decisions regarding proliferation/differentiation, differentiation/dedifferentiation, and sperm/oocyte fates, depending on the genetic context [25]. Racher and Hansen demonstrated that PUF-8 inhibits glp-1(ar202)-mediated tumorigenesis in the C. elegans germline [26]. Other PUF proteins (FBF-1/2 and PUF-3/11) also play a critical role in GSC maintenance [27].
- Syndecan: Gopal et al. identified SDN-1 (a syndecan transmembrane proteoglycan) as a positive regulator of GLP-1/Notch signaling. SDN-1 promotes GLP-1 expression and mitotic germ cell fate by controlling a somatic TRP calcium channel. The TRP enhances glp-1 expression by governing the calcium-dependent binding of an AP-2 transcription factor (APTF-2) [28].
3. GLD-1 Loss-Mediated Tumorigenesis: Meiotic Entry Failure
3.1. STAR Family of RNA-Binding Proteins
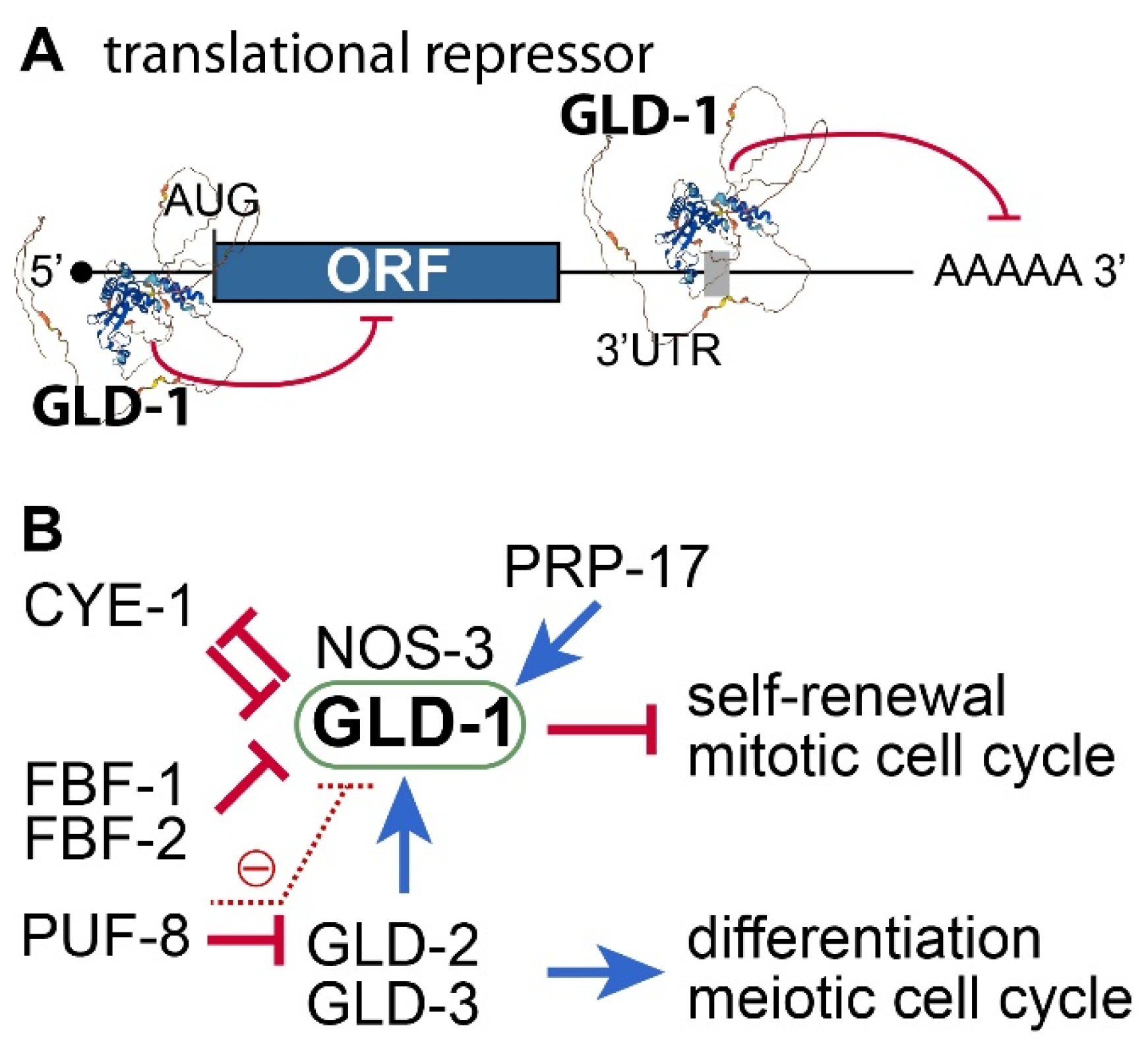
3.2. C. elegans gld-1 and Its Partners
3.4. Positive or Negative Regulators of GLD-1
- GLD-2 poly(A) polymerase (PAP): GLD-2 is a cytoplasmic poly(A) polymerase [52]. It plays a critical role in meiotic entry and progression [53,54]. Thus, no functional gametes are produced in the absence of GLD-2 [53]. Notably, the gld-1 mRNA is a direct target of GLD-2 [55]. GLD-2 poly(A) polymerase promotes meiotic entry, at least in part, by activating GLD-1 expression. Consequently, GLD-2 loss enhances the formation of germline tumors in gld-1 loss-of-function mutant worms [53].
- FBF/PUF RNA-binding protein: C. elegans FBF/PUF proteins play a crucial role in maintaining GSCs by regulating the expression of various target mRNAs, including the gld-1 mRNA [56]. Since GLD-1 is essential for inhibiting proliferation and maintaining the differentiation state of germ cells, FBF/PUF repression of gld-1 mRNAs is critical for GSC maintenance. In addition, C. elegans PUF-8 proteins negatively inhibit the expression of gld-1 mRNAs by inhibiting gld-2 mRNAs indirectly [57].
- CYE-1/CDK2: CYE-1/CDK2 influences the mitosis/meiosis balance through GLD-1 [15,58]. Specifically, GLD-1 has CDK2 phosphorylation sites and appears to be a direct substrate of CYE-1/CDK2. Functional analysis showed that CYE-1/CDK2 works with FBF-1 to maintain GSCs by inhibiting GLD-1. Notably, cye-1 mRNA is also a repressing target of GLD-1 [59]. Therefore, GLD-1 and CYE-1/CDK2 inhibit each other for the mitosis/meiosis balance.
- Pre-mRNA Splicing Factor (PRP-17): Kerins et al. reported that PRP-17 and other C. elegans splicing factor orthologs function to promote meiotic entry by positively regulating the splicing of mRNAs of genes in the GLD-1 pathway [60].
4. PUF-8 Loss-Mediated Tumorigenesis: Spermatogenic Dedifferentiation
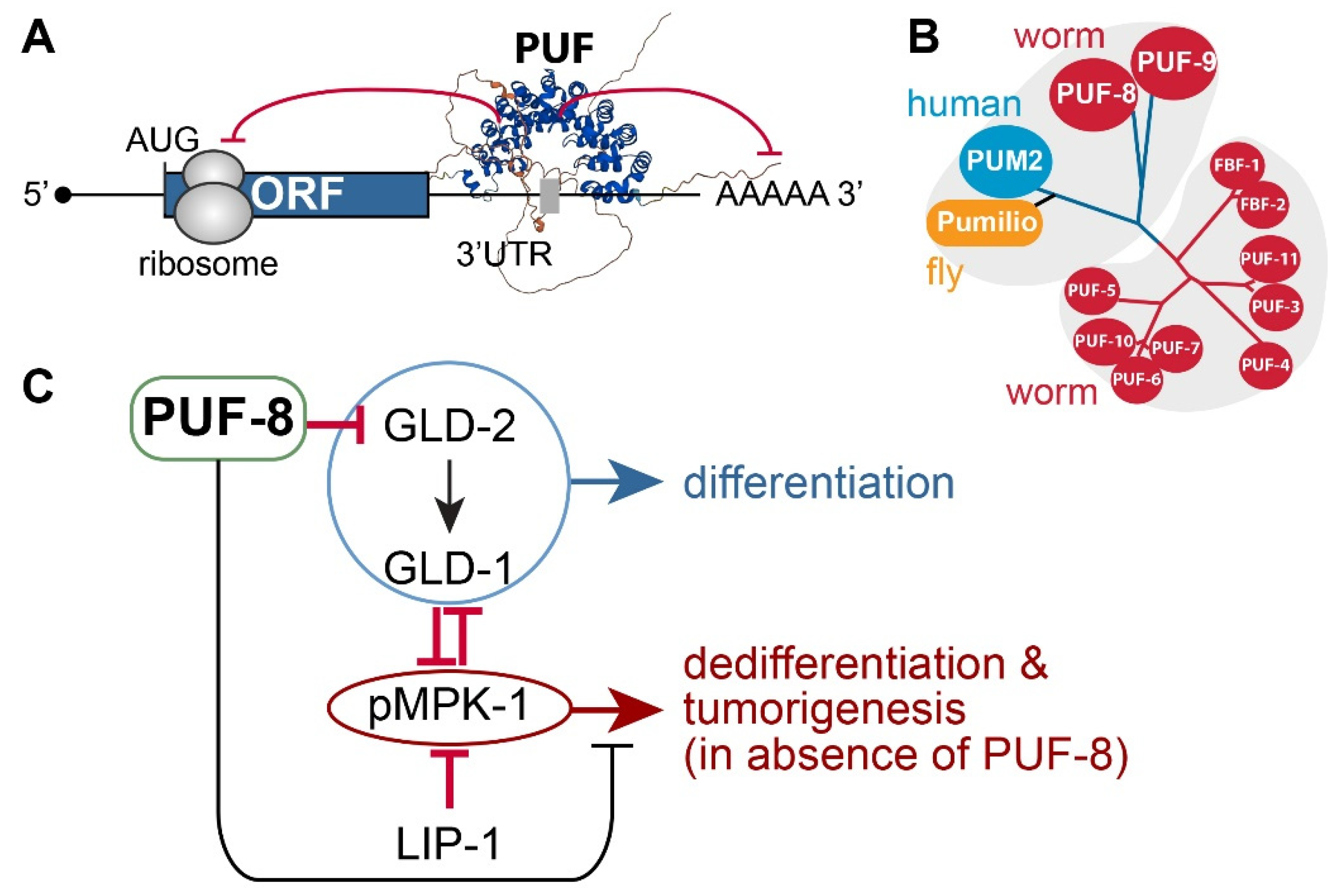
4.1. PUF RNA-Binding Proteins
4.2. C. elegans PUF-8
4.4. Positive or Negative Regulators of PUF-8
- LIP-1 dual-specificity phosphatase: puf-8(q725) mutants are self-fertile at 20°C. However, at 25°C, ~10% of 1-day adult puf-8(q725) mutants develop germline tumors [82]. Notably, the germline tumor phenotype of puf-8(q725) mutants is dramatically enhanced by the additional loss of LIP-1 [82]. This finding indicates that PUF-8 works with LIP-1 to inhibit dedifferentiation-mediated tumorigenesis by promoting the meiotic division of spermatocytes in the C. elegans germline [82].
- MPK-1/ERK MAPK: The Ras-ERK/MAP kinase signaling pathway governs many cellular processes, such as proliferation, differentiation, cell fate decision, and survival in most eukaryotes [69]. Components of the C. elegans Ras-ERK pathway, such as LET-60/Ras and MPK-1/ERK, are highly conserved and essential for germline development, including meiotic progression, sperm fate specification, and oocyte maturation [83]. Notably, the reduction of Ras-ERK MAPK signaling, either by mutation or chemical inhibition, blocked the initiation of dedifferentiation in puf-8(q725); lip-1(zh15) mutant germlines [82,84]. These findings indicate that MPK-1/ERK signaling pathways are critical for puf-8(q725) dedifferentiation-mediated tumorigenesis.
- GLD-1 and GLD-2: Park et al. recently reported that PUF-8 binds specifically to a PBE in gld-2 3’UTR and represses a GFP reporter gene carrying gld-2 3’UTR in the C. elegans mitotic germ cells [57]. Notably, the removal of both gld-2 and its activating target, gld-1, significantly increased puf-8(q725) dedifferentiation-mediated germline tumors [57]. These results indicate that GLD-1 and GLD-2 may inhibit dedifferentiation-mediated germline tumors in puf-8(q725) mutant germline by promoting germ cell differentiation.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Kirienko, N.V.; Mani, K.; Fay, D.S. Cancer models in Caenorhabditis elegans. Developmental dynamics: an official publication of the American Association of Anatomists 2010, 239, 1413–1448. [Google Scholar] [CrossRef] [PubMed]
- Kimble, J.; Crittenden, S.L. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annual review of cell and developmental biology 2007, 23, 405–433. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Lin, W.; Long, Y.; Yang, Y.; Zhang, H.; Wu, K.; Chu, Q. Notch signaling pathway: architecture, disease, and therapeutics. Signal transduction and targeted therapy 2022, 7, 95. [Google Scholar] [CrossRef] [PubMed]
- Greenwald, I. LIN-12/Notch signaling in C. elegans. WormBook: the online review of C. elegans biology, 2005; 1–16. [Google Scholar] [CrossRef]
- Henderson, S.T.; Gao, D.; Lambie, E.J.; Kimble, J. lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development 1994, 120, 2913–2924. [Google Scholar] [CrossRef]
- Tax, F.E.; Thomas, J.H. Cell-cell interactions. Receiving signals in the nematode embryo. Current biology: CB 1994, 4, 914–916. [Google Scholar] [CrossRef] [PubMed]
- Kobet, R.A.; Pan, X.; Zhang, B.; Pak, S.C.; Asch, A.S.; Lee, M.H. Caenorhabditis elegans: A Model System for Anti-Cancer Drug Discovery and Therapeutic Target Identification. Biomolecules & therapeutics 2014, 22, 371–383. [Google Scholar] [CrossRef] [PubMed]
- Kershner, A.M.; Shin, H.; Hansen, T.J.; Kimble, J. Discovery of two GLP-1/Notch target genes that account for the role of GLP-1/Notch signaling in stem cell maintenance. Proceedings of the National Academy of Sciences of the United States of America 2014, 111, 3739–3744. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.; Haupt, K.A.; Kershner, A.M.; Kroll-Conner, P.; Wickens, M.; Kimble, J. SYGL-1 and LST-1 link niche signaling to PUF RNA repression for stem cell maintenance in Caenorhabditis elegans. PLoS genetics 2017, 13, e1007121. [Google Scholar] [CrossRef]
- Kodoyianni, V.; Maine, E.M.; Kimble, J. Molecular basis of loss-of-function mutations in the glp-1 gene of Caenorhabditis elegans. Molecular biology of the cell 1992, 3, 1199–1213. [Google Scholar] [CrossRef]
- Austin, J.; Kimble, J. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell 1987, 51, 589–599. [Google Scholar] [CrossRef]
- Berry, L.W.; Westlund, B.; Schedl, T. Germ-line tumor formation caused by activation of glp-1, a Caenorhabditis elegans member of the Notch family of receptors. Development 1997, 124, 925–936. [Google Scholar] [CrossRef] [PubMed]
- Pepper, A.S.; Killian, D.J.; Hubbard, E.J. Genetic analysis of Caenorhabditis elegans glp-1 mutants suggests receptor interaction or competition. Genetics 2003, 163, 115–132. [Google Scholar] [CrossRef] [PubMed]
- Fox, P.M.; Vought, V.E.; Hanazawa, M.; Lee, M.H.; Maine, E.M.; Schedl, T. Cyclin E and CDK-2 regulate proliferative cell fate and cell cycle progression in the C. elegans germline. Development 2011, 138, 2223–2234. [Google Scholar] [CrossRef] [PubMed]
- Yoon, D.S.; Cha, D.S.; Alfhili, M.A.; Keiper, B.D.; Lee, M.H. Subunits of the DNA polymerase alpha-primase complex promote Notch-mediated proliferation with discrete and shared functions in C. elegans germline. The FEBS journal 2018. [Google Scholar] [CrossRef] [PubMed]
- Robinson-Thiewes, S.; Kershner, A.M.; Shin, H.; Haupt, K.A.; Kroll-Connor, P.; Kimble, J. A sensitized genetic screen to identify regulators of Caenorhabditis elegans germline stem cells. G3 (Bethesda) 2022, 12. [Google Scholar] [CrossRef] [PubMed]
- Greenblatt, E.; Casper, R.F. Endocrine changes after laparoscopic ovarian cautery in polycystic ovarian syndrome. American journal of obstetrics and gynecology 1987, 156, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Roy, D.; Kahler, D.J.; Yun, C.; Hubbard, E.J.A. Functional Interactions Between rsks-1/S6K, glp-1/Notch, and Regulators of Caenorhabditis elegans Fertility and Germline Stem Cell Maintenance. G3 (Bethesda) 2018, 8, 3293–3309. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Maine, E.M. The Bro1-domain protein, EGO-2, promotes Notch signaling in Caenorhabditis elegans. Genetics 2007, 176, 2265–2277. [Google Scholar] [CrossRef]
- Singh, R.; Smit, R.B.; Wang, X.; Wang, C.; Racher, H.; Hansen, D. Reduction of Derlin activity suppresses Notch-dependent tumours in the C. elegans germ line. PLoS genetics 2021, 17, e1009687. [Google Scholar] [CrossRef]
- Chi, C.; Ronai, D.; Than, M.T.; Walker, C.J.; Sewell, A.K.; Han, M. Nucleotide levels regulate germline proliferation through modulating GLP-1/Notch signaling in C. elegans. Genes & development 2016, 30, 307–320. [Google Scholar] [CrossRef] [PubMed]
- Brenner, J.L.; Jyo, E.M.; Mohammad, A.; Fox, P.; Jones, V.; Mardis, E.; Schedl, T.; Maine, E.M. TRIM-NHL protein, NHL-2, modulates cell fate choices in the C. elegans germ line. Developmental biology 2022, 491, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Gutnik, S.; Thomas, Y.; Guo, Y.; Stoecklin, J.; Neagu, A.; Pintard, L.; Merlet, J.; Ciosk, R. PRP-19, a conserved pre-mRNA processing factor and E3 ubiquitin ligase, inhibits the nuclear accumulation of GLP-1/Notch intracellular domain. Biology open 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Datla, U.S.; Scovill, N.C.; Brokamp, A.J.; Kim, E.; Asch, A.S.; Lee, M.H. Role of PUF-8/PUF protein in stem cell control, sperm-oocyte decision and cell fate reprogramming. Journal of cellular physiology 2014, 229, 1306–1311. [Google Scholar] [CrossRef]
- Racher, H.; Hansen, D. PUF-8, a Pumilio homolog, inhibits the proliferative fate in the Caenorhabditis elegans germline. G3 (Bethesda) 2012, 2, 1197–1205. [Google Scholar] [CrossRef] [PubMed]
- Haupt, K.A.; Law, K.T.; Enright, A.L.; Kanzler, C.R.; Shin, H.; Wickens, M.; Kimble, J. A PUF Hub Drives Self-Renewal in Caenorhabditis elegans Germline Stem Cells. Genetics 2020, 214, 147–161. [Google Scholar] [CrossRef] [PubMed]
- Gopal, S.; Amran, A.; Elton, A.; Ng, L.; Pocock, R. A somatic proteoglycan controls Notch-directed germ cell fate. Nature communications 2021, 12, 6708. [Google Scholar] [CrossRef]
- Hernandez, M.L.; Martinez, M.J.; Lopez de Heredia, M.; Ochoa, B. Protein phosphatase 1 and 2A inhibitors activate acyl-CoA:cholesterol acyltransferase and cholesterol ester formation in isolated rat hepatocytes. Biochimica et biophysica acta 1997, 1349, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Keene, J.D.; Lager, P.J. Post-transcriptional operons and regulons co-ordinating gene expression. Chromosome research: an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology 2005, 13, 327–337. [Google Scholar] [CrossRef]
- Ryder, S.P.; Massi, F. Insights into the structural basis of RNA recognition by STAR domain proteins. Advances in experimental medicine and biology 2010, 693, 37–53. [Google Scholar] [CrossRef]
- Artzt, K.; Wu, J.I. STAR trek: An introduction to STAR family proteins and review of quaking (QKI). Advances in experimental medicine and biology 2010, 693, 1–24. [Google Scholar] [PubMed]
- Zaffran, S.; Astier, M.; Gratecos, D.; Semeriva, M. The held out wings (how) Drosophila gene encodes a putative RNA-binding protein involved in the control of muscular and cardiac activity. Development 1997, 124, 2087–2098. [Google Scholar] [CrossRef] [PubMed]
- Ohno, G.; Hagiwara, M.; Kuroyanagi, H. STAR family RNA-binding protein ASD-2 regulates developmental switching of mutually exclusive alternative splicing in vivo. Genes & development 2008, 22, 360–374. [Google Scholar] [CrossRef] [PubMed]
- Sakers, K.; Liu, Y.; Llaci, L.; Lee, S.M.; Vasek, M.J.; Rieger, M.A.; Brophy, S.; Tycksen, E.; Lewis, R.; Maloney, S.E.; et al. Loss of Quaking RNA binding protein disrupts the expression of genes associated with astrocyte maturation in mouse brain. Nature communications 2021, 12, 1537. [Google Scholar] [CrossRef] [PubMed]
- Haroutunian, V.; Katsel, P.; Dracheva, S.; Davis, K.L. The human homolog of the QKI gene affected in the severe dysmyelination “quaking” mouse phenotype: downregulated in multiple brain regions in schizophrenia. The American journal of psychiatry 2006, 163, 1834–1837. [Google Scholar] [CrossRef]
- Francis, R.; Barton, M.K.; Kimble, J.; Schedl, T. gld-1, a tumor suppressor gene required for oocyte development in Caenorhabditis elegans. Genetics 1995, 139, 579–606. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.R.; Francis, R.; Schedl, T. GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Developmental biology 1996, 180, 165–183. [Google Scholar] [CrossRef]
- Carmel, A.B.; Wu, J.; Lehmann-Blount, K.A.; Williamson, J.R. High-affinity consensus binding of target RNAs by the STAR/GSG proteins GLD-1, STAR-2 and Quaking. BMC molecular biology 2010, 11, 48. [Google Scholar] [CrossRef] [PubMed]
- Ryder, S.P.; Frater, L.A.; Abramovitz, D.L.; Goodwin, E.B.; Williamson, J.R. RNA target specificity of the STAR/GSG domain post-transcriptional regulatory protein GLD-1. Nature structural & molecular biology 2004, 11, 20–28. [Google Scholar] [CrossRef]
- Doh, J.H.; Jung, Y.; Reinke, V.; Lee, M.H. C. elegans RNA-binding protein GLD-1 recognizes its multiple targets using sequence, context, and structural information to repress translation. Worm 2013, 2, e26548. [Google Scholar] [CrossRef]
- Lee, M.H.; Schedl, T. Translation repression by GLD-1 protects its mRNA targets from nonsense-mediated mRNA decay in C. elegans. Genes & development 2004, 18, 1047–1059. [Google Scholar] [CrossRef] [PubMed]
- Farley, B.M.; Ryder, S.P. POS-1 and GLD-1 repress glp-1 translation through a conserved binding-site cluster. Molecular biology of the cell 2012, 23, 4473–4483. [Google Scholar] [CrossRef] [PubMed]
- Mootz, D.; Ho, D.M.; Hunter, C.P. The STAR/Maxi-KH domain protein GLD-1 mediates a developmental switch in the translational control of C. elegans PAL-1. Development 2004, 131, 3263–3272. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, B.; Hanazawa, M.; Lee, M.H.; Nayak, S.; Volkmann, K.; Hofmann, E.R.; Hengartner, M.; Schedl, T.; Gartner, A. Translational repression of C. elegans p53 by GLD-1 regulates DNA damage-induced apoptosis. Cell 2005, 120, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Paulsen, J.; Yoo, Y.; Goodwin, E.B.; Strome, S. Caenorhabditis elegans MES-3 is a target of GLD-1 and functions epigenetically in germline development. Genetics 2001, 159, 1007–1017. [Google Scholar] [CrossRef] [PubMed]
- Scheckel, C.; Gaidatzis, D.; Wright, J.E.; Ciosk, R. Genome-wide analysis of GLD-1-mediated mRNA regulation suggests a role in mRNA storage. PLoS genetics 2012, 8, e1002742. [Google Scholar] [CrossRef] [PubMed]
- Crittenden, S.L.; Eckmann, C.R.; Wang, L.; Bernstein, D.S.; Wickens, M.; Kimble, J. Regulation of the mitosis/meiosis decision in the Caenorhabditis elegans germline. Philosophical transactions of the Royal Society of London. Series B, Biological sciences 2003, 358, 1359–1362. [Google Scholar] [CrossRef] [PubMed]
- Hansen, D.; Wilson-Berry, L.; Dang, T.; Schedl, T. Control of the proliferation versus meiotic development decision in the C. elegans germline through regulation of GLD-1 protein accumulation. Development 2004, 131, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Francis, R.; Maine, E.; Schedl, T. Analysis of the multiple roles of gld-1 in germline development: interactions with the sex determination cascade and the glp-1 signaling pathway. Genetics 1995, 139, 607–630. [Google Scholar] [CrossRef]
- Rutkowski, R.; Dickinson, R.; Stewart, G.; Craig, A.; Schimpl, M.; Keyse, S.M.; Gartner, A. Regulation of Caenorhabditis elegans p53/CEP-1-dependent germ cell apoptosis by Ras/MAPK signaling. PLoS genetics 2011, 7, e1002238. [Google Scholar] [CrossRef]
- Wang, L.; Eckmann, C.R.; Kadyk, L.C.; Wickens, M.; Kimble, J. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature 2002, 419, 312–316. [Google Scholar] [CrossRef] [PubMed]
- Kadyk, L.C.; Kimble, J. Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development 1998, 125, 1803–1813. [Google Scholar] [CrossRef] [PubMed]
- Eckmann, C.R.; Crittenden, S.L.; Suh, N.; Kimble, J. GLD-3 and control of the mitosis/meiosis decision in the germline of Caenorhabditis elegans. Genetics 2004, 168, 147–160. [Google Scholar] [CrossRef] [PubMed]
- Suh, N.; Jedamzik, B.; Eckmann, C.R.; Wickens, M.; Kimble, J. The GLD-2 poly(A) polymerase activates gld-1 mRNA in the Caenorhabditis elegans germ line. Proceedings of the National Academy of Sciences of the United States of America 2006, 103, 15108–15112. [Google Scholar] [CrossRef] [PubMed]
- Crittenden, S.L.; Bernstein, D.S.; Bachorik, J.L.; Thompson, B.E.; Gallegos, M.; Petcherski, A.G.; Moulder, G.; Barstead, R.; Wickens, M.; Kimble, J. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature 2002, 417, 660–663. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; O’Rourke, S.; Taki, F.A.; Alfhili, M.A.; Lee, M.H. Dose-Dependent Effects of GLD-2 and GLD-1 on Germline Differentiation and Dedifferentiation in the Absence of PUF-8. Frontiers in cell and developmental biology 2020, 8, 5. [Google Scholar] [CrossRef]
- Jeong, J.; Verheyden, J.M.; Kimble, J. Cyclin E and Cdk2 control GLD-1, the mitosis/meiosis decision, and germline stem cells in Caenorhabditis elegans. PLoS genetics 2011, 7, e1001348. [Google Scholar] [CrossRef]
- Biedermann, B.; Wright, J.; Senften, M.; Kalchhauser, I.; Sarathy, G.; Lee, M.H.; Ciosk, R. Translational repression of cyclin E prevents precocious mitosis and embryonic gene activation during C. elegans meiosis. Developmental cell 2009, 17, 355–364. [Google Scholar] [CrossRef]
- Kerins, J.A.; Hanazawa, M.; Dorsett, M.; Schedl, T. PRP-17 and the pre-mRNA splicing pathway are preferentially required for the proliferation versus meiotic development decision and germline sex determination in Caenorhabditis elegans. Developmental dynamics: an official publication of the American Association of Anatomists 2010, 239, 1555–1572. [Google Scholar] [CrossRef]
- Wang, X.; Voronina, E. Diverse Roles of PUF Proteins in Germline Stem and Progenitor Cell Development in C. elegans. Frontiers in cell and developmental biology 2020, 8, 29. [Google Scholar] [CrossRef]
- Nishanth, M.J.; Simon, B. Functions, mechanisms and regulation of Pumilio/Puf family RNA binding proteins: a comprehensive review. Molecular biology reports 2020, 47, 785–807. [Google Scholar] [CrossRef] [PubMed]
- Goldstrohm, A.C.; Hall, T.M.T.; McKenney, K.M. Post-transcriptional Regulatory Functions of Mammalian Pumilio Proteins. Trends in genetics: TIG 2018, 34, 972–990. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.H.; Hook, B.; Pan, G.; Kershner, A.M.; Merritt, C.; Seydoux, G.; Thomson, J.A.; Wickens, M.; Kimble, J. Conserved regulation of MAP kinase expression by PUF RNA-binding proteins. PLoS genetics 2007, 3, e233. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.H.; Wu, X.; Zhu, Y. RNA-binding protein PUM2 regulates mesenchymal stem cell fate via repression of JAK2 and RUNX2 mRNAs. Journal of cellular physiology 2020, 235, 3874–3885. [Google Scholar] [CrossRef]
- Goldstrohm, A.C.; Seay, D.J.; Hook, B.A.; Wickens, M. PUF protein-mediated deadenylation is catalyzed by Ccr4p. The Journal of biological chemistry 2007, 282, 109–114. [Google Scholar] [CrossRef]
- Friend, K.; Campbell, Z.T.; Cooke, A.; Kroll-Conner, P.; Wickens, M.P.; Kimble, J. A conserved PUF-Ago-eEF1A complex attenuates translation elongation. Nature structural & molecular biology 2012, 19, 176–183. [Google Scholar] [CrossRef]
- Kershner, A.M.; Kimble, J. Genome-wide analysis of mRNA targets for Caenorhabditis elegans FBF, a conserved stem cell regulator. Proceedings of the National Academy of Sciences of the United States of America 2010, 107, 3936–3941. [Google Scholar] [CrossRef]
- Whelan, J.T.; Hollis, S.E.; Cha, D.S.; Asch, A.S.; Lee, M.H. Post-transcriptional regulation of the Ras-ERK/MAPK signaling pathway. Journal of cellular physiology 2012, 227, 1235–1241. [Google Scholar] [CrossRef]
- Silva, I.L.Z.; Kohata, A.A.; Shigunov, P. Modulation and function of Pumilio proteins in cancer. Seminars in cancer biology 2022, 86, 298–309. [Google Scholar] [CrossRef]
- de la Roche, M.R.P.; Froats, M.; Bell, A.; McDonald, L.; Bolton, C.; Devins, R.; Hall, R.; Leclerc, J.; Istead, J.; Miron, M.; et al. Estimation of unregistered patients who left without being seen: At an urban mid-sized Canadian community emergency department. Canadian family physician Medecin de famille canadien 2021, 67, 114–120. [Google Scholar] [CrossRef]
- Vermani, L.; Kumar, R.; Senthil Kumar, N. GAPDH and PUM1: Optimal Housekeeping Genes for Quantitative Polymerase Chain Reaction-Based Analysis of Cancer Stem Cells and Epithelial-Mesenchymal Transition Gene Expression in Rectal Tumors. Cureus 2020, 12, e12020. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Liu, Z.; Yuan, Y.; Yang, Z.; Zhang, J.; Lu, Q.; Wang, W.; Fang, C.; Lin, H.; Liu, S. PUMILIO proteins promote colorectal cancer growth via suppressing p21. Nature communications 2022, 13, 1627. [Google Scholar] [CrossRef] [PubMed]
- Wickens, M.; Bernstein, D.S.; Kimble, J.; Parker, R. A PUF family portrait: 3’UTR regulation as a way of life. Trends in genetics: TIG 2002, 18, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Ariz, M.; Mainpal, R.; Subramaniam, K. C. elegans RNA-binding proteins PUF-8 and MEX-3 function redundantly to promote germline stem cell mitosis. Developmental biology 2009, 326, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Morgan, C.T.; Lee, M.H.; Kimble, J. Chemical reprogramming of Caenorhabditis elegans germ cell fate. Nature chemical biology 2010, 6, 102–104. [Google Scholar] [CrossRef]
- Bachorik, J.L.; Kimble, J. Redundant control of the Caenorhabditis elegans sperm/oocyte switch by PUF-8 and FBF-1, two distinct PUF RNA-binding proteins. Proceedings of the National Academy of Sciences of the United States of America 2005, 102, 10893–10897. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Zhao, J.; Hong, M.; Zeng, C.; Guang, S.; Shi, Y. Structural recognition of the mRNA 3’ UTR by PUF-8 restricts the lifespan of C. elegans. Nucleic acids research 2021, 49, 10082–10097. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Jiang, Y.; Sherrard, R.; Ikegami, K.; Conradt, B. PUF-8, a C. elegans ortholog of the RNA-binding proteins PUM1 and PUM2, is required for robustness of the cell death fate. Development 2023, 150. [Google Scholar] [CrossRef] [PubMed]
- Vaid, S.; Ariz, M.; Chaturbedi, A.; Kumar, G.A.; Subramaniam, K. PUF-8 negatively regulates RAS/MAPK signalling to promote differentiation of C. elegans germ cells. Development 2013, 140, 1645–1654. [Google Scholar] [CrossRef]
- Subramaniam, K.; Seydoux, G. Dedifferentiation of primary spermatocytes into germ cell tumors in C. elegans lacking the pumilio-like protein PUF-8. Current biology: CB 2003, 13, 134–139. [Google Scholar] [CrossRef]
- Cha, D.S.; Datla, U.S.; Hollis, S.E.; Kimble, J.; Lee, M.H. The Ras-ERK MAPK regulatory network controls dedifferentiation in Caenorhabditis elegans germline. Biochimica et biophysica acta 2012, 1823, 1847–1855. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.H.; Ohmachi, M.; Arur, S.; Nayak, S.; Francis, R.; Church, D.; Lambie, E.; Schedl, T. Multiple functions and dynamic activation of MPK-1 extracellular signal-regulated kinase signaling in Caenorhabditis elegans germline development. Genetics 2007, 177, 2039–2062. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Gaddy, M.; Hyun, M.; Jones, M.E.; Aslam, H.M.; Lee, M.H. Genetic and Chemical Controls of Sperm Fate and Spermatocyte Dedifferentiation via PUF-8 and MPK-1 in Caenorhabditis elegans. Cells 2023, 12. [Google Scholar] [CrossRef] [PubMed]
| Allele | CGC stock | Phenotype | Ref. |
|---|---|---|---|
| bn18 | DG2389 | Temperature-sensitive loss-of-function mutant | [11] |
| q224 | JK1107 | Temperature-sensitive loss-of-function mutant | [12] |
| oz112 | - | A ligand-independent gain-of-function mutant characterized by the formation of germline tumors. | [13] |
| ar202 | GC833 | A temperature-sensitive gain-of-function mutant characterized by the formation of proximal (Pro) germline tumors. This phenotype differs from that of the glp-1(oz112) mutants. The glp-1(ar202) mutants develop “Pro” germline tumors due to delayed initial meiotic entry during the L4 stage at the restrictive temperature. However, our genetic results revealed that additional mechanisms may induce the formation of germline tumors, even in the adult stage (Jones et al., unpublished results). | [14] |
| Allele | CGC stock | Phenotype | Ref. |
|---|---|---|---|
| op236 | TG34 | Fertile but hypersensitive to CEP-1/p53 mediated apoptosis. | [51] |
| q485 | JK3025 | Sterile with germline tumors | [37] [37] [37] [37] |
| q268 | JK3025 | Sterile with germline tumors | |
| q93 | JK3934 | Sterile with germline tumors | |
| q343 | JK1058 | Small abnormal oocytes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





