Submitted:
12 May 2024
Posted:
13 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Laboratory and Clinical Data
2.3. Gene Expression Analysis
2.3.1. RNA Extraction and cDNA Synthesis
2.3.2. Real-Time PCR Amplification
2.4. Statistical Analysis
3. Results
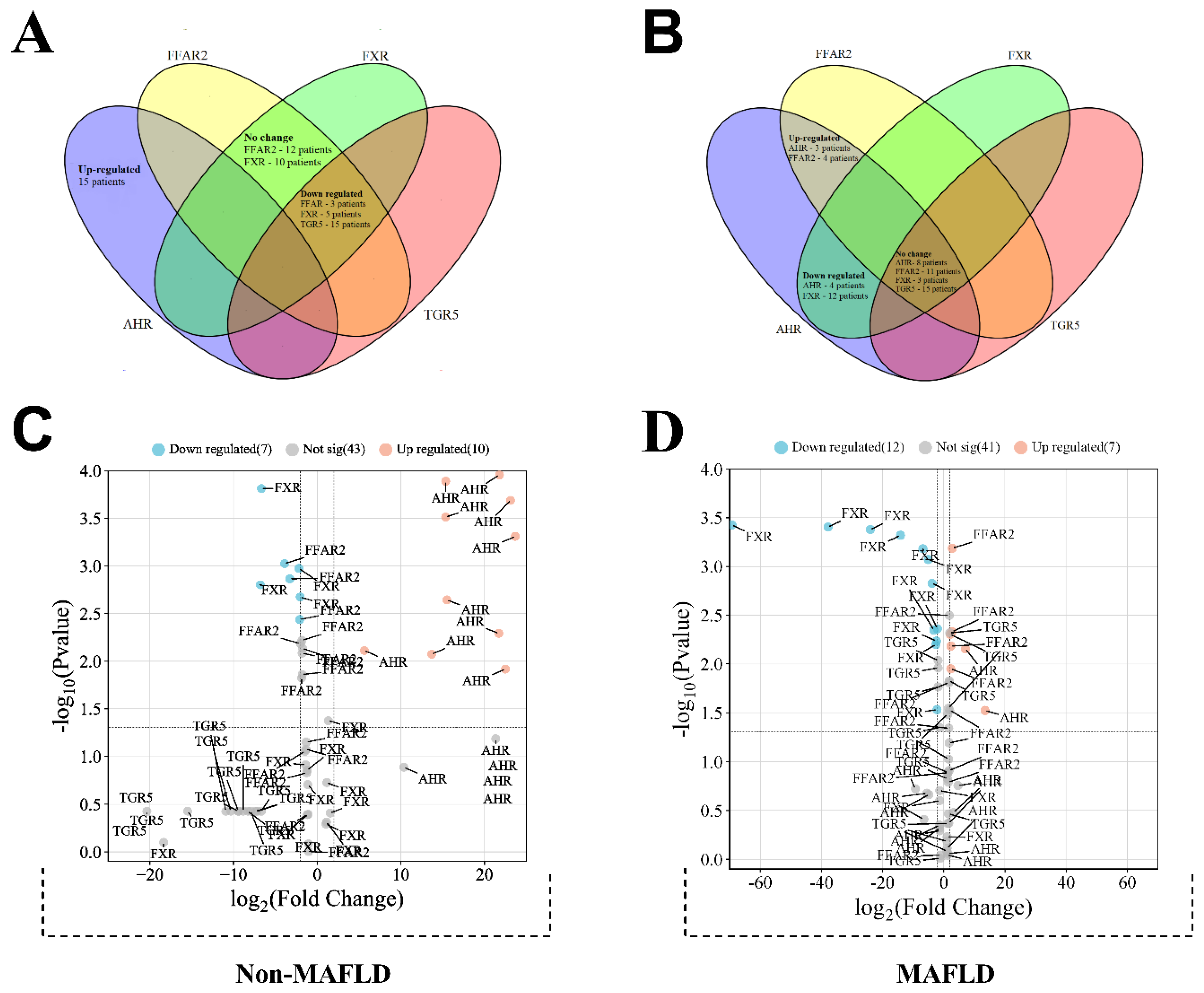
3.1. Сomparing Group Expression
| No-MAFLD cohort | ||||||
| Regulation of FXR gene expression | ||||||
| Down regulated (n = 5) | No change (n = 10) | p-Value a | ||||
| Band neutrophils, % (admission) | 9 (8.5–18) | 6 (4–10) | p = 0.045 | |||
| INR *, n (admission) | 1.1 (1.06–1.38) | 0.96 (0.865–1.06) | p = 0.038 | |||
| PT *, sec (admission) | 14.2 (13.5–16.6) | 12.5 (11.4–13.2) | p = 0.021 | |||
| GGT *, unit/l (admission) | 67 (49–127) | 31(19.1–42.5) | p = 0.008 | |||
| Albumin, g/l (admission) | 40 (35–40.5) | 45 (40–59.5) | p = 0.045 | |||
| Albumin, g/l (discharge) | 37 (33.5–40.5) | 47.5 (42.5–50) | p = 0.004 | |||
| Regulation of FFAR2 gene expression | ||||||
| No change (n = 12) | Down regulated (n = 3) | p-Value a | ||||
| Albumin, g/l (discharge) | 46 (41.5–49) | 35 (32–40) | p = 0.018 | |||
| Mafld cohort | ||||||
| Regulation of FFAR2 gene expression | ||||||
| No change (n = 11) | Up regulated (n = 4) | p-Value a | ||||
| Length of hospital stay, days | 11 (10–13) | 16.5 (15.3–17) | p = 0.001 | |||
| Leukocytes, 109/L (admission) | 7 (6.59–8.97) | 11.3 (10.4–14.6) | p = 0.010 | |||
| ALP *, mmol/L (admission) | 132 (107–271) | 89.5 (80.8–96.8) | p = 0.006 | |||
| ALP *, mmol/L (discharge) | 132 (106–195) | 90.5 (79–112) | p = 0.017 | |||
| Regulation of FXR gene expression | ||||||
| Down regulated (n = 12) | No change (n = 3) | p-Value a | ||||
| Length of hospital stay, days | 11.5 (10–13.8) | 17 (13–17) | p = 0.033 | |||
| ALP *, mmol/L (admission) | 130 (100–246) | 86 (79–103) | p = 0.031 | |||
| Regulation of AHR gene expression | ||||||
| Down regulated (n = 4) | No change (n = 8) | Up regulated (n = 3) | p-Value b | |||
| INR *, n (admission) | 1.17 (1.06–1.66) | 0.98 (0.9–1) | 0.99 (0.76–1.14) | p = 0.018 | ||
| PT *, sec (admission) | 14.8 (13.5–19.4) | 12.4(11.6–13.1) | 12.4 (10.12–14.9) | p = 0.033 | ||
3.2. Relative Expression of AHR, FFAR2, FXR and TGR5 in COVID-19 Patients with and without MAFLD
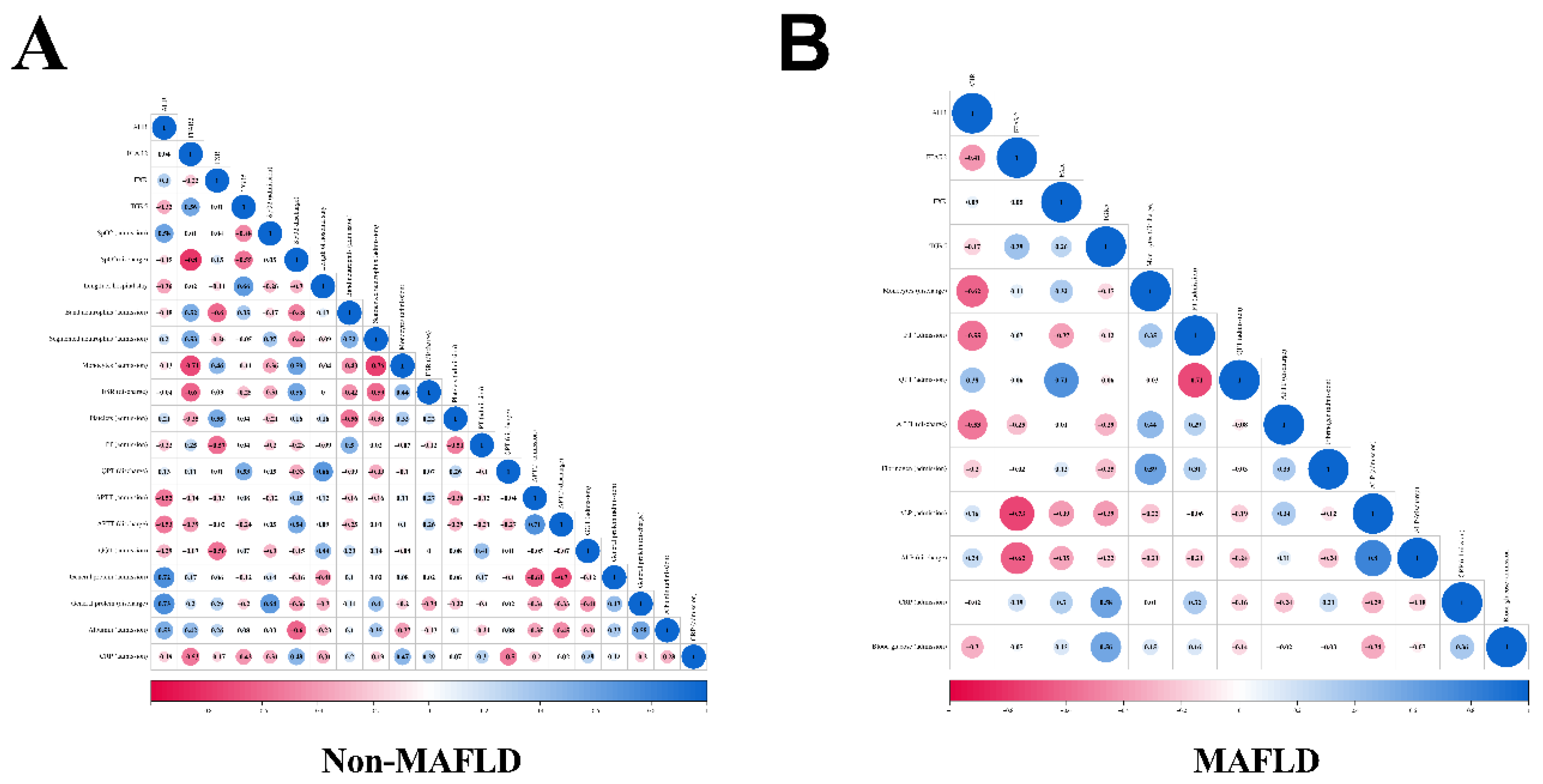
3.3. Correlation Analysis of Genes Normalized Expression
3.4. Principal Component Analysis
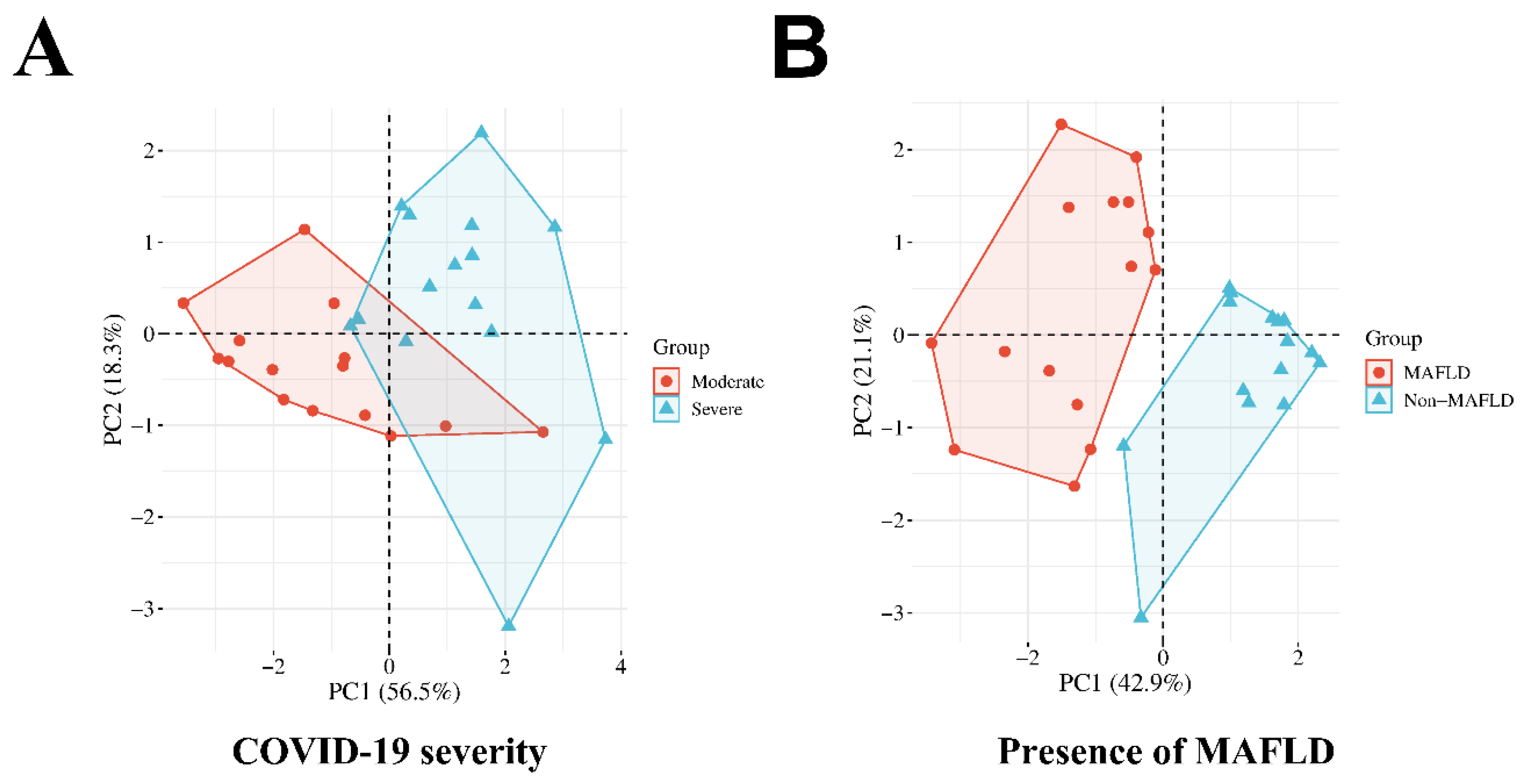
| Component | ||
| Factor 1 | Factor 2 | |
| BMI | 0.724 | 0.129 |
| FFAR2 Normalized Expression | 0.693 | 0.233 |
| FXR Normalized Expression | -0.635 | -0.085 |
| SpO2 (admission) | -0.18 | -0.914 |
| T2DM | 0.768 | 0.062 |
| The need for oxygen supply | 0.13 | 0.938 |
4. Discussion
5. Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- COVID - Coronavirus Statistics - Worldometer. Available online: https://www.worldometers.info/coronavirus/ (accessed on 29 April 2024).
- Buchynskyi, M.; Oksenych, V.; Kamyshna, I.; Kamyshnyi, O. Exploring Paxlovid Efficacy in COVID-19 Patients with MAFLD: Insights from a Single-Center Prospective Cohort Study. Viruses 2024, 16, 112. [Google Scholar] [CrossRef] [PubMed]
- Buchynskyi, M.; Kamyshna, I.; Lyubomirskaya, K.; Moshynets, O.; Kobyliak, N.; Oksenych, V.; Kamyshnyi, A. Efficacy of Interferon Alpha for the Treatment of Hospitalized Patients with COVID-19: A Meta-Analysis. Front. Immunol. 2023, 14, 1069894. [Google Scholar] [CrossRef] [PubMed]
- Kamyshnyi, A.; Koval, H.; Kobevko, O.; Buchynskyi, M.; Oksenych, V.; Kainov, D.; Lyubomirskaya, K.; Kamyshna, I.; Potters, G.; Moshynets, O. Therapeutic Effectiveness of Interferon-A2b against COVID-19 with Community-Acquired Pneumonia: The Ukrainian Experience. Int. J. Mol. Sci. 2023, 24, 6887. [Google Scholar] [CrossRef] [PubMed]
- Steenblock, C.; Schwarz, P.E.H.; Ludwig, B.; Linkermann, A.; Zimmet, P.; Kulebyakin, K.; Tkachuk, V.A.; Markov, A.G.; Lehnert, H.; de Angelis, M.H.; et al. COVID-19 and Metabolic Disease: Mechanisms and Clinical Management. Lancet Diabetes Endocrinol. 2021, 9, 786–798. [Google Scholar] [CrossRef] [PubMed]
- Hegyi, P.J.; Váncsa, S.; Ocskay, K.; Dembrovszky, F.; Kiss, S.; Farkas, N.; Erőss, B.; Szakács, Z.; Hegyi, P.; Pár, G. Metabolic Associated Fatty Liver Disease Is Associated With an Increased Risk of Severe COVID-19: A Systematic Review With Meta-Analysis. Front. Med. 2021, 8, 626425. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Hussain, S.; Antony, B. Non-Alcoholic Fatty Liver Disease and Clinical Outcomes in Patients with COVID-19: A Comprehensive Systematic Review and Meta-Analysis. Diabetes Metab. Syndr. Clin. Res. Rev. 2021, 15, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Tao, Z.; Li, Y.; Cheng, B.; Zhou, T.; Gao, Y. Risk of Severe COVID-19 Increased by Metabolic Dysfunction-Associated Fatty Liver Disease: A Meta-Analysis. J. Clin. Gastroenterol. 2021, 55, 830. [Google Scholar] [CrossRef] [PubMed]
- Buchynskyi, M.; Oksenych, V.; Kamyshna, I.; Vari, S.G.; Kamyshnyi, A. Genetic Predictors of Comorbid Course of COVID-19 and MAFLD: A Comprehensive Analysis. Viruses 2023, 15, 1724. [Google Scholar] [CrossRef] [PubMed]
- Buchynskyi, M.; Kamyshna, I.; Oksenych, V.; Zavidniuk, N.; Kamyshnyi, A. The Intersection of COVID-19 and Metabolic-Associated Fatty Liver Disease: An Overview of the Current Evidence. Viruses 2023, 15, 1072. [Google Scholar] [CrossRef]
- Veldhoen, M.; Hirota, K.; Westendorf, A.M.; Buer, J.; Dumoutier, L.; Renauld, J.-C.; Stockinger, B. The Aryl Hydrocarbon Receptor Links TH17-Cell-Mediated Autoimmunity to Environmental Toxins. Nature 2008, 453, 106–109. [Google Scholar] [CrossRef]
- Neavin, D.R.; Liu, D.; Ray, B.; Weinshilboum, R.M. The Role of the Aryl Hydrocarbon Receptor (AHR) in Immune and Inflammatory Diseases. Int. J. Mol. Sci. 2018, 19, 3851. [Google Scholar] [CrossRef] [PubMed]
- Anderson, G.; Carbone, A.; Mazzoccoli, G. Aryl Hydrocarbon Receptor Role in Co-Ordinating SARS-CoV-2 Entry and Symptomatology: Linking Cytotoxicity Changes in COVID-19 and Cancers; Modulation by Racial Discrimination Stress. Biology 2020, 9, 249. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, B.P.; Roberts, A.D.; Neumiller, J.J.; Cundiff, J.A.; Woodland, D.L. Aryl Hydrocarbon Receptor Activation Impairs the Priming but Not the Recall of Influenza Virus-Specific CD8+ T Cells in the Lung. J. Immunol. 2006, 177, 5819–5828. [Google Scholar] [CrossRef] [PubMed]
- Thomas, T.; Stefanoni, D.; Reisz, J.A.; Nemkov, T.; Bertolone, L.; Francis, R.O.; Hudson, K.E.; Zimring, J.C.; Hansen, K.C.; Hod, E.A.; et al. COVID-19 Infection Alters Kynurenine and Fatty Acid Metabolism, Correlating with IL-6 Levels and Renal Status. JCI insight 2020, 5. [Google Scholar] [CrossRef] [PubMed]
- Giovannoni, F.; Li, Z.; Remes-Lenicov, F.; Dávola, M.E.; Elizalde, M.; Paletta, A.; Ashkar, A.A.; Mossman, K.L.; Dugour, A.V.; Figueroa, J.M.; et al. AHR Signaling Is Induced by Infection with Coronaviruses. Nat. Commun. 2021, 12, 5148. [Google Scholar] [CrossRef] [PubMed]
- Morelli, A.E.; Zahorchak, A.F.; Larregina, A.T.; Colvin, B.L.; Logar, A.J.; Takayama, T.; Falo, L.D.; Thomson, A.W. Cytokine Production by Mouse Myeloid Dendritic Cells in Relation to Differentiation and Terminal Maturation Induced by Lipopolysaccharide or CD40 Ligation. Blood 2001, 98, 1512–1523. [Google Scholar] [CrossRef]
- Lee, J.H.; Wada, T.; Febbraio, M.; He, J.; Matsubara, T.; Lee, M.J.; Gonzalez, F.J.; Xie, W. A Novel Role for the Dioxin Receptor in Fatty Acid Metabolism and Hepatic Steatosis. Gastroenterology 2010, 139, 653–663. [Google Scholar] [CrossRef]
- Lu, P.; Yan, J.; Liu, K.; Garbacz, W.G.; Wang, P.; Xu, M.; Ma, X.; Xie, W. Activation of Aryl Hydrocarbon Receptor Dissociates Fatty Liver from Insulin Resistance by Inducing Fibroblast Growth Factor 21. Hepatology 2015, 61, 1908–1919. [Google Scholar] [CrossRef]
- Wang, C.; Xu, C.-X.; Krager, S.L.; Bottum, K.M.; Liao, D.-F.; Tischkau, S.A. Aryl Hydrocarbon Receptor Deficiency Enhances Insulin Sensitivity and Reduces PPAR-α Pathway Activity in Mice. Environ. Health Perspect. 2011, 119, 1739–1744. [Google Scholar] [CrossRef]
- Lusis, A.J.; Attie, A.D.; Reue, K. Metabolic Syndrome: From Epidemiology to Systems Biology. Nat. Rev. Genet. 2008, 9, 819–830. [Google Scholar] [CrossRef]
- Kamp, M.E.; Shim, R.; Nicholls, A.J.; Oliveira, A.C.; Mason, L.J.; Binge, L.; Mackay, C.R.; Wong, C.H.Y. G Protein-Coupled Receptor 43 Modulates Neutrophil Recruitment during Acute Inflammation. PLoS One 2016, 11, e0163750. [Google Scholar] [CrossRef] [PubMed]
- McNelis, J.C.; Lee, Y.S.; Mayoral, R.; van der Kant, R.; Johnson, A.M.F.; Wollam, J.; Olefsky, J.M. GPR43 Potentiates β-Cell Function in Obesity. Diabetes 2015, 64, 3203–3217. [Google Scholar] [CrossRef]
- Tolhurst, G.; Heffron, H.; Lam, Y.S.; Parker, H.E.; Habib, A.M.; Diakogiannaki, E.; Cameron, J.; Grosse, J.; Reimann, F.; Gribble, F.M. Short-Chain Fatty Acids Stimulate Glucagon-like Peptide-1 Secretion via the G-Protein-Coupled Receptor FFAR2. Diabetes 2012, 61, 364–371. [Google Scholar] [CrossRef] [PubMed]
- Henao-Mejia, J.; Elinav, E.; Jin, C.; Hao, L.; Mehal, W.Z.; Strowig, T.; Thaiss, C.A.; Kau, A.L.; Eisenbarth, S.C.; Jurczak, M.J.; et al. Inflammasome-Mediated Dysbiosis Regulates Progression of NAFLD and Obesity. Nature 2012, 482, 179–185. [Google Scholar] [CrossRef]
- Liang, Y.; Liang, S.; Zhang, Y.; Deng, Y.; He, Y.; Chen, Y.; Liu, C.; Lin, C.; Yang, Q. Oral Administration of Compound Probiotics Ameliorates HFD-Induced Gut Microbe Dysbiosis and Chronic Metabolic Inflammation via the G Protein-Coupled Receptor 43 in Non-Alcoholic Fatty Liver Disease Rats. Probiotics Antimicrob. Proteins 2019, 11, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Xie, J.; Zhao, L.; Fei, X.; Zhang, H.; Tan, Y.; Nie, X.; Zhou, L.; Liu, Z.; Ren, Y.; et al. Alveolar Macrophage Dysfunction and Cytokine Storm in the Pathogenesis of Two Severe COVID-19 Patients. EBioMedicine 2020, 57, 102833. [Google Scholar] [CrossRef]
- Pham, M.T.; Yang, A.J.; Kao, M.-S.; Gankhuyag, U.; Zayabaatar, E.; Jin, S.-L.C.; Huang, C.-M. Gut Probiotic Lactobacillus Rhamnosus Attenuates PDE4B-Mediated Interleukin-6 Induced by SARS-CoV-2 Membrane Glycoprotein. J. Nutr. Biochem. 2021, 98, 108821. [Google Scholar] [CrossRef]
- Kao, M.-S.; Yang, J.-H.; Balasubramaniam, A.; Traisaeng, S.; Jackson Yang, A.; Yang, J.J.; Salamon, B.P.; Herr, D.R.; Huang, C.-M. Colonization of Nasal Cavities by Staphylococcus Epidermidis Mitigates SARS-CoV-2 Nucleocapsid Phosphoprotein-Induced Interleukin (IL)-6 in the Lung. Microb. Biotechnol. 2022, 15, 1984–1994. [Google Scholar] [CrossRef]
- Kawamata, Y.; Fujii, R.; Hosoya, M.; Harada, M.; Yoshida, H.; Miwa, M.; Fukusumi, S.; Habata, Y.; Itoh, T.; Shintani, Y.; et al. A G Protein-Coupled Receptor Responsive to Bile Acids. J. Biol. Chem. 2003, 278, 9435–9440. [Google Scholar] [CrossRef]
- Mencarelli, A.; Renga, B.; Migliorati, M.; Cipriani, S.; Distrutti, E.; Santucci, L.; Fiorucci, S. The Bile Acid Sensor Farnesoid X Receptor Is a Modulator of Liver Immunity in a Rodent Model of Acute Hepatitis. J. Immunol. 2009, 183, 6657–6666. [Google Scholar] [CrossRef]
- Vavassori, P.; Mencarelli, A.; Renga, B.; Distrutti, E.; Fiorucci, S. The Bile Acid Receptor FXR Is a Modulator of Intestinal Innate Immunity. J. Immunol. 2009, 183, 6251–6261. [Google Scholar] [CrossRef] [PubMed]
- Biagioli, M.; Carino, A.; Fiorucci, C.; Marchianò, S.; Di Giorgio, C.; Bordoni, M.; Roselli, R.; Baldoni, M.; Distrutti, E.; Zampella, A.; et al. The Bile Acid Receptor GPBAR1 Modulates CCL2/CCR2 Signaling at the Liver Sinusoidal/Macrophage Interface and Reverses Acetaminophen-Induced Liver Toxicity. J. Immunol. 2020, 204, 2535–2551. [Google Scholar] [CrossRef] [PubMed]
- Fiorucci, S.; Carino, A.; Baldoni, M.; Santucci, L.; Costanzi, E.; Graziosi, L.; Distrutti, E.; Biagioli, M. Bile Acid Signaling in Inflammatory Bowel Diseases. Dig. Dis. Sci. 2021, 66, 674–693. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, R.; Takayama, T.; Yoneno, K.; Kamada, N.; Kitazume, M.T.; Higuchi, H.; Matsuoka, K.; Watanabe, M.; Itoh, H.; Kanai, T.; et al. Bile Acids Induce Monocyte Differentiation toward Interleukin-12 Hypo-Producing Dendritic Cells via a TGR5-Dependent Pathway. Immunology 2012, 136, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Valdés, A.; Moreno, L.O.; Rello, S.R.; Orduña, A.; Bernardo, D.; Cifuentes, A. Metabolomics Study of COVID-19 Patients in Four Different Clinical Stages. Sci. Rep. 2022, 12, 1650. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Yi, X.; Sun, Y.; Bi, X.; Du, J.; Zhang, C.; Quan, S.; Zhang, F.; Sun, R.; Qian, L.; et al. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell 2020, 182, 59–72.e15. [Google Scholar] [CrossRef] [PubMed]
- Yeoh, Y.K.; Zuo, T.; Lui, G.C.-Y.; Zhang, F.; Liu, Q.; Li, A.Y.; Chung, A.C.; Cheung, C.P.; Tso, E.Y.; Fung, K.S.; et al. Gut Microbiota Composition Reflects Disease Severity and Dysfunctional Immune Responses in Patients with COVID-19. Gut 2021, 70, 698–706. [Google Scholar] [CrossRef] [PubMed]
- Durairajan, S.S.K.; Singh, A.K.; Saravanan, U.B.; Namachivayam, M.; Radhakrishnan, M.; Huang, J.D.; Dhodapkar, R.; Zhang, H. Gastrointestinal Manifestations of SARS-CoV-2: Transmission, Pathogenesis, Immunomodulation, Microflora Dysbiosis, and Clinical Implications. Viruses 2023, 15, 1231. [Google Scholar] [CrossRef]
- Stutz, M.R.; Dylla, N.P.; Pearson, S.D.; Lecompte-Osorio, P.; Nayak, R.; Khalid, M.; Adler, E.; Boissiere, J.; Lin, H.; Leiter, W.; et al. Immunomodulatory Fecal Metabolites Are Associated with Mortality in COVID-19 Patients with Respiratory Failure. Nat. Commun. 2022, 13, 1–11. [Google Scholar] [CrossRef]
- Campbell, C.; McKenney, P.T.; Konstantinovsky, D.; Isaeva, O.I.; Schizas, M.; Verter, J.; Mai, C.; Jin, W.-B.; Guo, C.-J.; Violante, S.; et al. Bacterial Metabolism of Bile Acids Promotes Generation of Peripheral Regulatory T Cells. Nature 2020, 581, 475–479. [Google Scholar] [CrossRef]
- Fiorucci, S.; Urbani, G.; Biagioli, M.; Sepe, V.; Distrutti, E.; Zampella, A. Bile Acids and Bile Acid Activated Receptors in the Treatment of Covid-19. Biochem. Pharmacol. 2023, 115983. [Google Scholar] [CrossRef]
- Brevini, T.; Maes, M.; Webb, G.J.; John, B.V.; Fuchs, C.D.; Buescher, G.; Wang, L.; Griffiths, C.; Brown, M.L.; Scott, W.E.; et al. FXR Inhibition May Protect from SARS-CoV-2 Infection by Reducing ACE2. Nature 2022, 615, 134–142. [Google Scholar] [CrossRef]
- Biagioli, M.; Marchianò, S.; Roselli, R.; Di Giorgio, C.; Bellini, R.; Bordoni, M.; Distrutti, E.; Catalanotti, B.; Zampella, A.; Graziosi, L.; et al. GLP-1 Mediates Regulation of Colonic ACE2 Expression by the Bile Acid Receptor GPBAR1 in Inflammation. Cells 2022, 11, 1187. [Google Scholar] [CrossRef] [PubMed]
- Verbeke, L.; Mannaerts, I.; Schierwagen, R.; Govaere, O.; Klein, S.; Vander Elst, I.; Windmolders, P.; Farre, R.; Wenes, M.; Mazzone, M.; et al. FXR Agonist Obeticholic Acid Reduces Hepatic Inflammation and Fibrosis in a Rat Model of Toxic Cirrhosis. Sci. Rep. 2016, 6, 33453. [Google Scholar] [CrossRef]
- Inagaki, T.; Moschetta, A.; Lee, Y.-K.; Peng, L.; Zhao, G.; Downes, M.; Yu, R.T.; Shelton, J.M.; Richardson, J.A.; Repa, J.J.; et al. Regulation of Antibacterial Defense in the Small Intestine by the Nuclear Bile Acid Receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 3920–3925. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Xie, S.; Chi, Z.; Zhang, J.; Liu, Y.; Zhang, L.; Zheng, M.; Zhang, X.; Xia, D.; Ke, Y.; et al. Bile Acids Control Inflammation and Metabolic Disorder through Inhibition of NLRP3 Inflammasome. Immunity 2016, 45, 802–816. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-D.; Chen, W.-D.; Yu, D.; Forman, B.M.; Huang, W. The G-Protein-Coupled Bile Acid Receptor, Gpbar1 (TGR5), Negatively Regulates Hepatic Inflammatory Response through Antagonizing Nuclear Factor κ Light-Chain Enhancer of Activated B Cells (NF-ΚB) in Mice. Hepatology 2011, 54, 1421–1432. [Google Scholar] [CrossRef]
- Clinical Spectrum | COVID-19 Treatment Guidelines. Available online: https://www.covid19treatmentguidelines.nih.gov/overview/clinical-spectrum/ (accessed on 29 October 2023).
- Fouad, Y.; Waked, I.; Bollipo, S.; Gomaa, A.; Ajlouni, Y.; Attia, D. What’s in a Name? Renaming ‘NAFLD’ to ‘MAFLD.’ Liver Int. 2020, 40, 1254–1261. [Google Scholar] [CrossRef] [PubMed]
- Méndez-Sánchez, N.; Bugianesi, E.; Gish, R.G.; Lammert, F.; Tilg, H.; Nguyen, M.H.; Sarin, S.K.; Fabrellas, N.; Zelber-Sagi, S.; Fan, J.G.; et al. Global Multi-Stakeholder Endorsement of the MAFLD Definition. Lancet Gastroenterol. Hepatol. 2022, 7, 388–390. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, D.; Kim, H.J.; Lee, C.H.; Yang, J.I.; Kim, W.; Kim, Y.J.; Yoon, J.H.; Cho, S.H.; Sung, M.W.; et al. Hepatic Steatosis Index: A Simple Screening Tool Reflecting Nonalcoholic Fatty Liver Disease. Dig. Liver Dis. 2010, 42, 503–508. [Google Scholar] [CrossRef]
- Gadaleta, R.M.; Oldenburg, B.; Willemsen, E.C.L.; Spit, M.; Murzilli, S.; Salvatore, L.; Klomp, L.W.J.; Siersema, P.D.; van Erpecum, K.J.; van Mil, S.W.C. Activation of Bile Salt Nuclear Receptor FXR Is Repressed by Pro-Inflammatory Cytokines Activating NF-ΚB Signaling in the Intestine. Biochim. Biophys. Acta 2011, 1812, 851–858. [Google Scholar] [CrossRef] [PubMed]
- Massafra, V.; Ijssennagger, N.; Plantinga, M.; Milona, A.; Ramos Pittol, J.M.; Boes, M.; van Mil, S.W.C. Splenic Dendritic Cell Involvement in FXR-Mediated Amelioration of DSS Colitis. Biochim. Biophys. Acta 2016, 1862, 166–173. [Google Scholar] [CrossRef] [PubMed]
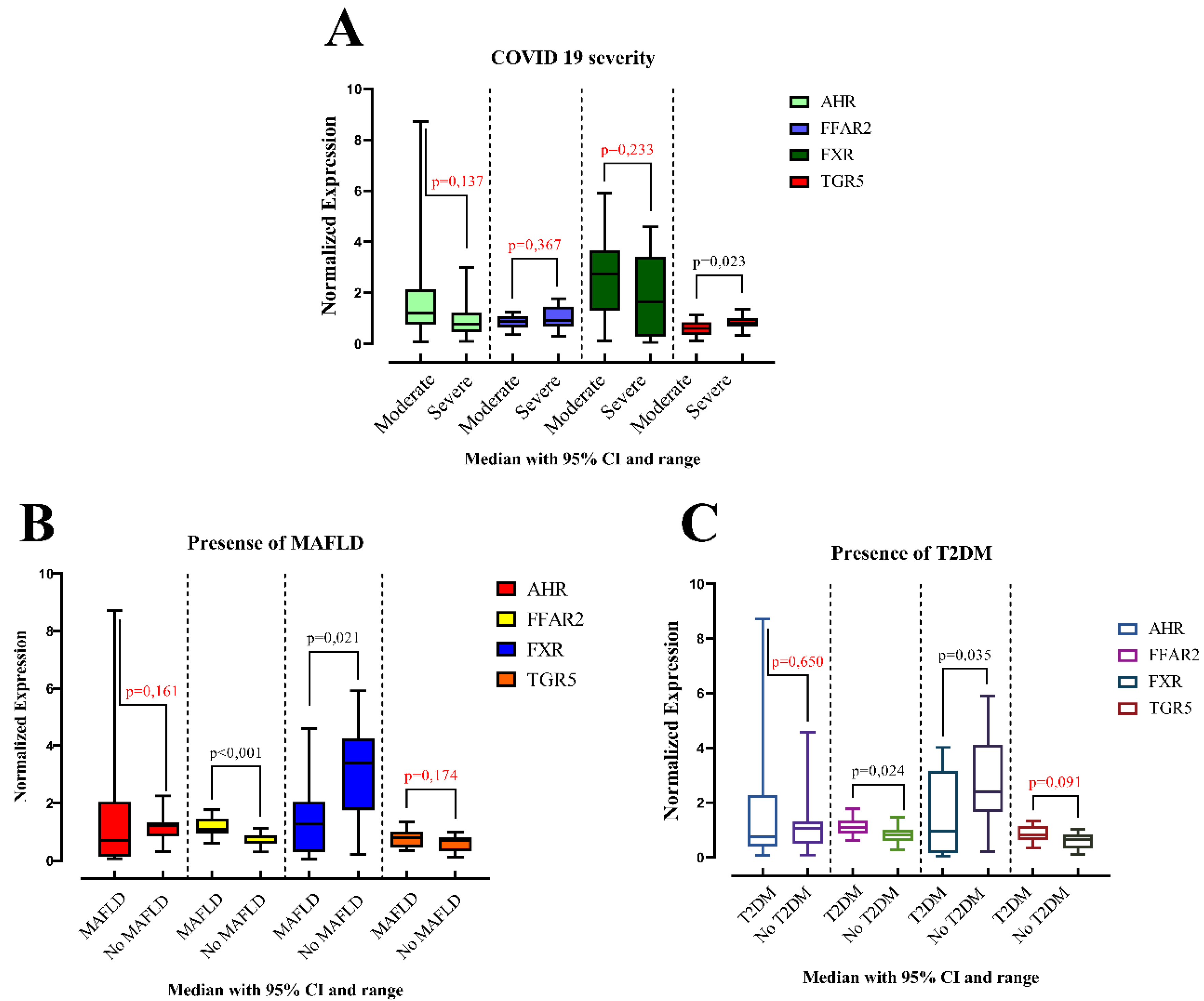
| Normalized Expression (median, IQR a) | |||||
| AHR | FFAR2 | FXR | TGR5 | ||
| Sex | Male (n = 17) | 0.871 (0.519–1.53) | 0.938 (0.621–1.14) | 1.88 (0.863–3.77) | 0.798 (0.404–0.999) |
| Female (n = 13) | 1.22 (0.494–1.72) | 0.876 (0.656–1.1) | 1.92 (0.430–3.54) | 0.681 (0.387–0.848) | |
| p-Value b | p = 0.837 | p = 1.000 | p = 0.650 | p = 0.509 | |
| MAFLD | Presence (n = 15) | 0.715 (0.133–2.04) | 1.11 (0.938–1.46) | 1.3 (0.298–2.04) | 0.801 (0.46–1.03) |
| Absence (n = 15) | 1.22 (0.863–1.33) | 0.646 (0.584–0.88) | 3.41 (1.75–4.27) | 0.702 (0.325–0.832) | |
| p-Value | p = 0.161 | p < 0.001 | p = 0.021 | p = 0.174 | |
| COVID 19 severity | Moderate (15) | 1.20 (0.77–2.14) | 0.876 (0.64–1.07) | 2.74 (1.30–3.66) | 0.606 (0.344–0.846) |
| Severe (15) | 0.774 (0.456–1.22) | 0.928 (0.665–1.46) | 1.65 (0.298–3.41) | 0.801 (0.681–1.01) | |
| p-Value | p = 0.137 | p = 0.367 | p = 0.233 | p = 0.023 | |
| Presence of pneumonia | Presence (n = 16) | 0.649 (0.351–1.13) | 0.997 (0.7–1.4) | 1.7 (0.364–2.87) | 0.815 (0.484–1) |
| Absence (n = 14) | 1.53 (0.863–2.44) | 0.871 (0.636–0.999) | 3.19 (1.04–4.08) | 0.65 (0.348–0.812) | |
| p-Value | p = 0.001 | p = 0.193 | p = 0.093 | p = 0.179 | |
| Obesity | Presence (n = 10) | 0.847 (0.375–2.67) | 1.05 (0.868–1.23) | 1.86 (0.539–3.17) | 0.763 (0.348–1.05) |
| Absence (20) | 1.04 (0.616–1.32) | 0.835 (0.628–1.09) | 2.31 (0.707–3.92) | 0.703 (0.436–0.856) | |
| p-Value | p = 0.846 | p = 0.109 | p = 0.530 | p = 0.373 | |
| T2DM | Presence (n = 10) | 0.772 (0.396–2.28) | 1.12 (0.868–1.35) | 0.971 (0.160–3.17) | 0.833 (0.634–1.16) |
| Absence (n = 20) | 1.06 (0.519–1.32) | 0.835 (0.620–1.02) | 2.39 (1.67–4.12) | 0.671 (0.345–0.842) | |
| p-Value | p = 0.650 | p = 0.024 | p = 0.035 | p = 0.091 | |
| Arterial hypertension | Presence (n = 20) | 0.891 (0.552–1.25) | 0.953 (0.7–1.13) | 1.98 (0.698–3.5) | 0.774 (0.569–0.986) |
| Absence (n = 10) | 1.08 (0.124–1.86) | 0.761 (0.614–1.08) | 1.47 (0.608–4.48) | 0.581 (0.308–0.850) | |
| p-Value | p = 0.948 | p = 0.328 | p = 0.983 | p = 0.169 | |
| Coronary heart disease | Presence (n = 13) | 0.863 (0.607–1.67) | 0.987 (0.754–1.14) | 3.14 (0.237–3.77) | 0.705 (0.598–1.03) |
| Absence (n = 17) | 0.919 (0.386–1.53) | 0.876 (0.601–1.08) | 1.80 (0.727–3.54) | 0.702 (0.336–0.855) | |
| p-Value | p = 0.967 | p = 0.133 | p = 1.000 | p = 0.229 | |
| Component | ||
| Factor 1 | Factor 2 | |
| Сommunity acquired pneumonia | 0.227 | 0.816 |
| SpO2 (admission) | -0.083 | -0.870 |
| Length of hospital stay (days) | 0.561 | 0 |
| Segmented neutrophils (discharge) | 0.868 | 0.411 |
| NLR (discharge) | 0.918 | 0.13 |
| Lymphocytes (discharge) | -0.903 | -0.307 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





