1. Introduction
Missing data is one of the critical problems encountered in scientific research. It is a typical problem that affects the availability of datasets necessary for drawing statistical inferences. Furthermore, several statistical analysis techniques require complete datasets. Therefore, researchers often struggle with imputing missing data or dropping the cases with missing values. However, dropping incomplete cases is inefficient and often unacceptable in many cases because it discards original information that could be beneficial in drawing more reliable conclusions [
1]. As a result, imputing is the only viable option when one does not wish to remove original cases from the dataset to conduct analysis.
The missing data problem has been studied in the traditional statistics literature [
2]. There exist some well-known methods for handling missing data. The easiest of them is the Case Wise Deletion method (CWD) which is based on merely omitting the missing sample points. [
2] reported problems with the use of CWD, especially for linear regression problems. Two other problems identified are bias and low efficiency. The alternative approach to CWD is the Mean Substitution (MS) method. The procedure involves replacing the missing points with the mean of available points. The technique is simple yet efficient with good results on most occasions.
[
3] provided a more refined approach called Multiple Imputation (MI) approaches. The method involves imputing missing values based on conditional non-missing values. The entire estimation process is repeated
t times (usually 3-5 times). Each time the imputation is done, the desired analysis is performed on the complete imputed datasets. The average parameter estimates and their standard errors are estimated using the
t repetitions.
Another well-established method to handle missing data, in comparison with MI, is the Expectation-Maximization algorithm introduced by [
4]. The EM algorithm, much like MI, comprises multiple iterative steps with an expectation step followed by a maximization phase. Specifically, during the expectation step, the algorithm computes a latent variable likelihood that assumes the missing data is present and incorporates them in the calculation. In the maximization step, in contrast, the EM algorithm maximizes the expected likelihood and computes new estimates of the covariance matrix and mean vector based on it. Through its foundation in probability theory, the EM algorithm has firmly established itself as one of the most effective imputation approaches due to its adopted methodology. Because it generates only one plausible value computation for missing data, it is often called a single imputation approach yet remains a heavily utilized methodology.
A novel robust methodology referred to as the Multiple Imputation by Chained Equations (MICE) or Full Conditional Specification (FCS) method has gained popularity in the recent past where the technique is used to address the missing data problems [
5]. Several researchers have suggested that MICE is a powerful tool used in imputing quantitative variables with missing values in a multivariate data setting, and the method performs better compared to the ad-hoc and single imputation methods [
6]. Despite its robustness and applications, other alternatives exist that incorporate the handling of missing data in their algorithms. For instance, the proximity imputation, highly adopted in the random forest algorithms, is an imputation approach that starts by imputing the missing values to fit a random forest, and then the initial imputed missing values are subsequently updated by the proximity of the data [
7]. Consequently, such processes achieve the desired results over several iterations.
The procedure explained in [
8], takes advantage of the so-called proximity matrix, which measures the proximity between pairs of observations in the forest, to estimate missing values. Data imputation based on random forests has further been explored by [
9], and extended to unsupervised classification by [
10]. [
11] also proposed an adaptive imputation approach for Random Survival forest. [
11] established the supremacy over the traditional proximity imputation in survival analysis studies.
Another comparison of some selected classification methods (k-nearest neighbours (kNN), C4.5 and support vector machines (SVM)) on data with inherent missing values with MICE algorithm by [
12] showed the supremacy of MICE over other imputation methods. Most of the datasets using C
do not benefit from the imputation technique and lead to an increase in the misclassification error rate. [
13] conducted a comprehensive simulation study on the application of the
k-nearest neighbours (kNN) imputation approach to Random Forest.
An approach proposed within the Bayesian paradigm is BARTm, introduced by [
14]. BARTm extends the Bayesian Additive Regression Trees (BART) by incorporating the statistical missingness of some of the covariates and, therefore, making BART less sensitive to missing data. Specifically, while traditional statistical approaches would require imputation or censoring of the missing data, BARTm adjusts the splitting scheme of decision trees to allow missing values to be allocated to nodes along with observations and then maximize the overall likelihood of the tree. A critical advantage of BARTm is that it treats missingness as a “valid” splitting criterion, that is, it allows the models to capture the “signal” in that data, which is especially useful when missing data is not missing at random, but there is a process influencing the response function which is related to the cause of missing regarding certain predictors. BARTm can work for linearity or any other transformation of the predictor, and it can model both continuous and nominal data. It works for selection and pattern-mixture models and does not have any requirements for the nature of missing data. By imputing missing data directly in the construction of the model, BARTm can automatically predict data with missing entries. Moreover, as the model produces Bayesian credible intervals, the credible intervals automatically account for the increased uncertainty due to imputation. Importantly, as [
14] note, the approach is computationally inexpensive, permitting automatic prediction on future points with missing entries.
Most methods for dealing with missing data discussed in the previous paragraphs were originally developed for low-to-moderate dimensional data. These methods are likely to fail in high dimensions and high-scale circumstances, such as microarray analysis, proteomics, neuroimaging, and many other high-throughput applications. It is recommended to impute all variables in multiple imputations to obtain unbiased correlation estimates [
15]. However, this practice is not ideal in high-dimensional cases, where the number of variables is significantly larger than the number of samples. Imputing all variables could lead to overparameterization in the model, which further results in non-convexity optimization procedures such as maximum likelihood-based methods, as well as Gaussian and all other similar models like the EM algorithm [
16]. Furthermore, the majority of the existing missing data methods were developed for continuous data types, such as gene expression data [
17,
18]. Therefore, they ignore the complexity of distinct interactions and nonlinearities among different variables. Standard MI procedures cannot adequately handle interaction effects and produce biased parameter estimates in such situations [
19]. Recently developed methods, such as FCS of covariates, are more promising [
20], but it is relatively challenging to implement these methods in practice, particularly when one is interested in complex interactions.
In this paper, we propose a novel approach that integrates the standalone MICE method with Bayesian random forest (BRF) techniques originally proposed by [
21,
22] into a cohesive framework. This hybridization is aimed at enhancing the imputation accuracy and predictive power of the BRF models. Our method extends the traditional BRF algorithms to handle both categorical and continuous response variables more effectively. The key innovation lies in preprocessing the data using MICE imputation before feeding it into the BRF model. By doing so, we capitalize on the strengths of both methodologies: MICE’s ability to handle missing data efficiently and BRF’s robustness in capturing complex relationships within the data. This integration offers a comprehensive solution for addressing missing data issues while leveraging the predictive capabilities of BRF.
3. Bayesian Random Forest for Missing Data
Suppose we let
,
represent
incomplete dataset with
assuming continuous or categorical values and
be the vector of
p covariates. Here
is the unequal sample size of each
j covariate in the covariate set
p. If we denote the complete sample size by
n, then
is the number of missing entries in each covariate
j and
is the estimate of the missing probability
. The first step of the proposed procedure is to apply the MICE imputation procedure in equation (
1) to achieve a complete dataset with dimension
. Subsequently, according to [
21,
22] we can define the sum of trees model
where
and
K is the total number of trees that make the forest. In the tree notation, we can rewrite (
2) as
where
is an estimate of
y in region
,
is a single regression tree,
is the number of branches of each tree,
is the random noise that occurs in estimating
and its assumed to be independent and identically Gaussian distributed with mean zero and constant variance
over all trees. If the response is continuous, we proceed with the two-stage BRF [
22] to estimate the target
y after imputation.
-
Variable splitting:
Bayesian Weighted Splitting
where
is the Bayesian weighted sum of squares splitting scores for trees,
is the probability that a specific covariate
is more relevant to
y than all other
.
is the cumulative density function of the
statistic of the effect of each variable
from a fitted Bootstrap Bayesian simple linear regression model [
29] defined below as
Correspondingly, the estimate of the statistic
for variable
is
where
and
are the posterior mean and standard deviation of bootstrap Bayesian distribution of the fitted model (
6). The next step involves estimating the target
y by averaging over all the trees
that makes the forest.
Parameter estimation:
Bootstrap Bayesian Estimation
where
is the prior predictive density of each observation
i in each regression tree
k which is distributed normal-invese-gamma (NIG) with prior parameters as defined in [
22]. On the other hand, if the response
y is categorical, after appropriate imputation using MICE we also proceed with the two-stage approach presented in [
21].
For the categorical response
y, instead of the averaging as in regression, the forest model is majority voting presented below as
where
is the proportion of target response for class
c in region
,
is
single classification tree. The process of estimating
proceed as
-
Variable splitting:
Bayesian Weighted Splitting
where
is the Bayesian weighted Gini impurity [
21] splitting scores for trees,
is the variable
relevant probability as defined above.
is the cumulative density function of the
statistic of each variable
from a fitted Bootstrap Bayesian ANOVA model [
30] given below as
Correspondingly, the estimate of the statistic
for variable
is
where
, are the posterior mean of bootstrap Bayesian distribution of the fitted model (
14),
and
are sums of squares regression and sum squares error respectively. The next step involves estimating the proportion of target response for class
c,
by majority voting over all the trees
that makes the forest.
Parameter estimation:
Bootstrap Bayesian Estimation
where
is the prior predictive density of each observation
i in each classification tree
k which is distributed Dirichlet-Multinomial (DM) with prior parameters as defined in [
21].
4. Simulation, Results and Discussion
The three missing mechanisms (MCAR, MAR and MNAR) were simulated for both regression and classification cases. For the regression case, we adopted the simulation strategies of [
22] for simulating the high-dimensional Friedman nonlinear Gaussian response model and [
7] for different missing mechanisms injection. Specifically, we set
and
with the nonlinear model
The model is high-dimensional in that only variables
are relevant and the remaining
are noise. For MCAR, the relevant independent variables
are missing randomly with Bernoulli probabilities
such that a case is missing if the Bernoulli random vector returned is 1. For MAR, variables
are missing if the probability of missing defined over the Probit link equation
is
. Here,
and
are defined such that
. Similarly, for MNAR the relevant variables
are missing if the probability of missing defined over the Probit link equation
is
. The proportion of missingness
was adapted from the studies of [
7,
31,
32,
33] that found up to
,
,
missing entries in simulation and real-life datasets used. Two other methods (RF [
7] and BART2: Bartmachine [
14]) were compared with BRF using the root mean square error
and average root mean square
as performance measures over 10-fold cross-validations. All simulations and analyses were carried out in the R package version (4.3.1).
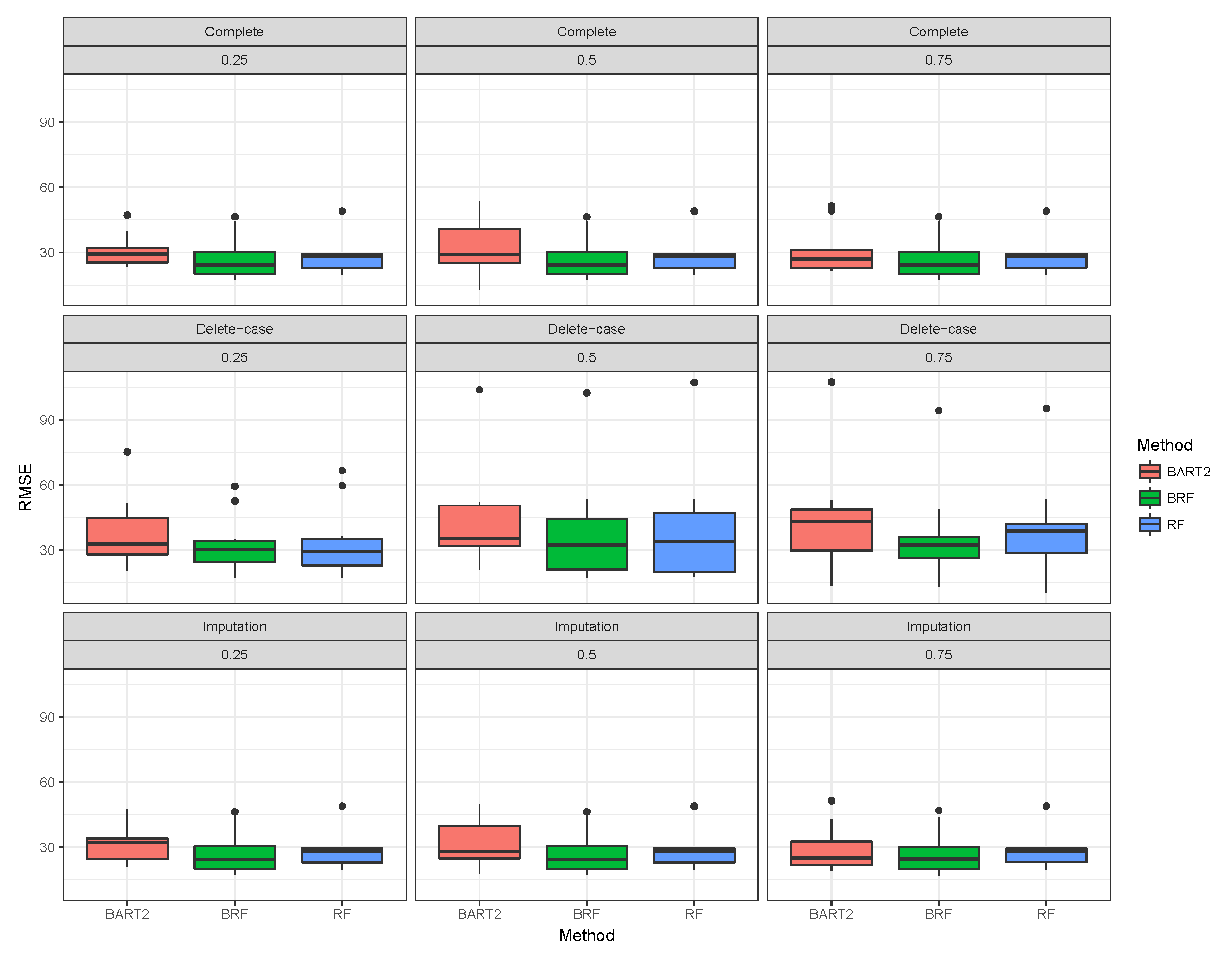
Table 1 presents the average test data RMSE of the three missing data mechanisms (MCAR, MAR and MNAR) with the proportion of missing observation
and
for Gaussian response. The first three rows gave the results when there was no missing observation, and it stands as the threshold for comparing the performances of the methods. A method is termed as robust if there is no significant increase in RMSE when missing observations are omitted or imputed. The RMSE results when there were no missing cases is constant as expected for BRF and likewise RF. The RMSE results for BART2 exhibit little changes at different simulation timestamps across the three missing mechanisms which arises as a result of MCMC simulation involved in the estimation technique of BART2. The second compartment of the table shows the results when the missing data have been imputed using missing data strategies by the various methods. Specifically, proximity imputation was used for RF while MICE was used for BRF and BARTm for BART2. For MCAR with imputed missing observations, BRF maintains the same value of RMSE as observed when there were no missing cases for the proportion of missingness
and
. A slight increase was observed when the proportion of missingness approached 0.75. A similar pattern was observed for RF except for larger RMSE when compared with BRF. The unstable behaviour of BART2 was also observed when the data were imputed using the BARTm strategy. On average, BRF maintains the lowest RMSE for MCAR at various levels of missingness. Similar behaviours were found for MAR and MNAR at different levels of missingness. The detrimental effect of deleting the missing entries before estimation can be observed in the third compartment of the table. The RMSE of the three methods significantly deviates from the results when there are no missing entries. Although, on average the effect is minimal on BRF when compared to RF and BART2. Therefore, for high-dimensional data with missing entries up to 75% arising from different missing mechanisms, BRF is the best among the three methods considered here.
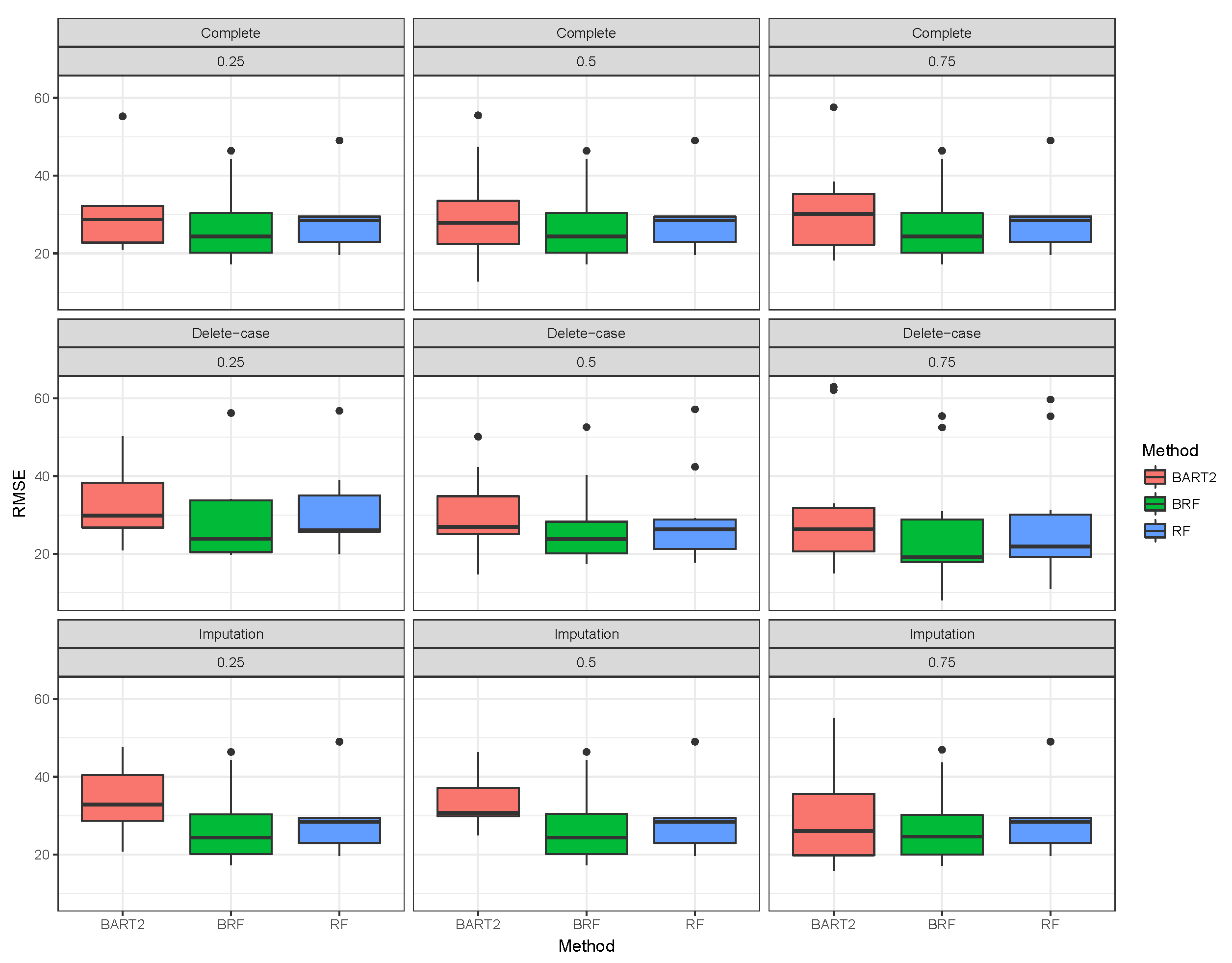
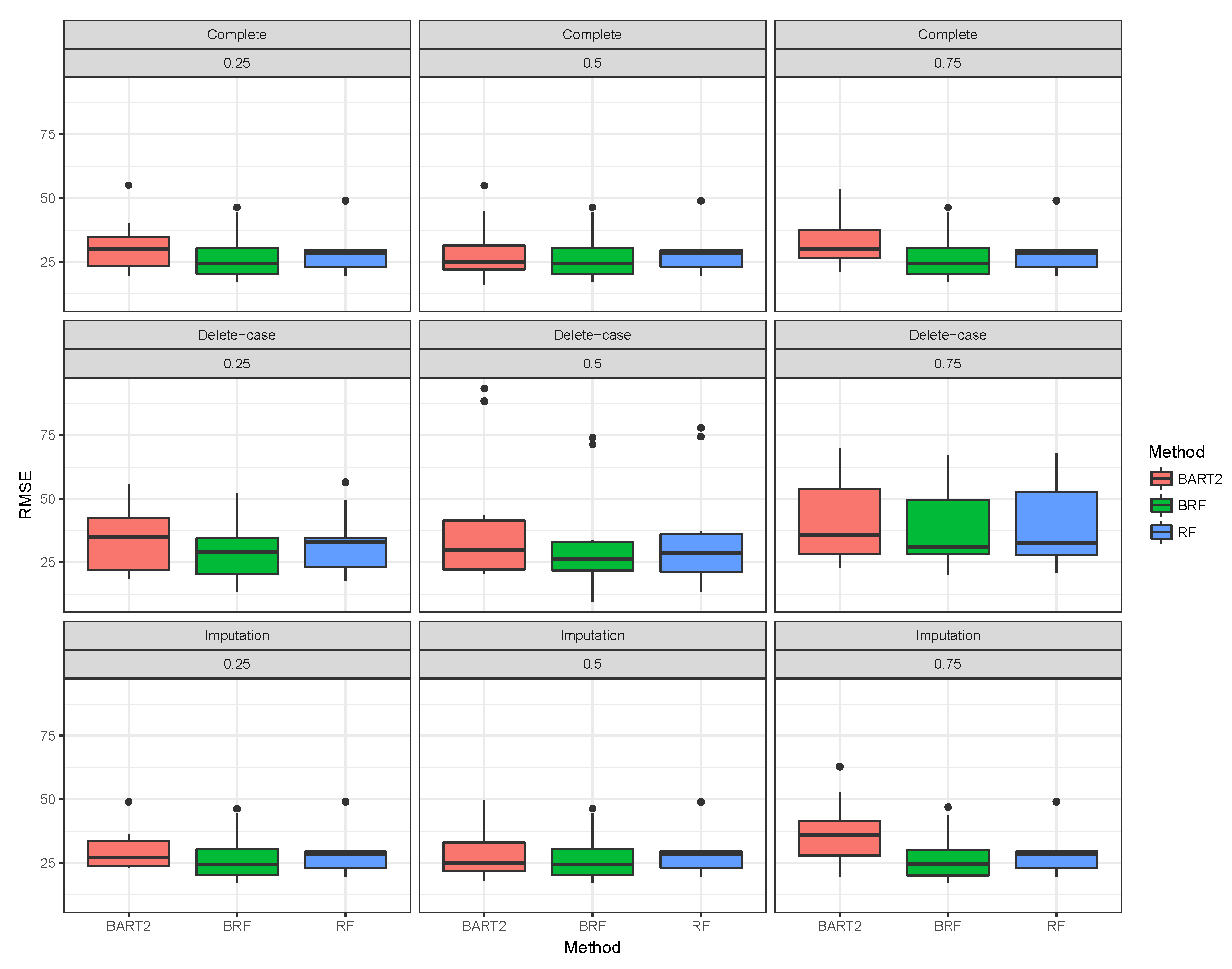
Figure 1,
Figure 2,
Figure 3 shows the visual behaviour over the folds. The median RMSE in
Figure 1–3 confirms that BRF with the MICE imputation technique is the best among the three methods for analysing high-dimensional data with missing data.
For the classification case, we adopted the simulation strategies of [
21] for simulating the high-dimensional classification model with data dimension as in the regression case. In addition, we also followed the approach of [
7] as in the regression case for different missing mechanisms injection. Two other methods (RF [
7] and BART2: Bartmachine [
14]) were compared with BRF using the misclassification error rate
and average misclassification error rate
as performance measures over 10-fold cross-validations. The MER and AMER were computed using a
confusion matrix [
34]
A given by;
where row elements of matrix
A represent the true classes and the column elements represent the predicted classes.
are the diagonal elements and they are interpreted as the number of true predicted classes and the off-diagonal elements
are the false predicted classes. Thus, the accuracy
of a method for predicting the classes of test samples correctly is defined as;
and correspondingly the
and
are defined as;
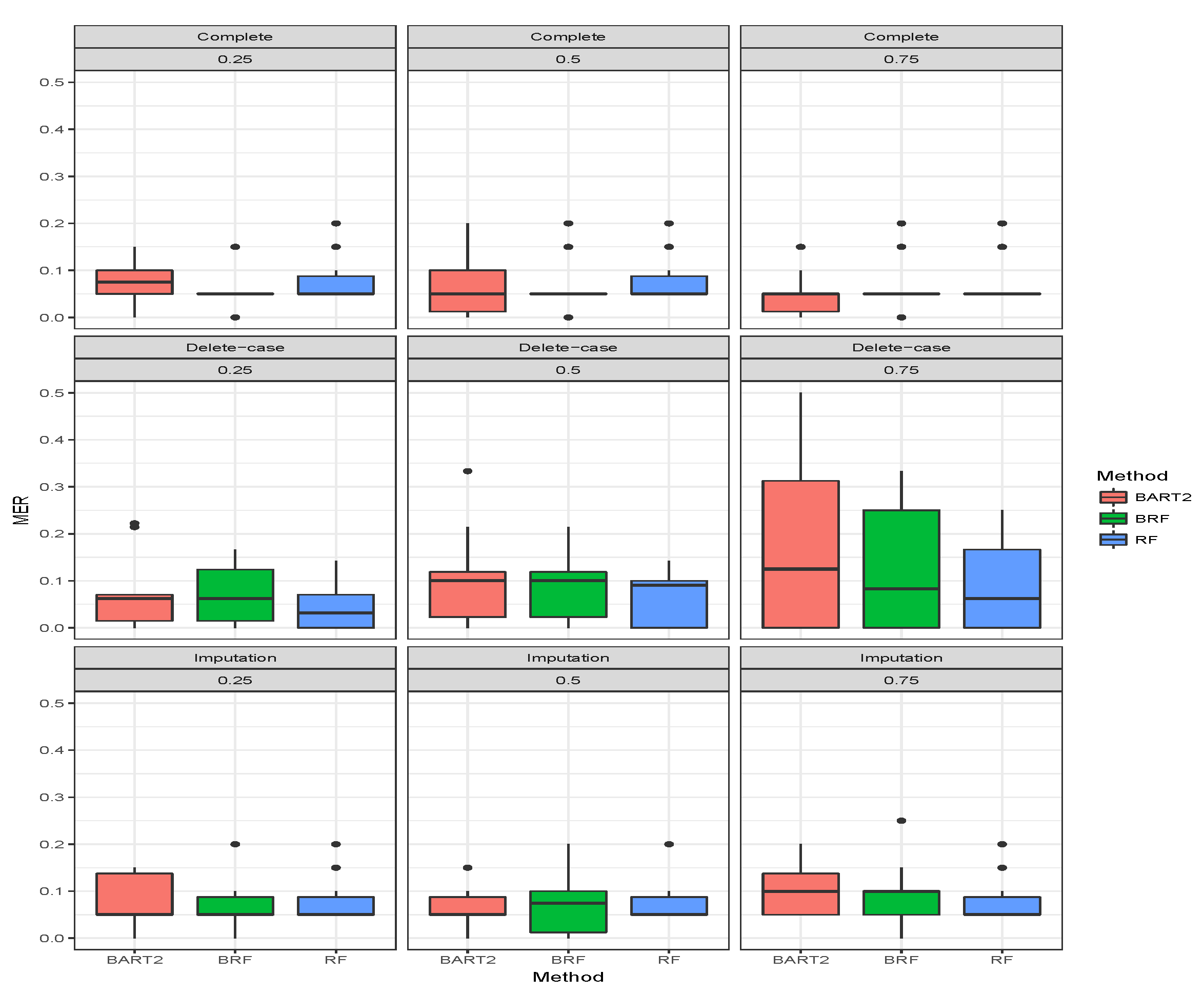
Table 2 presents the average MER of the three missing data mechanisms (MCAR, MAR and MNAR) with the proportion of missing observation
and
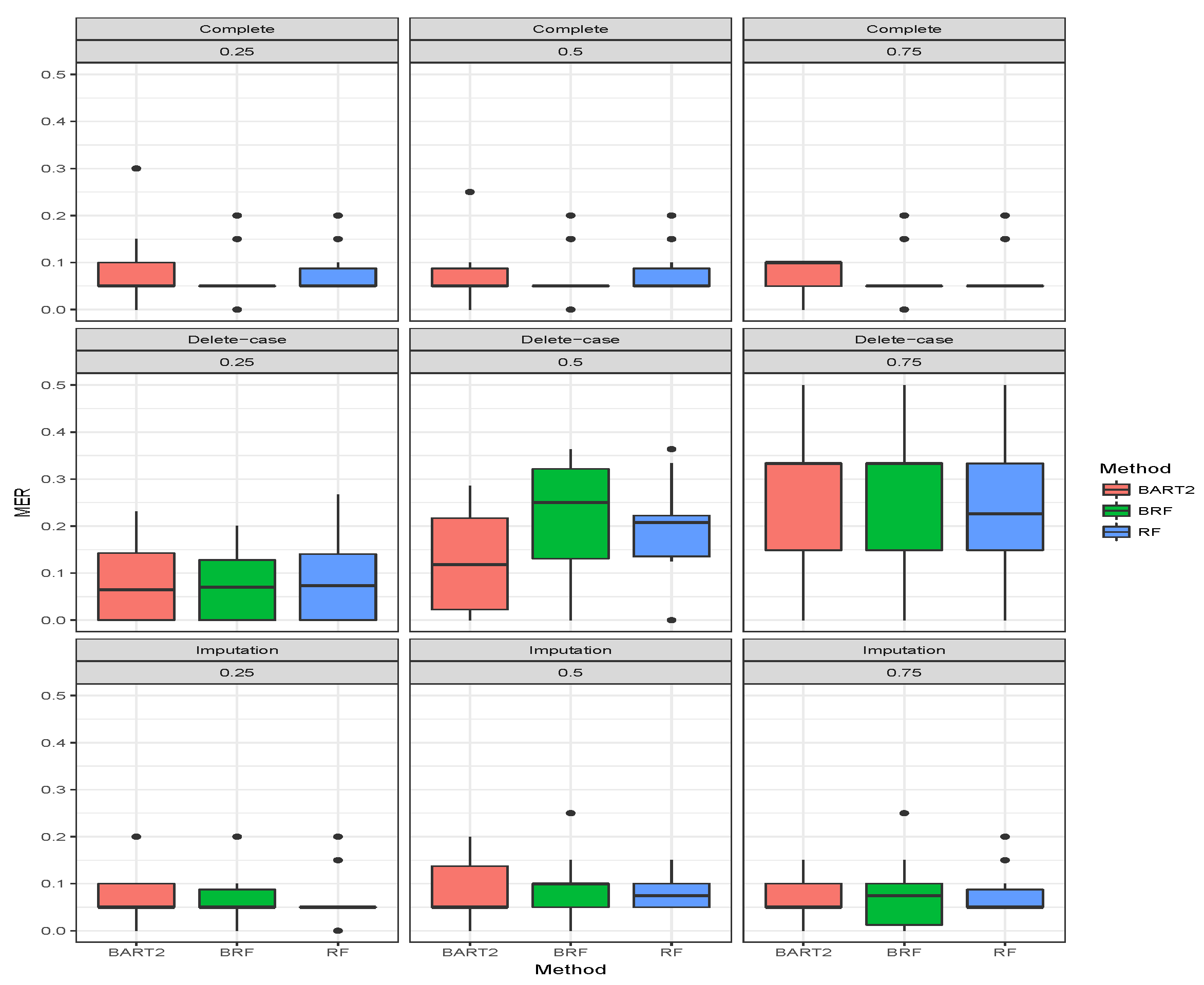
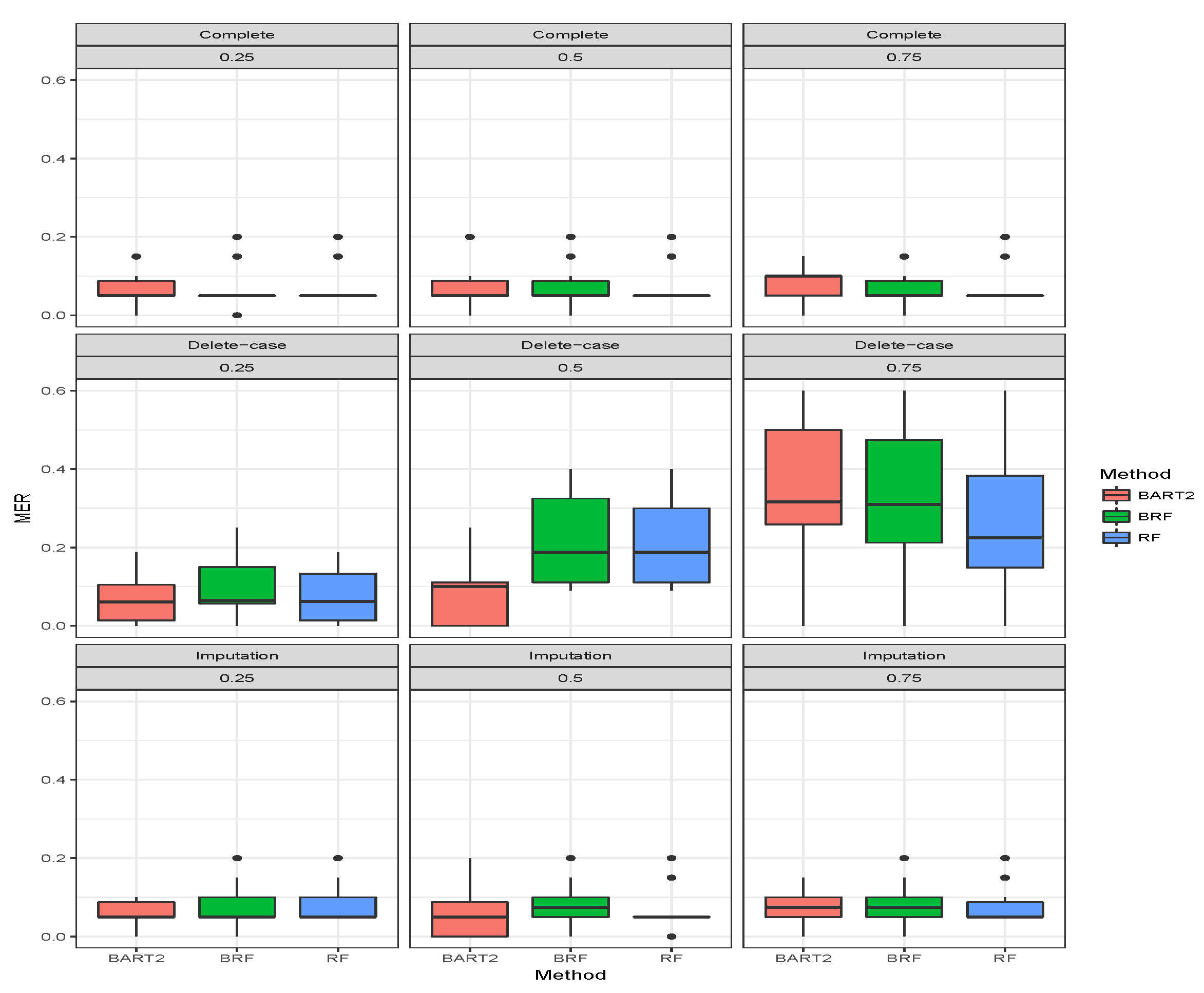
for the binary categorical response. The first three rows gave the results when there was no missing observation, and it stands as the threshold for comparing the performance of the methods. A method is termed as robust if there is no significant increase in MER when missing observations are omitted or imputed. The MER results when there were no missing cases are constant as expected for BRF and likewise RF. The MER results for BART2 exhibit little changes at different simulation timestamps across the three missing mechanisms which arises as a result of MCMC simulation involved in the estimation technique of BART2.
The second compartment of Table 2 shows the results when the missing data have been imputed using missing data strategies for the methods. For MCAR with imputed missing observations, BRF MER increases with an increase in the proportion of missing observations and the values differ from the case when there were no missing values. For RF the performance when imputed remains the same as the case when there were no missing values. The unstable behaviour of BART2 was also observed when the data were imputed using the BARTm strategy. On average, RF maintains the lowest MER for MCAR at and proportion of missingness. Similar behaviours were found for MAR and MNAR at different levels of missingness. The detrimental effect of deleting the missing entries before estimation can be observed in the third compartment of the table. The MER of the three methods significantly deviates from the results when there are no missing entries. The effect is on average minimal on RF when compared to BRF and BART2. Therefore, for high-dimensional data with missing entries up to arising from different missing mechanisms, RF is the best among the three methods considered here.
Figure 4,
Figure 5,
Figure 6 show the visual behaviour over the folds. The median MER in
Figure 4,
Figure 5,
Figure 6 confirm that BRF with the MICE imputation technique is better than BART2 for analyzing missing data while RF is the best among the three methods.
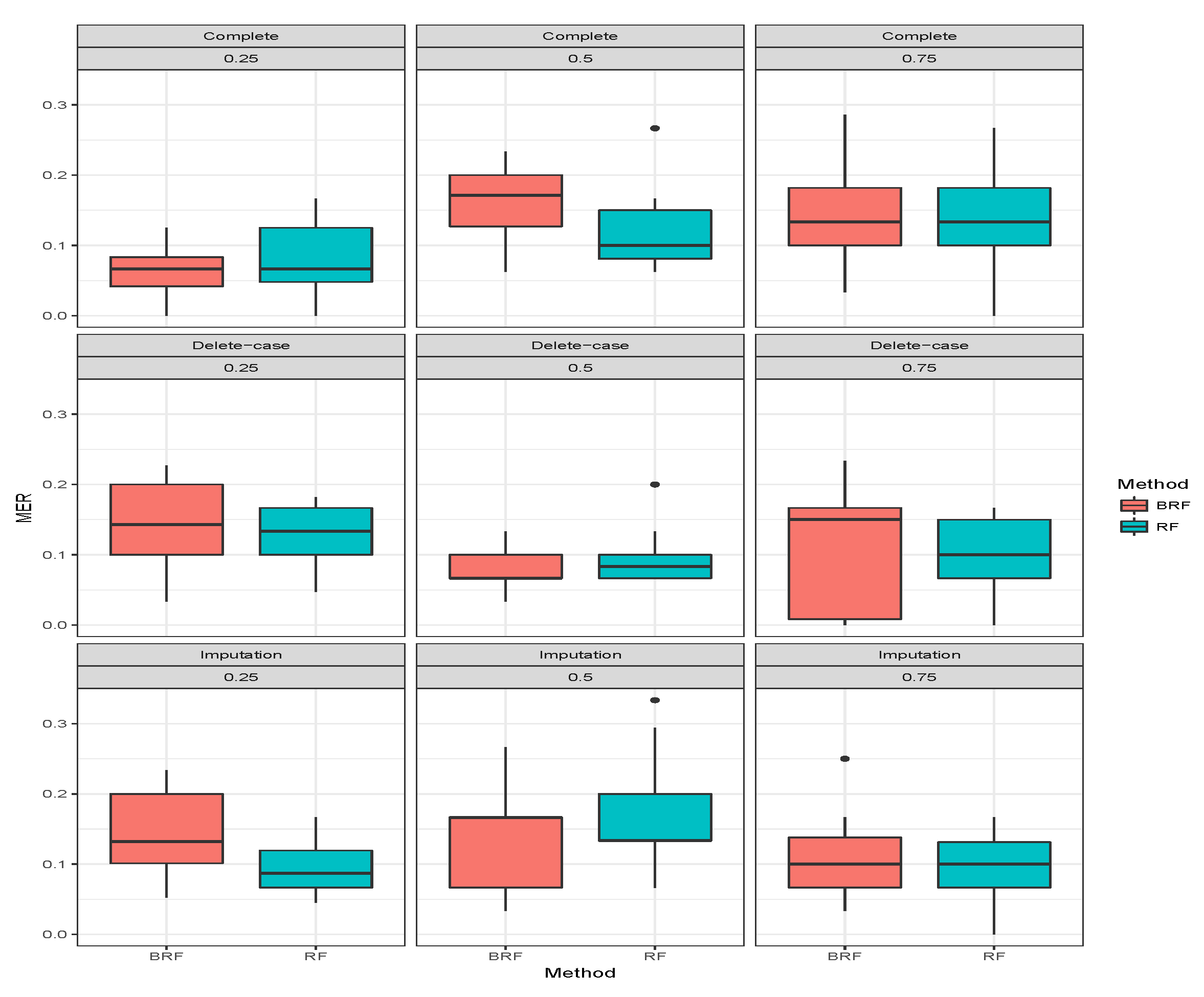
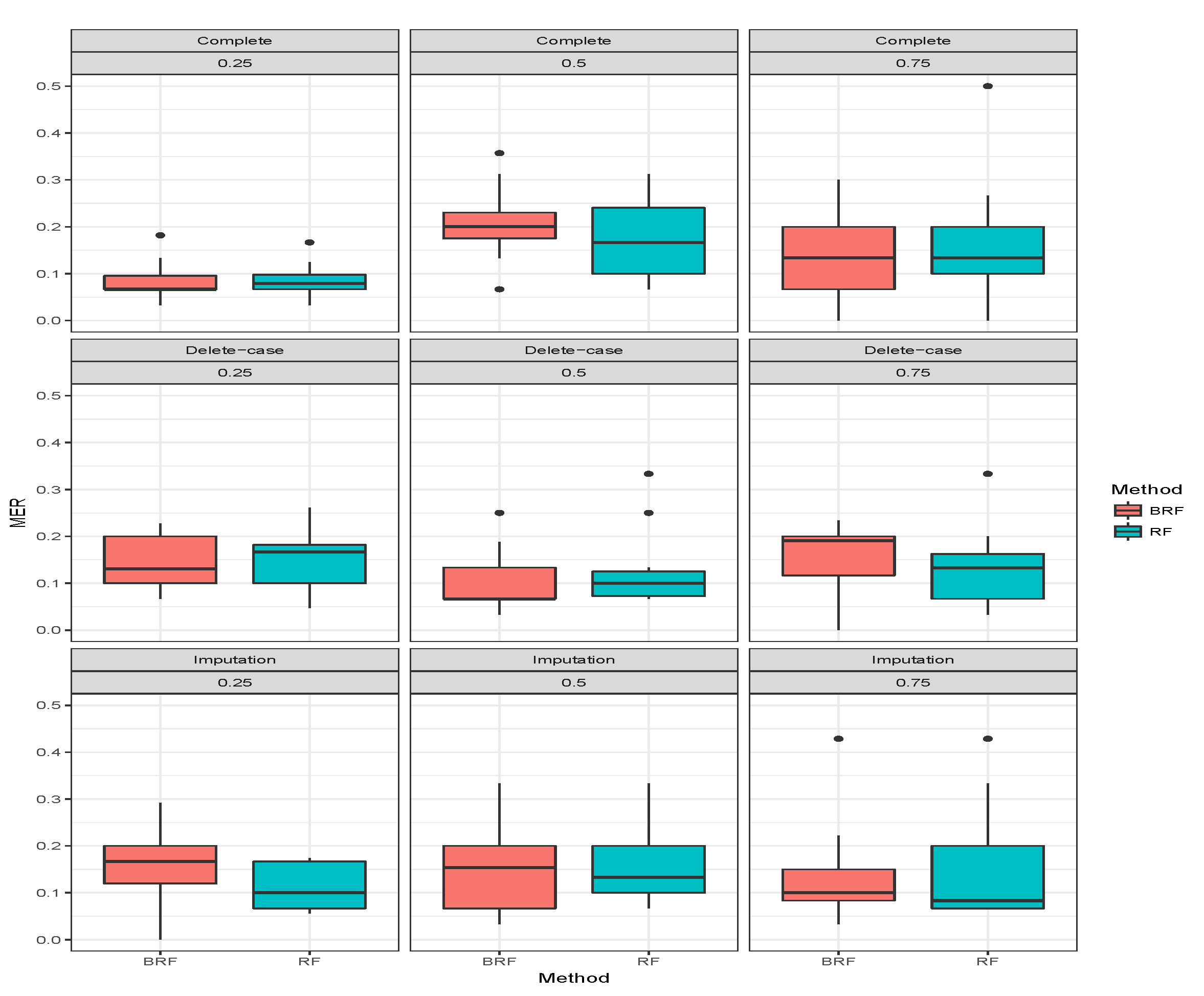
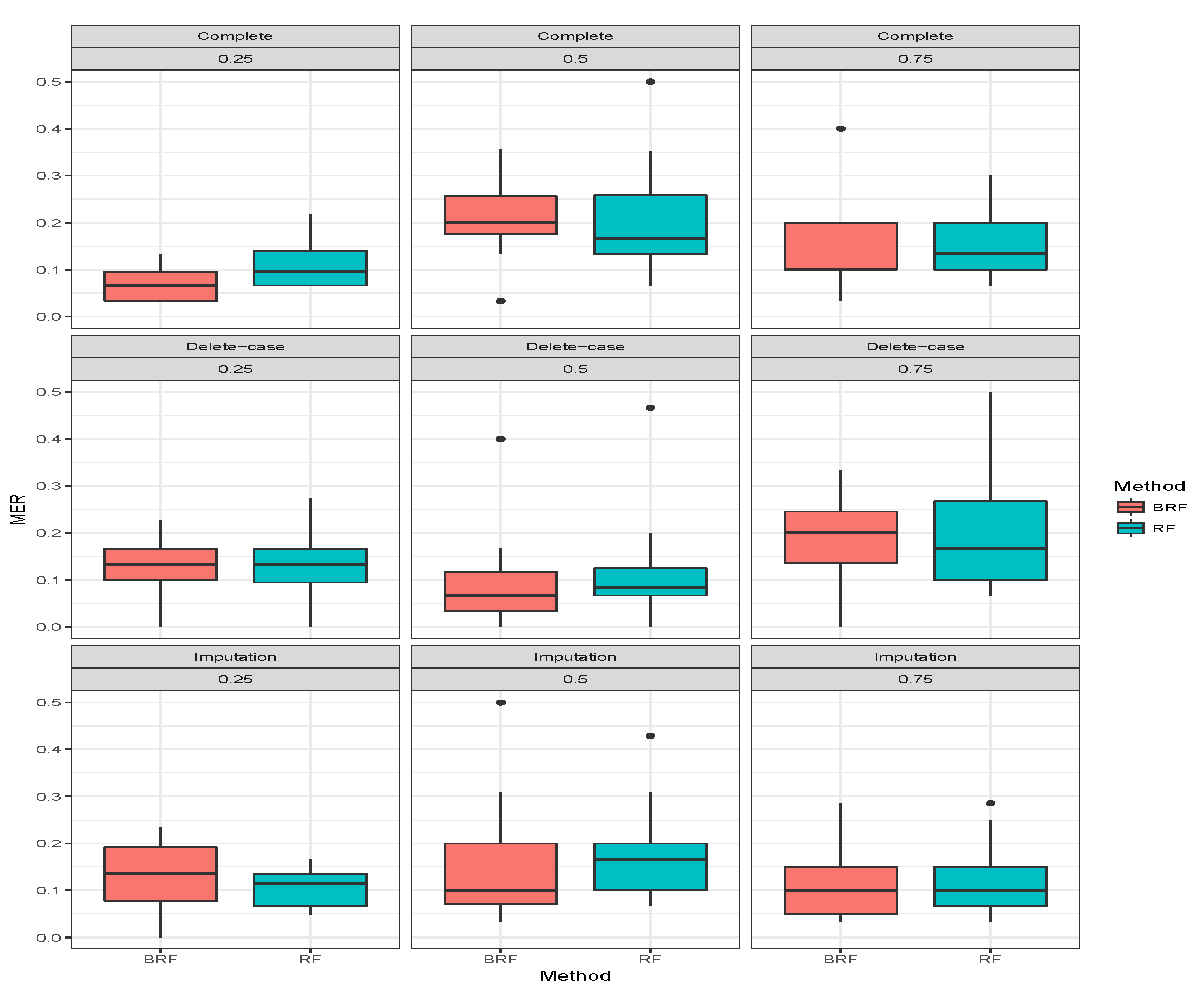
Table 3 presents the average MER of the three missing data mechanisms (MCAR, MAR and MNAR) with the proportion of missing observations 0.25, 0.5 and 0.75 for multiclass categorical response. The methods compared here are RF and BRF as BART2 has not been implemented for multi-class. Again, the first three rows show when there was no missing observation, and it stands as the threshold for comparing the performances of the methods. The second compartment of the table shows the results when the missing data have been imputed. For MCAR with imputed missing observations, BRF MER increases with an increase in the proportion of missing observations and the values differ from the case when there were no missing values. The MER of RF also increases with an increase in the proportion of missing but the performance is better than BRF at 0.5 and 0.75. Similar behaviours were found for MAR and MNAR at different levels of missingness. The detrimental effect of deleting the missing entries before estimation can be observed in the third compartment of the table. The MER of the two methods significantly deviates from the results when there are no missing entries. Overall, on average the effect of missing values is minimal on BRF when compared to RF.
Figure 7,
Figure 8,
Figure 9 show the visual behaviour over the folds.
Author Contributions
Conceptualization, O.R.O., A.R.R.A.; methodology, O.R.O.; software, O.R.O.; validation, O.R.O.; formal analysis, O.R.O.; investigation, O.R.O., A.R.R.A.; resources, A.R.R.A.; data curation, O.R.O.; writing—original draft preparation, O.R.O.; writing—review and editing, O.R.O., A.R.R.A.; visualization, O.R.O; supervision, O.R.O.; project administration, O.R.O. All authors have read and agreed to the published version of the manuscript.