1. Introduction
Rotavirus vaccines have been highly successful at reducing the burden of rotavirus-induced gastroenteritis in children in high income countries. Two oral vaccines were first approved in 2006 and 2008, developed from a human rotavirus strain (Rotarix, GSK) and a human-bovine rotavirus reassortant virus (Rotateq, Merck) [
1,
2]. A total of four live attenuated rotavirus vaccines are now pre-qualified with the World Health Organization, and a further two are country-specific [
3]. However, live attenuated vaccines have failed to protect infants in low- to middle-income countries, with vaccine efficacy often lower than 50% [
4,
5], leading to higher rates of gastroenteritis deaths in unprotected infants. As a consequence, there is a pressing need for the development of improved vaccines.
A major hurdle for vaccine development has been the absence of suitable pre-clinical models to test new vaccine candidates. Rotavirus strains are highly species-specific, meaning that human rotavirus strains are primarily pathogenic in humans, and murine rotavirus strains only cause disease in mice [
6,
7]. This hinders the study of disease pathology and immune responses to human rotavirus strains in mice, as in this species human rotavirus strains only replicate to low titres and cause minimal disease.
In this study, we aimed to develop a new strategy to study the immune response to human rotaviruses in a mouse model. To achieve this, we generated a chimeric rotavirus strain that can replicate well in mice
and contains key immunogenic proteins from a human rotavirus strain. This approach builds on previous work that showed reassortment of certain murine rotavirus genes with those from a non-murine rotavirus strain could still permit virus replication in mice [
6].
The advent of rotavirus reverse genetics has provided the field with the capacity to rapidly generate chimeric rotaviruses with relative ease [
8,
9]. Rotavirus has a triple-layered structure with an outer capsid composed of two proteins, VP4 and VP7, which become a major target of the adaptive immune response [
10]. Chimeric rotaviruses have previously been produced using reverse genetics with a murine backbone and human rotavirus VP4 protein [
11], and we have now extended this by successfully generating a chimeric rotavirus with VP4 and VP7 from a human rotavirus strain. As a control, we also generated a chimeric virus encoding the outer capsid proteins of a heterologous murine rotavirus strain. We demonstrated that both viruses could infect neonatal mice and replicate to comparable titres.
Antibody responses to human rotavirus proteins in chimeric rotaviruses have not previously been studied in mice. We aimed to determine the magnitude and specificity of the antibody response to human rotavirus outer capsid proteins using our small animal model. We observed strong germinal centre formation in the draining lymph nodes of mice infected with both chimeric viruses, which correlated with antibody production. Neutralisation and ELISpot assays were used to clearly demonstrate that the chimeric virus with human rotavirus outer capsid proteins induced antibodies with human rotavirus specificity. This novel approach to studying human rotaviruses in a small animal model will be valuable for pre-clinical evaluation of vaccine efficacy and therapeutics targeting the outer capsid of human rotaviruses.
2. Materials and Methods
2.1. Cells and Viruses
MA104 African green monkey kidney cells, provided by Dr. John Parker (Baker Institute for Animal Health, Cornell University, USA), were grown in Dulbecco’s Minimum Essential Media (DMEM) supplemented with 10% heat-inactivated fetal bovine serum, 100 IU/mL penicillin, and 100 μg/mL streptomycin (complete DMEM).
The pre-existing rotavirus strains used in this study were the Rotarix vaccine strain (G1P[
8]) and the primate strain SA11 (G3P[
2]). Two rotavirus chimeric strains were developed using a published plasmid-based reverse genetics system [
5]. Using the reassortant virus rD6/2-2g as the backbone, chimeric viruses were rescued with a human CDC-9 strain VP4 and VP7 (human outer capsid proteins), or a murine strain (ETD) VP4 and VP7. All viruses were propagated in MA104 cells following activation with 10 μg/mL TPCK-treated trypsin at 37°C for 30 minutes. Prior to
in vivo infection, viruses were diluted to the appropriate titre in sterile PBS without calcium chloride and magnesium chloride.
2.2. Virus Quantification by Fluorescent Focus Assay (FFA)
A fluorescent focus assay was used to determine viral titre (in fluorescent focus units, FFU) as previously described [
12]. Briefly, MA104 cells were infected with virus for 16-hours, then cells were fixed with 1:1 methanol:acetone at -20°C for 20 minutes. After blocking with PBS-2%FBS for 20 minutes at room temperature, 10 μg/mL rotavirus polyclonal antibody (sheep) diluted in PBS-2%FBS was added for 1 hour at room temperature. After three washes with PBS-T, 4 μg/mL Alexa Fluor 488 Donkey Anti-Sheep IgG and Hoechst 33342 diluted in PBS-2%FBS were added to each well, and the plate was incubated for 1 hour at room temperature. All plates were kept at 4-6°C coated with PBS prior to imaging. Quantification of rotavirus-infected cells was achieved using the BioTek Cytation 7 Cell Imaging Multimode Reader and Gen5 Image Prime (v3.13) software.
2.3. Rotavirus Infection of Mice
129S6/SvEvTac mice (Taconic Biosciences) were maintained by an in-house breeding colony housed at the Baker Institute for Animal Health. All mouse work was approved by the Cornell University Institutional Animal Care and Use Committee (IACUC), Protocol 2022-0152. Seven-day-old pups were infected with 1 x 104 FFU virus by oral gavage. Pups were monitored for the development of diarrhoea, and scored positive if mucus or liquid stool was observed. Stool samples were collected once daily post-infection from each litter, then pooled and diluted 1:10 in PBS. Diluted stool was centrifuged at 8,000 × g for 5 minutes to remove debris, and the supernatant was stored at -80°C. Blood samples were collected from the lateral saphenous vein at 4, 6 and 8 weeks old, and by terminal cardiac puncture at 10 weeks of age. All blood samples were centrifuged at 6,000 × g for 5 minutes, and sera was stored short-term at 4°C. In separate experiments, 14 days post-infection mice were humanely culled with terminal cardiac puncture samples collected, and the Peyer's patches (PPs), mesenteric lymph nodes (MLNs), and spleens harvested.
2.4. Quantification of Virus Shedding by RT-qPCR
RNA extraction from a clarified stool suspension was achieved using the Monarch Total RNA Miniprep Kit, according to manufacturer’s instructions. Each sample was eluted in a total volume of 50 μL nuclease-free water, followed by denaturation of dsRNA at 95°C for 5 minutes. RT-qPCR was performed using the Luna Universal One-Step RT-qPCR Kit, per the manufacturer’s instructions, with 5 μL of RNA in a total reaction volume of 20 μL using NSP5 forward primer CTGCTTCAAACGATCCACTCAC 400 nM, NSP5 reverse primer TGAATCCATAGACACGCC 400 nM, NSP5 TaqMan probe FAM-TCAAATGCAGTTAAGACAAATGCAGACGCT-TAMRA 200 nM. The reaction was carried out on a QuantStudio 3 thermocycler (Applied Biosystems), under the cycling conditions of 55°C for 10 minutes, 95°C for 1 minute, and 40 cycles of 95°C for 10 seconds and 60°C for 30 seconds. A 10-fold serial dilution of SA11 total RNA was included on each plate to quantify rotavirus genome copies per mL of stool supernatant using QuantStudio Design & Analysis Software (v1.5.1). A lower limit of quantification of 100 genome copy numbers was set and assigned to samples with no detectable virus.
2.5. ELISAs
ELISAs were performed to detect IgG-specific anti-rotavirus antibodies using an in-house method as previously described [
12]. MA104 cells were infected with rotavirus to produce infected cell lysate, or phosphate-buffered saline (PBS) to produce control cell lysate. Cells were collected and resuspended in Radio-Immunoprecipitation Assay (RIPA) buffer supplemented with protease inhibitors. The Bicinchoninic Acid Kit for protein determination was used to measure protein concentration following manufacturer’s instructions, and lysate stocks were diluted to 1 mg/mL in PBS.
Plates were washed three times using 0.1% Tween-20 (PBS-T) between each step. High-binding 96-well plates (Greiner) were coated with 5 μg/mL rotavirus-specific polyclonal antibody (sheep) in PBS and incubated at 4-6°C for 16 hours. Plates were then blocked with 5% milk-PBS-T at room temperature for 1 hour. The purified cell culture lysates (Rotarix and SA11 virus infected lysate or mock-infected control lysate) were diluted to 10 μg/mL in PBS, and incubated at 37°C for two hours. Sera was diluted 1:200 in 5% milk-PBS-T and added in duplicate and incubated at 37°C for 2 hours. Positive and negative control sera from known infected and uninfected mice were included on each plate. The anti-mouse IgG HRP secondary antibody was diluted 1:1000 in 5% milk-PBS–T and added before plates were incubated at 37°C for 1 hour. To detect the bound antibody, 3,3',5,5'-tetramethylbenzidine (TMB) was incubated at room temperature for 10 minutes. The reaction was stopped with 1M sulfuric acid (H2SO4) and the optical density (OD) read at 450 nm using the BioTek Cytation 7 Cell Imaging Multimode Reader. The OD was normalised by subtracting the OD of the mock-infected control cell lysate well from the OD of the virus-infected cell lysate well.
2.6. Extracellular Neutralisation Assay
MA104 cells were seeded in a 96-well black sided plate (Corning 3340) at 2 x 104 per well in complete DMEM, and incubated at 37°C for 4 hours to allow cells to adhere. Eight two-fold serial dilutions of serum in serum free media (SFM) were incubated with trypsin-activated rotavirus at 37°C for 1 hour. The serum-virus mixture was then added in triplicate to seeded cells. After 1 hour at 37°C, 50 μL complete DMEM was added to each well and the plate was incubated at 37°C for 16 hours. Rotavirus neutralisation was quantified by FFA.
2.7. Intracellular Neutralisation Assay
A previously published intracellular neutralisation assay protocol was applied to sera from mice infected with the human or mouse outer capsid chimeric viruses [
9]. Using a Neon Transfection System Kit, sera was diluted 1:3 in PBS and mixed with 2 x 10
5 MA104 cells suspended in Resuspension Buffer R. Sera were then electroporated with two pulses at 1400 V and a 20-pulse width using the Neon ® Transfection System (Thermo Fisher Scientific). Electroporated cells were resuspended in complete DMEM and plated onto a 96-well black sided plate in duplicate. After incubation at 37°C for 4 hours, wells were washed once with PBS, and trypsin-activated rotavirus in SFM was added to each well. Infection and virus quantification then proceeded as for the extracellular neutralisation assay, described above.
2.8. Isolation of Single Cells from Lymph Nodes and Spleens
Peyer's patches (PPs), mesenteric lymph nodes (MLNs), and spleens were harvested from infected mice 14-days post-infection or control uninfected mice. The tissues were homogenised through 70 μm mesh cell strainers to obtain single-cell suspensions, and then washed with RPMI supplemented with 2%FCS (RPMI- 2%FCS) by centrifugation at 300 × g for 5 minutes. The PP and MLN cell pellets were resuspended in 1 mL cold staining buffer (PBS- 1%FCS), then filtered through 70 μm mesh, washed and resuspended in 100 μL staining buffer. Spleen cell pellets were resuspended in red blood cell lysis buffer and incubated at room temperature for 3 minutes. These were then washed with PBS by centrifugation, and resuspended in RPMI - 2%FCS.
2.9. Flow Cytometry
PP and MLN single-cell suspensions were incubated with Fc Block (1:100) in staining buffer for 30 minutes at 4°C, then cells were incubated with viability dye and fluorescently conjugated antibodies targeting B220, GL7 Antigen, CD95, and CD45 in staining buffer for 30 minutes at 4°C. Single-color controls were included using PP cells or compensation beads. Cells were washed twice with staining buffer by centrifugation, and resuspended in 100 μL 4%PFA-PBS at 4°C for 15 minutes to fix. Cells were then washed twice by centrifugation, and the cell pellets were resuspended in 300 μL staining buffer. Cells were analyzed using a BD LSRFortessa X-20 (BD Biosciences) and BD FACSDiva Software (v9.0) and data analysis was performed in FlowJo (v10.9.0).
2.10. ELISpot Assay
Spleen single-cell suspensions were analysed by ELISpot to identify rotavirus-specific B cell responses. PVDF-based membrane plates (MSIP white, Mabtech) were activated with 35% ethanol, followed by five washes with sterile water. All further washing steps were performed five times with sterile PBS. Plates were coated with 2.7 x 104 FFU Rotarix per well and incubated at 4°C for 16 hours. Wells were washed to remove excess antigen, and incubated with RPMI supplemented with 10%FCS, 100 IU/mL penicillin, and 100 μg/mL streptomycin for 30 minutes at room temperature. Media was removed, and 3 x 105 spleen cells were incubated in triplicate at 37°C for 16 hours. The plate was then washed, and 1 μg/mL anti-mouse IgG biotin in PBS- 0.5%FCS was incubated for 2 hours at room temperature. After washing, 1:1000 streptavidin-ALP diluted in PBS- 0.5%FCS was incubated for 1 hour at room temperature. The plate was washed a final time before BCIP/NBT-plus for ALP was added to develop until distinct spots emerged. Colour development was then stopped by rinsing the plate with water, and plates were left to dry before imaging on the upright microscope of a BioTek Cytation 7 Cell Imaging Multimode Reader.
2.11. Statistics
Statistical analysis was performed using GraphPad Prism (v10.2.0). Immune responses by ELISA, neutralisation assay, flow cytometry and ELISpot assay were analysed using unpaired two-tailed T-tests. Statistical differences were considered significant at P-values < 0.05 for all comparisons. Error bars indicate the standard error of the mean.
Table 1.
Reagent details.
Table 1.
Reagent details.
| Reagent |
Source |
Identifier |
| Antibodies and Dyes |
| Alexa Fluor 488 Anti-Sheep IgG (Donkey) |
Invitrogen |
CAT#A-11015 |
| Anti-Mouse IgG Biotin (Goat) |
Mabtech |
CAT#3825-6 |
| Anti-Mouse IgG HRP (Goat) |
Sigma Aldrich |
CAT#A0168 |
| CD45 (BUV395 Rat Anti-Mouse) |
BD Biosciences |
CAT#564279; Clone: 30-F11 |
| CD45R (PerCP/Cyanine 5.5 Anti-Mouse/Human CD45R/B220) |
BioLegend |
CAT#103236; Clone: RA3-6B2 |
| CD95 (APC Anti-Mouse (Fas)) |
BioLegend |
CAT#152603; Clone: SA367H8 |
| Fc Block (TruStain FcX Anti-Mouse CD16/32) |
BioLegend |
CAT#101320; Clone: 93 |
| Fixable Viability Dye eFlour 780 |
Invitrogen |
CAT#65-0865 |
| GL7 Antigen (Pacific Blue Anti-Mouse/Human) |
BioLegend |
CAT#144614; Clone: GL7 |
| Hoechst 33342 |
Invitrogen |
CAT#H3570 |
| Rotavirus Polyclonal Antibody (Sheep) |
Invitrogen |
CAT#PA1-85845 |
| Streptavidin-ALP |
Mabtech |
CAT#3310-10 |
| Chemicals |
| BCIP/NBT-plus for ALP |
Mabtech |
CAT#3650-10 |
| Dulbecco’s Modified Eagle’s Medium (DMEM) |
Corning |
CAT#10-013 |
| Ethidium Bromide |
VWR |
CAT#E406-5ML |
| Fetal Bovine Serum (FBS) |
Corning |
CAT#28622001 |
| Pierce Protease Inhibitor Mini Tablets |
Thermo Fisher Scientific |
CAT#A32953 |
| RBC Lysis Buffer (10X) |
BioLegend |
CAT#420301 |
| RIPA Lysis and Extraction Buffer |
Thermo Fisher Scientific |
CAT#89900 |
| TPCK-Treated Trypsin |
Worthington Biochemical |
CAT#LS003740 |
| 2X RNA Loading Dye |
Thermo Fisher Scientific |
CAT#R0641 |
| 3,3',5,5'-Tetramethylbenzidine (TMB) Membrane Peroxidase Substrate Plus |
Avantor |
CAT#K830 |
| Commercial Assays |
| BCA Protein Assay Kit |
Thermo Fisher Scientific |
CAT#23227 |
| Luna Universal One-Step RT-qPCR Kit |
New England Biolabs |
CAT#E3006 |
| Monarch Total RNA Miniprep Kit |
New England Biolabs |
CAT#T2010S |
| Neon Transfection System 100 μL Kit |
Invitrogen |
CAT#MPK10025 |
3. Results
3.1. Construction and Characterisation of Chimeric Rotaviruses Expressing Murine or Human Strain Outer Capsid Proteins
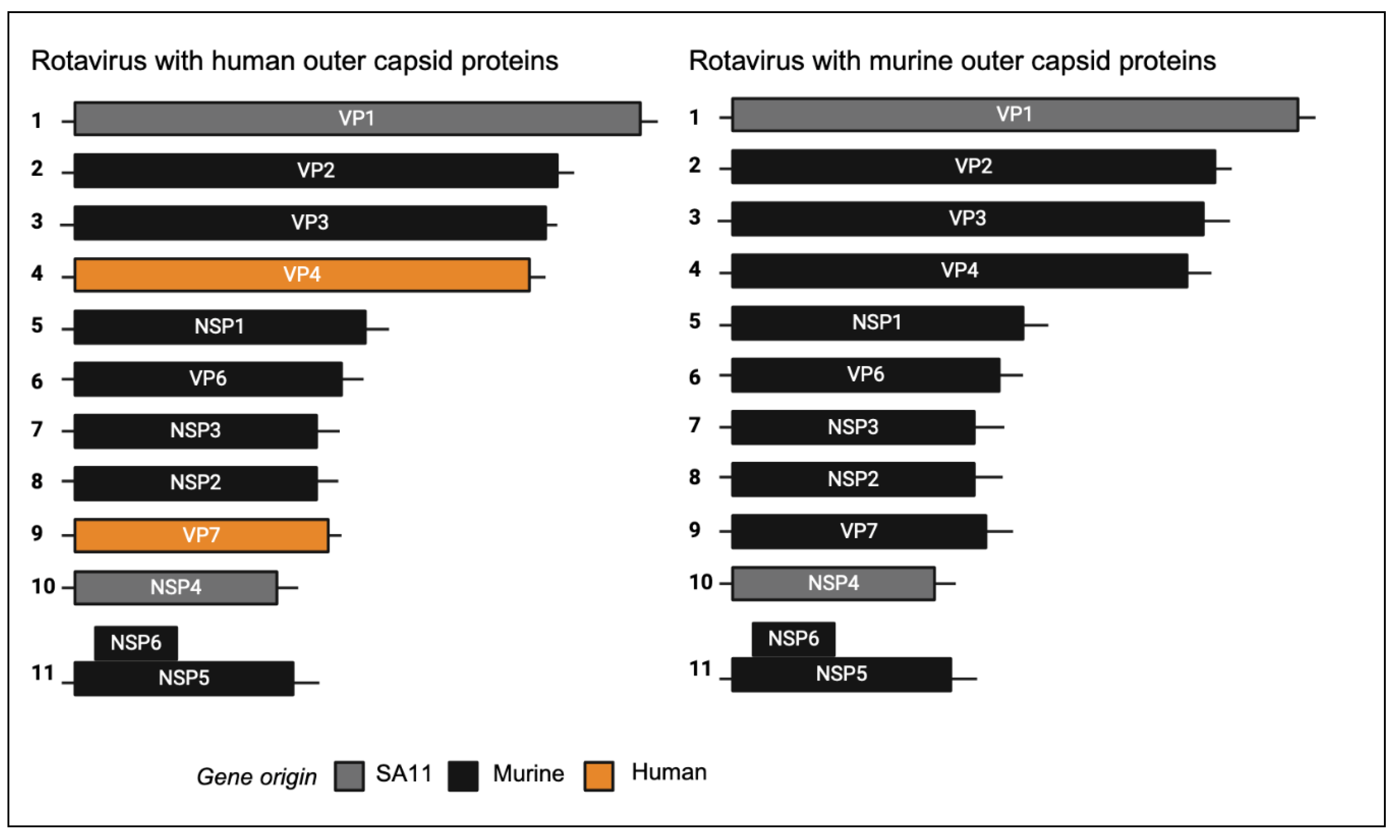
We previously generated a chimeric virus named rD6/2-2g, which has a murine rotavirus backbone of the non-tissue culture adapted wild-type murine EW strain with gene segment 4 (VP4) of simian rotavirus RRV strain, and gene segments 1 (VP1) and 10 (NSP4) of simian rotavirus strain SA11. In addition, we generated a murine reassortant virus with gene segment 4 (encoding VP4) from the human rotavirus strain CDC9 (G1P[
8]) that replicates in mice [
11]. To generate a murine-like virus with an entirely human outer capsid composition, we inserted both gene segment 4 and gene segment 9 (encoding VP7) into the murine-like rD6/2-2g backbone. As a control, we also produced a chimeric virus encoding the VP4 of the cell culture adapted murine ETD strain. This generated two different chimeric viruses that were identical except one encoded a human rotavirus outer capsid and the other a murine rotavirus outer capsid.
Figure 1.
Schematic diagram of segmented dsRNA genome of chimeric rotaviruses. Reverse genetics was used to generate chimeric viruses encoding either human or murine outer capsid proteins.
Figure 1.
Schematic diagram of segmented dsRNA genome of chimeric rotaviruses. Reverse genetics was used to generate chimeric viruses encoding either human or murine outer capsid proteins.
3.2. Chimeric Rotaviruses with Human Outer Capsid Proteins Replicate in Mice
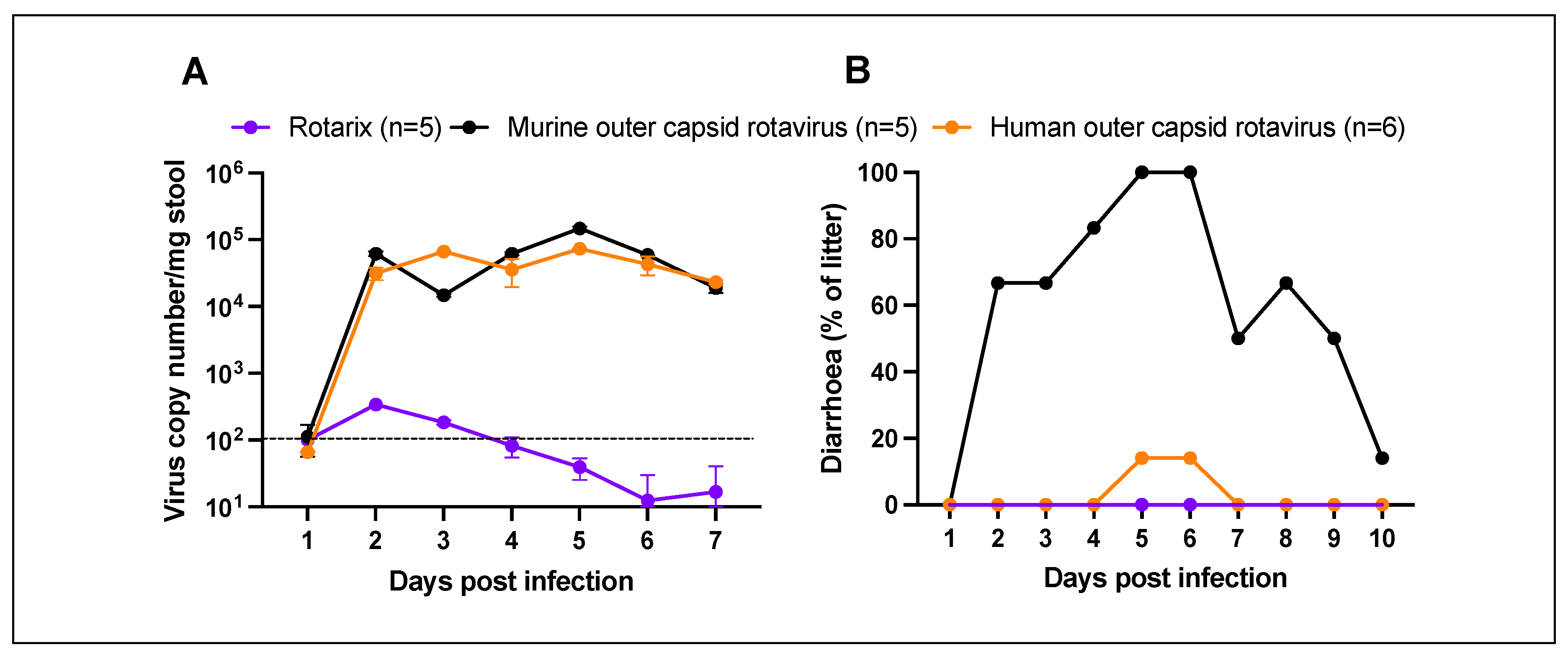
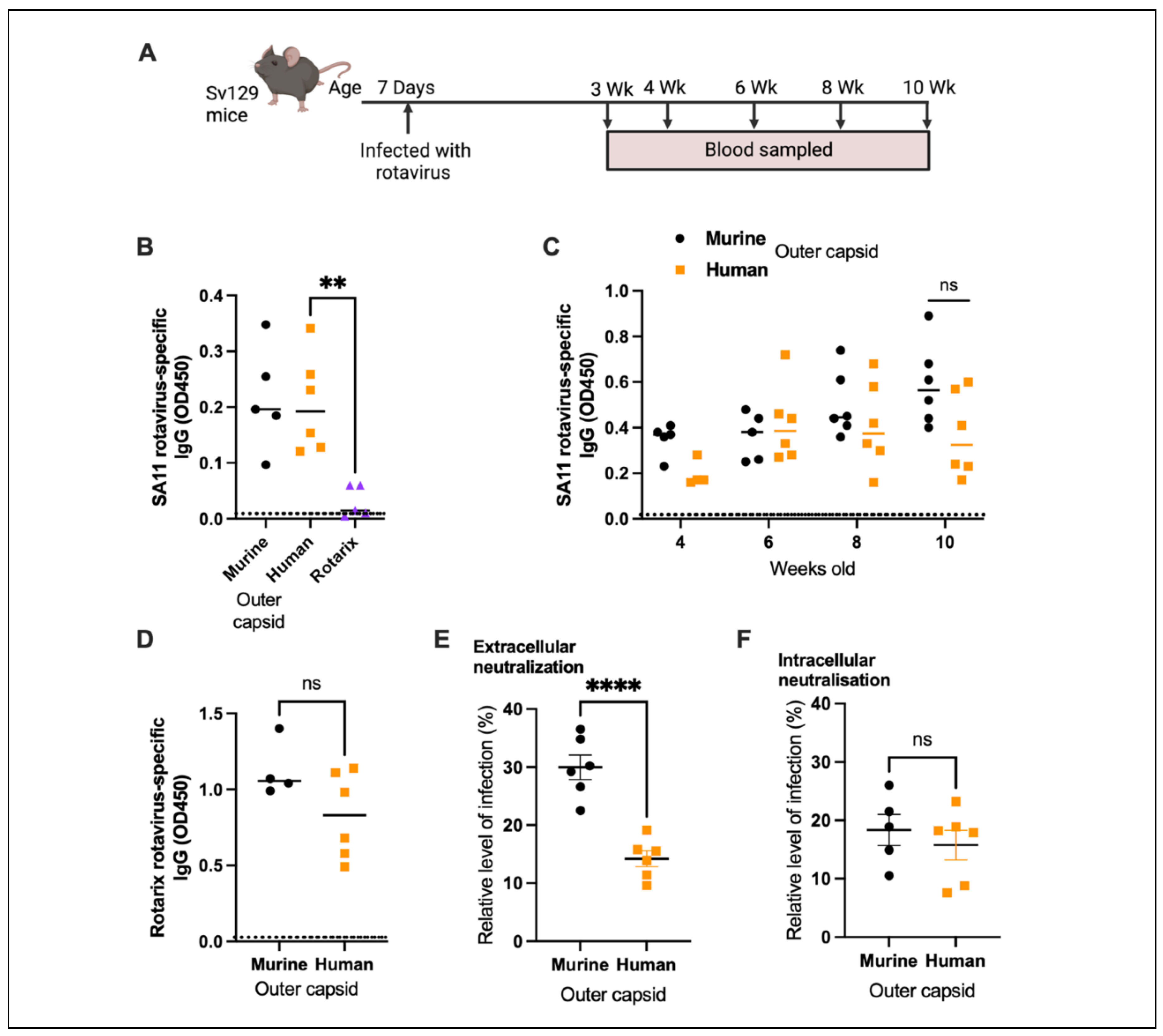
To examine the ability of chimeric rotaviruses to replicate and cause disease in mice, two separate litters of five pups (mouse outer capsid rotavirus) or six pups (human outer capsid rotavirus) were infected with virus by oral gavage at seven days old. For comparison with an entirely human rotavirus strain, an additional litter of five pups were infected with the human vaccine strain (Rotarix) at the same viral titre. All pups in each litter were monitored daily for the development of diarrhoea, and stool samples were collected for quantification by qPCR.
As expected, Rotarix did not robustly replicate and was detectable at only low copy numbers in pup stool (
Figure 2A). In contrast, both chimeric rotaviruses showed strong evidence of viral amplification from day 2 onwards. Viral shedding was detectable for at least seven days. As shown in
Figure 2B, diarrhoea was only detected in the litter of pups infected with chimeric virus expressing the murine rotavirus outer capsid proteins. The chimeric virus with human rotavirus outer capsid proteins was significantly attenuated in comparison. Given that viral loads were comparable, this suggests that the outer capsid proteins are important in the pathogenesis of diarrhoea in this model, but the mechanism remains unclear.
3.3. Human Rotavirus-Specific Antibody Responses Are Generated in Mice Infected with Chimeric Rotaviruses
Antibody responses following infection of seven-day-old mice with different rotaviruses were analysed to determine the immunogenicity of the chimeric strains. Serum samples were collected from mice infected with chimeric rotaviruses at 2 weeks post infection, and compared with samples from mice infected with Rotarix. An in-house sandwich ELISA based on the SA11 primate rotavirus strain was used to show that rotavirus-specific IgG were readily detected in mice infected with the chimeric rotaviruses, but no antibody response was evident in mice infected with Rotarix (
Figure 3B).
Next, we wanted to determine if there were any differences between longitudinal antibody responses in mice infected with either chimeric virus. We demonstrated that antibody responses were detected for over two months following infection (
Figure 3C). Despite heterogeneity in antibody responses within litters, there were no significant differences between IgG titres in mice infected with chimeric viruses expressing the murine or human outer capsid proteins. To determine whether use of a different antigen in our sandwich ELISA could differentiate between the antibody responses of the two litters, we generated a lysate of cells infected with the Rotarix strain. We analysed samples collected at the 10-week timepoint and, as shown in
Figure 3D, there was no significant difference between IgG responses detected using the Rotarix based ELISA. This likely reflects the fact that the infected cell lysate used in the sandwich ELISA contains all rotavirus proteins, and the chimeric viruses are identical except for VP4 and VP7. This supports previous work that has shown this type of ELISA predominantly detects antibodies specific for the inner capsid protein VP6 [
13], an immunodominant antigen that is identical between both chimeric virus strains.
To evaluate whether antibodies specifically targeting the human rotavirus capsid were induced by the human chimeric strain, we next performed serum neutralisation assays. Sera from 10-week-old mice were incubated with Rotarix for 1 hour, and then the resulting complexes were added to MA104 cells and infection allowed to proceed overnight (
Figure 3E). Whereas sera from mice infected with entirely murine chimeric virus could neutralise to a mean of 30.0% relative to no-sera control, sera from mice infected with virus containing human outer capsid proteins neutralised to 14.2% (
p <0.0001). This provides clear evidence that a human outer capsid-specific antibody response was induced by this chimeric virus.
To verify that this functional response was specific for the outer capsid and not for other rotavirus proteins, we also performed an intracellular neutralisation assay. This assay evaluates the activity of VP6-specific antibodies inside cells, achieved when serum antibodies are electroporated into the cytoplasm of MA104 cells [
12]. As the VP6 sequence was identical for both chimeric viruses, we hypothesised that intracellular neutralisation by sera from infected mice would be very similar. As shown in
Figure 3F, there was indeed no significant difference in intracellular neutralisation induced by the antibodies raised to both chimeric viruses.
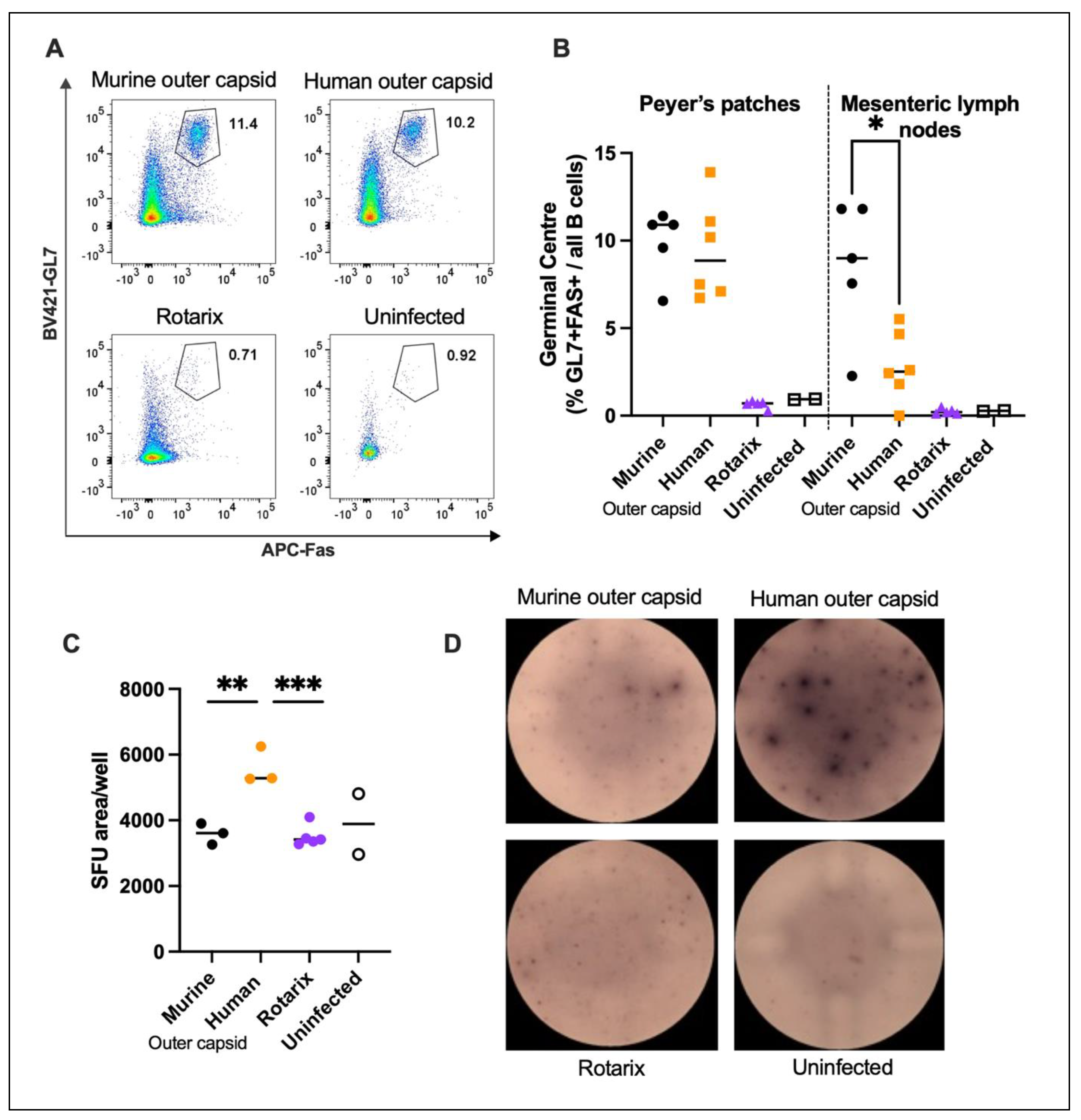
3.4. Chimeric Viruses Induced Human Rotavirus-Specific B Cell Responses in Mice
To complement and extend the results obtained from the serum antibody analysis of mice infected with chimeric viruses, we also characterised B cell responses to infection. Two litters of mice were infected with chimeric viruses, and 14 days post infection, cells from the spleen and draining lymph nodes (PP and MLN) were analysed. A third litter of mice were infected with an equal titre of the Rotarix vaccine strain to verify the inability of an entirely human rotavirus strain to induce a detectable B cell response in mice. Two age-matched uninfected mice were also included as controls. Flow cytometry on 50,000 cells was used to identify the germinal centre B cells present in PP and MLN of each litter of mice (
Figure 4 A, B). No germinal centre formation was observed in pups infected with Rotarix, in accordance with
Figure 3A, and in line with the lack of virus replication measured in
Figure 2A. In contrast, germinal centre formation in draining lymph nodes was readily observed in both litters of mice infected with the two chimeric rotavirus strains. Interestingly there was a small but significant difference (
p = 0.0128) between the number of germinal centre B cells measured in the MLN. This indicates that the chimeric strain with the human rotavirus outer capsid induces fewer germinal centre B cells than the entirely murine strain.
Whilst germinal centre B cell quantification clearly shows a strong B cell response, this approach does not identify antigen specificity of the B cells. To investigate this we performed ELISpot assays, using Rotarix as the antigen coated onto wells of an ELISpot plate. Splenocytes from three mice from each litter, plus two uninfected control mice were incubated in the antigen coated wells overnight, then staining for IgG production was completed the following day.
Figure 4C/D shows that mice infected with chimeric virus encoding the human outer capsid generated B cells that produced significantly more human rotavirus-specific antibody than the other viruses. This therefore confirms that a human rotavirus-specific B cell response can be readily induced and detected by infection with a chimeric virus.
4. Discussion
The species-specificity of rotaviruses has made development of a small animal model to study human rotavirus strains a significant challenge. This has hampered efforts to characterise the immune responses to human rotavirus by experimental infections, and meant that a robust pre-clinical system to analyse the efficacy of vaccine candidates and therapeutics has been lacking. To address this, we have successfully generated a novel approach to permit replication of rotaviruses encoding human rotavirus proteins in mice, and shown these mice generate robust antibody responses to human rotavirus proteins. This was achieved using reverse genetics to create a chimeric rotavirus that encodes the outer capsid proteins of a human rotavirus. We verified that whereas human rotavirus stains replicate poorly in mice and are not immunogenic, a chimeric rotavirus replicates to high titre and enables characterisation and quantification of human-rotavirus specific antibody responses.
A common alternative approach to studying non-murine viruses in mouse models is to use immunocompromised mice [
14,
15]. This has been used to permit replication of non-murine rotaviruses in mouse models e.g. STAT1 knockout mice [
6,
16], but this does not provide a comprehensive overview of the interactions between the innate and adaptive immune responses. Use of our chimeric virus system therefore has an advantage over use of immunocompromised mice as an immune response that more closely resembles the complexity and functionality seen in humans is induced. This generates a more representative picture of the response to the outer capsid epitopes that occurs in a human rotavirus infection. It is acknowledged that use of humanised mice could be a means of advancing this model further, but use of the widely available wild type 129S6/SvEvTac mouse strain makes our approach more accessible.
We propose a number of different situations where this chimeric rotavirus could be valuable for advancing our understanding of how to control human rotavirus infections. Firstly, the ability to study antibody responses to human rotavirus outer capsid proteins could be valuable for investigating a number of factors that have been associated with reduced rotavirus vaccine efficacy in low-middle income countries. For example, maternal antibodies have been correlated with a reduced ability of infants to seroconvert following vaccination in a number of vaccine clinical trials, yet the target of these interfering maternal antibodies is unclear. Infection of female mice with one strain, and then infection of their pups with another would determine whether antibodies targeting the outer capsid protein are responsible for interference. A second situation where these viruses could be useful is in pre-clinical vaccine trials. Whereas immune responses to mice vaccinated with new strategies targeting human rotaviruses e.g. recently described rotavirus VP8* mRNA vaccines [
17] can be readily studied in mice, the ability of these immune responses to protect against human rotavirus infection is not possible using standard strains. A chimeric virus infection would provide a solution for this. Finally, chimeric viruses could be used to test new therapeutic strategies targeting the human outer capsid protein. This would be especially useful for testing monoclonal antibodies specific for human rotavirus strains [
18,
19].
One potential limitation of our model is the inability of the human chimeric virus to recapitulate gastrointestinal disease seen in a natural species-specific rotavirus infections. Current rotavirus vaccines do not induce sterilising immunity, but instead reduce the severity of clinical signs following infection. The ideal model system would therefore induce gastroenteritis in mice in order to enable testing of vaccine candidates or therapeutics that aim to reduce the severity of clinical disease. Whilst viral replication in mice infected with rotaviruses with murine or human outer capsid proteins was similar, there was an interesting decrease in pathogenicity when switching from mouse to human. We found that the incidence of diarrhoea in pups infected with the rotavirus with a human capsid was significantly reduced. It is known that the pathogenesis of diarrhoea in rotavirus infection is multi-factorial, with reduced epithelial absorption, NSP4 enterotoxin and activation of the nervous system all reported to be involved [
20]. As the only difference between our two chimeric viruses was the outer capsid protein, this eliminates a possible role for NSP4, and instead suggests that interaction of the outer capsid proteins and the murine intestinal tract is important.
An additional limitation of our outer capsid chimera approach is that this model does not enable study of a human rotavirus VP6-specific immune response. This is an issue as the middle capsid protein VP6 is highly immunogenic and known to be the target of many rotavirus-specific antibodies in humans [
21,
22]. Furthermore, VP6-specific antibodies have been shown to be protective in mouse models [
12,
23]. The absence of human rotavirus-specific VP6 in our chimeric virus system means the repertoire of antibodies induced by chimeric viruses will not fully recapitulate that induced by natural human rotavirus strains. One possible solution to this issue could be to generate a chimeric rotavirus with human VP4, VP7 and VP6 on a murine backbone. However, this is predicted to be technically challenging as VP6 plays a crucial role in the structure and function of rotavirus, and therefore a human strain VP6 may not be compatible with a murine rotavirus backbone.
5. Conclusions
In conclusion, we have shown that using reverse genetics to manipulate rotavirus can be an effective strategy to study human rotaviruses in an immunocompetent pre-clinical model. This approach has the potential to facilitate future vaccine and therapeutic development against a childhood pathogen whose global disease burden is still unacceptably high.
Author Contributions
Conceptualization, Caitlin Williams, Sallie Permar and Sarah Caddy; Funding acquisition, Caitlin Williams, Sallie Permar and Sarah Caddy; Investigation, Sarah Woodyear, Tawny Chandler, Takahiro Kawagishi, Tom Lonergan and Vanshika Patel; Methodology, Siyuan Ding; Resources, Siyuan Ding and Sallie Permar; Writing – original draft, Sarah Woodyear and Sarah Caddy; Writing – review & editing, Tawny Chandler, Takahiro Kawagishi and Siyuan Ding. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a Multi-Investigator Seed Grant (MISG) from Cornell University, New York, USA.
Data Availability Statement
All the relevant data are provided in this paper.
Acknowledgments
We thank the staff at the Baker Institute for Animal Health for helping to care for our mice, in particular Julie Reynolds, and Dr. John Parker for kindly gifting MA104 cells.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study, collection, analyses, interpretation of data, manuscript writing, or in the decision to publish the results.
References
- Ruiz-Palacios, G.M.; Pérez-Schael, I.; Velázquez, F.R.; Abate, H.; Breuer, T.; Clemens, S.C.; Cheuvart, B.; Espinoza, F.; Gillard, P.; Innis, B.L.; et al. Safety and Efficacy of an Attenuated Vaccine against Severe Rotavirus Gastroenteritis. N. Engl. J. Med. 2006, 354, 11–22. [Google Scholar] [CrossRef]
- Vesikari, T.; Matson, D.O.; Dennehy, P.; Van Damme, P.; Santosham, M.; Rodriguez, Z.; Dallas, M.J.; Heyse, J.F.; Goveia, M.G.; Black, S.B.; et al. Safety and Efficacy of a Pentavalent Human–Bovine (WC3) Reassortant Rotavirus Vaccine. N. Engl. J. Med. 2006, 354, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Cates, J.E.; Tate, J.E.; Parashar, U. Rotavirus Vaccines: Progress and New Developments. Expert Opin. Biol. Ther. 2022, 22, 423–432. [Google Scholar] [CrossRef]
- Desselberger, U. Differences of Rotavirus Vaccine Effectiveness by Country: Likely Causes and Contributing Factors. Pathogens 2017, 6, 65. [Google Scholar] [CrossRef]
- Caddy, S.; Papa, G.; Borodavka, A.; Desselberger, U. Rotavirus Research: 2014–2020. Virus Res. 2021, 304, 198499. [Google Scholar] [CrossRef] [PubMed]
- Feng, N.; Yasukawa, L.L.; Sen, A.; Greenberg, H.B. Permissive Replication of Homologous Murine Rotavirus in the Mouse Intestine Is Primarily Regulated by VP4 and NSP1. J. Virol. 2013, 87, 8307–8316. [Google Scholar] [CrossRef] [PubMed]
- Cook, N. The Zoonotic Potential of Rotavirus. J. Infect. 2004, 48, 289–302. [Google Scholar] [CrossRef]
- Kanai, Y.; Komoto, S.; Kawagishi, T.; Nouda, R.; Nagasawa, N.; Onishi, M.; Matsuura, Y.; Taniguchi, K.; Kobayashi, T. Entirely Plasmid-Based Reverse Genetics System for Rotaviruses. Proc. Natl. Acad. Sci. 2017, 114, 2349–2354. [Google Scholar] [CrossRef]
- Kanai, Y.; Kobayashi, T. Rotavirus Reverse Genetics Systems: Development and Application. Virus Res. 2021, 295, 198296. [Google Scholar] [CrossRef]
- Desselberger, U. Rotaviruses. Virus Res. 2014, 190, 75–96. [Google Scholar] [CrossRef]
- Sánchez-Tacuba, L.; Kawagishi, T.; Feng, N.; Jiang, B.; Ding, S.; Greenberg, H.B. The Role of the VP4 Attachment Protein in Rotavirus Host Range Restriction in an In Vivo Suckling Mouse Model. J. Virol. 2022, 96, e00550-22. [Google Scholar] [CrossRef] [PubMed]
- Caddy, S.L.; Vaysburd, M.; Wing, M.; Foss, S.; Andersen, J.T.; O‘Connell, K.; Mayes, K.; Higginson, K.; Iturriza-Gómara, M.; Desselberger, U.; et al. Intracellular Neutralisation of Rotavirus by VP6-Specific IgG. PLOS Pathog. 2020, 16, e1008732. [Google Scholar] [CrossRef] [PubMed]
- Ishida, S.; Feng, N.; Gilbert, J.M.; Tang, B.; Greenberg, H.B. Immune Responses to Individual Rotavirus Proteins Following Heterologous and Homologous Rotavirus Infection in Mice. J. Infect. Dis. 1997, 175, 1317–1323. [Google Scholar] [CrossRef] [PubMed]
- Winkler, C.W.; Peterson, K.E. Using Immunocompromised Mice to Identify Mechanisms of Zika Virus Transmission and Pathogenesis. Immunology 2018, 153, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Kutle, I.; Dittrich, A.; Wirth, D. Mouse Models for Human Herpesviruses. Pathogens 2023, 12, 953. [Google Scholar] [CrossRef] [PubMed]
- Vancott, J.L.; McNeal, M.M.; Choi, A.H.C.; Ward, R.L. The Role of Interferons in Rotavirus Infections and Protection. J. Interferon Cytokine Res. 2003, 23, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Roier, S.; Mangala Prasad, V.; McNeal, M.M.; Lee, K.K.; Petsch, B.; Rauch, S. mRNA-Based VP8* Nanoparticle Vaccines against Rotavirus Are Highly Immunogenic in Rodents. Npj Vaccines 2023, 8, 190. [Google Scholar] [CrossRef] [PubMed]
- Langel, S.; Steppe, J.; Chang, J.; Travieso, T.; Webster, H.; Otero, C.; Williamson, L.; Crowe, J.; Greenberg, H.; Wu, H.; et al. Protective Transfer: Maternal Passive Immunization with a Rotavirus-Neutralizing Dimeric IgA Protects against Rotavirus Disease in Suckling Neonates; Immunology, 2021.
- Zha, M.; Yang, J.; Zhou, L.; Wang, H.; Pan, X.; Deng, Z.; Yang, Y.; Li, W.; Wang, B.; Li, M. Preparation of Mouse Anti-human Rotavirus VP7 Monoclonal Antibody and Its Protective Effect on Rotavirus Infection. Exp. Ther. Med. 2019. [Google Scholar] [CrossRef]
- Crawford, S.E.; Ramani, S.; Tate, J.E.; Parashar, U.D.; Svensson, L.; Hagbom, M.; Franco, M.A.; Greenberg, H.B.; O’Ryan, M.; Kang, G.; et al. Rotavirus Infection. Nat. Rev. Dis. Primer 2017, 3, 17083. [Google Scholar] [CrossRef]
- Svensson, L.; Sheshberadaran, H.; Vene, S.; Norrby, E.; Grandien, M.; Wadell, G. Serum Antibody Responses to Individual Viral Polypeptides in Human Rotavirus Infections. J. Gen. Virol. 1987, 68, 643–651. [Google Scholar] [CrossRef]
- Johansen, K.; Granqvist, L.; Karlén, K.; Stintzing, G.; Uhnoo, I.; Svensson, L. Serum IgA Immune Response to Individual Rotavirus Polypeptides in Young Children with Rotavirus Infection. Arch. Virol. 1994, 138, 247–259. [Google Scholar] [CrossRef] [PubMed]
- Burns, J.W.; Siadat-Pajouh, M.; Krishnaney, A.A.; Greenberg, H.B. Protective Effect of Rotavirus VP6-Specific IgA Monoclonal Antibodies That Lack Neutralizing Activity. Science 1996, 272, 104–107. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).