1. Introduction
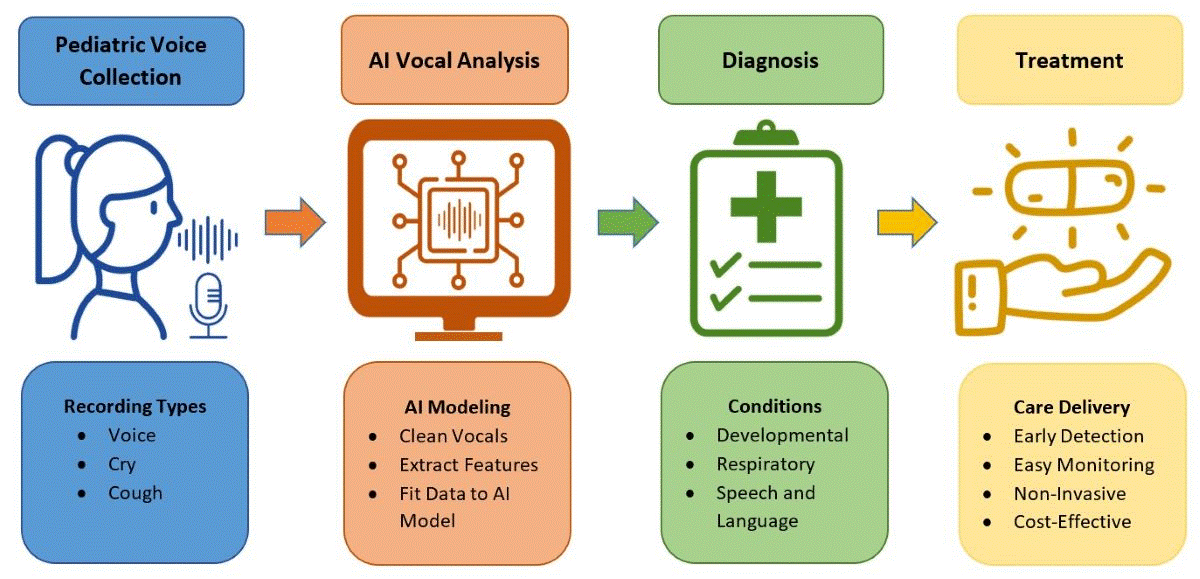
The human voice is often referred to as a unique print for each individual. It contains biomarkers that have been linked in the adult literature to various diseases ranging from Parkinson's disease [
66] to dementia, mood disorders, and cancers [
64,
65,
67]. The voice contains complex acoustic markers that depend on respiration, phonation, articulation, and prosody coordination. Recent advances in acoustic analysis technology, especially when coupled with machine learning, have shed new insights into the detection of diseases. As a biomarker, the voice is cost-effective, easy, and safe to collect in low-resource settings. Moreover, the human voice contains not only speech, but other acoustic biomarkers such as cry, cough, and other respiratory sounds. The objective of this scoping review is to synthesize existing knowledge on the application of Artificial Intelligence (AI) in the analysis of pediatric voice as a biomarker for health to foster a deeper understanding of its potential use as an investigative or diagnostic tool within the pediatric clinical setting.
2. Materials Methods
2.1. Registration and Funding
This scoping review was registered with the Open Science Framework (OSF) to enhance transparency and reproducibility. The review was registered on July 24, 2023 under the OSF registration DOI 10.17605/OSF.IO/SC6MG. The full registration details, including the review protocol and objectives can be accessed at
https://osf.io/sc6mg. All phases of this study were supported by National Institutes of Health grant number: 1OT20D032720-01.
2.2. Search Strategy
Precise searches were conducted to identify relevant keywords and controlled vocabulary for the following concepts: artificial intelligence, voice, pediatrics, and disorders. Controlled vocabulary terms were combined logically by a medical librarian using Boolean logic, with keywords searched in the title and abstract to form a sensitive search strategy. The final search strategy utilized 217 keywords, including 91 related to "artificial intelligence," 45 related to "voice," 20 related to "pediatric," and 61 related to "disorder,” as shown in
Appendix I. The original PubMed search was translated into the following databases: Embase, Web of Science Core Collection, and the Cochrane database. Google Scholar and ClinicalTrials.gov were searched in order to pull in grey literature. All searches were run in May 2023 and deduplicated in EndNote using the validated deduplication method put forth by Bramer et al. [
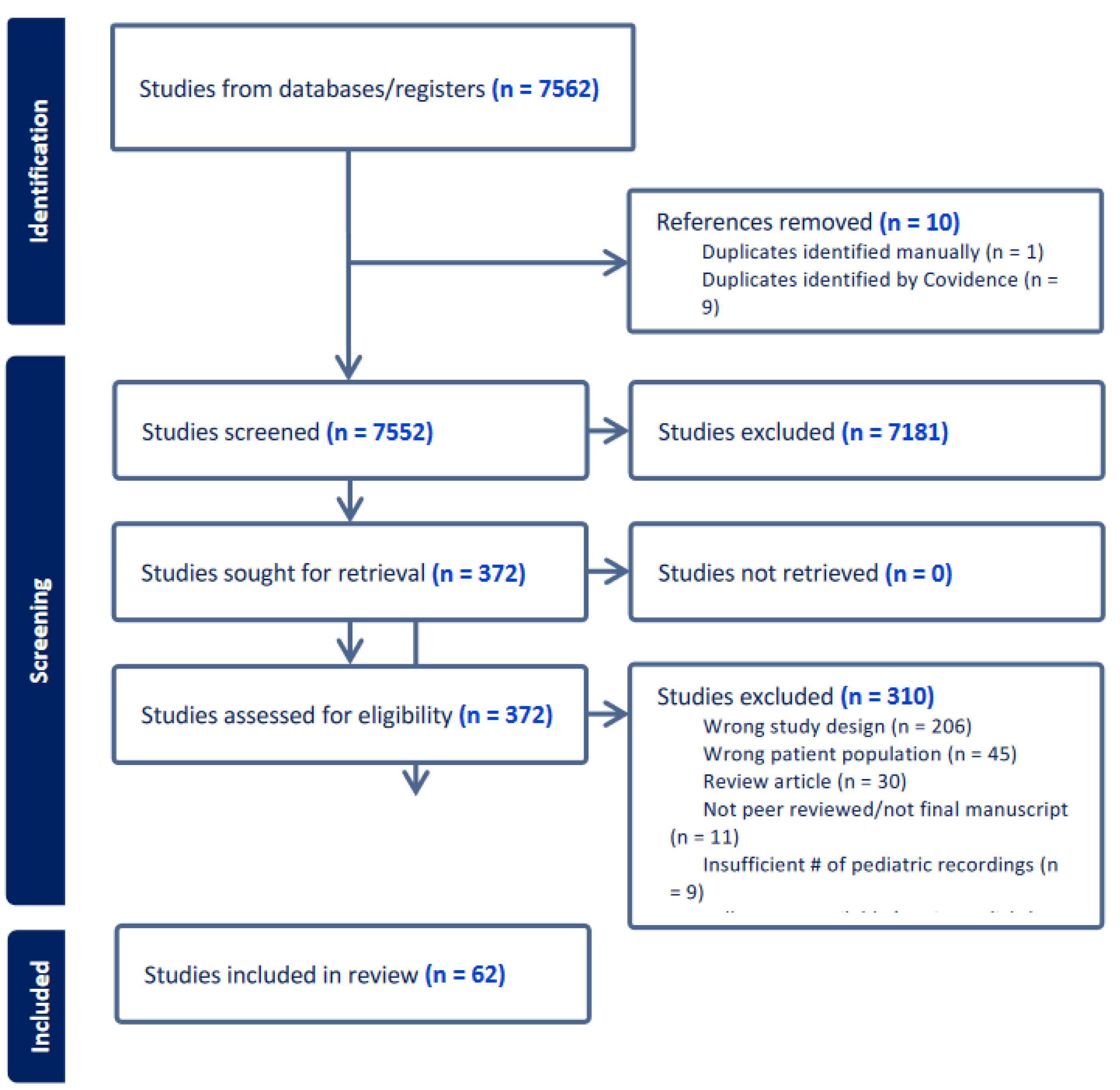
13]. Results were imported into Covidence, a systematic review software. Titles and abstracts were independently reviewed by two reviewers against pre-defined inclusion criteria. Relevant texts were moved to the full-text review, whereby the same process evaluated PDFs of eligible citations. Conflicting votes were resolved via discussion until the two original reviewers reached a consensus. The PRISMA flow chart of article inclusion is shown in
Figure 1.
2.3. Inclusion Criteria
Each study was required to include voice recordings in pediatric populations aged 0-17 years. Studies involving both pediatric and adult cohorts were considered on the basis that pediatric data were collected and analyzed separately from adult data. A minimum of 10 pediatric participants were required in each study. All pediatric health conditions were considered except for newborn or infant cry to detect hunger, discomfort, pain, or sleepiness. Studies were limited to peer-reviewed prospective or retrospective research studies written originally in English and excluded scoping reviews, literature reviews, and meta-analyses. Studies were required to utilize one or more feature extraction methods to produce a vocal dataset and required an analysis of pathological biomarkers contained in voice, cry, or respiratory sounds using one or more machine learning or artificial intelligence models.
2.4. Data Extraction
At the final stage, 62 studies met the inclusion criteria (
Figure 1). A study was eligible for data extraction after two independent reviewers reached a consensus on its inclusion in the title, abstract, and full-text review phases. Utilizing the data extraction template in Covidence, we customized a tool to collect general study information, study characteristics, participant characteristics, recording sources and data, feature extraction methods, and machine learning or artificial intelligence model types.
3. Results
3.1. Global Representation
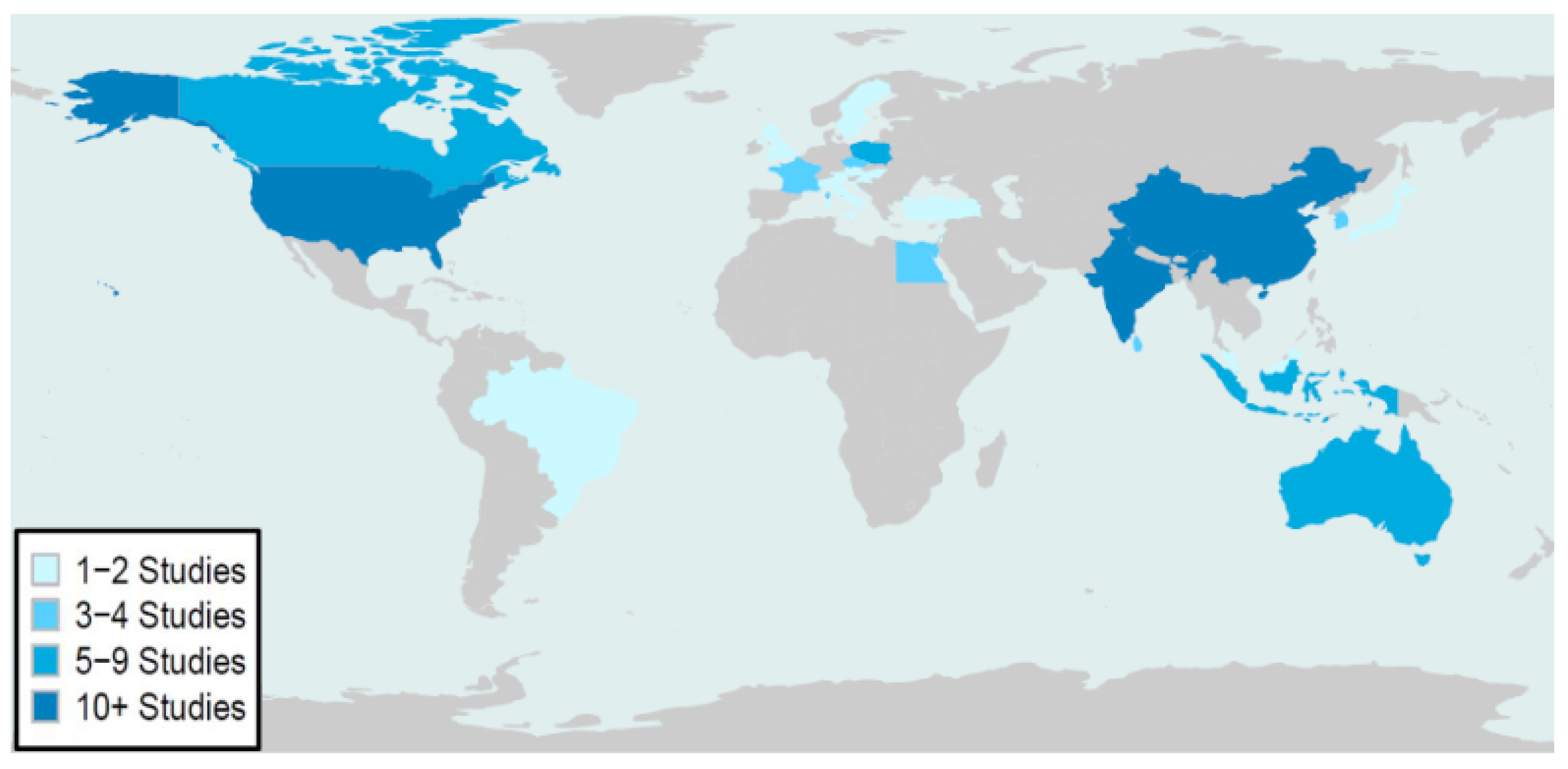
Across 62 studies, 25 countries were represented (
Appendix II). The global distribution and frequency of publication are shown in
Figure 2. Pediatric populations from the United States, India, and China were the most frequently studied. Data primarily represented pediatric populations from North America, Asia, Europe, and Oceania and less representative of Central and South America, Africa, and the Middle East.
3.2. Studies by Year
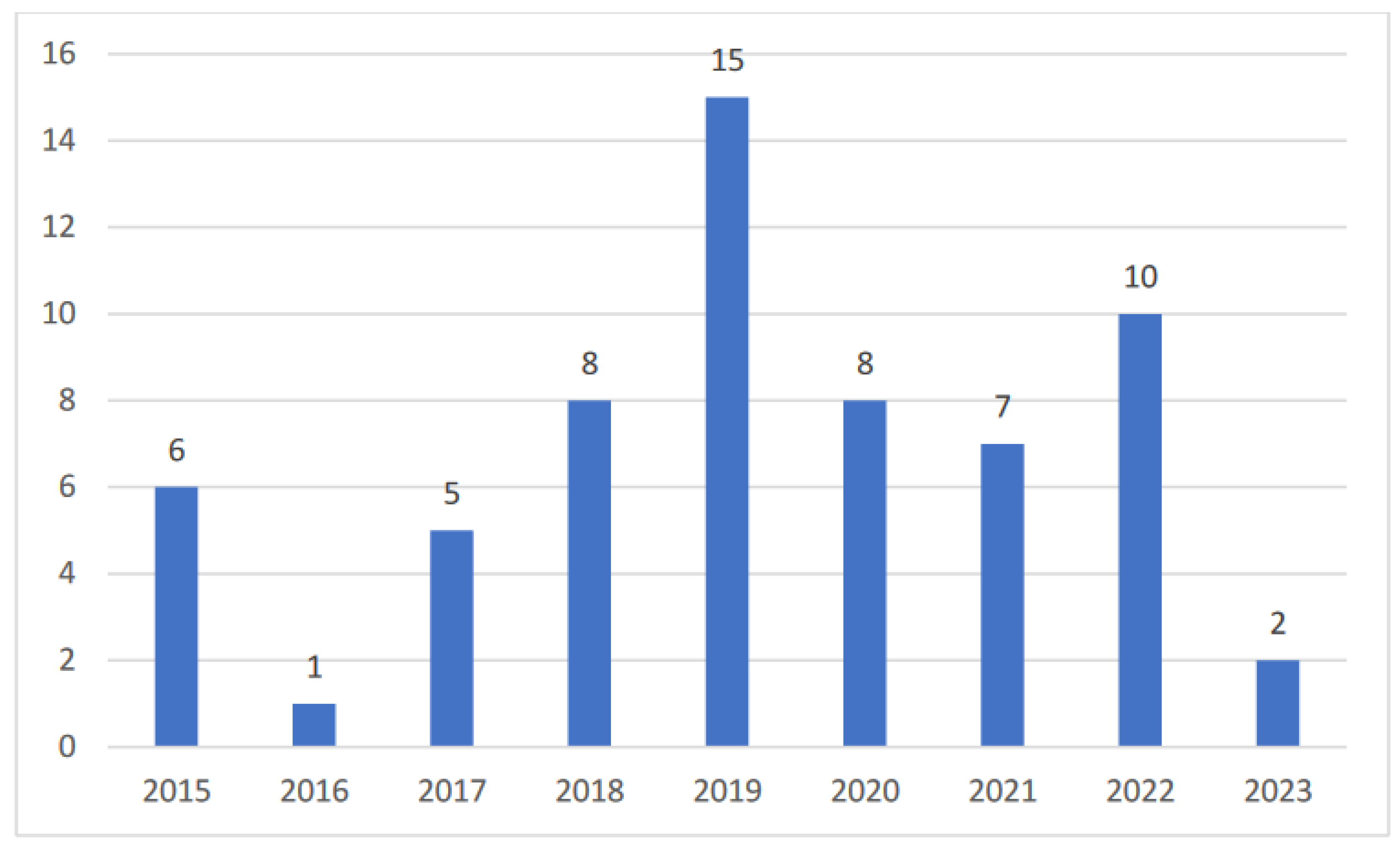
This review identified studies published between 2015 and 2023, and data was extracted on May 25, 2023. The number of studies per year is shown in
Figure 3, with an average of 7 pediatric voice studies per year between 2015 and 2023 and a peak of 15 publications in 2019.
3.3. Funding Sources
Research funding supported 29 studies (46.7%), and 56 different funding sources were represented (
Appendix III). Organizations that provided funding to two or more studies included the National Natural Science Foundation of China, Manipal University Jaipur (India), SMART Innovation Centre (USA), Austrian National Bank, National Institute on Deafness and Other Communication Disorders (USA), Austrian Science Fund, Natural Sciences and Engineering Research Council of Canada, and the Bill & Melinda Gates Foundation (USA). Most funding came from public and private organizations from the United States, China, India, and Austria.
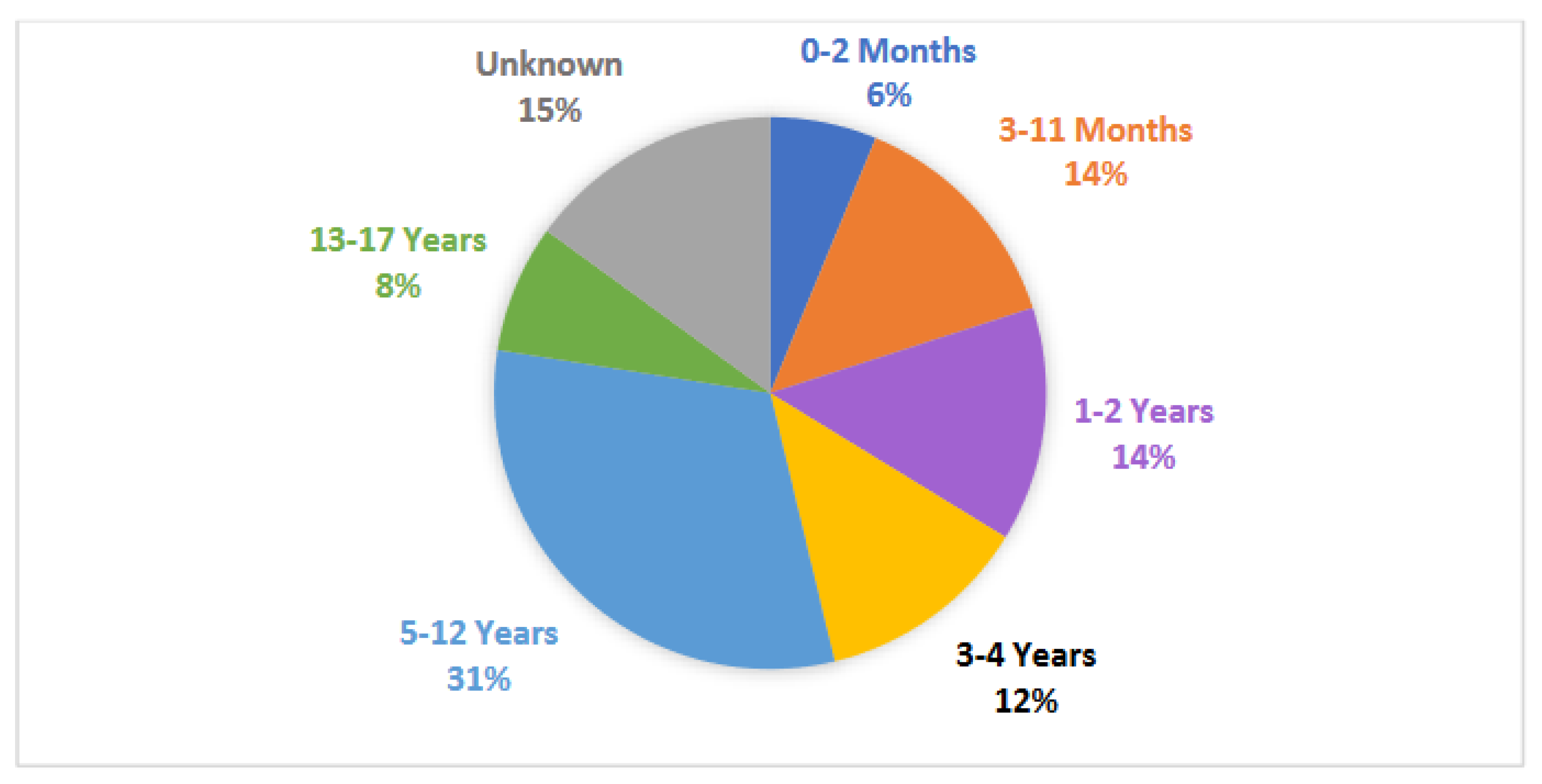
3.4. Participant Age
Each study had, on average, 202 participants [range: 12-2268], with a median of 76 participants. 27% of participants (n = 3347) were distinguished by sex, of which 61% were male. School-aged children (ages 5-12 years) were the most commonly studied (25 studies). Newborn (ages 0-2 months), infant (ages 3-11 months), toddler (ages 1-2 years), preschool (ages 3-4 years), school-aged (ages 5-12 years), and teenage (ages 13-17) groups were also represented in at least 5 studies each as shown in
Figure 4. The specific pediatric age group being studied was not defined for 12 studies.
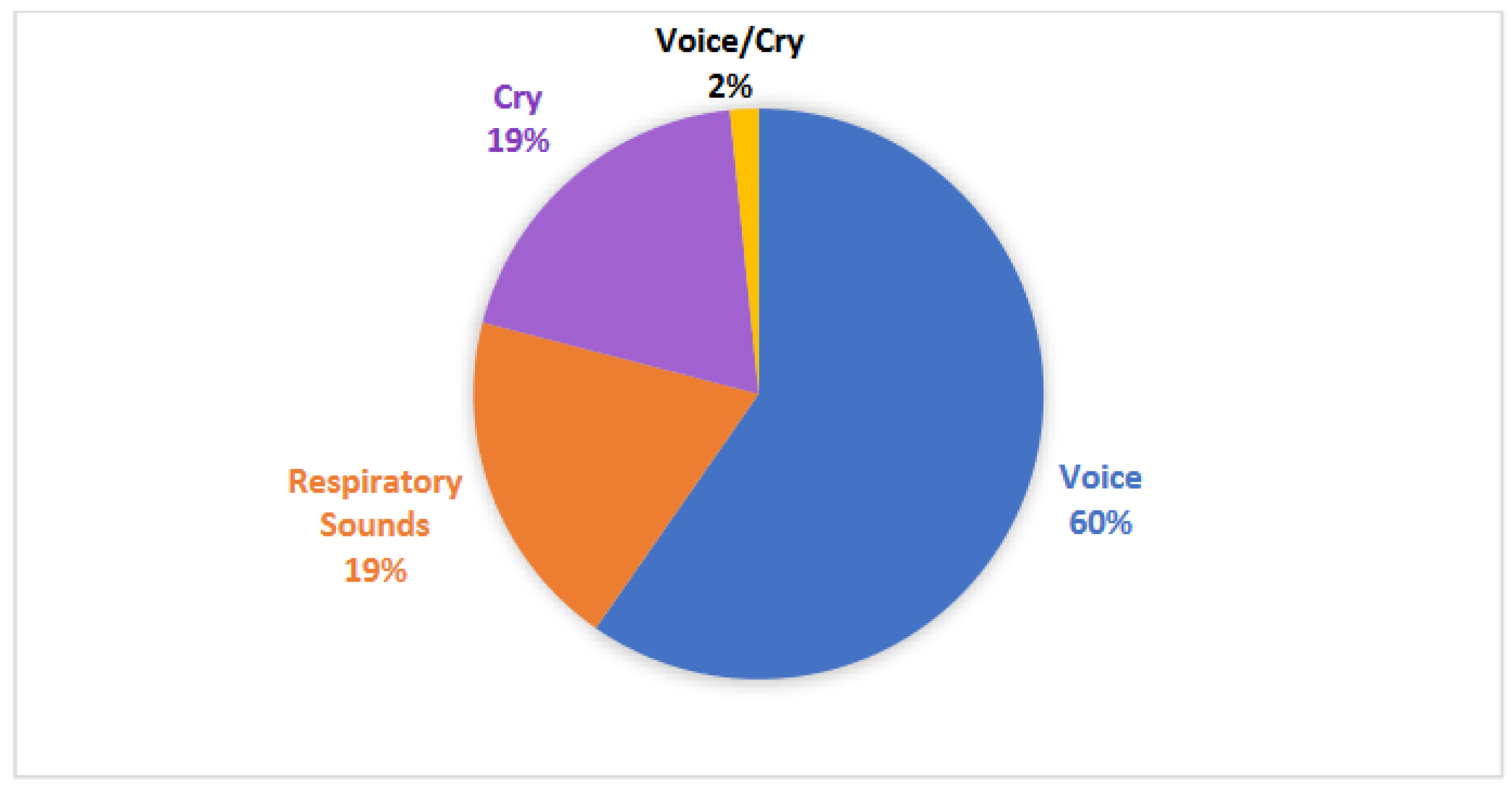
3.5. Recording Characteristics
As shown in
Figure 5, studies included three types of vocal recordings: voice (38 studies), cry (13 studies), and respiratory sounds (12 studies). The majority of studies (45 studies) collected unique vocal data, while 17 studies utilized 13 different existing datasets to conduct their studies, of which recordings from the Baby Chillanto Infant Cry Database (Mexico) and the LANNA Research Group Child Speech Database (Czech Republic) were the most commonly studied.
3.5. Clinical Conditions
Vocal recordings were analyzed using AI as a biomarker for 31 clinical conditions, represented in IV. Among these conditions, developmental conditions (21 studies), respiratory conditions (21 studies), speech and language conditions (13 studies), and non-respiratory conditions (7 studies) were represented. The most frequently studied conditions included Autism Spectrum Disorder (ASD) (12 studies), intellectual disabilities (7 studies), asphyxia (7 studies), and asthma (5 studies).
3.6. Feature Extraction Methods
Among 62 studies, 33 feature extraction methods were utilized (
Appendix V). Mel Frequency Cepstral Coefficients were the most utilized feature extraction method (43 studies), followed by Spectral Components (10 studies), Cepstral Coefficients (10 studies), Pitch and Fundamental Frequency (9 studies), and Linear Predictive Coefficients (9 studies).
3.7. Artificial Intelligence and Machine Learning Models
Across studies, 33 artificial intelligence or machine learning models were utilized (
Appendix VI). The most common AI/ML models were Support Vector Machine (SVM) (34 studies), Neural Network (31 studies), Random Forest (9 studies), Linear Discriminant Analysis (LDA) (7 studies), and K-Nearest Neighbor (KNN) (5 studies).
4. Discussion
The human voice contains unique, complex acoustic markers that vary depending on one's coordination between respiration, phonation, articulation, and prosody. As technology progresses, especially in artificial intelligence and acoustic analysis, voice is emerging as a cost-effective, non-invasive, and accessible biomarker for the detection of pathologies. Our primary objective was to determine what is currently known about using pediatric voice paired with AI models for the early detection, diagnosis, and monitoring of pediatric conditions. This review identified 62 studies that met the inclusion criteria, utilizing pediatric voice, cry, or respiratory sounds for the detection of 31 pediatric conditions among 4 condition groups, representing pediatric populations from 25 countries.
4.1. Developmental Conditions
Twenty-one of the included studies trained and evaluated machine learning algorithms using voice data to classify children with developmental disorders. Speech was the predominantly utilized feature, with studies considering various aspects of speech, including vocal, acoustic, phonetic, and language features. Acoustic features [
1,
2,
3] and phonetic features [
14] were extracted to train machine learning algorithms in classifying children with intellectual disability. A majority of the included studies centered on training machine learning algorithms to classify children with autism spectrum disorder (and Down Syndrome [
22]) using acoustic features [
16,
17,
22,
29,
30,
61], vocal features [
8,
31], voice prosody features [
38], prelinguistic vocal features [
41], and speech features [
15,
59]. In particular, Wu et al. (2019) [
61] focused on acoustic features of crying sounds in children of 2 to 3 years of age, while Pokorny et al. (2017) [
41] concentrated on prelinguistic vocal features in 10-month-old babies. Speech features were also utilized in training machine learning algorithms to classify children with developmental language disorders [
63], specific language impairment [
50,
51], and dyslexia [
25,
43].
4.2. Respiratory Conditions
Twenty-one of the included studies focused on the unintentional air movement across vocal cords by cry, cough, or breath. Machine learning techniques characterized infant cries in the setting of asphyxia [
9,
23,
24,
39,
46]. Spontaneous pediatric coughs are rigorously described through AI methodology [
5,
7,
42,
47] and analyzed to detect specific clinical entities such as croup [
47,
48,
49], pertussis [
49], asthma [
21], and pneumonia [
6]. Asthma, a common childhood illness, has also been studied through AI analysis of pediatric breath sounds [
12,
33].
4.3. Speech and Language Conditions
The detection and evaluation of voice and speech disorders in children is uniquely challenging due to the intricate nature of speech production and the variability inherent in children's speech patterns. To address these challenges, researchers have explored a variety of computational approaches leveraging machine learning, neural networks, and signal processing techniques aimed toward early identification of speech delay [
44,
52]. Several studies highlight promising methodologies to identify stuttering and specific language impairment (SLI) using acoustic and linguistic features [
4,
11]. Feature extraction techniques and convolutional neural networks can help to detect hypernasality in children with cleft palates [
18,
58]. Voice acoustic parameters have been developed to identify dysphonia and vocal nodules in children [
54,
55]. Automatic acoustic analysis can also be used to differentiate typically developing children from those who are hard of hearing, language-delayed, and autistic [
56]. Other notable research has utilized deep learning models and computer-aided systems to identify SLI and sigmatism, also known as lisping [
28,
35,
60].
4.4. Other Non-Respiratory Conditions
Researchers have explored using voice recordings and AI to identify other non-respiratory genetic or medical conditions, usually based on known characteristics affecting cry, voice, or speech that can lead to a clinical suspicion that a diagnosis is present. A Voice Biometric System was developed using recordings of 15 two-syllable words to identify whether a child has cerebral palsy and the severity of the condition, with potential usefulness to evaluate therapeutic benefit [
36]. A hierarchical machine learning model using voice recordings of the standardized PATA speech test was able to identify and grade the level of severity of dysarthria associated with ataxia [
54]. Early detection of anxiety and depression using a 3-minute Speech Task in 3 to 8-year-olds showed reasonable accuracy when recordings were high quality [
34], and multimodal text and audio data was able to discriminate adolescents with depression based on recorded interviews [
62]. Recordings of cry sounds have also been evaluated using machine learning and have shown reasonable accuracy in detecting life-threatening sepsis in neonates [
26,
27] and neonatal opioid withdrawal syndrome [
32].
4.5. Limitations
This review was restricted to studies published in English, which may not capture the full scope of research in non-English-speaking regions. Additionally, the inclusion criteria required a sample size of at least 10 pediatric participants in each study. These studies may offer valuable insights, but they did not meet inclusion criteria within this review.
5. Conclusions
This scoping review highlights the current and potential application of AI in analyzing pediatric voice as a biomarker for health. So far, pediatric voice has been paired with AI models for the early detection, diagnosis, and monitoring of 32 pediatric conditions, primarily observing Autism Spectrum Disorder (ASD), intellectual disabilities, asphyxia, and asthma. While most applications of using pediatric voice as a biomarker have been for the diagnosis of developmental, respiratory, and speech and language conditions, this review highlights the application of pediatric voice analysis for the detection of non-respiratory conditions such as anxiety and depression, sepsis, and jaundice. Research thus far has demonstrated the enormous potential to use voice recordings to detect and monitor diseases and conditions in children. While most research thus far has recorded voice in more clinical settings, one can imagine a future where recordings are used as a biomarker in non-clinical settings where children are more comfortable, such as at home or school. Further development of this field could lead to innovative, new diagnostic tools and interventions for pediatric populations globally.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org. Appendix I: A table showing the Boolean search strategy employed to compile studies from PubMed, Cochrane Database, Embase, Web of Science, ClinicalTrials.Gov, and Google Scholar. Appendix II: A table listing the country associated with each study and their reference number. Appendix III: A table listing the funding source, funding country, study, and reference number for each study that stated a funding source within their respective publication. Appendix IV: A table listing the condition group and condition type being analyzed by each study included in the scoping review and their associated reference number. Appendix V: A table listing the feature extraction method utilized by each respective study and their associated reference number. Appendix VI: A table listing the artificial intelligence or machine learning model utilized by each respective study and their associated reference number. Appendix VII: Bridge2AI- Voice Consortium List of Authors.
Author Contributions
Hannah Paige Rogers conducted the comprehensive scoping review, including the design, data collection, analysis, and writing of the manuscript, with contributions from all team members. Stacy Jo contributed to the data collection and analysis of the scoping review. Dr. Elizabeth Silberholz participated in the development of the search strategy, review of articles, and writing of manuscript. Drs Jung Kim and Anne Hseu participated in the review of articles and writing of manuscript. Anna Dorste conceptualized and executed the search strategies and wrote the methodology portion of the paper concerning the searches and databases. Dr. Kathy Jenkins participated in developing the search strategy and the review of articles, data interpretation, and presentation, and writing of manuscript.
Funding
This research was funded by National Institutes of Health, grant number 1OT20D032720-01.
Institutional Review Board Statement
Not Applicable
Informed Consent Statement
Not Applicable
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors on request.
Acknowledgments
We would like to acknowledge Yaël Bensoussan, MD MSc, Chief of Laryngology at the University of South Florida Morsani College Of Medicine, and Olivier Elemento PhD, Professor of Physiology and Biophysics at Weill Cornell Medicine, for acquiring funding from the National Institutes of Health, allowing this project to be possible. We would also like to acknowledge Jessily Ramirez-Mendoza, EPHM IMPH, for her contribution to the design and implementation of the scoping review.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Aggarwal G, Monga R, Gochhayat SP. A Novel Hybrid PSO Assisted Optimization for Classification of Intellectual Disability Using Speech Signal. Wireless Personal Communications. 2020;113(4):1955-1971. [CrossRef]
- Aggarwal G, Singh L. Evaluation of Supervised Learning Algorithms Based on Speech Features as Predictors to the Diagnosis of Mild to Moderate Intellectual Disability. 3d Research. 2018;9(4):11. [CrossRef]
- Aggarwal G, Singh L. Comparisons of Speech Parameterisation Techniques for Classification of Intellectual Disability Using Machine Learning. International Journal of Cognitive Informatics and Natural Intelligence. 2020;14(2):16-34. [CrossRef]
- Alharbi S, Hasan M, Simons AJH, et al. A Lightly Supervised Approach to Detect Stuttering in Children's Speech. 19th Annual Conference of the International-Speech-Communication-Association (INTERSPEECH 2018). 2018:3433-3437. [CrossRef]
- Amrulloh Y, Abeyratne U, Swarnkar V, et al. Cough Sound Analysis for Pneumonia and Asthma Classification in Pediatric Population. 6th International Conference on Intelligent Systems, Modelling and Simulation (ISMS). 2015:127-131. [CrossRef]
- Amrulloh YA, Abeyratne UR, Swarnkar V, Triasih R, Setyati A. Automatic cough segmentation from non-contact sound recordings in pediatric wards. Biomedical Signal Processing and Control. 2015;21:126-136. [CrossRef]
- Amrulloh YA, Priastomo IH, Wahyuni ES, et al. Optimum Features Computation Using Genetic Algorithm for Wet and Dry Cough Classification. 2nd International Conference on Biomedical Engineering (IBIOMED). 2018:111-114.
- Asgari M, Chen L, Fombonne E. Quantifying Voice Characteristics for Detecting Autism. Front Psychol. 2021;12:665096. [CrossRef]
- Badreldine OM, Elbeheiry NA, Haroon ANM, et al. Automatic Diagnosis of Asphyxia Infant Cry Signals Using Wavelet Based Mel Frequency Cepstrum Features. 14th International Computer Engineering Conference (ICENCO). 2018:96-100.
- Balamurali BT, Hee HI, Kapoor S, et al. Deep Neural Network-Based Respiratory Pathology Classification Using Cough Sounds. Sensors (Basel). 2021;21(16). [CrossRef]
- Barua PD, Aydemir E, Dogan S, et al. Novel favipiravir pattern-based learning model for automated detection of specific language impairment disorder using vowels. Neural Comput Appl. 2023;35(8):6065-6077. [CrossRef]
- Bokov P, Mahut B, Delclaux C. Automatic wheezing recognition algorithm using recordings of respiratory sounds at the mouth: Methodology and development in peadiatric population. Acta Physiologica. 2015;214(S700):76. [CrossRef]
- Bramer WM, Giustini D, de Jonge GB, Holland L, Bekhuis T. De-duplication of database search results for systematic reviews in EndNote. J Med Libr Assoc. Jul 2016;104(3):240-3. [CrossRef]
- Chen Y, Ma S, Yang X, Liu D, Yang J. Screening Children's Intellectual Disabilities with Phonetic Features, Facial Phenotype and Craniofacial Variability Index. Brain Sci. 2023;13(1). [CrossRef]
- Chi NA, Washington P, Kline A, et al. Classifying Autism From Crowdsourced Semistructured Speech Recordings: Machine Learning Model Comparison Study. JMIR Pediatr Parent. 2022;5(2):e35406. [CrossRef]
- Cho S, Liberman M, Ryant N, Cola M, Schultz RT. Automatic Detection of Autism Spectrum Disorder in Children Using Acoustic and Text Features from Brief Natural Conversations. Interspeech. 2019; [CrossRef]
- Deng J, Cummins N, Schmitt M, et al. Speech-based Diagnosis of Autism Spectrum Condition by Generative Adversarial Network Representations. 7th International Conference on Digital Health (DH). 2017:53-57. [CrossRef]
- Dubey AK, Prasanna SRM, Dandapat S, et al. Pitch-Adaptive Front-end Feature for Hypernasality Detection. 19th Annual Conference of the International-Speech-Communication-Association (INTERSPEECH 2018). 2018:372-376. [CrossRef]
- Gouda A, El Shehaby S, Diaa N, et al. Classification Techniques for Diagnosing Respiratory Sounds in Infants and Children. 9th IEEE Annual Computing and Communication Workshop and Conference (CCWC). 2019:354-360.
- Hariharan M, Sindhu R, Vijean V, et al. Improved binary dragonfly optimization algorithm and wavelet packet based non-linear features for infant cry classification. Comput Methods Programs Biomed. 2018;155:39-51. [CrossRef]
- Hee HI, Balamurali BT, Karunakaran A, et al. Development of Machine Learning for Asthmatic and Healthy Voluntary Cough Sounds: A Proof of Concept Study. Applied Sciences-Basel. 2019;9(14):14. [CrossRef]
- Jayasree T, Shia SE. Combined Signal Processing Based Techniques and Feed Forward Neural Networks for Pathological Voice Detection and Classification. Sound and Vibration. 2021;55(2):141-161. [CrossRef]
- Ji CY, Pan Y, Ieee, et al. Infant Vocal Tract Development Analysis and Diagnosis by Cry Signals with CNN Age Classification. 11th International Conference on Speech Technology and Human-Computer Dialogue (SpeD). 2021:37-41. [CrossRef]
- Ji CY, Xiao XL, Basodi S, et al. Deep Learning for Asphyxiated Infant Cry Classification Based on Acoustic Features and Weighted Prosodic Features. IEEE Int Congr on Cybermat / 12th IEEE Int Conf on Cyber, Phys and Social Comp (CPSCom) / 15th IEEE Int Conf on Green Computing and Communications (GreenCom) / 12th IEEE Int Conf on Internet of Things (iThings) / 5th IEEE Int Conf on Smart Data. 2019:1233-1240. [CrossRef]
- Kariyawasam R, Nadeeshani M. Pubudu: Deep learning based screening and intervention of dyslexia, dysgraphia and dyscalculia. 2019 IEEE 14th Conference on Industrial and Information Systems (ICIIS). [CrossRef]
- Khalilzad Z, Hasasneh A, Tadj C. Newborn Cry-Based Diagnostic System to Distinguish between Sepsis and Respiratory Distress Syndrome Using Combined Acoustic Features. Diagnostics (Basel). 2022;12(11). [CrossRef]
- Khalilzad Z, Kheddache Y, Tadj C. An Entropy-Based Architecture for Detection of Sepsis in Newborn Cry Diagnostic Systems. Entropy (Basel). 2022;24(9). [CrossRef]
- Kotarba K, Kotarba M, Ieee, et al. Efficient detection of specific language impairment in children using ResNet classifier. 24th IEEE Conference on Signal Processing: Algorithms, Architectures, Arrangements, and Applications (IEEE SPA). 2020:169-173.
- Lee JH, Lee GW, Bong G, Yoo HJ, Kim HK. Deep-Learning-Based Detection of Infants with Autism Spectrum Disorder Using Auto-Encoder Feature Representation. Sensors (Basel). 2020;20(23). [CrossRef]
- Lee JH, Lee GW, Bong G, Yoo HJ, Kim HK. End-to-End Model-Based Detection of Infants with Autism Spectrum Disorder Using a Pretrained Model. Sensors (Basel). 2022;23(1). [CrossRef]
- MacFarlane H, Salem AC, Chen L, Asgari M, Fombonne E. Combining voice and language features improves automated autism detection. Autism Res. 2022;15(7):1288-1300. [CrossRef]
- Manigault AW, Sheinkopf SJ, Silverman HF, Lester BM. Newborn Cry Acoustics in the Assessment of Neonatal Opioid Withdrawal Syndrome Using Machine Learning. JAMA Netw Open. 2022;5(10):e2238783. [CrossRef]
- Mazić I, Bonković M, Džaja B. Two-level coarse-to-fine classification algorithm for asthma wheezing recognition in children's respiratory sounds. Biomedical Signal Processing and Control. 2015;21:105-118. [CrossRef]
- McGinnis EW, Anderau SP, Hruschak J, et al. Giving Voice to Vulnerable Children: Machine Learning Analysis of Speech Detects Anxiety and Depression in Early Childhood. IEEE J Biomed Health Inform. 2019;23(6):2294-2301. [CrossRef]
- Miodonska Z, Krecichwost M, Szymanska A, Silesian Tech Univ FACE, Comp Sci GP, Silesian Tech Univ FBEZP. Computer-Aided Evaluation of Sibilants in Preschool Children Sigmatism Diagnosis. 5th International Conference on Information Technologies in Biomedicine (ITIB). 2016;471:367-376. [CrossRef]
- Moharir M, Sachin MU, Nagaraj R, et al. Identification of Asphyxia in Newborns using GPU for Deep Learning. 2nd International Conference for Convergence in Technology (I2CT). 2017:236-239.
- Nafisah S, Effendy N. Voice Biometric System: The Identification of the Severity of Cerebral Palsy using Mel-Frequencies Stochastics Approach. International Journal of Integrated Engineering. 2019;11(3):194-206.
- Nakai Y, Takiguchi T, Matsui G, Yamaoka N, Takada S. Detecting Abnormal Word Utterances in Children With Autism Spectrum Disorders: Machine-Learning-Based Voice Analysis Versus Speech Therapists. Percept Mot Skills. 2017;124(5):961-973. [CrossRef]
- Onu CC, Lebensold J, Hamilton WL, et al. Neural Transfer Learning for Cry-based Diagnosis of Perinatal Asphyxia. Interspeech Conference. 2019:3053-3057. [CrossRef]
- Pokorny FB, Schmitt M, Egger M, et al. Automatic vocalisation-based detection of fragile X syndrome and Rett syndrome. Sci Rep. 2022;12(1):13345. [CrossRef]
- Pokorny FB, Schuller BW, Marschik PB, et al. Earlier Identification of Children with Autism Spectrum Disorder: An Automatic Vocalisation-based Approach. 18th Annual Conference of the International-Speech-Communication-Association (INTERSPEECH 2017). 2017:309-313. [CrossRef]
- Porter P, Abeyratne U, Swarnkar V, et al. A prospective multicentre study testing the diagnostic accuracy of an automated cough sound centred analytic system for the identification of common respiratory disorders in children. Respir Res. 2019;20(1):81. [CrossRef]
- Ribeiro FM, Pereira AR, Paiva DMB, et al. Early Dyslexia Evidences using Speech Features. 22nd International Conference on Enterprise Information Systems (ICEIS). 2020:640-647. [CrossRef]
- Sadeghian R, Zahorian SA, Isca-Int Speech Commun A, et al. Towards an Automated Screening Tool for Pediatric Speech Delay. 16th Annual Conference of the International-Speech-Communication-Association (INTERSPEECH 2015). 2015:1650-1654.
- Salehian Matikolaie F, Tadj C. On the use of long-term features in a newborn cry diagnostic system. Biomedical Signal Processing and Control. 2020;59. [CrossRef]
- Satar M, Cengizler C, Hamitoglu S, Ozdemir M. Investigation of Relation Between Hypoxic-Ischemic Encephalopathy and Spectral Features of Infant Cry Audio. J Voice. 2022; [CrossRef]
- Sharan RV, Abeyratne UR, Swarnkar VR, Porter P. Cough sound analysis for diagnosing croup in pediatric patients using biologically inspired features. Annu Int Conf IEEE Eng Med Biol Soc. 2017;2017:4578-4581. [CrossRef]
- Sharan RV, Abeyratne UR, Swarnkar VR, Porter P. Automatic Croup Diagnosis Using Cough Sound Recognition. IEEE Trans Biomed Eng. 2019;66(2):485-495. [CrossRef]
- Sharan RV, Berkovsky S, Navarro DF, Xiong H, Jaffe A. Detecting pertussis in the pediatric population using respiratory sound events and CNN. Biomedical Signal Processing and Control. 2021;68. [CrossRef]
- Sharma G, Prasad D, Umapathy K, Krishnan S. Screening and analysis of specific language impairment in young children by analyzing the textures of speech signal. Annu Int Conf IEEE Eng Med Biol Soc. 2020;2020:964-967. [CrossRef]
- Sharma Y, Singh BK. One-dimensional convolutional neural network and hybrid deep-learning paradigm for classification of specific language impaired children using their speech. Comput Methods Programs Biomed. 2022;213:106487. [CrossRef]
- Suthar K, Yousefi Zowj F, Speights Atkins M, He QP. Feature engineering and machine learning for computer-assisted screening of children with speech disorders. PLOS Digit Health. 2022;1(5):e0000041. [CrossRef]
- Szklanny K, Wrzeciono P. The Application of a Genetic Algorithm in the Noninvasive Assessment of Vocal Nodules in Children. IEEE Access. 2019;7:44966-44976. [CrossRef]
- Tartarisco G, Bruschetta R, Summa S, et al. Artificial Intelligence for Dysarthria Assessment in Children With Ataxia: A Hierarchical Approach. IEEE Access. 2021;9:166720-166735. [CrossRef]
- Tulics MG, Vicsi K. Automatic classification possibilities of the voices of children with dysphonia. Infocommunications Journal. 2018;10(3):30-36.
- VanDam M, Oller DK, Ambrose SE, et al. Automated Vocal Analysis of Children With Hearing Loss and Their Typical and Atypical Peers. Ear and hearing. 2015;36(4):e146-e152. [CrossRef]
- Wang X, Yang S, Tang M, Yin H, Huang H, He L. HypernasalityNet: Deep recurrent neural network for automatic hypernasality detection. Int J Med Inform. 2019;129:1-12. [CrossRef]
- Wang XY, Tang M, Yang S, Yin H, Huang H, He L. Automatic Hypernasality Detection in Cleft Palate Speech Using CNN. Circuits Systems and Signal Processing. 2019;38(8):3521-3547. [CrossRef]
- Wijesinghe A, Samarasinghe P, Seneviratne S, et al. MACHINE LEARNING BASED AUTOMATED SPEECH DIALOG ANALYSIS OF AUTISTIC CHILDREN. 11th International Conference on Knowledge and Systems Engineering (KSE). 2019:163-167.
- Woloshuk A, Krecichwost M, Miodonska Z, et al. CAD of Sigmatism Using Neural Networks. 6th International Conference on Information Technology in Biomedicine (ITIB). 2018;762:260-271. [CrossRef]
- Wu K, Zhang C, Wu XP, et al. Research on Acoustic Feature Extraction of Crying for Early Screening of Children with Autism. 34th Youth Academic Annual Conference of Chinese-Association-of-Automation (YAC). 2019:295-300.
- Zhang L, Fan Y, Jiang J, Li Y, Zhang W. Adolescent Depression Detection Model Based on Multimodal Data of Interview Audio and Text. Int J Neural Syst. 2022;32(11):2250045. [CrossRef]
- Zhang X, Qin F, Chen Z, Gao L, Qiu G, Lu S. Fast screening for children's developmental language disorders via comprehensive speech ability evaluation-using a novel deep learning framework. Ann Transl Med. 2020;8(11):707. [CrossRef]
- Faurholt-Jepsen M, Rohani DA, Busk J, Vinberg M, Bardram JE, Kessing LV. Voice analyses using smartphone-based data in patients with bipolar disorder, unaffected relatives and healthy control individuals, and during different affective states. Int J Bipolar Disord. 2021 Dec 1;9(1):38. [CrossRef] [PubMed] [PubMed Central]
- Kim H, Jeon J, Han YJ, Joo Y, Lee J, Lee S, Im S. Convolutional Neural Network Classifies Pathological Voice Change in Laryngeal Cancer with High Accuracy. J Clin Med. 2020 Oct 25;9(11):3415. [CrossRef] [PubMed] [PubMed Central]
- Moro-Velazquez L, Gomez-Garcia JA, Godino-Llorente JI, Grandas-Perez F, Shattuck-Hufnagel S, Yagüe-Jimenez V, Dehak N. Phonetic relevance and phonemic grouping of speech in the automatic detection of Parkinson's Disease. Sci Rep. 2019 Dec 13;9(1):19066. [CrossRef] [PubMed] [PubMed Central]
- Xue, C., Karjadi, C., Paschalidis, I.C. et al. Detection of dementia on voice recordings using deep learning: a Framingham Heart Study. Alz Res Therapy 13, 146 (2021). [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).