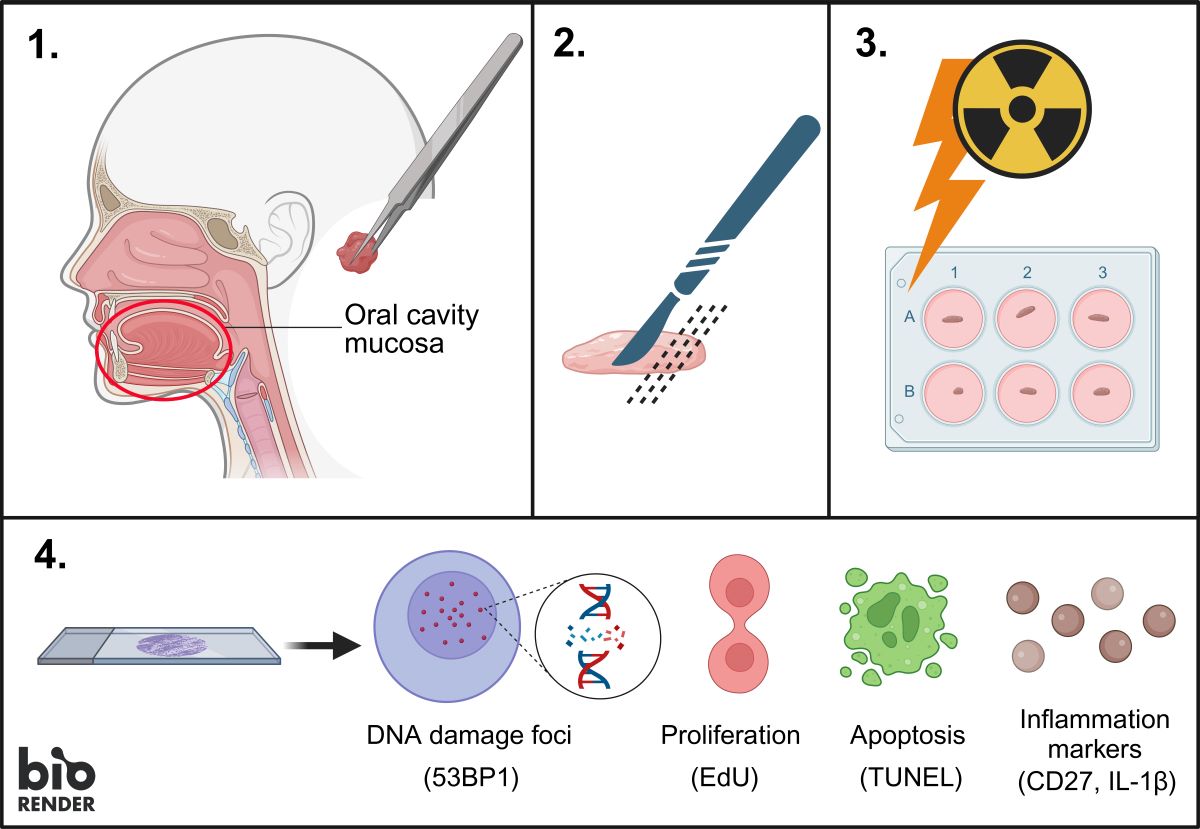
1. Introduction
Oral mucositis is a common side effect of radiotherapy in the head-and-neck area, affecting up to 80% of patients treated with curative chemo-radiotherapy [
1]. Although mucositis is an acute, largely self-limiting side effect of treatment, it can seriously impact the quality of life of patients, as it is associated with severe pain. Furthermore, it may cause decreased oral intake, malnutrition, and consequently hospitalization or discontinuation of treatment. The probability of developing oral mucositis upon irradiation is described by the normal tissue complication probability (NTCP) model. However, it has been demonstrated that dosimetric parameters of a treatment plan account only partly for the variability in the development of normal tissue complications. Even though patient-specific factors, including individual radiosensitivity, play an important role in determining normal tissue response to irradiation [
2,
3,
4], this factor is not accounted for in NTCP models. Therefore, pre-treatment identification of patients with an increased risk of developing severe mucositis may help to optimize clinical decision making and improve personalized treatment, guiding treatment choices where possible, or adjusting radiotherapy treatment planning.
Mucositis development starts with the initial induction of DNA double strand breaks and oxidative DNA damage, resulting from reactive oxygen species (ROS) caused by chemo- or radiotherapy. This DNA damage can lead to apoptosis or necrosis of cells in the mucosa. Increased tissue cell death induces immune cell activation and pro-inflammatory signaling [
5,
6]. A major player of this pro-inflammatory response is nuclear factor-Kappa B (NF-κB), which is, next to other factors, activated by TNF receptor (TNFR) superfamily members. Activation of NF-κB induces the expression of pro-inflammatory cytokines. This initial inflammatory response initiates a positive feedback loop, sustaining NF-kB activity and inducing the expression of more pro-inflammatory cytokines, which in turn promote cell death of epithelial basal cells and damage to endothelium and connective tissue [
5,
6,
7].
Several assays predicting normal mucosa response to radiation have been proposed, but so far none of them has reached clinical validation. That is, on the one hand, due to the fact that many of the proposed assays, such as levels of pro-inflammatory cytokines, are non-specific, measuring simply the tissues response to injury [
6,
8,
9]. On the other hand, more specific assays using primary human keratinocytes, or 3D engineered mucosa models, are too labor- and time-intensive to fit the timeframe of the clinical process [
10,
11,
12,
13]. Moreover, assays using only keratinocytes might be an oversimplification of the complex process of radiation-induced injury and inflammation, in which various other cell types are involved [
6].
Assays using thin slices of the complete tissue have the advantage of including all cell types present in oral mucosa, including fibroblasts and immune cells. Furthermore, this approach allows visualization of radiation response in the various mucosal cell layers, including the basal epithelial cell layer, which comprises the proliferation capacity of the mucosa. Clinically, irradiation of this epithelial progenitor cell layer can result in the temporary loss of regenerative potential of this tissue and development of mucositis [
14]. Radiation response may additionally differ between different types of irradiation, which can be directly compared using thin tissue slices of the same patient. Proton radiotherapy is offered for various tumor sites, since it has the advantage of better dose distribution and decreased excess radiation in healthy tissue compared to photons (such as X-rays; [
15,
16,
17]).
To be able to detect the effects of irradiation in ex vivo patient tissue, we have recently developed an ex vivo assay for head-and-neck squamous cell carcinoma (HNSCC). This assay sustains cancer cell viability and proliferation over several days, allowing ex vivo treatment and assessment of functional outcomes of treatment, such as induction of apoptosis, decrease in proliferation, and DNA repair kinetics [
18]. Moreover, the minimal tissue processing allows a quick read out of the radiation response (in under two weeks).
Expanding from this tumor tissue-response evaluation tool, we here report development of an ex vivo functional assay that is suitable for evaluating the effect of irradiation in the healthy oral mucosa on an individual patient level and can be adapted towards prediction of normal tissue damage in the clinical setting.
2. Materials and Methods
2.1. Collection of oral Mucosa Tissue
Fresh oral mucosa tissue was obtained from HNSCC patients undergoing surgery at the Erasmus University Medical Center (Erasmus MC), Rotterdam, The Netherlands. The resection material was directly transported to the pathology department for macroscopic inspection and determination of tumor areas for diagnostic purposes. The regions of healthy mucosa were subsequently identified by a dedicated head and neck pathologist and a sample was acquired. The healthy oral mucosa tissue was used for research purposes according to the code of proper secondary use of human tissue in the Netherlands established by the Dutch Federation of Medical Scientific Societies, and approved by the Erasmus MC Medical Ethical Committee (number MEC-2017-1049). Research samples were kept at 4 °C and transported in culture medium (see section below). Specimens were anonymized such that patient information could not be traced by the research personnel.
2.2. Tissue Slice Preparation and Culture
Oral mucosa tissue preparation methods were adapted from Capala et al. [
18]. Mucosa specimens were embedded in 4% low gelling temperature agarose and cut manually with a scalpel in a thickness between 500 and 1000 µm under semi-sterile conditions. Mucosa tissue slices were cultured in medium consisting of aDMEM-F12 (ThermoFischer Scientific (Waltham, MA, USA), catalog number 12634028) supplemented with 1% penicillin-streptomycin, 0.1% primocin (50 mg/mL stock solution; InvivoGen (San Diego, CA, USA), cat. code ant-pm-1), and freshly added 20 ng/mL epidermal growth factor (EGF; Sigma-Aldrich, cat. nr SRP3027) and 20 ng/mL basic fibroblast growth factor (bFGF; Sigma-Aldrich, Saint Louis, MO, USA, cat. nr F3685), within 4 hours after surgical resection. Culturing was performed at 37°C, 5% CO
2, and atmospheric oxygen levels under continuous rotation at 60 rpm on a Stuart SSM1 mini orbital shaker placed in the incubator. Proliferating cells were labeled using 30 μmol/L EdU (Invitrogen, Waltham, MA, USA, cat. nr C10086) two hours before fixation. Tissue slices were fixed in 10% neutral buffered formalin for at least 24 hours at room temperature. Subsequently, tissue slices were embedded in paraffin and 4 μm sections were generated for microscopy analysis.
2.3. Tissue Irradiation
X-ray irradiation was performed directly after placing the mucosa tissue slices in the culture media using the X-Strahl RS320 X-ray cabinet with 0.5 mm Cu filter at 195 keV and a dose rate of 1.6 Gy/min.
All proton irradiations were performed at Holland PTC (Delft, Netherlands) in the R&D beam line, simultaneously with X-ray irradiation at Erasmus MC with the same physical dose of 5 Gy (not RBE weighted). The proton beam line of HollandPTC is fixed and horizontal, therefore the samples were positioned vertically on a proper target station [
19]. The same setup was used for respective samples at Erasmus MC. Details of the proton experimental setup can be found in the supplementary Materials and Methods.
From a therapeutic proton pencil beam, a passive scattering system produces a field of 8x8 cm
2, with a uniformity of 98% over the whole area, irradiating the whole sample with a dose rate of 9 Gy/min [
19]. To irradiate the sample evenly over its depth (beam direction), a Spread-out-Bragg peak (SOBP) is produced with a 2D range energy modulator, with an initial energy of 150 MeV, creating a SOBP of 25 mm with 98% ±1% uniformity. Tissue samples were placed into 4 wells of a 6-well plate, and positioned in the middle of the SOBP using a solid plastic phantom of RW3 material [
19].
After irradiation, plates were immediately put back in the incubator.
2.4. Immunofluorescence (IF) Staining
Histological mucosa architecture was examined by hematoxylin-eosin (H&E) staining. Samples that did not contain healthy (non-cancerous) epithelium were excluded from analysis. To be able to analyze the progenitor and stem cell population in all samples, p63 (isoform ΔNp63) as a marker of epithelial progenitor cells was combined with EdU or 53BP1 staining. Immunostaining was performed as described previously [
20].
2.5. TUNEL Assay
TUNEL assay was performed using In Situ Cell Death Detection Kit (Roche Life Sciences) as described previously [
20].
2.6. Immunohistochemistry (IHC) Staining
All IHC stainings were automated by using the Ventana Benchmark ULTRA (Ventana Medical Systems Inc.). Sequential 4 µm thick (FFPE) sections were stained for the antibody of interest (
Table S1) using optiview (OV) (#760-700, Ventana). In brief, following deparaffinization and heat-induced antigen retrieval with CC1 (#950-500, Ventana) for 32 minutes the tissue samples were incubated with the antibody of interest for 32 minutes at 37˚C. Incubation was followed by optiview detection and hematoxylin II counter stain for 8 minutes followed by a blue coloring reagent for 8 minutes according to the manufactures instructions (Ventana).
2.7. Image Acquisition
H&E, CD45, CD27 and IL-1β IHC stainings were visualized using an Olympus BX40 F4 System microscope. For the TUNEL assay, from each mucosa slice section multiple images (at least three fields of view (FoV) per sample) were acquired using a Leica DM4000 B fluorescent microscope with a Leica DFC300 FX camera. 53BP1/p63 immunostaining (at least 50 nuclei per sample) and EdU incorporation (at least three FoV per sample) were imaged using a Leica Stellaris 5 LIA confocal microscope.
2.8. Image Analysis of 53BP1 DNA Damage Foci
For the image analysis of the 53BP1 foci and p63-positive nuclei, the same method as previously described was used [
18,
21]. Data is presented as violin plots showing the median of all values and 1st and 3rd quartiles depicted.
2.9. Image Analysis of Proliferation Assay
To calculate proliferation, nuclei were segmented on the p63 images and on the corresponding EdU images. For the p63 images, the same U-net model was used as explained in the image analysis DNA damage section. Afterwards, the segmentations were improved using the ImageJ hole filling procedure. The EdU images were segmented using a threshold of 90. Subsequently, the percentage of proliferation was calculated using the following formula:
In this formula, N_(EdU∩p63) is the number of pixels that is both EdU and p63 positive and N_p63is the number of pixels that is p63 positive. Image segmentation and proliferation calculations were performed using Python 3.8. Data is shown as mean with standard error of the mean (SEM).
2.10. Image Analysis of Apoptosis Assay
To analyze apoptosis, nuclei were segmented on the DAPI images and on the TUNEL images. Segmentation was performed using the StartDist module in Python 3.8, which is a deep-learning-based method for nucleus detection [
22]. A pre-trained model was used (2D_versatile_flou) and the normalized DAPI and TUNEL imaged were used as input. The probability threshold was set to 0.5 and 0.7 for the DAPI and TUNEL images respectively to decrease the number of false positive TUNEL segmentations. For the other parameters, the standard settings were used.
TUNEL images were cut to only contain the same cell layer that is stained with p63. Furthermore, the first two cell layers at the tissue border were cut, because high TUNEL signals can sometimes be observed at the edges of the tissue slices, which is usually not a treatment effect. Subsequently, the images excluding the edges were used to calculate the percentage of apoptosis according to the following formula:
In this formula, N_(TUNEL∩DAPI) is the number of pixels that is both TUNEL and DAPI positive and N_DAPI is the number of pixels that is DAPI positive. Image segmentation and apoptosis calculations were performed using Python 3.8. Data is shown as mean with SEM.
2.11. IMAGE analysis of CD45, CD27 and IL-1β IHC Staining
With the ImageJ function of “Deconvolution”, different channels of the IHC staining were separated automatically. A threshold was set for nuclei channel (205) and for DAB channel (210) and images were inverted. Area was measured with the function “Analyze particles” and percentage of CD27-positive or IL-1β-positive area relative to total nuclear area was calculated. Data is shown as mean with SEM.
2.12. Statistical Analysis
Statistical analysis and generation of graphs was performed using GraphPad Prism 8.0. Differences were tested, depending on number of compared groups, with Kruskal Wallis test with Dunn’s comparison or Mann Whitney test and p-values <0.05 were considered significant.
3. Results
3.1. Healthy Mucosa Tissue Slices Maintain Viability for Several Days in Culture
In order to be able to assess treatment response
ex vivo, it is crucial to ensure viability of analyzed tissue during the assay. In the initial assessment of tissue survival in culture, morphological mucosa structure was histologically investigated with H&E staining, demonstrating normal morphology over three days in culture (
Figure 1A). Apoptosis was measured using TUNEL staining as another evaluation of viability in culture. Here we observed that mucosa samples showed low baseline apoptosis levels, which did not increase after one or three days in culture (
Figure 1B and C,
Figure S1A).
Furthermore, EdU incorporation of p63 positive epithelial progenitor cells was measured to compare proliferation capacity over the course of culture. Although there was a decrease after 24 hours, proliferation measurements did not differ significantly between two hours and three days in culture (
Figure 1D and E,
Figure S1B), indicating that normal mucosa remained viable throughout the culture period.
3.2. Healthy Mucosa Displays Dose-Dependent Response to X-ray Irradiation
To assess the effect of irradiation on healthy oral mucosa, morphology, proliferation capacity, and induction of apoptosis were investigated. Consistent with degrading morphology after irradiation (
Figure 2A), we observed a dose-dependent decrease in proliferation in the basal cell layer after a single dose of 5 Gy (p = 0.0081) and 10 Gy (p = 0.0001;
Figure 2B and C). This decrease in proliferation was heterogeneous between samples, which was most evident at 5 Gy (
Figure S2A). While two samples did not show a decrease in proliferation, in four samples proliferation was decreased to various extent (n = 6; 65% - 1.8% of control value). All 10 Gy irradiated slices (n = 5) showed a decrease in proliferation (53% - 4% of control value).
Apoptosis increased after 5 Gy (p = 0.1338) and 10 Gy (p = 0.0003) of X-ray irradiation (
Figure 2D and E). All 5 Gy (n = 6) irradiated slices displayed an increase in apoptosis (
Figure S2B), although for most conditions this increase was small (0.3 - 13%). 10 Gy (n = 5) irradiated slices showed on average higher apoptosis values (1.6 - 52%). While no significant differences were observed between the two irradiation doses, a decreasing trend in viability can be seen in the higher dose in all but one sample.
Taken together, we conclude that the ex vivo model can be used to assess responses to varying doses of X-ray irradiation.
3.3. Residual DNA Damage Foci after Irradiation of Normal Mucosa
Sensitivity to irradiation may be associated with DNA repair capacity, which can be deduced from the formation and subsequent dissolution of protein accumulations (foci) at DNA breaks. Therefore, we quantified 53BP1 DNA damage foci at 24 hours post-irradiation. Significantly higher levels of residual 53BP1 foci were measured after 5 Gy (p < 0.0001) and 10 Gy (p < 0.0001) X-ray irradiation than in the unirradiated controls in a dose-dependent manner (
Figure 3A and B). All individual irradiated slices exhibited significantly more residual 53BP1 foci than control conditions (n = 6). One analyzed 10 Gy condition was classified as necrotic by a pathologist and therefore excluded from analysis. Similarly to apoptosis and proliferation measurements, the increased number of foci after irradiation differed between samples (
Figure S3A). These results indicate that residual DNA damage is quantifiable with our setup, showing a dose-dependent increase in DNA double strand breaks upon irradiation.
Interestingly, a slight increase in foci numbers was still visible in irradiated slices after 3 days, (n = 3;
Figure 3C and D;
Figure S3B). Additionally, when looking at 53BP1 foci size, we observed a significant increase (p < 0.0001) in both 5 Gy and 10 Gy irradiated slices compared to control after three days (
Figure 3E;
Figure S3C and S3D).
3.4. Proton Irradiation of Mucosa Tissue Slices
Due to the clinical significance of proton radiotherapy for HNSCC, we adapted our setup for this treatment modality. We analyzed DNA damage induction, as visualized by 53BP1 foci, after 5 Gy proton irradiation in normal mucosa tissue slices. X-ray irradiation was performed simultaneously to provide reference for the induced damage in a pre-developed setup (
Figure 4A). We observed a significant increase in 53BP1 foci number (p < 0.0001) and size (p < 0.0001) in both X-ray- and proton irradiated samples compared to untreated control (n = 3;
Figure 4B - E). We did not observe a significantly different 53BP1 foci number or size in proton irradiated samples compared to X-ray irradiation. Individual samples showed heterogeneity in the residual 53BP1 foci number and size 24 hours after treatment (
Figure S4A and S4B), suggesting that this may be an interesting parameter to include in ex vivo predictive assays for normal tissue response.
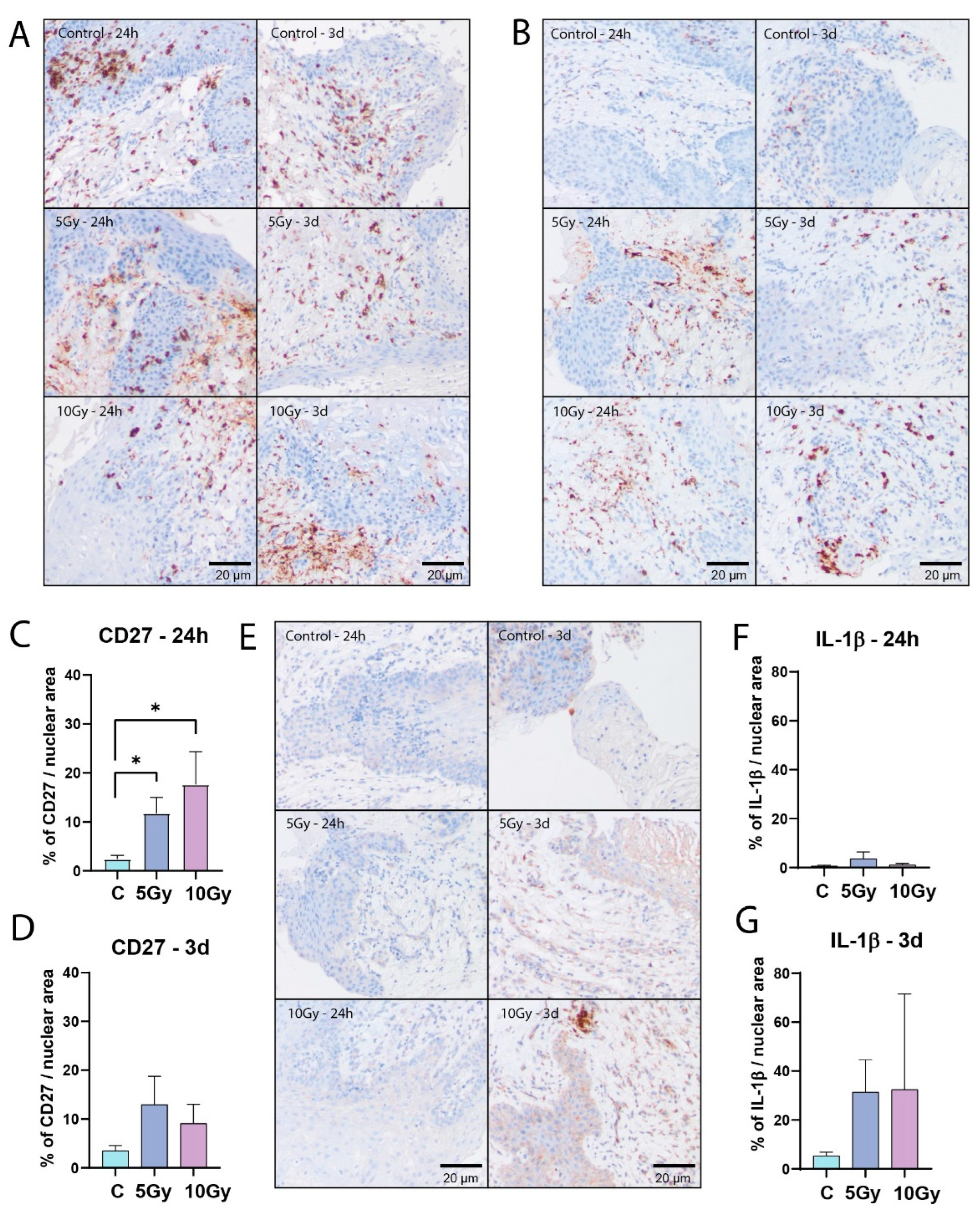
3.5. Immune Cells Express Inflammation Inducing Factors upon Irradiation Ex Vivo
One of the advantages of tissue slices is that they preserve the composition of the tissue, including various stromal cell types. Accordingly, lymphocytes could be detected by CD45 immunohistochemistry staining in all analyzed samples. They were mainly located in the lamina propria, but also in the stratified epithelium (
Figure 5A). To detect lymphocyte immunological function after irradiation induced cell damage, we analyzed expression levels of CD27, a member of the tumor necrosis factor family, and Interleukin 1 beta (IL-1β), a pro-inflammatory cytokine. Twenty-four hours after irradiation, the mean CD27 signal showed a dose-dependent increase (5 Gy, p = 0.0239; 10Gy, p = 0.0185), which could still be measured three days after treatment, albeit at lower levels (
Figure 5B - D). We did not observe an increase in IL-1β levels after 24 hours in most samples. However, after three days the average IL-1β signal displayed an increasing trend in irradiated conditions (
Figure 5E - G). Both CD27 and IL-1β signals in individual samples were heterogeneous, suggesting that this parameter may be useful for a radiation sensitivity prediction model (
Figure S5A – S5D).
4. Discussion
In this study, we developed an ex vivo culture system for healthy oral mucosa tissue slices, that maintained normal tissue morphology and viability for at least three days. Moreover, automated image analysis allowed comparison between various treatment conditions. We measured a clear dose-dependent response to X-ray irradiation and could compare X-ray and proton irradiation in the same healthy mucosa sample. Furthermore, we detected increased levels of inflammation inducing factors, which are major drivers in mucositis development. The irradiation responses were heterogeneous in the various samples, suggesting that the assay can be used to stratify patients based on ex vivo radiation sensitivity.
Adaptation of the HNSCC culture system for mucosa tissue was met with some challenges. Unlike HNSCC tumor tissue, the partly spongy texture of mucosa tissue necessitated manual cutting of samples with a scalpel. This process could be standardized by keeping the mucosa in agarose, so that samples were positioned to obtain similar mucosal layers in comparable thickness of approximately 500-1000 µm. This technical bottleneck had no influence on viability since no difference between the beginning and day 3 in culture was detected. The minor decrease in proliferation at 24 hours can probably be explained as an adaptation effect at the start of the culture. Healthy mucosa was macroscopically identified in the resection specimens by a dedicated head and neck pathologist. However, after slicing epithelium could not be distinguished macroscopically. To increase the chances of finding back epithelial tissue in every slice, we used more than one slice per condition.
Previously established models for radiosensitivity include measurement of inflammatory factors, such as pro-inflammatory cytokines, before and after treatment in patient saliva, blood, or oral cytological smears [
6,
8,
12]. Furthermore, γH2AX DNA damage foci, which similarly to 53BP1 DNA damage foci mark double strand breaks, have been investigated to develop a predictive model for healthy tissue sensitivity using blood lymphocytes and tumor cells. However, these approaches have not been clinically validated yet [
23]. Moreover, in most studies, only one factor or cell type was investigated and not the combination of different factors and cell types [
6,
8,
12]. While 3D engineered tissue models that include different patient cell types can be used for studying the mechanisms of mucositis, the high costs for personalization and long development time render them unsuitable for clinical treatment planning [
9,
11,
24]. The same can be said about large sequencing studies, which did not lead to the identification of one or multiple factors correlating strongly enough to mucositis risk for clinical implementation [
25,
26,
27,
28].
Our assay can be set up relatively easily and analyzed in under two weeks, facilitating clinical implementation. Furthermore, it contains all cell layers and cell types of healthy mucosa tissue, including immune cells. Therefore, it can be used for observing the effects of radiation on the whole mucosa microenvironment and every individual cell type. It could be utilized to test preventative intervention options for mucositis in patients. Moreover, more fundamental investigations into the process of mucositis and risk-predicting factors can be undertaken with a relatively large sample number due to the simple and quick procedures involved.
Furthermore, the effect of different types of irradiation on normal mucosa can be explored. Proton radiotherapy is an alternative to X-ray irradiation for HNSCC patients, that provides improved dose distribution [
15,
29,
30,
31]. Consequently, several studies observed that acute healthy tissue toxicity was decreased after proton irradiation compared to X-ray radiotherapy. However, reduction of normal tissue complications differed between patients of the same treatment group and between different treatment groups [
16,
30,
32,
33,
34]. This indicates the need to investigate these differences on a patient-specific level for both tumor and healthy mucosa tissue. For this purpose, we developed a proton irradiation setup, where exact positioning of tissue slices relative to the SOBP is possible. This precise tissue positioning allows investigation of several aspects of proton therapy, such as the effect of positioning in various parts of the Bragg peak, or ultra-high dose irradiation (FLASH) [
35,
36,
37,
38]. Furthermore, as our automated analysis methods allow analysis of a large number of treated nuclei, it is well suited to study differences in DNA damage response to different types of irradiation. In our experiments, we have not observed a significant difference between X-ray and proton irradiation, while at least a 10% increase in the effect of proton irradiation could be expected given non-RBE corrected delivered dose. The absence of a difference between protons and photons may be explained by low LET values in the proton SOBP and additionally higher LET in the orthovoltage X-ray than for conventional patient treatment, but may also be connected to the exact measurement time point or a too subtle difference for our assay to pick up.
To capture the full complexity of radiation responses, it is important to investigate the mechanisms of mucositis development in the context of the complete oral mucosa microenvironment. Our assay offers this opportunity, measuring irradiation response of the broad array of cell types in the oral mucosa. CD27 as an activator of NF-κB, showed a pro-inflammatory reaction 24 hours after irradiation that is still visible after 3 days. Similarly, we observed an average increasing trend in abundance of IL-1β pro-inflammatory cytokines in irradiated samples after three days. This demonstrates the presence of functioning immune cells in our ex vivo system that react to damage similarly to in vivo observations in literature [
39,
40,
41,
42]. We conclude that our setup enables investigation of the separate initial stages of mucositis functionally
ex vivo, including immune reactions.
Mucositis is one of the dose-limiting side effects in radiotherapy for HNSCC. Marked heterogeneity in the occurrence and severity of mucositis can be observed among patients. Therefore, development of an ex vivo model for normal mucosa irradiation damage is crucial to not only predict individual patient’s radiosensitivity but also investigate the underlying mechanisms of mucositis so that interventions limiting this side effect can be developed. Furthermore, this assay may be easily combined with HNSCC biopsies for testing individual tumor radiosensitivity, which we are currently investigating in a clinical study (PMID: 35584883), ultimately allowing personalized radiotherapy treatment.
5. Conclusions and Future Perspectives
We developed an ex vivo normal mucosa model for assessing radiation responses, representing epithelial cells in their natural environment containing immune components, fibroblasts and other mucosal cell types. Our findings should be confirmed in a larger cohort in which in vivo radiation responses can directly be compared to the ex vivo outcomes. Parameters from this analysis could then be combined with NTCP modeling, leading to the establishment of a predictive model for oral mucositis.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org:, Supplementary Materials and Methods, Table S1. Immunohistochemistry antibody information, Figure S1: Healthy mucosa tissue slices maintain viability for several days in culture, Figure S2: Healthy mucosa displays dose-dependent response to X-ray irradiation, Figure S3: Residual DNA damage foci after irradiation of normal mucosa, Figure S4: Proton irradiation of mucosa tissue slices, Figure S5: Immune cells express inflammation inducing factors upon irradiation ex vivo.
Author Contributions
Conceptualization, M.E.C. and D.C.v.G.; investigation, K.S.P., M.E.C., I.L. and N.S.V.; resources, M.R., E.v.d.W., H.M., B.P.J., A.S., J.A.H., S.K. (Stijn Keereweer), D.M., B.K., S.K. (Sjors Koppes) and T.v.d.B.; writing—original draft preparation, K.S.P.; writing—review and editing, K.S.P., M.E.C., I.L., N.S.V., M.R., E.v.d.W., S.K. (Stijn Keereweer), S.K. (Sjors Koppes), G.M.V., S.P., B.S.S and D.C.v.G.; visualization, K.S.P., M.E.C. and I.L.; supervision, M.E.C. and D.C.v.G.; funding acquisition, D.C.v.G., S.P., M.E.C. and G.M.V.; All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by HollandPTC-Varian consortium-confined call 2019, project number 2019011 and by KWF Dutch Cancer Society, grant number 12141.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committee of Erasmus MC (protocol code MEC-2017-1049 (2017)).
Informed Consent Statement
Patient consent was waived for surgical material due to laws in the Netherlands for opting out. All use of human material was approved by the Erasmus MC Medical Ethics commission.
Data Availability Statement
All data can be found in the manuscript.
Acknowledgments
Special thank you to Wouter van Burik, Soraya Nait Abdellah and Eva van Oosten for helping with proton treatment of the tumor material.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Trotti, A.; Bellm, L. A.; Epstein, J. B.; Frame, D.; Fuchs, H. J.; Gwede, C. K.; Komaroff, E.; Nalysnyk, L.; Zilberberg, M. D. , Mucositis incidence, severity and associated outcomes in patients with head and neck cancer receiving radiotherapy with or without chemotherapy: a systematic literature review. Radiotherapy and Oncology 2003, 66(3), 253–262. [Google Scholar] [CrossRef] [PubMed]
- Turesson, I.; Nyman, J.; Holmberg, E.; Odén, A. , Prognostic factors for acute and late skin reactions in radiotherapy patients. Int J Radiat Oncol Biol Phys 1996, 36(5), 1065–75. [Google Scholar] [CrossRef] [PubMed]
- Johansen, J.; Bentzen, S. M.; Overgaard, J.; Overgaard, M. , Relationship between the in vitro radiosensitivity of skin fibroblasts and the expression of subcutaneous fibrosis, telangiectasia, and skin erythema after radiotherapy. Radiother Oncol 1996, 40(2), 101–9. [Google Scholar] [CrossRef] [PubMed]
- Subedi, P.; Gomolka, M.; Moertl, S.; Dietz, A. Ionizing Radiation Protein Biomarkers in Normal Tissue and Their Correlation to Radiosensitivity: A Systematic Review. Journal of Personalized Medicine 2021, 11 (2), 140.
- Liu, S.; Zhao, Q.; Zheng, Z.; Liu, Z.; Meng, L.; Dong, L.; Jiang, X. Status of Treatment and Prophylaxis for Radiation-Induced Oral Mucositis in Patients With Head and Neck Cancer. Front Oncol 2021, 11.
- Pulito, C.; Cristaudo, A.; Porta, C.; Zapperi, S.; Blandino, G.; Morrone, A.; Strano, S. Oral mucositis: the hidden side of cancer therapy. J Exp Clin Cancer Res 2020, 39 (1), 210.
- Liu, T.; Zhang, L.; Joo, D.; Sun, S. C., NF-kappaB signaling in inflammation. Signal Transduct Target Ther 2017, 2, 10.1038/sigtrans.2017.23.
- Palmier, N. R.; Leme, A. F. P.; De Rossi, T.; Telles, G. P.; Morais-Faria, K.; Kowalski, L. P.; Marta, G. N.; Brandao, T. B.; Arany, P. R.; Migliorati, C. A.; Santos-Silva, A. R.; Prado-Ribeiro, A. C., Salivary alpha-1-antitrypsin and macrophage migration inhibitory factor may be potential prognostic biomarkers for oncologic treatment-induced severe oral mucositis. Support Care Cancer 2021, 29 (6), 2939-2946.
- Tschachojan, V.; Schroer, H.; Averbeck, N.; Mueller-Klieser, W. Carbon ions and X-rays induce pro-inflammatory effects in 3D oral mucosa models with and without PBMCs. Oncol Rep 2014, 32 (5), 1820-1828.
- Rakhorst, H. A.; Tra, W. M.; Posthumus-Van Sluijs, S. T.; Hovius, S. E.; Levendag, P. C.; Kanaar, R.; Hofer, S. O. , Quantitative Analysis of Radiation-Induced DNA Break Repair in a Cultured Oral Mucosal Model. Tissue Engineering, 2006,12 (12), 3395-3403.
- Lambros, M. P.; DeSalvo, M. K.; Mulamalla, H. C.; Moreno, J.; Kondapalli, L. , Genome wide expression after different doses of irradiation of a three-dimensional (3D) model of oral mucosal. Genomics Data 2016, 7, 137–139. [Google Scholar] [CrossRef] [PubMed]
- Tobita, T.; Izumi, K.; Feinberg, S. E. , Development of an in vitro model for radiation-induced effects on oral keratinocytes. International Journal of Oral and Maxillofacial Surgery 2010, 39(4), 364–370. [Google Scholar] [CrossRef] [PubMed]
- Donetti, E.; Bedoni, M.; Capone, P.; Gualerzi, A.; Tartaglia, G.; Sforza, C. An in vitro model of human oral explants to study early effects of radiation mucositis. Eur J Oral Sci 2009, 117 (2), 169-74.
- Duncan, M.; Grant, G. Oral and intestinal mucositis - causes and possible treatments. Aliment Pharmacol Ther 2003, 18 (9), 853-74.
- Prasanna, P. G.; Rawojc, K.; Guha, C.; Buchsbaum, J. C.; Miszczyk, J. U.; Coleman, C. N. Normal Tissue Injury Induced by Photon and Proton Therapies: Gaps and Opportunities. Int J Radiat Oncol Biol Phys 2021, 110 (5), 1325-1340.
- Chen, Z.; Dominello, M. M.; Joiner, M. C.; Burmeister, J. W. Proton versus photon radiation therapy: A clinical review. Front Oncol 2023, 13.
- Alan Mitteer, R.; Wang, Y.; Shah, J.; Gordon, S.; Fager, M.; Butter, P. P.; Jun Kim, H.; Guardiola-Salmeron, C.; Carabe-Fernandez, A.; Fan, Y. Proton beam radiation induces DNA damage and cell apoptosis in glioma stem cells through reactive oxygen species. Sci Rep 2015, 5.
- Capala, M. E.; Pachler, K. S.; Lauwers, I.; de Korte, M. A.; Verkaik, N. S.; Mast, H.; Jonker, B. P.; Sewnaik, A.; Hardillo, J. A.; Keereweer, S.; Monserez, D.; Koljenovic, S.; Mostert, B.; Verduijn, G. M.; Petit, S.; van Gent, D. C., Ex Vivo Functional Assay for Evaluating Treatment Response in Tumor Tissue of Head and Neck Squamous Cell Carcinoma. Cancers (Basel) 2023, 15 (2).
- Rovituso, M.; Groenendijk, C. F.; Van der Wal, E.; Van Burik, W.; Ibrahimib, A.; Rituerto Prieto, H.; Brown, J. M. C.; Weber, U.; Simeonov, Y.; Fontana, M.; Lathouwers, D.; Van Vulpena, M.; Hoogeman, M., Characterisation of the HollandPTC R&D proton beamline for physics and radiobiology studies. arXiv 2023.
- Naipal, K. A. T.; Verkaik, N. S.; Sánchez, H.; van Deurzen, C. H. M.; den Bakker, M. A.; Hoeijmakers, J. H. J.; Kanaar, R.; Vreeswijk, M. P. G.; Jager, A.; van Gent, D. C., Tumor slice culture system to assess drug response of primary breast cancer. BMC Cancer 2016, 16 (1), 78.
- Lauwers, I.; Pachler, K. S.; Capala, M. E.; Sijtsema, N. D.; Van Gent, D. C.; Rovituso, M.; Hoogeman, M. S.; Verduijn, G. M.; Petit, S. F., Ex vivo radiation sensitivity assessment for individual head and neck cancer patients using deep learning-based automated nuclei and DNA damage foci detection. Clin Transl Radiat Oncol 2024, 45.
- Uwe Schmidt, M. W., Coleman Broaddus, and Gene Myers, Cell Detection with Star-convex Polygons. arXiv 2018.
- Habash, M.; Bohorquez, L. C.; Kyriakou, E.; Kron, T.; Martin, O. A.; Blyth, B. J., Clinical and Functional Assays of Radiosensitivity and Radiation-Induced Second Cancer. Cancers (Basel) 2017, 9 (11),.
- Rakhorst, H. A.; Posthumus-Van Sluijs, S. J.; Tra, W. M.; Van Neck, J. W.; Van Osch, G. J.; Hovius, S. E.; El Ghalbzouri, A.; Hofer, S. O., Fibroblasts accelerate culturing of mucosal substitutes. Tissue Eng 2006, 12 (8), 2321-31.
- Wardill, H. R.; Sonis, S. T.; Blijlevens, N. M. A.; Van Sebille, Y. Z. A.; Ciorba, M. A.; Loeffen, E. A. H.; Cheng, K. K. F.; Bossi, P.; Porcello, L.; Castillo, D. A.; Elad, S.; Bowen, J. M.; Mucositis Study Group of the Multinational Association of Supportive Care in Cancer/International Society of Oral, O., Prediction of mucositis risk secondary to cancer therapy: a systematic review of current evidence and call to action. Support Care Cancer 2020, 28 (11), 5059-5073.
- Venkatesh, G. H.; Manjunath, V. B.; Mumbrekar, K. D.; Negi, H.; Fernandes, D. J.; Sharan, K.; Banerjee, S.; Bola Sadashiva, S. R., Polymorphisms in radio-responsive genes and its association with acute toxicity among head and neck cancer patients. PLoS One 2014, 9 (3).
- Mlak, R.; Powrozek, T.; Brzozowska, A.; Homa-Mlak, I.; Mazurek, M.; Malecka-Massalska, T. RRM1 gene expression evaluated in the liquid biopsy (blood cfRNA) as a non-invasive, predictive factor for radiotherapy-induced oral mucositis and potential prognostic biomarker in head and neck cancer patients. Cancer Biomark 2018, 22 (4), 657-667.
- Reyes-Gibby, C. C.; Melkonian, S. C.; Wang, J.; Yu, R. K.; Shelburne, S. A.; Lu, C.; Gunn, G. B.; Chambers, M. S.; Hanna, E. Y.; Yeung, S. J.; Shete, S., Identifying novel genes and biological processes relevant to the development of cancer therapy-induced mucositis: An informative gene network analysis. PLoS One 2017, 12 (7).
- Sio, T. T.; Lin, H. K.; Shi, Q.; Gunn, G. B.; Cleeland, C. S.; Lee, J. J.; Hernandez, M.; Blanchard, P.; Thaker, N. G.; Phan, J.; Rosenthal, D. I.; Garden, A. S.; Morrison, W. H.; Fuller, C. D.; Mendoza, T. R.; Mohan, R.; Wang, X. S.; Frank, S. J., Intensity Modulated Proton Therapy Versus Intensity Modulated Photon Radiation Therapy for Oropharyngeal Cancer: First Comparative Results of Patient-Reported Outcomes. Int J Radiat Oncol Biol Phys 2016, 95 (4), 1107-14.
- Romesser, P. B.; Cahlon, O.; Scher, E.; Zhou, Y.; Berry, S. L.; Rybkin, A.; Sine, K. M.; Tang, S.; Sherman, E. J.; Wong, R.; Lee, N. Y., Proton beam radiation therapy results in significantly reduced toxicity compared with intensity-modulated radiation therapy for head and neck tumors that require ipsilateral radiation. Radiother Oncol 2016, 118 (2), 286-92.
- Hutchinson, M. N. D.; Mierzwa, M.; D’Silva, N. J., Radiation resistance in head and neck squamous cell carcinoma: dire need for an appropriate sensitizer. Oncogene 2020, 39 (18), 3638-3649.
- Jakobi, A.; Bandurska-Luque, A.; Stutzer, K.; Haase, R.; Lock, S.; Wack, L. J.; Monnich, D.; Thorwarth, D.; Perez, D.; Luhr, A.; Zips, D.; Krause, M.; Baumann, M.; Perrin, R.; Richter, C., Identification of Patient Benefit From Proton Therapy for Advanced Head and Neck Cancer Patients Based on Individual and Subgroup Normal Tissue Complication Probability Analysis. Int J Radiat Oncol Biol Phys 2015, 92 (5), 1165-1174.
- Jakobi, A.; Stutzer, K.; Bandurska-Luque, A.; Lock, S.; Haase, R.; Wack, L. J.; Monnich, D.; Thorwarth, D.; Perez, D.; Luhr, A.; Zips, D.; Krause, M.; Baumann, M.; Perrin, R.; Richter, C., NTCP reduction for advanced head and neck cancer patients using proton therapy for complete or sequential boost treatment versus photon therapy. Acta Oncol 2015, 54 (9), 1658-64.
- Alterio, D.; D’Ippolito, E.; Vischioni, B.; Fossati, P.; Gandini, S.; Bonora, M.; Ronchi, S.; Vitolo, V.; Mastella, E.; Magro, G.; Franco, P.; Ricardi, U.; Krengli, M.; Ivaldi, G.; Ferrari, A.; Fanetti, G.; Comi, S.; Tagliabue, M.; Verri, E.; Ricotti, R.; Ciardo, D.; Jereczek-Fossa, B. A.; Valvo, F.; Orecchia, R., Mixed-beam approach in locally advanced nasopharyngeal carcinoma: IMRT followed by proton therapy boost versus IMRT-only. Evaluation of toxicity and efficacy. Acta Oncol 2020, 59 (5), 541-548.
- Bourhis, J.; Montay-Gruel, P.; Gonçalves Jorge, P.; Bailat, C.; Petit, B.; Ollivier, J.; Jeanneret-Sozzi, W.; Ozsahin, M.; Bochud, F.; Moeckli, R.; Germond, J.-F.; Vozenin, M.-C., Clinical translation of FLASH radiotherapy: Why and how? Radiotherapy and Oncology 2019, 139.
- Bourhis, J.; Sozzi, W. J.; Jorge, P. G.; Gaide, O.; Bailat, C.; Duclos, F.; Patin, D.; Ozsahin, M.; Bochud, F.; Germond, J.-F.; Moeckli, R.; Vozenin, M.-C., Treatment of a first patient with FLASH-radiotherapy. Radiotherapy and Oncology 2019, 139, 18-22.
- Bohlen, T. T.; Germond, J. F.; Bourhis, J.; Vozenin, M. C.; Ozsahin, E. M.; Bochud, F.; Bailat, C.; Moeckli, R., Normal Tissue Sparing by FLASH as a Function of Single-Fraction Dose: A Quantitative Analysis. Int J Radiat Oncol Biol Phys 2022, 114 (5), 1032-1044.
- Ruan, J. L.; Lee, C.; Wouters, S.; Tullis, I. D. C.; Verslegers, M.; Mysara, M.; Then, C. K.; Smart, S. C.; Hill, M. A.; Muschel, R. J.; Giaccia, A. J.; Vojnovic, B.; Kiltie, A. E.; Petersson, K., Irradiation at Ultra-High (FLASH) Dose Rates Reduces Acute Normal Tissue Toxicity in the Mouse Gastrointestinal System. Int J Radiat Oncol Biol Phys 2021, 111 (5), 1250-1261.
- Chen, C.; Zhang, Q.; Yu, W.; Chang, B.; Le, A. D., Oral Mucositis: An Update on Innate Immunity and New Interventional Targets. J Dent Res 2020, 99 (10), 1122-1130.
- Han, G.; Bian, L.; Li, F.; Cotrim, A.; Wang, D.; Lu, J.; Deng, Y.; Bird, G.; Sowers, A.; Mitchell, J.; Gutkind, J. S.; Zhao, R.; Raben, D.; Dijke, P.; Refaeli, Y.; Zhang, Q.; Wang, X.-J., Preventive and therapeutic effects of Smad7 on radiation-induced oral mucositis. Nature medicine 2013, 19.
- Kanarek, N.; Grivennikov, S.; Leshets, M.; Lasry, A.; Alkalay-Snir, I.; Horwitz, E.; Shaul, Y.; Stachler, M.; Voronov, E.; Apte, R.; Pagano, M.; Pikarsky, E.; Karin, M.; Ghosh, S.; Ben-Neriah, Y., Critical role for IL-1 in DNA damage-induced mucositis. Proceedings of the National Academy of Sciences of the United States of America 2014, 111.
- Hintzen, R. Q.; de Jong R Fau - Lens, S. M.; Lens Sm Fau - van Lier, R. A.; van Lier, R. A., CD27: marker and mediator of T-cell activation? Immunol Today 1994, 15 (7), 307-311.
Figure 1.
Healthy mucosa tissue slices maintain viability for several days in culture. Representative H&E staining (A) and fluorescent microscopy images of TUNEL staining (B) at day 0 (2 hours), day 1 (24 hours) and day 3 of ex vivo culture, showing DAPI (nuclei; blue) and TUNEL (apoptosis; green) channel. (C) Percentage of apoptotic cells of all Fields of View (FoVs) in the untreated tissue slices at day 0, day 1 and day 3 of culture. (n = 7 samples) (D) Representative confocal microscopy images of EdU and p63 staining at day 0, day 1 and day 3 of ex vivo culture. DAPI channel depicts cell nuclei (blue), p63 epithelial progenitor cells (green), and EdU proliferative cells (red). The fourth row shows all channels merged. (E) Percentage of proliferating cells of all FoVs in the untreated slices at day 0, day 1 and day 3 of culture (n = 7 samples). The bars represent the mean of all FoVs of all control samples (≥3 FoV per sample) per timepoint and error bars depict SEM. Kruskal-Wallis and Dunn’s multiple comparison test were used for significance. ** p < 0.01, **** p < 0.0001.
Figure 1.
Healthy mucosa tissue slices maintain viability for several days in culture. Representative H&E staining (A) and fluorescent microscopy images of TUNEL staining (B) at day 0 (2 hours), day 1 (24 hours) and day 3 of ex vivo culture, showing DAPI (nuclei; blue) and TUNEL (apoptosis; green) channel. (C) Percentage of apoptotic cells of all Fields of View (FoVs) in the untreated tissue slices at day 0, day 1 and day 3 of culture. (n = 7 samples) (D) Representative confocal microscopy images of EdU and p63 staining at day 0, day 1 and day 3 of ex vivo culture. DAPI channel depicts cell nuclei (blue), p63 epithelial progenitor cells (green), and EdU proliferative cells (red). The fourth row shows all channels merged. (E) Percentage of proliferating cells of all FoVs in the untreated slices at day 0, day 1 and day 3 of culture (n = 7 samples). The bars represent the mean of all FoVs of all control samples (≥3 FoV per sample) per timepoint and error bars depict SEM. Kruskal-Wallis and Dunn’s multiple comparison test were used for significance. ** p < 0.01, **** p < 0.0001.
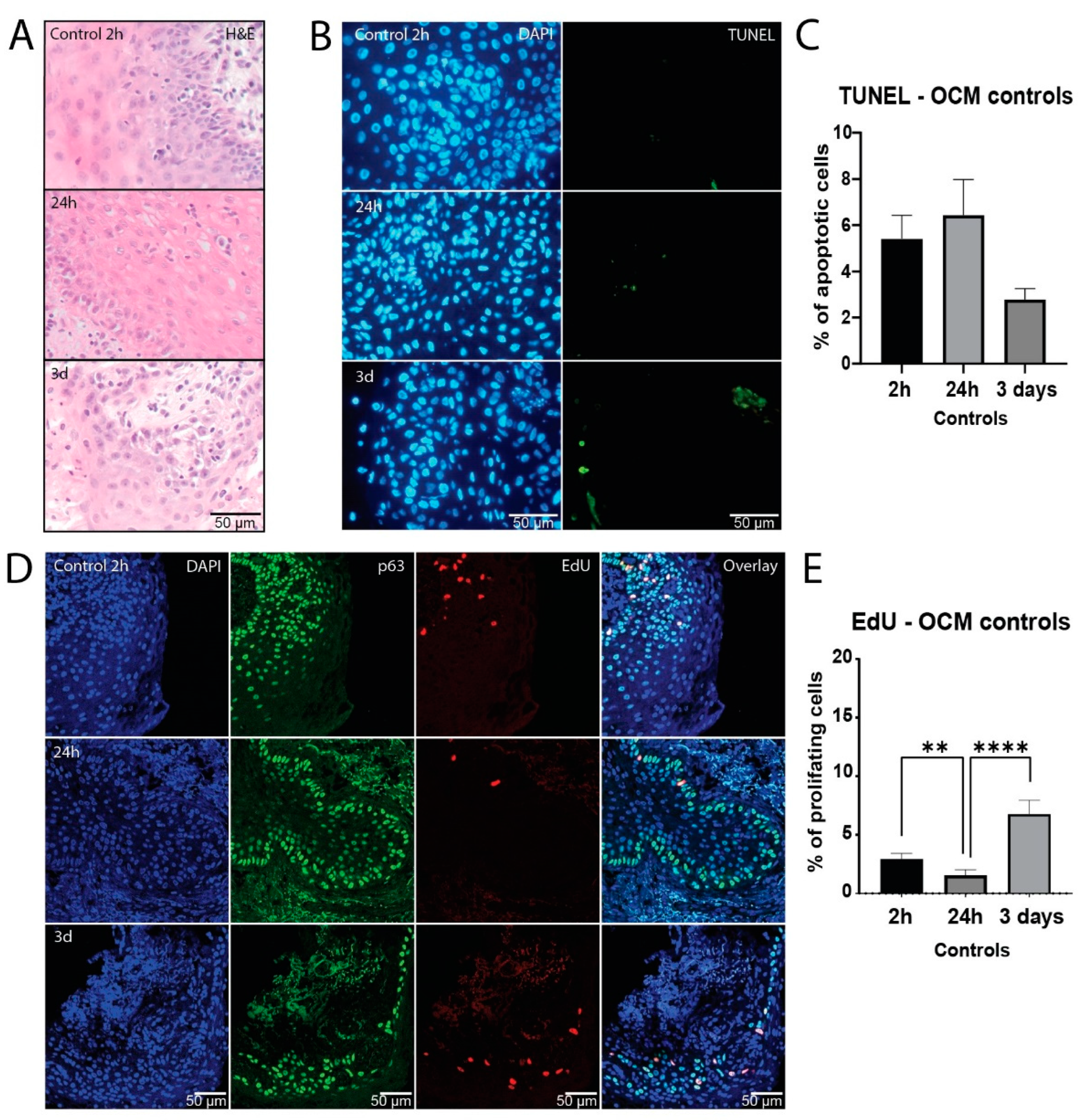
Figure 2.
Healthy mucosa displays dose-dependent response to X-ray irradiation. (A) Representative bright-field microscopy images of H&E staining of untreated, 5 and 10 Gy irradiated samples after 3 days of culture. Arrows depict degradation after irradiation. (B) Representative confocal microscopy images of EdU and p63 staining of untreated, 5 and 10 Gy irradiated samples after 3 days of culture. DAPI channel depicts cell nuclei (blue), p63 epithelial progenitor cells (green), and EdU proliferative cells (red). The fourth row shows all channels merged. (C) Percentage of proliferating cells of all FoVs per treatment condition on day 3 after irradiation (n = 7 samples). (D) Representative fluorescent microscopy images of TUNEL staining of untreated, 5 and 10 Gy irradiated samples on day 3 after irradiation, showing DAPI (nuclei; blue) and TUNEL (apoptosis; green) channel. (E) Percentage of apoptotic cells of all FoVs per treatment condition on day 3 after irradiation (n = 7 samples). All graphs represent the mean of all FoVs of all samples (≥3 FoV per sample), and error bars depict SEM. Kruskal-Wallis and Dunn’s multiple comparison test were used for significance. ** p < 0.01, *** p < 0.001.
Figure 2.
Healthy mucosa displays dose-dependent response to X-ray irradiation. (A) Representative bright-field microscopy images of H&E staining of untreated, 5 and 10 Gy irradiated samples after 3 days of culture. Arrows depict degradation after irradiation. (B) Representative confocal microscopy images of EdU and p63 staining of untreated, 5 and 10 Gy irradiated samples after 3 days of culture. DAPI channel depicts cell nuclei (blue), p63 epithelial progenitor cells (green), and EdU proliferative cells (red). The fourth row shows all channels merged. (C) Percentage of proliferating cells of all FoVs per treatment condition on day 3 after irradiation (n = 7 samples). (D) Representative fluorescent microscopy images of TUNEL staining of untreated, 5 and 10 Gy irradiated samples on day 3 after irradiation, showing DAPI (nuclei; blue) and TUNEL (apoptosis; green) channel. (E) Percentage of apoptotic cells of all FoVs per treatment condition on day 3 after irradiation (n = 7 samples). All graphs represent the mean of all FoVs of all samples (≥3 FoV per sample), and error bars depict SEM. Kruskal-Wallis and Dunn’s multiple comparison test were used for significance. ** p < 0.01, *** p < 0.001.
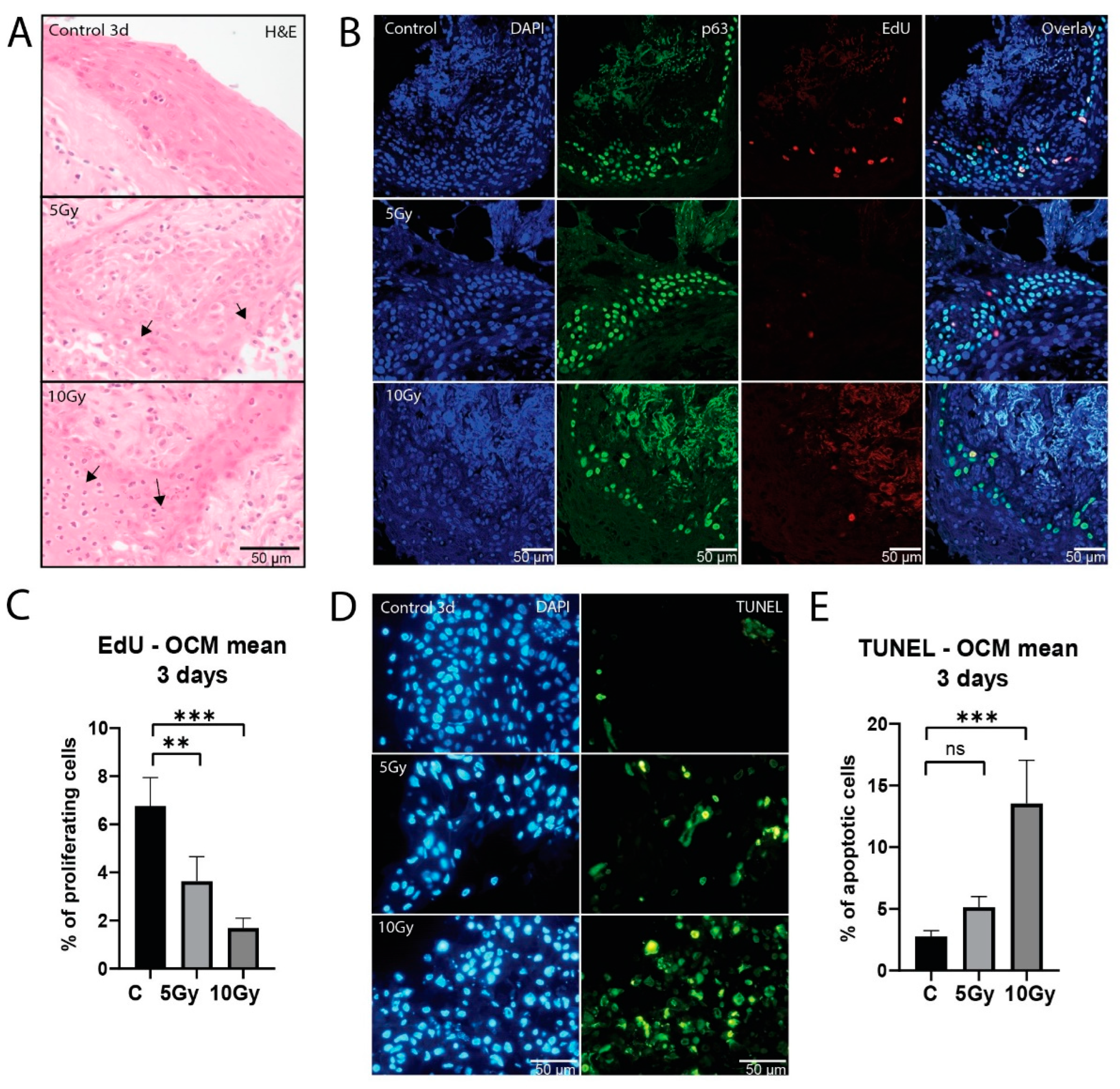
Figure 3.
Residual DNA damage foci after irradiation of normal mucosa. (A) Representative confocal images of 53BP1 DNA damage foci of control, 5 Gy and 10 Gy X-ray irradiated conditions after 24 hours of ex vivo culture. Depicted are DAPI (cell nuclei), p63 (epithelial progenitor cells) and 53BP1 foci channel. The fourth row shows segmentation of both p63 and 53BP1 channels combined. Number of 53BP1 foci per µm3 (B) in control, 5 Gy and 10 Gy X-ray irradiated conditions after 24 hours of culture (n = 6, all nuclei combined). (C) Representative confocal images of 53BP1 DNA damage foci of control, 5 Gy and 10 Gy X-ray irradiated conditions after three days of ex vivo culture. Depicted are DAPI (cell nuclei), p63 (epithelial progenitor cells) and 53BP1 foci channel. The fourth row shows segmentation of both p63 and 53BP1 channels combined. 53BP1 foci per µm3 in 3-days samples (n = 3; D) and foci size (E) in control, 5 Gy and 10 Gy X-ray irradiated conditions. All images were cropped for foci to be visible in print format. All graphs represent the median of all nuclei analyzed (≥100 nuclei per sample for foci number, ≥75 foci per sample for foci size) and 1st and 3rd quantile (dashed lines). Kruskal-Wallis and Dunn’s multiple comparison test were used for significance. ** p < 0.01, **** p < 0.0001.
Figure 3.
Residual DNA damage foci after irradiation of normal mucosa. (A) Representative confocal images of 53BP1 DNA damage foci of control, 5 Gy and 10 Gy X-ray irradiated conditions after 24 hours of ex vivo culture. Depicted are DAPI (cell nuclei), p63 (epithelial progenitor cells) and 53BP1 foci channel. The fourth row shows segmentation of both p63 and 53BP1 channels combined. Number of 53BP1 foci per µm3 (B) in control, 5 Gy and 10 Gy X-ray irradiated conditions after 24 hours of culture (n = 6, all nuclei combined). (C) Representative confocal images of 53BP1 DNA damage foci of control, 5 Gy and 10 Gy X-ray irradiated conditions after three days of ex vivo culture. Depicted are DAPI (cell nuclei), p63 (epithelial progenitor cells) and 53BP1 foci channel. The fourth row shows segmentation of both p63 and 53BP1 channels combined. 53BP1 foci per µm3 in 3-days samples (n = 3; D) and foci size (E) in control, 5 Gy and 10 Gy X-ray irradiated conditions. All images were cropped for foci to be visible in print format. All graphs represent the median of all nuclei analyzed (≥100 nuclei per sample for foci number, ≥75 foci per sample for foci size) and 1st and 3rd quantile (dashed lines). Kruskal-Wallis and Dunn’s multiple comparison test were used for significance. ** p < 0.01, **** p < 0.0001.
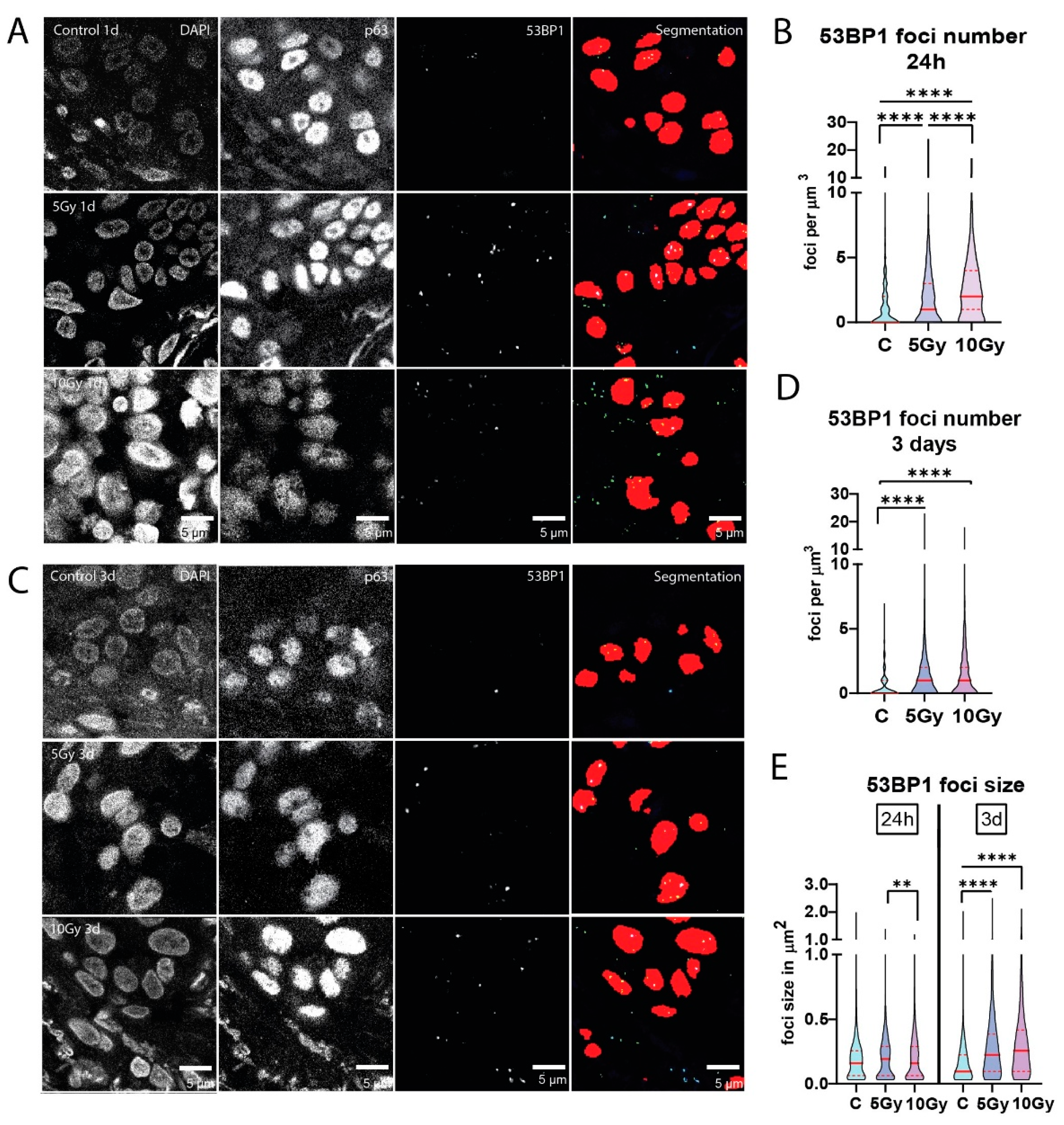
Figure 4.
Proton irradiation of mucosa tissue slices. (A) Schematic image of tissue slice setup for proton irradiation. A physical dose of 5 Gy was given with both X-ray and protons (not RBE weighted). (B) Representative confocal images of 53BP1 DNA damage foci in control, X-ray and proton irradiated conditions after 24 hours of ex vivo culture. Depicted are DAPI (cell nuclei), p63 (epithelial progenitor cells) and 53BP1 foci channel. The fourth row shows segmentation of both p63 and 53BP1 channels combined. (C) A zoom-in view of the 53BP1 channel and segmentation is shown. (D) Number of 53BP1 foci per µm3 and (E) foci size in control, 5 Gy X-ray and 5 Gy proton irradiated conditions after 24 hours of ex vivo culture. C = control, X = X-ray, and P = proton irradiation. All images were cropped for foci to be visible in print format. All graphs represent the median of all nuclei analyzed (≥50 nuclei per sample, ≥75 foci per sample for foci size) and 1st and 3rd quantile (dashed lines). Kruskal-Wallis and Dunn’s multiple comparison test was used for significance. **** p < 0.0001.
Figure 4.
Proton irradiation of mucosa tissue slices. (A) Schematic image of tissue slice setup for proton irradiation. A physical dose of 5 Gy was given with both X-ray and protons (not RBE weighted). (B) Representative confocal images of 53BP1 DNA damage foci in control, X-ray and proton irradiated conditions after 24 hours of ex vivo culture. Depicted are DAPI (cell nuclei), p63 (epithelial progenitor cells) and 53BP1 foci channel. The fourth row shows segmentation of both p63 and 53BP1 channels combined. (C) A zoom-in view of the 53BP1 channel and segmentation is shown. (D) Number of 53BP1 foci per µm3 and (E) foci size in control, 5 Gy X-ray and 5 Gy proton irradiated conditions after 24 hours of ex vivo culture. C = control, X = X-ray, and P = proton irradiation. All images were cropped for foci to be visible in print format. All graphs represent the median of all nuclei analyzed (≥50 nuclei per sample, ≥75 foci per sample for foci size) and 1st and 3rd quantile (dashed lines). Kruskal-Wallis and Dunn’s multiple comparison test was used for significance. **** p < 0.0001.

Figure 5.
Immune cells express inflammation inducing factors upon irradiation ex vivo. (A) Representative bright field images of CD45 staining. (B) Representative bright field images of CD27 staining. Area measurement of CD27 signal per nuclear area of all FoVs per condition after 24 hours (C; n = 6 samples) and 3 days (D; n = 6 samples) of ex vivo culture. (E) Representative bright field images of IL-1β staining. Area measurement of IL-1β signal per nuclear area of all FoVs per condition after 24 hours (F; n = of 6 samples) and 3 days (G; n = 7 samples) of ex vivo culture. All graphs represent mean + SEM. Kruskal-Wallis and Dunn’s multiple comparison test was used for significance. * p < 0.05.
Figure 5.
Immune cells express inflammation inducing factors upon irradiation ex vivo. (A) Representative bright field images of CD45 staining. (B) Representative bright field images of CD27 staining. Area measurement of CD27 signal per nuclear area of all FoVs per condition after 24 hours (C; n = 6 samples) and 3 days (D; n = 6 samples) of ex vivo culture. (E) Representative bright field images of IL-1β staining. Area measurement of IL-1β signal per nuclear area of all FoVs per condition after 24 hours (F; n = of 6 samples) and 3 days (G; n = 7 samples) of ex vivo culture. All graphs represent mean + SEM. Kruskal-Wallis and Dunn’s multiple comparison test was used for significance. * p < 0.05.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).