1. Introduction
Viral diseases in viticulture lead to serious reductions in yield quantity and quality. With the development of high-throughput sequencing technology, new viruses infecting grapevines have been identified. To date, more than 80 different viruses from 17 families and 34 genera have been identified in grapevines [
1,
2]. Grapevine geminivirus A (GGVA) was first reported in 2017 using high-throughput sequencing [
3]. Since its initial characterization, GGVA has also been reported in China [
4], South Korea [
5], New Zealand [
6], and India [
7]. GGVA is a monoparticle, single-stranded circular DNA virus belonging to the genus
Maldovirus in the family
Geminiviridae. The complete genome of GGVA ranges from 2903 to 2907 nucleotides (nt) in length [
8], including two open reading frames (ORFs) on the viral sense strand encoding V1 (coat protein) and V2 (putative movement protein), and four ORFs on the complementary strand encoding C1 (replication-associated protein), C2 (transcriptional activator protein), C3 (replication enhancer), and C4 (host-activated protein) [
3].
Infectious clones of viruses are important research tools for the analysis of viral biology and pathogenic mechanisms. Infectious cDNA (DNA) clones of some grapevine viruses [
8,
9,
10,
11,
12,
13,
14,
15,
16,
17,
18] have already been constructed. By using GGVA infectious clones, the pathogenicity of different GGVA isolates has been analyzed in
Nicotiana benthamiana [
8,
16] and
grapevine plants [
16]
. Those authors found that GGVA infection induces
dwarfing and upward leaf curling symptoms in N. benthamiana plants, whereas grapevine plants agroinfiltrated with GGVA were asymptomatic [
16]
.
Grapevine and other woody perennials are recalcitrant to mechanical inoculation. The main approaches for delivering infectious clones into grapevine plants include vacuum-assisted agroinfiltration [
2,
9,
16] and agro-drenching [
10,
15,
18]. However, these methods are complex and time-consuming, especially for the latter method which requires more than 20 days to establish a viral infection. In addition, the two methods result in relatively low survival and infection rates of the treated plants. Therefore, development of an efficient and simple experimental system to launch grapevine infections is needed.
In the present study, we constructed an infectious DNA clone of GGVA isolate QN (GGVAQN), which was isolated from a grapevine ‘Queen Nina’ plant with severe disease symptoms, including malformation, upward curling of the leaf margins, vein clearing, and reduced leaf size. The biological activity of infectious clone of GGVAQN (pXT-GGVAQN) was analyzed in N. benthamiana and grapevine plants. In addition, we provide a simple, time-saving and highly efficient inoculation method to deliver an infectious clone of a virus into woody perennial plants, including grapevine.
2. Results
2.1. Construction of an Infectious Clone of GGVA Isolate QN
The recombinant plasmid pCE3-GGVAQN, containing the full genome of GGVAQN, was used as template. The full genome of GGVAQN (2905 nt) was 100% identical to the isolate we obtained from grapevine ‘Marselan’ (GenBank accession no. PP397181), and shared 96.58% to 99.70% sequence identity with previous reported isolates.
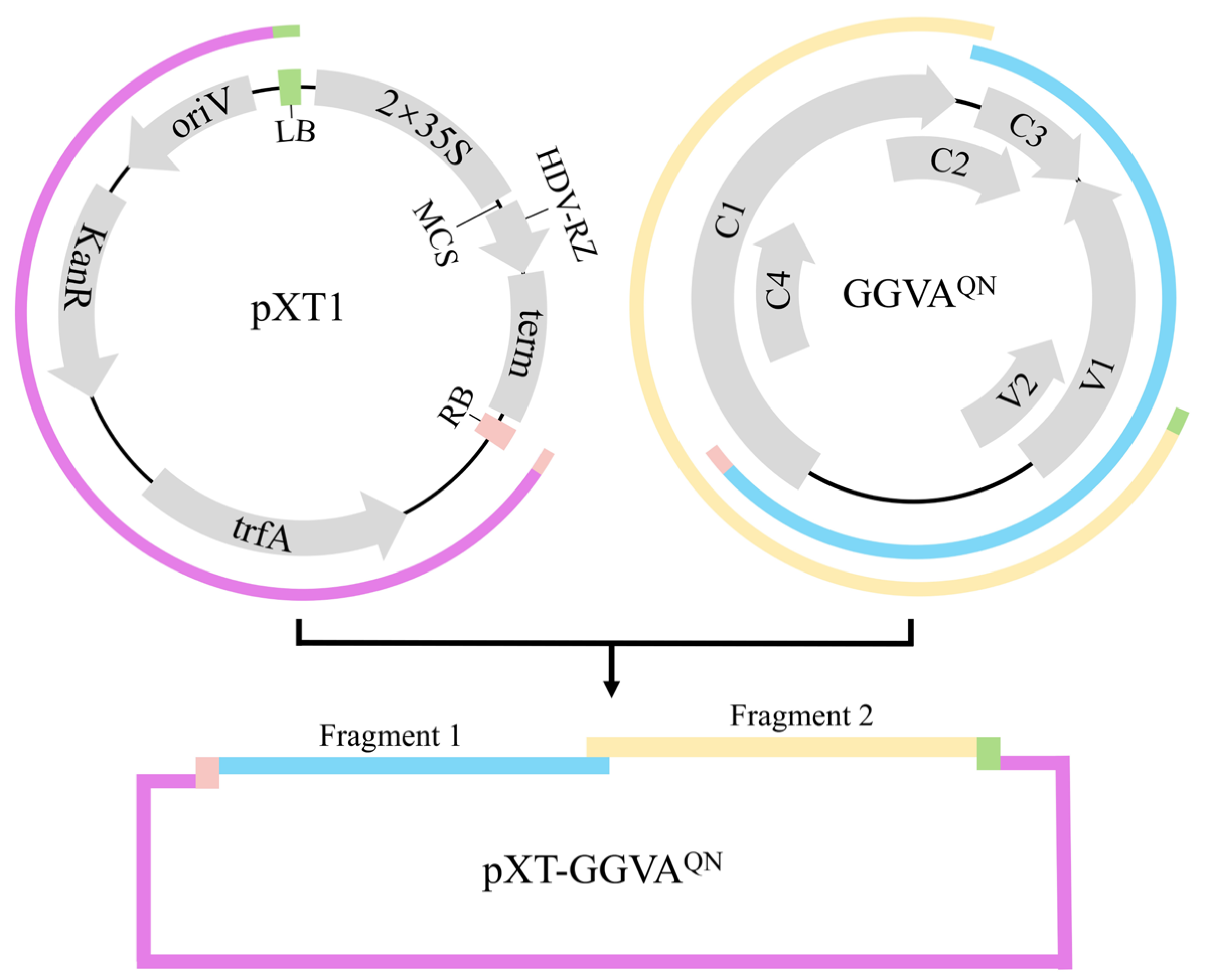
To construct an infectious clone of GGVA
QN, two fragments covering 1.2× viral genome sequence amplified from pCE3-GGVA
QN and the linearized vector pXT1 (JN029690) [
19] were assembled into circular plasmids (
Figure 1), which were then transformed into competent
Escherichia coli DH5a cells. Positive clones were identified by PCR, enzyme digestion and sequencing. The recombinant plasmid (pXT-GGVA
QN) was transformed into
Agrobacterium tumefaciens GV3101.
2.2. Infectivity of GGVAQN in N. benthamiana Plants
Six-week old
N. benthamiana plants were agroinfiltrated with the infectious clone of pXT-GGVA
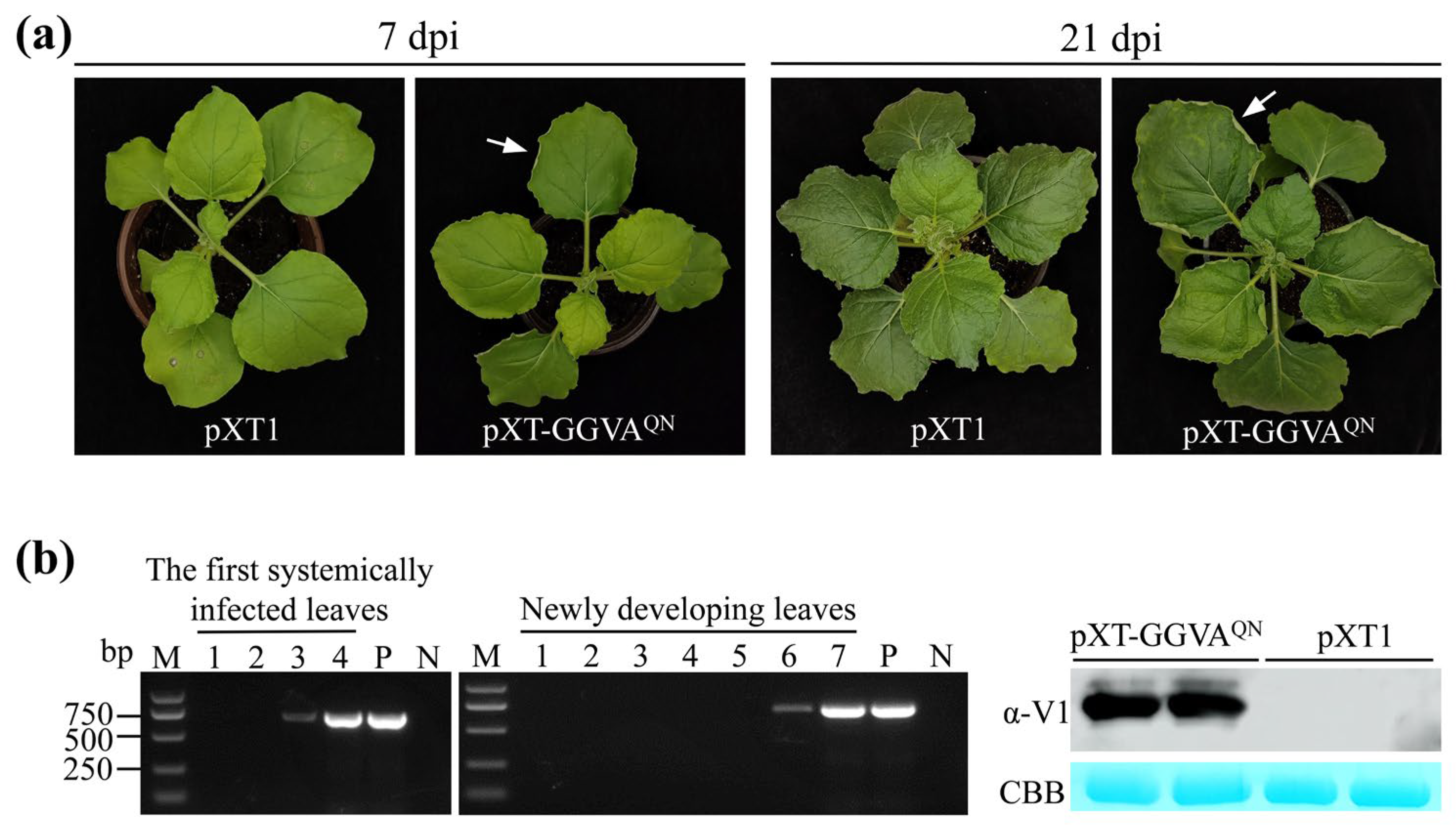
QN, and agroinfiltration of the empty vector pXT1 served as a control. At 7 days postinfiltration (dpi), the pXT-GGVA
QN-infiltrated leaves exhibited upward curling of the leaf edge, and symptoms of upward leaf curling together with chlorotic spots were observed on upper systemically infected leaves 21 dpi, whereas the control plants were asymptomatic (
Figure 2a). All 37
N. benthamiana plants (10–13 plants, three repeats) were systemically infected with pXT-GGVA
QN, as evidenced by PCR and western blot assays showing the presence of GGVA
QN in the first systemically infected and upper new developing leaves of all GGVA
QN-inoculated plants at 3 and 6 dpi, respectively (
Figure 2b). These results indicated that pXT-GGVA
QN can successfully infect
N. benthamiana plants and elicits upward leaf curling and chlorotic spot symptoms.
2.3. Infectivity of GGVAQN in Grapevine Plants
Before infection assays, the virological condition of the in vitro-grown ‘Red Globe’ grapevine plantlets, which had tested negative for 19 grapevine viruses, as well as grapevine leafroll-associated virus 4 (GLRaV-4) strains GLRaV-5, GLRaV-6, GLRaV-9, GLRaV-De, and GLRaV-Pr, were resubjected to RT-PCR and PCR assays. They were all free of the targeted viruses (
Supplementary Figure S1).
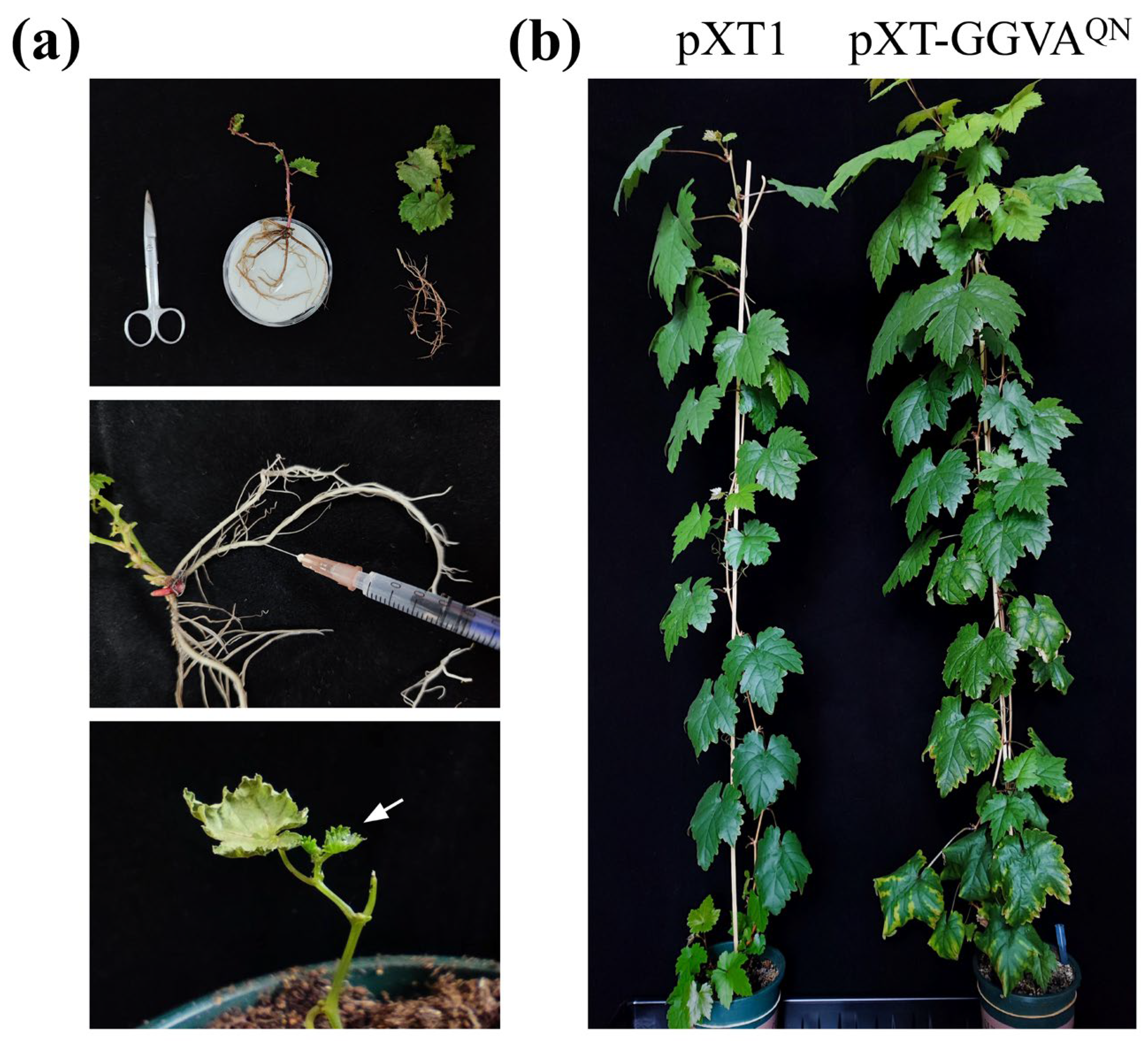
We then used a very simple and efficient method to deliver pXT-GGVA
QN into grapevine plants. Roots of each in vitro-grown grapevine plantlet were directly injected with 0.5 mL of
Agrobacterium tumefaciens cells carrying pXT-GGVA
QN by syringe and then transplanted into a pot (
Figure 3a). Empty vector pXT1 served as a control. A total of 18 plants were inoculated with GGVA
QN in two repeat experiments (9 plants each), and 15 plants survived after transplanting. As shown in
Figure 3b and
Figure 4a, about 2 month later, ‘Red Globe’ plants inoculated with GGVA
QN showed chlorotic mottling on the upper leaves; symptoms of downward curling and necrosis of the leaf margins, and interveinal yellowing were also observed on the lower leaves. Five months later, the color of interveinal tissues of the lower leaves of a few plants changed from yellow to red. All of these symptoms were absent in the corresponding control ‘Red Globe’ plants. All 15 surviving plants were systemically infected with GGVA
QN, as confirmed by the presence of GGVA
QN in the upper leaves detected by PCR and western blot assays 1 month after inoculation (
Figure 4b).
3. Discussion
GGVA was first found in 2017 in samples from both grapevine ‘Nagano purple’ plants with leaf chlorotic ringspot symptoms and asymptomatic ‘Black Beet’ plants [
3]. Fan et al. [
4] reported that most grapevine samples that were positive for GGVA were asymptomatic, except for a few samples with foliar ringspot symptoms. Similarly, Jo et al. [
5] reported the detection of GGVA in all 16 individual grapevine plants representing 12 different cultivars, but only the cultivar Shine Muscat displayed obvious symptoms (leaf malformation, vein clearing, and yellowing). Infectious clones of three GGVA isolates—GGVA-17YM1 [
8] and GGVA-76 and GGVA-93 [
16]—all induced upward leaf curling and stunting symptoms in
N. benthamiana plants, but grapevine (cultivars Colombard, Salt Creek, Cabernet Sauvignon, and Vaccarèse) plants infected with GGVA-76 or GGVA-93 did not display any obvious symptoms [
16]. Therefore, our knowledge of the pathogenicity of GGVA to grapevine plants remains largely unclear.
In this study, we constructed an infectious clone containing the full genome of GGVA
QN to investigate its pathogenicity.
N. benthamiana plants that were systemically infected with GGVA
QN showed symptoms of upward leaf curling and chlorotic mottling (
Figure 2a) which were somewhat different from the upward leaf curling and stunting symptoms observed by Sun et al. [
8] and Kuo et al. [
16]. GGVA
QN shares 99.52%, 97.26%, and 98.45% sequence identity with GGVA-17YM1, GGVA-76, and GGVA-93, respectively. The inconsistency may not be due to the sequence differences between GGVA
QN and the three GGVA isolates; rather, other factors, such as the different ages of the
N. benthamiana plants used for inoculation and fertilization, may be responsible.
The pathogenicity of GGVA
QN on grapevine was also investigated using in vitro-grown ‘Red Globe’ rooted plantlets. Upon GGVA
QN infection, all ‘Red Globe’ plants exhibited chlorotic mottling symptoms on their upper leaves, and symptoms of downward curling and necrosis of the leaf margins, and interveinal yellowing on the lower leaves 2 months after inoculation and transplanting (
Figure 3b and
Figure 4a). In contrast, all control ‘Red Globe’ plants which were inoculated with empty vector pXT1 were asymptomatic. The in vitro-grown ‘Red Globe’ rooted plantlets used for infection assays were negative for GGVA, GLRaV-4 and its strains GLRaV-5, GLRaV-6, GLRaV-9, GLRaV-De, and GLRaV-Pr, and another 17 viruses reported in China. Therefore, the symptoms on ‘Red Globe’ plants were closely associated to GGVA
QN infection. Our results differ from those reported by Kuo et al. [
16], where neither GGVA-76 nor GGVA-93 induced obvious symptoms on grapevine ‘Colombard’, ‘Salt Creek’, ‘Cabernet Sauvignon’, and ‘Vaccarèse’ plants, although the virological condition of these plants was unclear. Lovato et al. [
12] reported that grapevine plants infiltrated with the infectious clone of grapevine Algerian latent virus exhibited different symptoms in different cultivars. Therefore, the inconsistency may be due to the different grapevine cultivars used in the two studies. Future research investigating the pathogenicity of GGVA
QN in different grapevine cultivars is warranted.
One important factor that hinders the research on viruses of woody perennial plants, including grapevine, is the lack of a reliable experimental system for delivering infectious clones. Two main approaches, vacuum-agroinfiltration and agro-drenching, are currently used to deliver infectious clones into grapevine plants. However, they are complex and time-consuming, and the achieved rate of infection is relatively low. In this study, we directly injected A. tumefaciens cells carrying the infectious clone pXT-GGVAQN into roots of grapevine plantlets by syringe followed by transplanting the plantlets into pots, and all surviving plants (15 out of 18) were systemically infected with GGVAQN. We also successfully launched the infection of rooted grapevine plantlets with infectious clones of grapevine Pinot gris virus (GPGV) and GLRaV-2 with this method (data not shown). Therefore, we provide a very simple, quick, and efficient method for delivering infectious clones into grapevine plants, with no vacuuming operation, taking only a few minutes to complete the infiltration, and resulting in 100% infection rate. To our knowledge, the experimental protocol reported herein is the most effective system for achieving grapevine infection with infectious clones of a virus, potentially facilitating grapevine virology research.
In conclusion, an infectious clone of GGVAQN was constructed and its pathogenicity in N. benthamiana and grapevine plants was investigated. The results presented here provide important insights into GGVA pathogenesis, as well as a very simple and highly effective method for launching grapevine infection with the infectious clone of a virus.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
Grapevine cv. Red Globe in vitro-grown rooted plantlets used in the infection assays were previously obtained from treatments of thermotherapy combined with meristem tip culture, and kept in our laboratory. The virological condition of these plantlets were investigated three times and were negative for GGVA and other viruses reported in China, including GLRaV-1, GLRaV-2, GLRaV-3, GLRaV-4 (including GLRaV-4 strains GLRaV-5, GLRaV-6, and GLRaV-9, GLRaV-De, and GLRaV-Pr), GLRaV-7, GLRaV-13, grapevine rupestris stem pitting-associated virus (GRSPaV), grapevine fleck virus (GFkV), grapevine fanleaf virus (GFLV), grapevine virus A (GVA), GVB, GVE, GPGV, grapevine fabavirus (GFabV), grapevine red blotch virus (GRBV), grapevine Syrah virus-1 (GSyV-1), grapevine berry inner necrosis virus (GINV), and grapevine red globe virus (GRGV).
In vitro-grown rooted grapevine plantlets were grown on woody plant medium supplemented with 30 mg mL-1 sucrose and 1 mg mL-1 indole-3-butyric acid. N. benthamiana plants were grown in individual pots containing commercial soil, and 6-week-old plants were used. All plant materials were grown at 26–31 °C with a 16-h light regime.
4.2. Construction of Infectious Clone of GGVA
The full genome of GGVA
QN isolate was PCR amplified from a sample collected from a grapevine ‘Queen Nina’ plant in Guangxi Province in Southwest China using a TOPO-Blunt Cloning Kit (Vazyme, China) with the primer pair GGVA-1678-F/GGVA-1678-R previously described [
8]. The PCR products were cloned into the pCE3 Blunt Vector provided by the kit, and the recombinant plasmid pCE3-GGVA
QN was used as the template to construct the infectious clone of GGVA.
Construction of the infectious clone of GGVA
QN was performed essentially as described previously [
16] with modifications. A fragment (the linearized pXT1 without the sequence of the 35S promoter, multiple cloning site region, ribozyme, and NOS terminator) (
Figure 1a) was amplified using the binary vector pXT1 as template with primer pair F27/R27. Two fragments, each covering 0.6× viral genome sequence, were amplified from pCE3-GGVA
QN using primer pairs F25/R25 and F26/R26, with F25 and R26 containing a 30-bp sequence homologous to the T-DNA right border (RB) and left border (LB) on pXT1, respectively. The three fragments were assembled into circular plasmids using NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs, Ipswich, MA, USA). Positive clones were identified by
Hind III digestion (resulting in a 7197-bp fragment), and PCR with the primer pairs F25/R25 and F26/R26 for amplifying fragment 1 (1886 bp) and fragment 2 (1934 bp), respectively. Three positive clones were then sequenced to verify the correctness of the GGVA genome sequence. The recombinant plasmid pXT-GGVA
QN was delivered into competent
A. tumefaciens strain GV3101 using the freeze–thawing method.
4.3. Inoculation
A. tumefaciens GV3101 carrying the infectious clone pXT-GGVA
QN was cultured to an optical density of 1.0 at 600 nm.
Agrobacterium cells were harvested by centrifugation and resuspended in the infiltration buffer (10 mM MES/NaOH pH 5.8, 10 mM MgCl
2, 150 µM acetosyringone). Agroinfiltration of
N. benthamiana plants was performed essentially as described previously [
20].
For inoculation of in vitro-grown grapevine plantlets, rooted ‘Red Globe’ plantlets were removed from the culture jar and washed to remove the agar adhering to their base. Plantlets with pruned roots and leaves were placed on moist filter paper. A 0.5-mL aliquot of
A. tumefaciens GV3101 cells carrying pXT-GGVA
QN (for each grapevine plantlet) was injected into the main root bark with a syringe, and the plantlet was transplanted in a pot (10 cm diameter) containing commercial soil (
Figure 3a). These pots were placed in plates filled with a shallow layer of water. About 3 days after transplanting, new buds were observed, indicating that they had survived. We used 8–13 plants or plantlets for each treatment in each experiment.
All inoculated plants were fertilized once a week with water-soluble fertilizer (Peters Professional).
4.4. RT-PCR and PCR
The virological condition of the in vitro-grown grapevine plantlets was detected by RT-PCR and PCR, and the presence of GGVA in inoculated plants was assessed by PCR.
Primers used in the RT-PCR and PCR assays are listed in
Supplementary Table S1. Primer pairs F6/R6, F19/R19, F20/R20, F25/R25, F26/R26 and F27/R27 were designed in this study, and others were described previously [
21,
22,
23,
24,
25,
26,
27,
28,
29,
30,
31,
32,
33]. RNA and DNA were extracted from
N. benthamiana leaves, or phloem scrapings of grapevine petioles or leaves of in vitro-grown grapevine plantlets using the CTAB method. Synthesis of cDNA (at 37 °C) was primed with a mixture of random primers using 500 ng total RNA with HiScript III 1
st Strand cDNA Synthesis Kit (Vazyme, Nanjing, China). A 1-μL aliquot of cDNA or DNA was used as a template in a 10 μL PCR, and PCR amplification was conducted as follows: DNA denaturation at 95 °C for 5 min; 35 cycles at 95 °C for 30 s, 52–60 °C (depending on the specific primers used) for 30 s, and 72 °C for 30 s, and a final extension step at 72 °C for 5 min. A negative control (ddH
2O instead of cDNA or DNA) was included in the PCR assays. Positive PCR products were randomly selected for sequencing to confirm their identity.
4.5. Western Blot Analysis
Plant protein extraction and western blot analysis were performed as detailed previously [
20]. The dilution of anti-V1 antiserum (noncommercial) used in the western blot analysis was 1:2000. The hybridization signals were detected with High-sig ECL Western Blotting Substrate (Beyotime, China) according to the manufacturer’s instructions. Coomassie brilliant blue staining confirmed equal protein contents in each lane.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on
Preprints.org.
Author Contributions
Conceptualization, Y.-Q.C., C.L. and S.-Z.Y.; Construction of GGVA clone and inoculation, C.L. and S.-Z.Y.; Virus detection, J.-Y. W. and Y.-S.X.; supplying the plant materials, X.-Q.Z., C.-L.F. and W.-H.Z.; Western blot analysis, H.-W.L.; Symptom observation and photography, C.L., X.-Q.Z., C.-L.F. and W.-H.Z.; writing—original draft preparation, Y.-Q.C.; writing—review and editing, Y.-Q.C., C.L., J.-Y. W. and Y.-S.X. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by National Natural Science Foundation of China (No. 32272646) and the Construction of Beijing Science and Technology Innovation and Service Capacity in Top Subjects (CEFF-PXM2019_014207_000032).
Acknowledgments
The authors thank Xiaorong Tao (Nanjing Agriculture University, China) for kindly providing plasmid pXT1.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Fuchs, M. Grapevine viruses: A multitude of diverse species with simple but overall poorly adopted management solutions in the vineyard. J. Plant Pathol. 2020, 102, 643–653. [Google Scholar] [CrossRef]
- Shabanian, M.; Li, C.; Ebadi, A.; Dolja, V.; Meng, B. Optimization of a protocol for launching grapevine infection with the biologically active cDNA clones of a virus. Pathogens 2023, 12, 1314. [Google Scholar] [CrossRef] [PubMed]
- Al Rwahnih, M.; Alabi, O.J.; Westrick, N.M.; Golino, D.; Rowhani, A. Description of a novel monopartite geminivirus and its defective subviral genome in grapevine. Phytopathology 2017, 107, 240–251. [Google Scholar] [CrossRef]
- Fan, X.D.; Zhang, Z.P.; Ren, F.; Hu, G.J.; Li, Z.N.; Dong, Y.F. First report of Grapevine geminivirus A from grapevines in China. Plant Dis. 2017, 101, 1333–1333. [Google Scholar] [CrossRef]
- Jo, Y.; Choi, H.; Song, M.K.; Park, J.S.; Lee, J.W.; Cho, W.K. First report of Grapevine geminivirus A in diverse Vitis species in Korea. Plant Dis. 2018, 102, 255. [Google Scholar] [CrossRef]
- Veerakone, S.; Blouin, A.G.; Barrero, R.A.; Napier, K.R.; MacDiarmid, R.M.; Ward, L.I. First report of Grapevine geminivirus A in Vitis in New Zealand. Plant Dis. 2020, 104, 600–600. [Google Scholar] [CrossRef]
- Sidharthan, V.K.; Sevanthi, A.M.; Jaiswal, S.; Baranwal, V.K. Robust virome profiling and whole genome reconstruction of viruses and viroids enabled by use of available mRNA and sRNA-seq datasets in grapevine (Vitis vinifera L.). Front. Microbiol. 2020, 11, 1232. [Google Scholar] [CrossRef]
- Sun, S.; Hu, Y.; Jiang, G.; Tian, Y.; Ding, M.; Yu, C.; Zhou, X.; Qian, Y. Molecular characterization and genomic function of Grapevine geminivirus A. Front. Microbiol. 2020, 11, 555194. [Google Scholar] [CrossRef] [PubMed]
- Kurth, E.G.; Peremyslov, V.V.; Prokhnevsky, A.I.; Kasschau, K.D.; Miller, M.; Carrington, J.C.; Dolja, V.V. Virus-derived gene expression and RNA interference vector for grapevine. J. Virol. 2012, 86, 6002–6009. [Google Scholar] [CrossRef]
- Muruganantham, M.; Moskovitz, Y.; Haviv, S.; Horesh, T.; Fenigstein, A.; Preez, J. D.; Stephan, D.; Burger, J.T.; Mawassi, M. Grapevine virus A-mediated gene silencing in Nicotiana benthamiana and Vitis vinifera. J. Virol. Methods 2009, 155, 167–174. [Google Scholar] [CrossRef]
- Meng, B.; Venkataraman, S.; Li, C.; Wang, W.; Dayan-Glick, C.; Mawassi, M. Construction and biological activities of the first infectious cDNA clones of the genus Foveavirus. Virology 2013, 435, 453–462. [Google Scholar] [CrossRef] [PubMed]
- Lovato, A.; Faoro, F.; Gambino, G.; Maffi, D.; Bracale, M.; Polverari, A.; Santi, L. Construction of a synthetic infectious cDNA clone of Grapevine Algerian latent virus (GALV-Nf) and its biological activity in Nicotiana benthamiana and grapevine plants. Virol. J. 2014, 11, 186. [Google Scholar] [CrossRef] [PubMed]
- Jarugula, S.; Gowda, S.; Dawson, W.O.; Naidu, R.A. Development of infectious cDNA clones of Grapevine leafroll-associated virus 3 and analyses of the 5′ non-translated region for replication and virion formation. Virology 2018, 523, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Yepes, L.M.; Cieniewicz, E.; Krenz, B.; McLane, H.; Thompson, J.R.; Perry, K.L.; Fuchs, M. Causative role of Grapevine red blotch virus in red blotch disease. Phytopathology 2018, 108, 902–909. [Google Scholar] [CrossRef] [PubMed]
- Tarquini, G.; Zaina, G.; Ermacora, P.; De Amicis, F.; Franco-Orozco, B.; Loi, N.; Martini, M.; Bianchi, G.L.; Pagliari, L.; Firrao, G.; De Paoli, E.; Musetti, R. Agroinoculation of Grapevine Pinot Gris Virus in tobacco and grapevine provides insights on viral pathogenesis. PLoS ONE 2019, 14, e0214010. [Google Scholar] [CrossRef] [PubMed]
- Kuo, Y.W.; Bednarska, A.; Al Rwahnih, M.; Falk, B.W. Development of Agrobacterium tumefaciens infiltration of infectious clones of Grapevine geminivirus A directly into greenhouse-grown grapevine and Nicotiana benthamiana plants. Phytopathology 2022, 112, 1603–1609. [Google Scholar] [CrossRef]
- Liu, Y.P.; Peremyslov, V.V.; Medina, V.; Dolja, V.V. Tandem leader proteases of Grapevine leafroll-associated virus-2: Host-specific functions in the infection cycle. Virology 2009, 383, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Zhang, Z.; Ren, F.; Hu, G.; Li, C.; Zhang, B.; Dong, Y. Development of a full-length infectious cDNA clone of the Grapevine berry inner necrosis virus. Plants 2020, 9, 1340. [Google Scholar] [CrossRef] [PubMed]
- Yao, M.; Zhang, T.; Tian, Z.; Wang, Y.; Tao, X. Construction of Agrobacterium-mediated Cucumber mosaic virus infectious cDNA clones and 2b deletion viral vector. Sci. Agric. Sin. 2011, 44, 3060–3068. [Google Scholar]
- Li, M.; Zhang, J.; Feng, M.; Wang, X.; Luo, C.; Wang, Q.; Cheng, Y. Characterization of silencing suppressor p24 of Grapevine leafroll-associated virus 2. Mol. Plant Pathol. 2018, 19, 355–368. [Google Scholar] [CrossRef]
- Fan, X.D.; Zhang, Z.P.; Ren, F.; Hu, G.J.; Zhang, M.Y.; Li, C.; Zhang, B.D.; Dong, Y.F. Construction and screening of a yeast two-hybrid cDNA library from Beta grapevine infected by grapevine berry inner necrosis virus. Acta Phytopathol. Sin. 2021, 51, 210–216. [Google Scholar]
- Wang, X.Y.; Zhang, C.W.; Huang, W.T.; Yue, J.; Dou, J.J.; Wang, L.Y.; Wang, Q.; Cheng, Y.Q. Crude garlic extract significantly inhibits replication of grapevine viruses. Plant Pathol. 2020, 69, 149–158. [Google Scholar] [CrossRef]
- Zhou, J.; Fan, X.; Dong, Y.; Zhang, Z.P.; Ren, F.; Hu, G. Detection and genetic variation analysis of grapevine fanleaf virus (GFLV) isolates in China. Arch. Virol. 2015, 160, 2661–2667. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.J.; Dong, Y.F.; Zhang, Z.P.; Fan, X.D.; Fang, R.; Zhu, H.J. Detection and sequence analysis of grapevine virus B isolates from China. Acta Virol. 2014, 58, 180–184. [Google Scholar] [CrossRef] [PubMed]
- Maliogka, V.I.; Dovas, C.I.; Katis, N.I. Evolutionary relationships of virus species belonging to a distinct lineage within the Ampelovirus genus. Virus Res. 2008, 135, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.D.; Dong, Y.F.; Zhang, Z.P.; Ren, F.; Hu, G.J.; Li, Z.N.; Zhou, J. First report of Grapevine Red Globe Virus (GRGV) in grapevines in China. Plant Dis. 2016, 100, 2340. [Google Scholar] [CrossRef]
- Ahmed, I.; Fan, X.D.; Zhang, Z.P.; Ren, F.; Hu, G.J.; Li, Z.N.; Khaskheli, M.I.; Dong, Y.F. First report of Grapevine Syrah Virus-1 in grapevines in China. Plant Dis. 2018, 102, 466. [Google Scholar] [CrossRef]
- Fan, X.D.; Dong, Y.F.; Zhang, Z.P.; Ren, F.; Hu, G.J.; Zhu, H.J. First report of Grapevine Virus E from grapevines in China. J. Plant Pathol. 2013, 95, 659–668. [Google Scholar]
- Yu, S.; Kan, Q.; Huang, H.; Wang, J.; Xie, Y.; Li, H.; Zhang, X.; Liu, C.; Cheng, Y. Grapevine cultivar Shine Muscat in China: Occurrence of viruses and attempts to produce certified propagation material. J. Plant Pathol. 2023, 105, 1609–1616. [Google Scholar] [CrossRef]
- Krenz, B.; Thompson, J.R.; McLane, H.L.; Fuchs, M.; Perry, K.L. Grapevine Red Blotch-Associated Virus is widespread in the United States. Phytopathology 2014, 104, 1232–1240. [Google Scholar] [CrossRef]
- Ito, T.; Nakaune, R. Molecular characterization of a novel putative ampelovirus tentatively named grapevine leafroll-associated virus 13. Arch. Virol. 2016, 161, 2555–2559. [Google Scholar] [CrossRef] [PubMed]
- Lyu, M.D.; Li, X.M.; Guo, R.; Li, M.J.; Liu, X.M.; Wang, Q.; Cheng, Y.Q. Prevalence and distribution ofGrapevine Leafroll-associated Virus 7 in China detected by an improved reverse transcription polymerase chain reaction assay. Plant Pathol. 2014, 63, 1168–1176. [Google Scholar] [CrossRef]
- Osman, F.; Leutenegger, C.; Golino, D.; Rowhani, A. Real-time RT-PCR (TaqMan®) assays for the detection of Grapevine Leafroll associated viruses 1–5 and 9. J. Virol. Methods 2007, 141, 22–29. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).