1. Introduction
Skeletal muscle (SkM) is a highly plastic tissue capable of functional and structural remodeling in response to numerous physiological stimuli. Particularly during endurance training, skeletal myocytes undergo various challenges from neuronal, hormonal, mechanical, and metabolic stressors, leading to genetic and biochemical adaptations that ultimately determine the beneficial phenotypes evoked by chronic exercise [
1].
A predominant stressor on contracting muscle fibers is reactive oxygen species (ROS). ROS at high levels can react with biomolecules to oxidize proteins, lipids, and nucleic acids. This results in cell damage and organ dysfunction, a pathological redox state referred to as “oxidative stress”, which underlies the pathogenesis of numerous diseases. Indeed, in skeletal muscle, it is well-known that excessive ROS contribute to muscle fatigue during exhaustive exercise [
2], doxorubicin (DOX)-induced muscle wasting [
3], and age-related progressive loss of muscle mass and strength [
4]. On the other hand, ROS at low- to moderate- levels function as essential intracellular signaling molecules, playing a crucial role in many biological processes under normal conditions. Physiological ROS can induce posttranslational modifications (PTMs) on specific target proteins by a reversible oxidation of sulfur groups and metal centers [
5], therefore altering protein activity, localization, and interactions that result in orchestration of various biological events in cells and organs, such as cell proliferation, differentiation, migration, and angiogenesis [
6]. Physiological ROS or “normal redox signaling” is essential to maintain normal metabolism and function of cells and organs [
7]. Indeed, studies have demonstrated that selective depletion of ROS from normal skeletal muscle decreases force generation8. Accordingly, there is an optimal ROS concentration in skeletal myocytes which determines the maximal force generation during contraction. Deviations of redox status away from this point, either via an increase or decrease in ROS concentration, will impair muscle contractility forming a classic bell-shaped hormesis dose-response curve [
9].
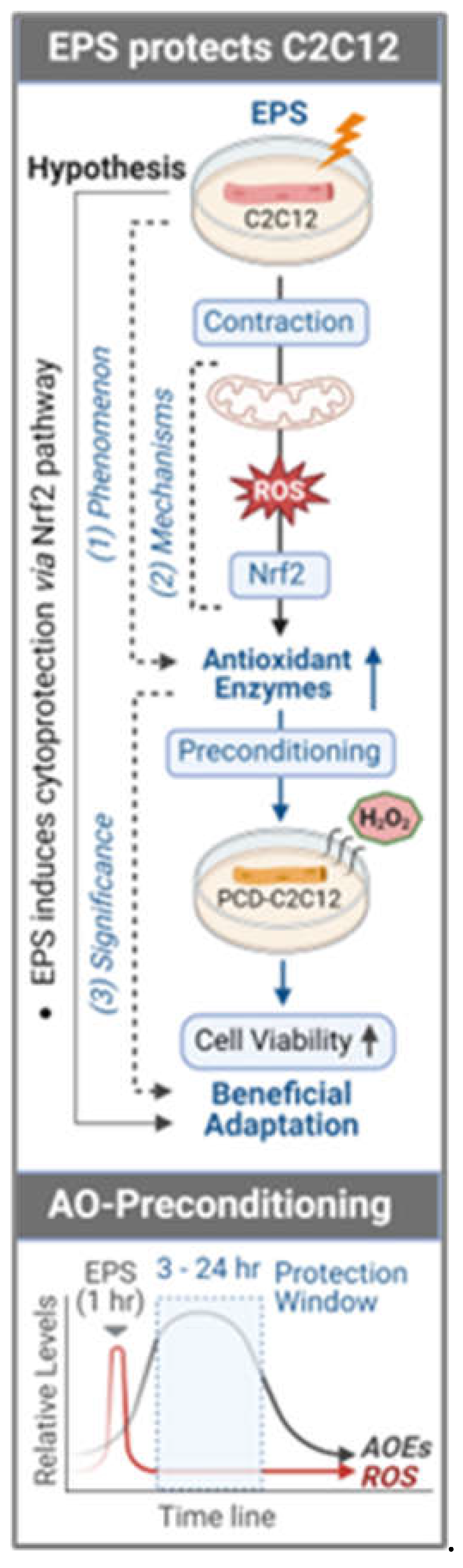
While there is no clear boundary of ROS levels to induce oxidative stress and function as physiological redox signaling, there is no doubt that these free radicals can activate intracellular antioxidant defense under both conditions via compensatory mechanisms to upregulate enzymatic and non-enzymatic antioxidants. This enhanced antioxidant mechanisms not only supports the cells to overcome instant oxidative stress, but also provides cells with an extended antioxidant capability to counteract subsequent similar insults, a phenomenon we term “antioxidant preconditioning”. This process has been proposed as a foundation of redox adaptation of skeletal muscle to endurance exercise [
9].
A well-known sensor-effector apparatus in response to a redox disturbance is the Keap1-Nrf2 system. Keap1 acts as a ROS sensor and Nrf2 repressor while Nrf2 is responsible for activating multiple antioxidant genes thus playing a critical role in maintaining intracellular redox homeostasis [
10]. Employing muscle-specific Nrf2 deletion and overexpression transgenic mouse models, iMS-Nrf2flox/flox and iMS-Keap1flox/flox, we previously identified over 200 cytoprotective and regulative proteins governed by Nrf2 in skeletal muscle, which are involved in several intracellular signaling pathways. These include antioxidant defense and mitochondrial function [
11]. We further demonstrated that these Nrf2-orchestrated molecular networks contribute to chronic exercise-induced enhancement of contractility and muscle antioxidant adaptatioin [
12]. However, it remains to be determined if short-term endurance training (STET) can evoke similar beneficial effects on skeletal myocytes to protect the cells against oxidative stress-associated injury.
In the present study, we used electrical pulse stimulation (EPS) on differentiated C2C12 myotubes, a validated in vitro and widely used acute muscle preparation [13, 14] to address the hypothesis that a short-term contraction can evoke an antioxidant preconditioning in skeletal myocytes via a mitochondrial-ROS-Nrf2 pathway. Furthermore, we provide evidence to demonstrate that this antioxidant adaptation protects cells against H2O2-induced toxicity.
2. Materials and Methods
2.1. Cell Culture
The C2C12 cell line (CRL-1722, ATCC, Manassas, VA) was used in these experiments. Cells were seeded in a 6-well plate at ~50,000 or 100,000 cells per well and grown for one day in complete media (CpM) (Dulbecco's Modification of Eagle's Medium (DMEM; Sigma-Aldrich) supplemented with 10% fetal bovine serum (FBS; Gibco/ThermoFisher Scientific) and 1% penicillin/streptomycin (P/S)). After ~24 hours, the media was changed from CpM to differentiation media (DM) (DMEM supplemented with 5% horse serum (HS; Gibco/ThermoFisher Scientific) and 1% P/S), where the cells differentiated from myoblasts to myotubes after four days in culture at 37 ˚C, 5% CO2, 20% O2, and 95% humidity. On the fourth day, the media was replaced by fresh DM and the cells underwent electrical pulse stimulation (EPS).
2.2. Electrical Pulse Stimulation (EPS) and Cell Collection
Electrical current of EPS was delivered to the C2C12 myotubes via stimulation electrodes designed and fabricated in our lab. These electrodes were built on the lid of standard 6-well plates, similar to what has previously been reported15. On the first day of EPS, the regular 6-well plate lids were replaced by the EPS-lids containing the electrodes, which were connected to a single channel pulse generator (A310 Accupulse, WPI, Sarasota, FL). The EPS lids were cleaned before and after each study using the following protocol: The EPS chamber was flooded with 70% ethanol for 10 minutes. The electrodes and EPS lid were washed with ~3 mL of sterile PBS. Finally, the EPS lid was exposed to UV light for 10 – 30 minutes in the cell-culture laminar flow hood. The C2C12 myotubes underwent EPS at 10 v, 50 Hz, 10 ms, 0.3 sec/3 sec for 1h/day for 4 days. At the end of EPS, the cells were rinsed with 200 µL of sterile PBS, followed by trypsinizing with 200 µL of 0.05% trypsin for 2 minutes in 37 ºC. The cells were then collected and transferred to 1.5 mL EP tubes, which were stored in -80 ºC for future analysis.
2.3. Western Blotting
2.3.1. Sample Preparation (Protein Extraction).
Cells were defrosted from -80 ºC, incubated on ice in 100 µL of 100:1 RIPA:PIC for 15 minutes, and then homogenized utilizing a sonicator. The soluble proteins of cell lysate were then extracted by centrifugation for 20 minutes at 13,800 x g and 4 ºC. Protein concentration was measured via BSA assay and a Multiskan Sky Microplate Spectrometer. Samples were then diluted to 2 µg/µL with RIPA. Equal volume of 2X SDS was added to each sample to further dilute the samples to 1 µg/µL and 1X SDS. Samples were then boiled in a 70 ºC water bath for 2 minutes and immediately returned to ice.
2.3.2. Gel Electrophoresis.
The protein samples (15 μg in 15 μL/each sample) were loaded on a precast polyacrylamide gel (NW00105BOX, Invitrogen) along with 2 μL of Invitrogen MagicMark™ Western Standard and 4 μL of Bio Rad Precision Plus Protein Dual Color Standards in separate wells. Electrophoresis was performed using Mini Gel Tank and Blot Module Set (NW2000, Invitrogen), initially at 70 V, for approximately 15 minutes, and then 120 V, until the desired separation had been reached.
2.3.3. Membrane transfer (iBlot2).
The fractionated protein on the gel was electrically transferred onto a nitrocellulose membrane by employing Thermo Fischer Scientific iBlot2 (IB21001, Thermo Fisher Scientific) with the preset program P0 (20 V for 1 minute, then 23 V for 4 minutes, followed by 25 V for 2 minutes). Membranes were gently rinsed with Milli-Q water and then stained with Ponceau S for 5 minutes to visualize total protein. Once ideal bands of total protein were obtained, Ponceau S on the membrane was removed by rinsing it in 10 mL of 1X phosphate-buffered saline-Tween 20 solution (PBST) for 5 minutes.
2.3.4. Immunodetection (Antibodies)
The membranes were then blocked for 30 minutes in 5% milk in 1X PBST at room temperature; this was followed by two 5-minute washes in 10 mL of 1X PBST at room temperature. The membranes were then incubated in primary antibody in 4% BSA overnight at 4 °C. The primary antibodies used in the present experiment were purchased from Abcam (ab-), ABclonal (A-), Santa Cruz Biotechnology (sc-), and Proteintech (-AB/-ig), including NQO1 (ab80588), GSTA2 (ab232833), GSTA4 (ab231601), GPX1 (ab22604), PRX Pathway (TRX, TXNRD1, and PRX1; ab184868), total OXPHOS (ab110413), calmodulin 1/2/3 (A4885), MFN1 (A9880), MFF (A8700), DRP1 (A2586), FIS1 (A21527), SOD1 (sc-8637), SOD2 (sc-30080), catalase (sc-50508), MyoG (sc-576), Nrf2 (A0674 and 16396-1-AP), and mTOR (66888-1-ig). The following day, membranes were washed three times for 5 minutes in 10 mL of 1X PBST; this was followed by incubation in anti-rabbit IgG horseradish peroxidase secondary antibody (1:5000) in 5% milk in 1X PBST for 30 minutes at room temperature. After this, the membrane was again washed three times for 5 minutes in 10 mL of 1X PBST. Membranes were developed in chemiluminescence detection reagent (SuperSignal West Dura or SuperSignal Femto; ThermoScientific). The films were scanned using the G:Box Syngene and Genesys. The intensity of the bands was quantified using the ImageJ software (NIH) and normalized for loading using Ponceau S staining.
2.4. ROS Measurements and NAC Application
2.4.1. Electron Paramagnetic Resonance (EPR) Spectroscopy
C2C12 myoblasts were seeded in six-well plates (100,000 cells/well) followed by differentiation and EPS processes, as described above. After the final EPS, the old DM was replaced with fresh DM, and the cells were processed for EPR spectroscopy analysis as described below at their specified time points (0, 3, 12, or 24 hours after EPS, or NS).
The media was removed from the wells, and the cells were then washed twice with 1 mL of KDD buffer (Krebs-HEPES buffer [pH 7.4]; 99 mM NaCl, 4.69 mM KCl, 2.5 mM CaCl2, 1.2 mM MgSO4, 25 mM NaHCO3, 1.03 mM KH2PO4, 5.6 mM d-glucose, 20 mM HEPES, 5 μM diethyldithiocarbamic acid sodium salt [DETC], 25 μM deferoxamine). Then, the cells were mixed with 200 μM cell-permeable ROS-sensitive spin probe 1-hydroxy-3-methoxycarbonyl-2,2,5,5-tetramethyl pyrrolidine (CMH; Noxygen Science Transfer and Diagnostics, Elzach, Germany) and incubated for 1 hour and 15 minutes at 37 °C. After incubation, 900 µL of KDD buffer was removed from the wells, and the cells were gently scraped into the remaining 100 µL. 50 µL of the cell suspension was pipetted into an EPR glass capillary tube, and inserted into a Bruker e-scan EPR spectrometer with the following settings: field sweep width, 60.0 G; microwave frequency, 9.75 kHz; microwave power, 21.90 mW; modulation amplitude, 2.37 G; conversion time, 10.24 ms; time constant, 40.96 ms. The remaining cell suspension was utilized to count the number of cells to normalize the EPR spectrum amplitude.
2.4.2. Confocal Imaging - MitoSOX Red (MSR, Mitochondrial ROS Indicator) and CM-H2DCFDA (Cytoplasmic ROS Indicator)
C2C12 myoblasts were seeded in six-well plates (5,000 cells/well) containing polylysine coated slides for cell growth, followed by differentiation and EPS processes, as described above. Immediately, after the final day of EPS, the well was washed with 1 mL of HBSS, and the coverslips were then transferred to a 24-well plate, where 2 mL of warm 1 µM MSR working solution or 1 mL of 5 µM CM-H2DCFDA was added, followed by incubation in at 37 °C and 5% CO2 for 30 minutes. The cover slips were then transferred into clean black boxes and gently washed 3 times with warm HBSS. Cells on the coverslips were fixed with cold 4% paraformaldehyde (PFA) for 10 minutes, washed 3 times with PBS, and then mounted onto a glass slide using mounting medium (sc-24941 with DAPI) and clear nail polish. The fluorescence was analyzed via confocal microscopy at the following wavelengths: Mitochondrial ROS: Ex 488 nm/Em 550 – 650 nm, Total ROS: Ex 488 nm/Em 520 nm, and Cellular Nuclear: Ex 360 nm/Ex 460 nm.
2.4.3. N-Acetylcysteine (NAC) Treatment
Three six-well plates were seeded at 100,000 cells per well and underwent differentiation as described above. On the fourth day of differentiation, 40.80 mg of NAC was dissolved in 1 mL of complete media, which was then further diluted to a final working concentration of 5 mM NAC in 50 mL of differentiation media. The pH of this media was neutralized to ~7.5 with 0.5 M NaOH, and then filtered through a 0.2 µm syringe filter. After 15 hours pretreatment with this differentiation media containing 5 mM NAC, the cells underwent EPS for 1 hour at 50 Hz, 10 V, 10 ms, and 0.3 sec/3 sec. 8 hours after EPS, the cells underwent a second treatment of NAC following the protocol outlined above. This was repeated daily for four days. After the fourth, and final day of EPS, the cells were collected as described above.
2.5. Mitochondrial Function Assays
2.5.1. Seahorse – Mito Stress Test
After undergoing 4 days EPS, the cells were collected as described above and replated in 96-well plates at 10,000 cells per well in 100 μL differentiation media (DM). The sensor cartridge was hydrated in calibration buffer at 37 °C in a non-CO2 incubator overnight. Sterile filtered 10 mM glucose, 2 mM glutamate, and 1 mM pyruvate were added to the Seahorse media, whose pH was then corrected to ~7.4 with 1 M HCl or 0.5 M NaOH. Next, 80 μL of DM and was removed from each well of the XF96-well plate, followed by washing with 100 μL of Seahorse media. 160 μL of fresh Seahorse media was added to each well, bringing the final volume to 180 μL. The cells were incubated in the Seahorse media in a non-CO2 incubator for 1 hour. Working solutions of Oligomycin, FCCP, and Rotenone/Antimycin A were made and then added to the injection ports of the sensor cartridge in the respective order. The plate was run on a 96-well plate Seahorse instrument using Wave programming. The Seahorse XF Mito Stress Test Report Generator was used to retrieve the data for later analysis.
2.5.2. Oroboros– High-Resolution Respirometry
After undergoing 4 days of EPS or no-stimulation (controls), approximately 300,000 cells suspended in DM were centrifuged at 1500 x g for 5 minutes, and the cell pellet was resuspended in 2 mL of mitochondrial respiration medium [MiR05, containing 0.5 mM EGTA, 3 mM MgCl2, 60mM lactobionic acid, 20 mM taurine, 10 mM KH2PO4, 20 mM HEPES, 110 mM D-Sucrose, 1 g/L BSA, pH 7.1] in a high-resolution respirometer (Oxygraph-2k, Oroboros, Innsbruck, Austria) to measure mitochondrial respiratory function. The chamber block maintained a constant temperature of 37 °C and magnetic stir bars mixed the solution continuously at 750 RPM. The chambers were hyper-oxygenated to 300 μM O2 via syringe with pure oxygen from an Oxia device (Oxia, Innsbruck, Austria). After the O2 respiration rate stabilized, the cells were permeabilized with digitonin (4.05 μM) for 20 minutes. After permeabilization, substrates were administered in the following order: 1) malate (2 mM) & glutamate (10 mM), 2) adenosine diphosphate (ADP, 5mM), 3) succinate (10 mM), 4) cytochrome c (10 μM), 5) rotenone (0.5 μM), 5) oligomycin (5 nM), 6) antimycin-A (2.5 μM), and 7) ascorbate (2 mM) & N,N,N’,N’-tetramethyl-p-phenylenediamine dihydrochloride (TMPD, 0.5 mM)16, 17. Mitochondrial complexes were assessed as follows: 1) complex I state 2 (LEAK respiration) was assessed after malate & glutamate, 2) complex I state 3 was assessed after malate & glutamate + ADP, 3) complex I+II state 3 was assessed after malate & glutamate + ADP + succinate, 4) membrane integrity was evaluated after the addition of cytochrome c, 5) complex II state 3 was assessed after the addition of rotenone, and 6) complex IV respiration was assessed after ascorbate & TMPD17. The respiration rate for each substrate or inhibitor was recorded after O2 flux reached a stable value (~5 min for substrates and ~10 min for inhibitors). The O2 respiration rates were measured as picomoles of O2 per second per mL and were converted to attomoles of O2 per second per cell (amol·s-1·cell).
2.6. H2O2 Treatment and CCK-8 Assay
After undergoing 4-day EPS, the cells were collected as described above and replated in 96-well plates at 5,000 cells per well in 100 μL CpM. The cells were allowed to adhere to the plate overnight. After cells had adhered to the plate, the CpM was removed and replaced with 100 μL of PBS in CpM (control) or 2 or 4 mM H2O2 in CpM. Cells were incubated in H2O2 at 5% CO2 and 37 °C for 5 hours. After 5 hours, 10 μL of CCK-8 was added to each well. Cells were incubated in CCK-8 at 5% CO2 and 37 °C for 1 hour. The absorbance was measured by a Multiskan Sky Microplate Spectrometer at 450 nm to evaluate cell viability.
2.7. Statistical Analyses
Data are expressed as means ± SD. A t test was used for analyzing the differences in protein expression between EPS and control group using GraphPad Prism 8 software. A two-way ANOVA was used to analyze the EPS-NAC data shown in panels C and D of
Figure 4. A P value of < 0.05 was taken as indicative of statistical significance..
4. Discussion
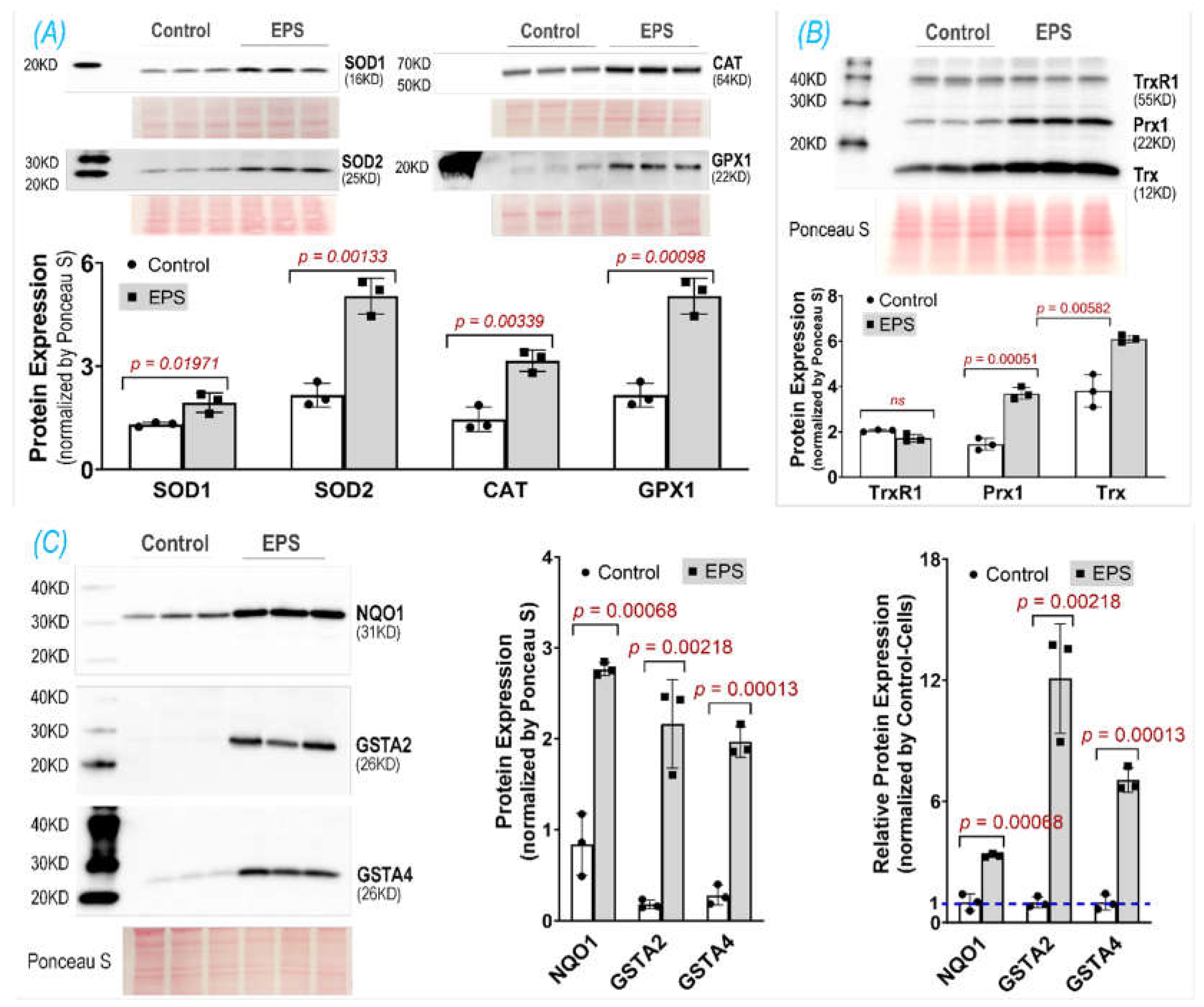
Using EPS on cultured C2C12 myotubes to investigate the effects of contraction on the intracellular antioxidant system in SkM, we showed that EPS for up to 4 days induced an obvious time- and dose- dependent upregulation of NQO1 and GSTA2 protein expression. We found that EPS markedly activated Nrf2 and multiple antioxidant systems, including the first line antioxidant system, the thioredoxin system, and the glutathione system by upregulating a large group of antioxidant enzymes and antioxidant-associated proteins. Among these systems, the glutathione system displayed the largest response to EPS, with approximately a 10-fold increase in GSTA2 and GSTA4 protein expression (Panel C,
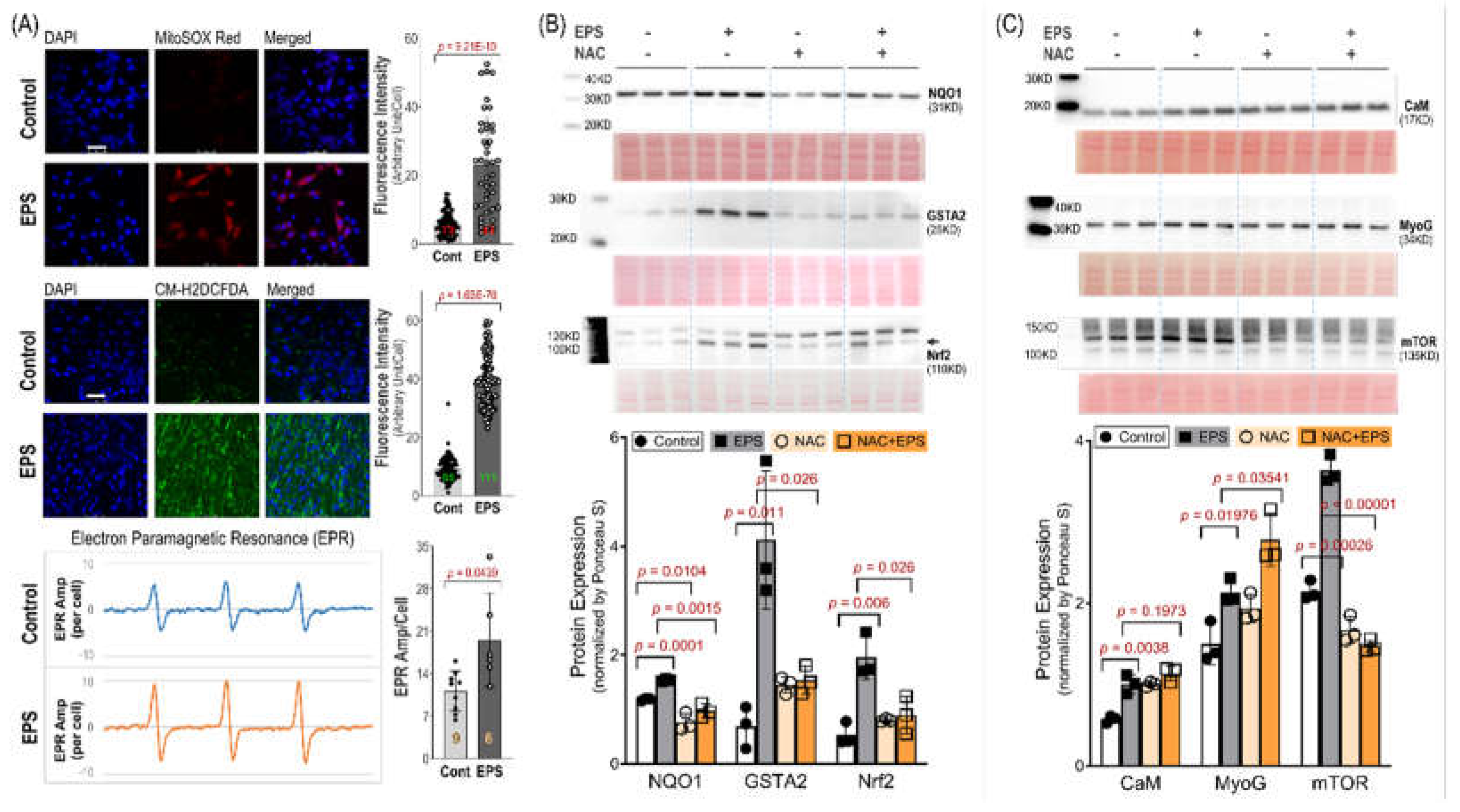
Figure 3). We further demonstrated that EPS-mediated increase in ROS played a critical role in the activation of Nrf2/antioxidant systems, since EPS-mediated enhancements in antioxidant capacity were abolished when the C2C12 myotubes were pre-treated with the ROS scavenger, NAC (Panel B,
Figure 4).
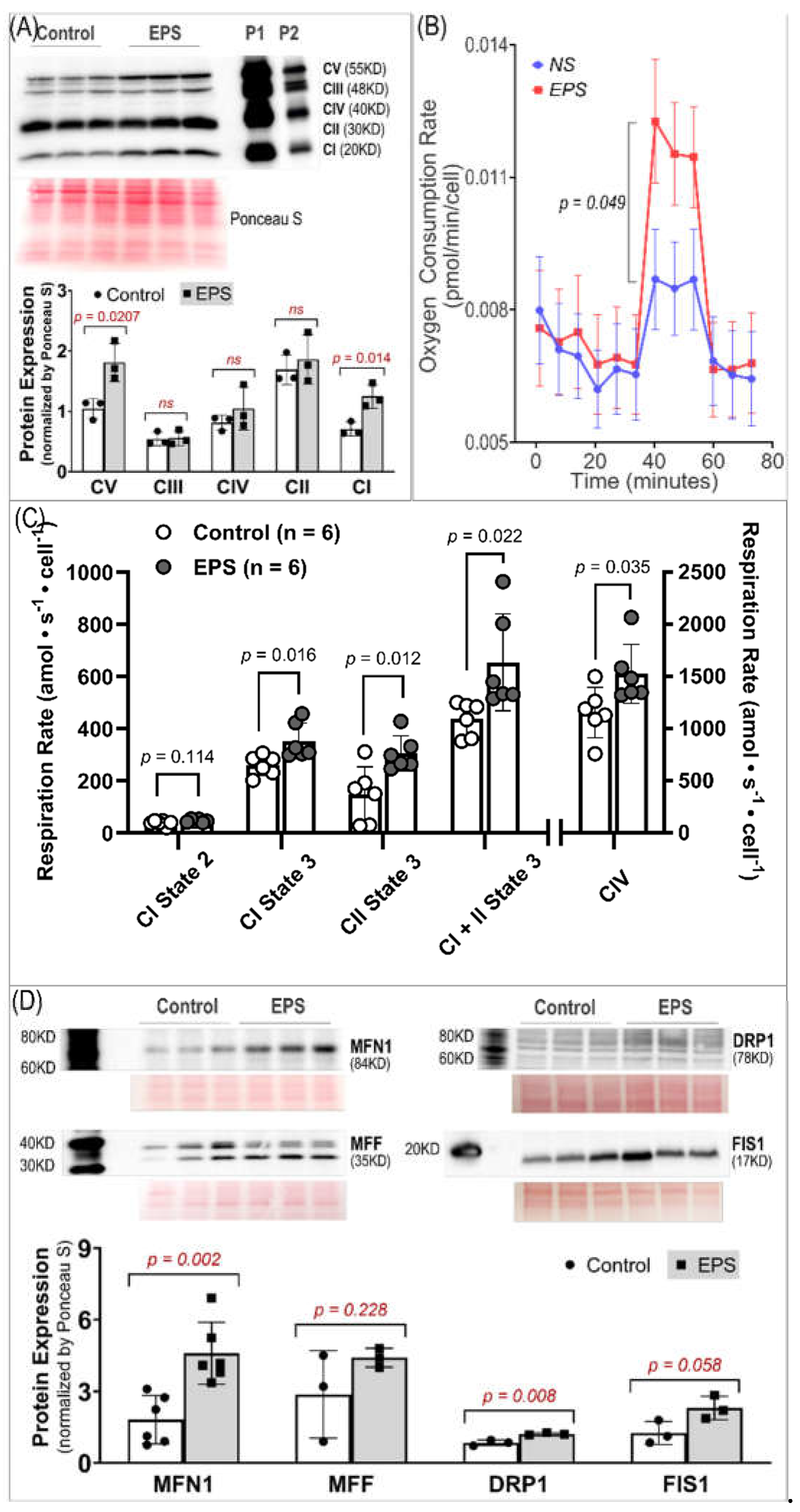
Importantly, our data, of substantially elevated mitochondrial-derived superoxide after EPS, supports the view that mitochondria may be one of the major sources of ROS and responsible for the activation of Nrf2/antioxidant pathways (Panel A,
Figure 4). Furthermore, our data suggest that EPS-induced mitochondrial adaptations may promote EPS-induced augmentation in antioxidant capacity. Of note, complex IV respiration was significantly increased after 4 days of EPS, compared to non-stimulated cells (Panels B and C,
Figure 5). Furthermore, ADP-stimulated respiration (state 3) through complex I, complex II, and complex I+II, were all elevated after 4-days of EPS (Panel C,
Figure 5). In addition, these adaptations were paralleled by increased protein expression of complexes I, the main mitochondrial site for ROS production [
28], and V (Panel A,
Figure 5), and upregulated fission/fusion factors (Panel D,
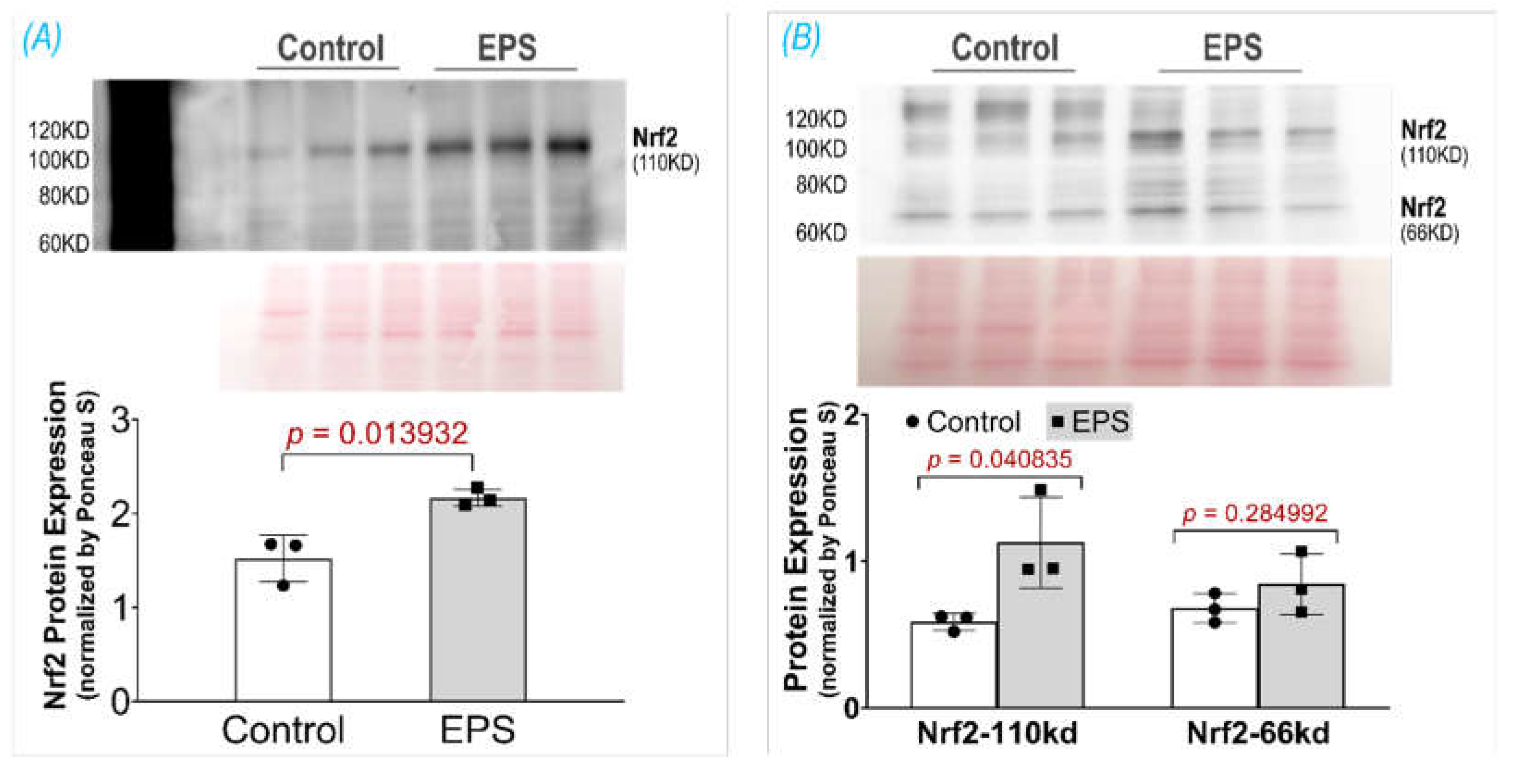
Figure 5). Altogether, these data indicate that EPS leads to substantial adaptations to mitochondrial function and elevations in OXPHOS capacity, which is consistent with known adaptations in human SkM mitochondria after aerobic training [29, 30]. Importantly, the stepwise elevation in OXPHOS capacity due to repeated EPS exposure may also lead to elevations in mitochondrial ROS production during high-intensity EPS [31-34], which may provide an incremental stimulus necessary to induce cumulative increases in antioxidant capacity through Nrf2 as observed in the present study (Panel A,
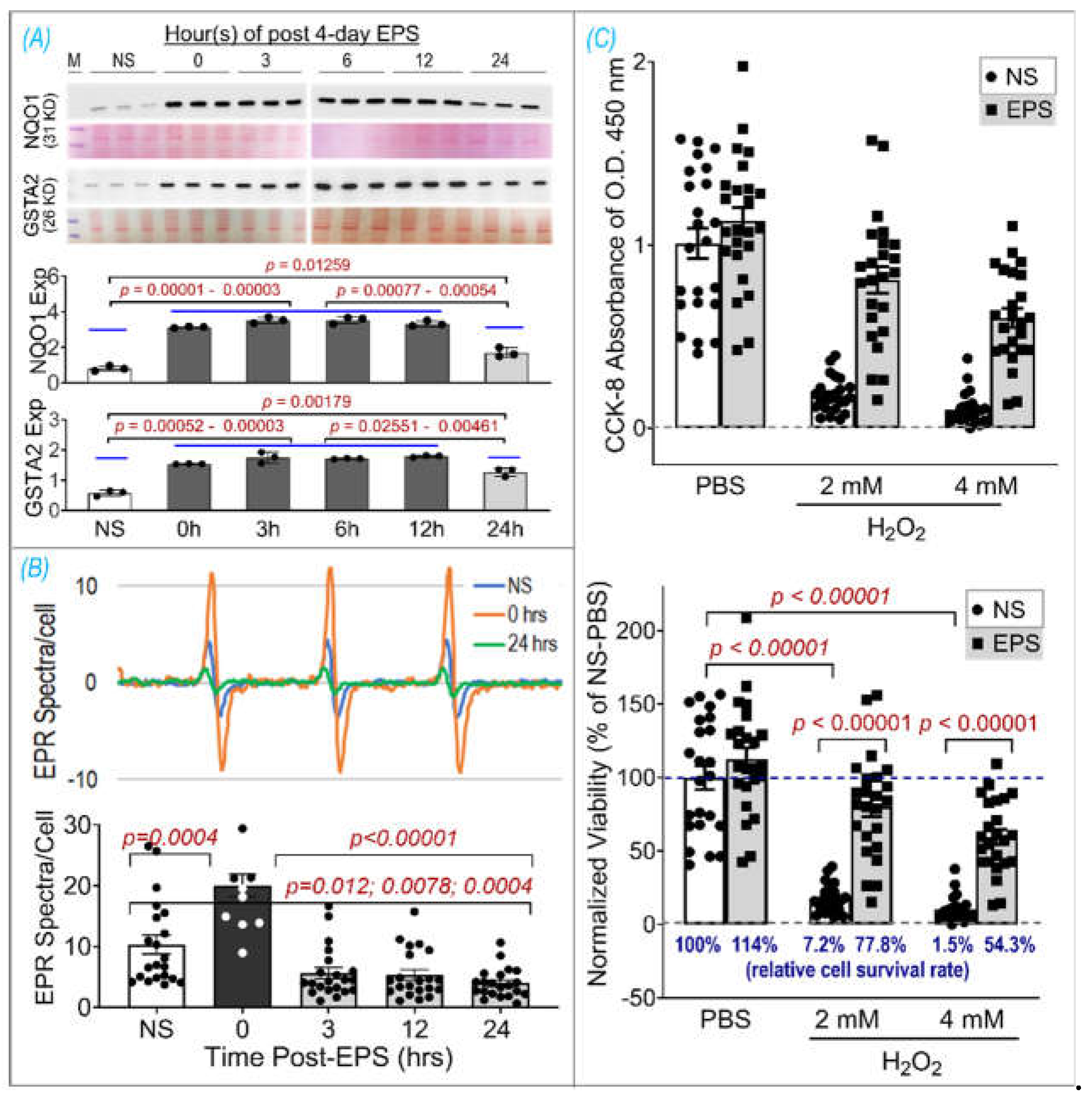
Figure 1). Furthermore, our data support the notion that ROS is transiently elevated during EPS (Panel B,
Figure 6) while antioxidant capacity remains elevated up to 24 hours after EPS (Panel A,
Figure 6), indicating an overcompensation of antioxidant synthesis and the development of an antioxidant reserve. Importantly, we demonstrated that “antioxidant preconditioning” induced by EPS protects C2C12 myotubules from future oxidative injury, since EPS-cells were more resistant to H
2O
2-induced cytotoxicity than non-stimulated control cells. Overall, our data indicate that mitochondrial ROS during EPS activates Nrf2/antioxidant pathways and may lead to an antioxidant reserve that protects cells from future oxidative injury.
C2C12 is a frequently used cell line to investigate the physiology and pathology of skeletal myocytes. After exposure to a differential media, C2C12 are transformed from primitive myoblasts into mature myotubes, which have the primary characteristics of rodent SkM, including the ability to contract and generate force. Indeed, it has been demonstrated that EPS evokes actual contraction of C2C12-myotubes, making this cell line a convenient in vitro model suitable to investigate the physiological significance and molecular responses of skeletal myocytes to stimulated exercise [
14]. A single-bout of EPS at 1 Hz for 1, 3, or 6 hour duration has been demonstrated to upregulate Nrf2/antioxidant system in C2C12 myotubes [
35]. We employed EPS at 50 Hz and 1 hour per day for 4 consecutive days in the present experiment, which may better simulate SkM response to chronic short-term exercise. Importantly, our time-course data clearly show a significantly higher protein expression of NQO1 and GSTA2 in 4-day-EPS-cells compared to those that undergo 1 day of EPS. These findings suggest cumulative effects of repeated contraction on the activation of the muscle antioxidant system and differential responses of SkM to a single bout of exercise compared to multiple sets of a short-term exercise training.
Intracellular redox homeostasis is crucial for maintaining normal muscle structure and function. However, this balance tends to be disturbed during exercise due to increased ROS production by membrane NADPH oxidases and mitochondria where the oxidative phosphorylation process increases dramatically to meet the increased ATP demand. As shown in Panel A of
Figure 4, we detected a profound increase in ROS in mitochondria, cytosol, and whole cells receiving 4 days of EPS. The exact mechanisms responsible for this increased ROS remain to be determined. However, given the data in Panels A and B of
Figure 5, a significant upregulation of complexes I/V and increased mitochondrial oxygen consumption rate were detected. We believe that mitochondria are, at least, one of the major sources for the increased ROS in these EPS cells [
36].
To preserve redox homeostasis in response to oxidative stress, intracellular antioxidant defenses must be mobilized to remove the excessive ROS. Antioxidant protein expression is primarily orchestrated by the Nrf2-Keap1 system, a thiol-based sensor-effector apparatus, where ROS oxidizes three key cysteine residues (Cys151, Cys273, and Cys288) on the Keap1 molecule, resulting in dissociation of Nrf2 from Keap1 and translocation from cytosol to the nucleus. In the nucleus, Nrf2 binds to antioxidant response elements (AREs) on DNA to upregulate hundreds of proteins involved in antioxidant defense, anti-inflammation, detoxification, and metabolism [
10]. This ROS-trigger and Nrf2-Keap1-dependent activation of antioxidant defense in SkM has been characterized by our laboratory employing transgenic mouse models with muscle-specific Nrf2/Keap1 deficiency, iMS-Nrf2flox/flox and iMS-Keap1flox/flox [
11], and in a chronic exercise training model in C57BL/6 mice [
12]. In the present study using 4 days of EPS on C2C12 myotubes, we describe a useful in vitro model whose Nrf2/antioxidant system can be activated to a similar level as observed in animal models. This is particularly true for the expression of NQO1 and GSTA2 proteins.
One of the most important findings from the present study, we believe, is the prolonged elevated protein expression of antioxidant enzymes for up to 24 hours after ceasing 4-day-EPS (Panel A,
Figure 6). This phenomenon suggests potential cumulative effects on protein expression induced by consecutive multiple sets of EPS when the interval of any two chronologically adjacent stimulations is less than 24 hours. This cumulative effect may represent a mechanism for the gradual increased protein content of NQO1 and GSTA2 observed in the time course experiment shown in Panel B of
Figure 1. Translation of this in vitro experimental data into a strategy to optimize long-term exercise points to the significance of intervals between two sets of exercise-induced muscle antioxidant adaptation. While the molecular mechanisms underlying this prolonged antioxidant protein overexpression remain to be elucidated, we postulate that they are involved in an extended alteration of DNA transcription, protein translation, and/or post-translational modifications.
In contrast to the expression of antioxidant enzymes, EPS-induced increase in ROS did not last more than 3 hours after ceasing stimulation (Panel B,
Figure 6). This chronological dissociation between antioxidant enzymes and ROS tilts the redox balance to the antioxidant side and confers cells with excess and chronic antioxidant capacity, a state we term as “antioxidant preconditioning” (Panel B,
Figure 7), thus protecting cells against subsequent oxidative injury. Indeed, as the data shows in Panel C of
Figure 6, EPS-cells have 11- and 36-fold higher viability than control-cells when they are exposed to the medium containing 2 mM and 4 mM H
2O
2.