1. Introduction
Xuanwei ham, as a traditional famous speciality product of Yunnan Province, has a his-tory of more than two hundred and seventy years [
1], and is favoured by consumers for its delicious taste, rich nutrition and strong aroma [
2]. Xuanwei ham is mainly made from Wujin pigs in the Xuanwei area, which are prepared by trimming, salting and air-drying under the unique geographical and natural conditions of the Wumeng Mountain area of the Yunnan-Guizhou Plateau. Unique fermentation conditions and a dry-curing process have created the unique flavour and texture of Xuanwei ham [
3,
4].
Microorganisms play an important role in flavour formation and preservation of fer-mented meat products. Li [
5] et al. explored the relationship between microbial communi-ties and VOCs in dry-cured boneless hams and found that molds may be the main con-tributors to the development and differentiation of flavour qualities of dry-cured boneless hams from Chinese hams. During fermentation of meat products, microorganisms induce a series of biochemical reactions such as protein hydrolysis, amino acid degradation, li-polysis and lipid oxidation [
6] that promote the production and accumulation of aromatic compounds to enhance meat product flavour and quality [
7,
8]. During processing, fat is hydrolyzed to form free fatty acids under the action of phosphatases and endogenous en-zymes, leading to an increase in free fatty acid content [
9]. Xuanwei ham, as a typical fermented meat product, is often evaluated by maturity period to assess quality, and it is generally believed that the longer the maturity period the better the quality, with greater accumulation of flavour components [
10]. Li [
11] et al. in revealing the intrinsic relation-ship between microbial communities and physicochemical properties during Xuanwei ham maturation showed that hams with a long maturity period scored the highest marks in colour, texture, flavour, aroma, and acceptability.
It is well known that microorganisms and endogenous enzymes are important ham qual-ity and flavour determinants [
12,
13]. In fermented fish, Wang [
14] et al. explored the po-tential influence of microorganisms on flavour formation by exploring the relationships between microbial diversity and changes in flavour components. In sour meat, Zhong [
15] et al. found that Lactobacillus and Staphylococcus were associated with changes in most flavour compounds. In Jinhua ham, Deng [
16] et al. found that Saccharomyces, Aspergil-lus and Staphylococcus were significantly and positively correlated with post-ripening flavor through the correlation between microbial diversity and flavour component chang-es. Currently, microbial community studies in Xuanwei ham are relatively simple, and mainly focused on analyzing its microbial species. However, the relationship between Xuanwei ham flavor changes after ripening and the microbial community remains un-clear.
We used gas chromatograph-mass spectrometry (GC-MS) and high-throughput sequenc-ing to determine the VOCs and microbial community structure of Xuanwei ham from dif-ferent years (1, 2, 3, and 4 years), and the relationships between dominant bacterial groups and flavour substances, to explore microbial influence on flavor. It could be significant for improving ham stability and quality during production to better understand the relation-ships between microorganisms and ham flavor.
2. Materials and Methods
2.1. Materials and Reagents
Three pieces of Xuanwei ham aged 1, 2, 3 and 4 years, processed from Wujin pigs reared from the same litter of the same age and similar weight were purchased from Xuanwei Yi-ji Foods Co. The hind legs of six pigs were taken as raw materials after slaughtering, and the average leg weight was 17.62±0.66 kg. Processing was carried out according to the technical regulations of Xuanwei ham production including: trimming and shaping, salting and curing, stacking and turning, washing and sunshine shaping, hanging and air-drying, and fermentation management. Processing time was from December 2019 to December 2023, during which 3 Xuanwei hams were randomly selected each year. The biceps femoris (BF) was taken as a sample, which was vacuum-packed after sampling and stored in a refrigerator at -80◦C for the determination of VOCs; o-phthalaldehyde (OPA) (analytically pure, sigma); FMOC (analytically pure, sigma); 3-mercaptopropionic acid (analytically pure, sigma); concentrated hydrochloric acid (analytically pure, Guangzhou Chemical Reagent Factory); and Boric acid (analytically pure, Guangzhou Chemical Reagent Factory); Sodium hydroxide (analytically pure, Guangzhou Chemical Reagent Factory); Sodium dihydrogen phosphate dihydrate (analytically pure, Guang-zhou Chemical Reagent Factory); Disodium hydrogen phosphate dodecahydrate (analyt-ically pure, Guangzhou Chemical Reagent Factory); 17 kinds of amino acid mixed stand-ard (2.5 umol/mL, sigma); asparagine, glutamine, citrulline, n-valine; Tryptophan, 21-hydroxyproline, sarcosine standard (analytical purity, sigma); methanol (chromato-graphic purity, CNW); 2.5 mg/mL FMOC-Cl acetonitrile solution; and o-phthalaldehyde (OPA) solution.
2.2. Instruments and Equipment
Constant temperature magnetic stirrer (08-2T, Shanghai Meiyinpu Instrumentation Man-ufacturing Co., Ltd.); solid-phase microextraction device (57330-U, supelco); 50/30um DVB/CAR/PDMS solid-phase microextraction needles (57348-U, supelco) gas chromatog-raphy-mass spectrometry (Agilent 6890N-5973 GC-MS); gas chromatography column; HP-INNOWax (60 m x 250 μm x 0 25 μm); Agilent 1100 liquid chromatograph. ); GC column; HP-INNOWax (60 m x 250 μm x 0.25 μm); Agilent 1100 liquid chromatograph.
2.3. Experimental Methods
2.3.1. Sample Processing
The Xuanwei ham BF portion (biceps femoris) was used as a sample after mold washing and trimming (
Figure 1).
2.3.2. Free Amino Acid Content Determination
We followed the method of Qiu et al [
17]. Briefly, the ham was stirred, a 0.5 g sample was weighed and placed in a 10 mL centrifuge tube, and 5 mL of 0.01 mol/L hydrochloric acid (or purified water) added. It was mixed and placed in a boiling water bath for 30 min, then centrifuged at 10,000 rpm for 10 min. The supernatant was retrieved, and the precipitate was added with another 4 mL of 0.01 mol/L hydrochloric acid suspension, ultrasonified for 5 min, centrifuged, combined with the supernatant, and fixed. It was then concentrated to 10 mL and measured by membrane. The resulting solution was derivatised with o-phthalaldehyde (OPA) for primary amino acids and fluorene-methoxycarbonyl chloride (FMOC) for secondary amino acids using Agilent's automatic on-line derivatization method [
18]. Chromatographic conditions: ZORBAX Eclipse AAA (4.6 x 150 mm, 3.5 μm); detection signals: UV 338 nm (0~19 min), 266 nm (19.01~25 min); mobile phase A: 40 mM sodium dihydrogen phosphate (pH 7.8); mobile phase B: acetonitrile/methanol/water = 45/45/10 flow rate: 1.0 mL/min; column temperature 45°C.
2.3.3. Taste Active Value (TAV) calculation
The TAV value can indicate the degree of contribution of the taste substance to the overall taste of the sample [
19].TAV>1 means that the substance has an important effect on the taste, and the larger the value, the larger the contribution; TAV<1 means that it does not have an important effect on the taste. The calculation formula is as follows:
where, 𝐶 is the content of the taste substance, mg/100 g; 𝑇 is the threshold value of the taste substance, mg/100 g.
2.3.4. Volatile Flavour Content Determination
We followed the method of Cao [
20] et al. Extraction conditions: A 5 g sample was weighed and placed into a 20mL extraction vial, 100uL of 2,4,6-trimethylpyridine (0.05 mg/mL) was added as the internal standard, sealed, and placed in a water bath at 85°C with magnetic stirring speed of 500 rpm, and equilibrated for 20min, then inserted into an extraction needle and extracted for 30 min. Before using the extraction needle, it was activated at the gas injection port for 20 min (250°C).
GC-MS conditions: inlet temperature 250°C, gas interface temperature 250°C, carrier gas flow rate 1.5 mL/min, no shunt injection. Temperature increase procedure: initial 40°C, hold for 5 min, 5°C /min increase to 250°C hold for 10 min. Ion source temperature 230°C, four-stage rod temperature 150°C, EI ionisation 70 eV, full scan 35~550 da.
Data analysis: Qualitative analysis: The mass spectrometry data were compared and searched in the NIST 17 spectral library, and compounds with a match of > 80% were retained. Quantitative analysis: 2,4,6-trimethylpyridine was used as the internal standard for the relative quantification of the compounds. The formulae were as follows:
where Cx is the content of the unknown flavour substance (ug/100 g); Sx is the chromatographic peak area of the unknown flavour substance; Ci is the concentration of the internal standard (mg/mL); Vi is the volume of the internal standard added (uL); Si is the chromatographic peak area of the internal standard; and Mx is the mass of the sample (g).
2.3.5. Electronic Nose Odour Fingerprints Extraction
Pre-treatment of ham samples: 1d before the experiment, the ham samples were thawed at 4℃. For electronic nose detection, a knife was used to scrape off the sample surface material (mainly some molds and dust), the inner core of the peeled Xuanwei ham samples was divided into 0.6×0.6×0.6 cm small pieces. About 75 g of each piece was divided into three 25 g parts, and each placed into 40mL headspace sampling bottles, caps screwed tightly on, placed in an oven (set at 50°C), and left for 30min. Then, according to the principles of solid-gas equilibrium and solid-liquid equilibrium, The gas component is volatilized, and the upper gas in the sample bottle reaches a stable state, and electronic nose detection is carried out.
Odour fingerprint data acquisition: cNose-10 electronic nose produced by Shanghai Baosheng Technology Company was used for the determination. Through the pre-test, the following conditions were set: carrier gas was dry air, gas flow rate was 1.0 L/min, test time was 120 s, injection time was 90 s, and then odour fingerprint data were collected from all the samples.
2.4. Experimental Procedures of Metagenomic Sequencing
Genomic DNA was extracted from the ham samples using the CTAB method. The concentration, integrity and purity of DNA were detected using Agilent5400.After the DNA samples passed the test, the samples were fragmented to a size of 350bp using a Covaris ultrasonic crusher, and then the whole library preparation was completed by the steps of end repair, addition of A-tail, addition of sequencing junction, fragment screening, PCR amplification and purification. Then further PCR amplification was performed, the PCR products were purified and the library quality was assessed by Agilent5400 system (Agilent,USA), and finally the library concentration was quantified by QPCR (1.5 nM). Based on the effective library concentration and target data volume, the eligible libraries were up-sequenced on the Illumina platform using the PE150 strategy. Macrogenomic sequencing was performed using the Illumina NovaSeq high-throughput sequencing platform to obtain raw macrogenomic data (Raw Data) of ham samples. The Raw sequencing data were preprocessed using Kneaddata software to ensure the reliability of the data,. [
21,
22,
23].
For comparative analyses, we used Kraken2 and custom databases for taxonomic delineation of species, and then predicted the actual relative abundance of species in the samples using Bracken. [
24,
25,
26,
27].
2.5. Data processing
Data were collated using Excel 2016. Data are presented as mean ± standard deviation (N=3). Statistical analyses were performed using IBM SPSS Statistics 26 software and general linear model variable analysis was performed using Duncan's test, with P < 0.05 indicating significant differences. Microbial flora mapping was performed using the Biotech Cloud platform
https://www.bioincloud.tech/. The analysis was performed according to Gao et al [
28] with slight modification for data visualisation .
3. Results
3.1. Free Amino Acid Content and TAV Value of Differently-Aged Xuanwei Hams
Free amino acid content, as the main contributor to ham flavour and texture, is related to protein degradation as well as amino acid degradation [
29], and can be classified as fresh, sweet, bitter, and tasteless amino acids according to flavoring characteristics [
30]. A total of 25 free amino acids were detected in differently-aged Xuanwei hams (
Table 1), which initially decreased and then increased, reaching a peak content in W4 (4328.70 mg/100 g), which was 65.45% higher than W3.
TAV is the taste activity value, and free amino acids with TAV>1 are considered to have a strong contribution to Xuanwei ham flavor, and the higher the TAV, the greater the contribution. Aspartic acid and glutamic acid are fresh flavour substances, and the TAV values of glutamic acid in differently-aged Xuanwei hams were higher than aspartic acid, and glutamic acid content in W3 was significantly lower than W1, W2, and W4 (P<0.05). Moreover, glutamic acid not only helps to improve the fresh taste, but also provides the amino receptor α-ketoglutaric acid when transamination of branched-chain amino acids occurs, which promotes characteristic flavor generation [
31]. Alanine is the main sweet amino acid in Xuanwei ham because it has the lowest threshold among sweet amino acids and can be converted to aldehydes in later stages to promote ham flavor [
32]. Alanine content was significantly lower in W4 than W1, W2, and W3 (P<0.05), and the glutamic acid TAV in W3 (1.06) was significantly lower than W1, W2, and W4 (P<0.05). W4 had the highest arginine content (TAV value of 5.59), which had a bitter flavor with a weak sweetness, which could be masked by NaCl or glutamic acid [
33]. Similarly, lysine, which was bitter in W4, also had a high TAV (6.90). A high bitter amino acid content can adversely affect ham quality, however, appropriate bitterness can increase the complexity of the flavor presentation, which can enhance overall ham flavor [
34].
3.2. VOCs in Xuanwei Ham of Different Vintages
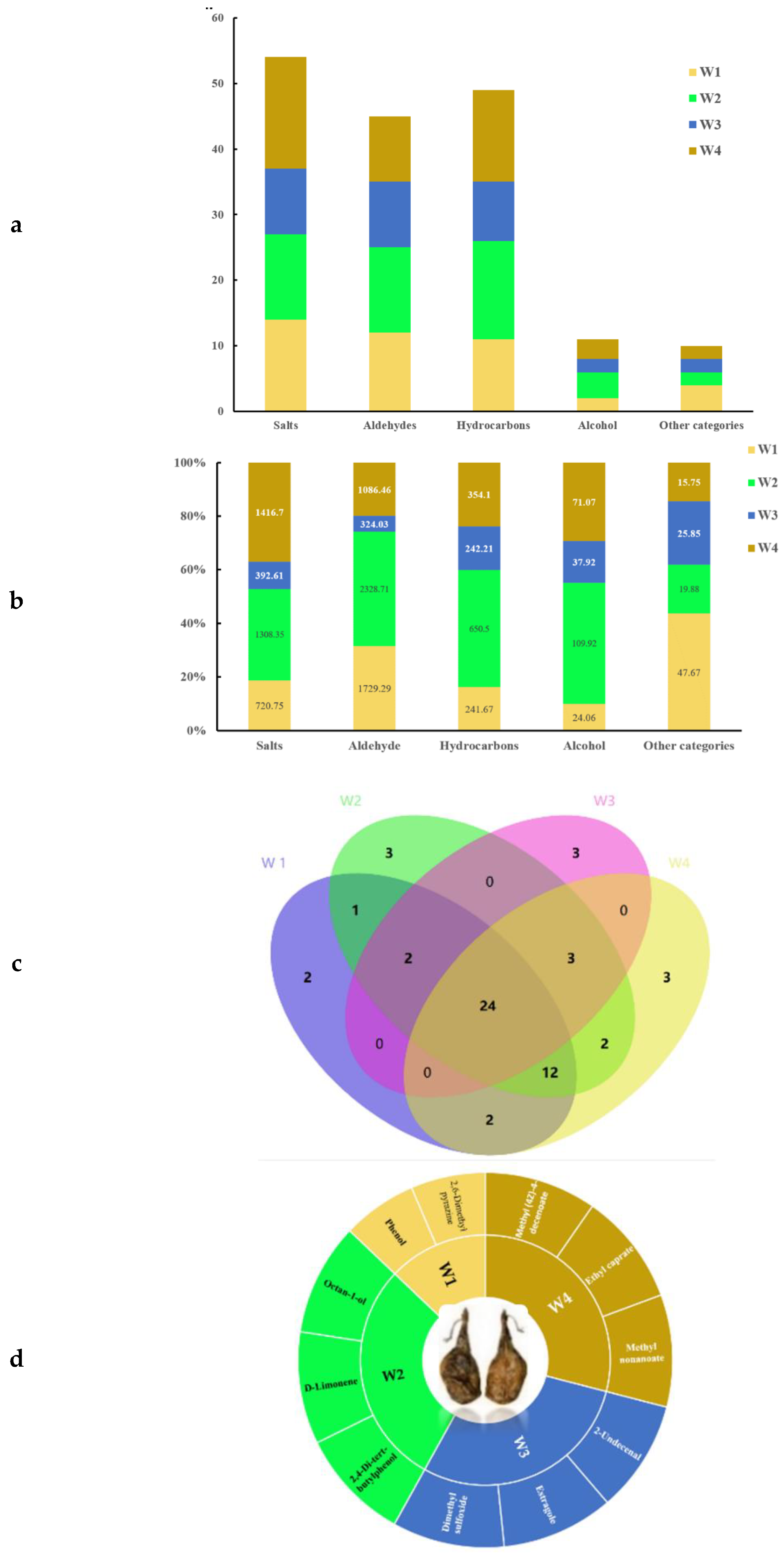
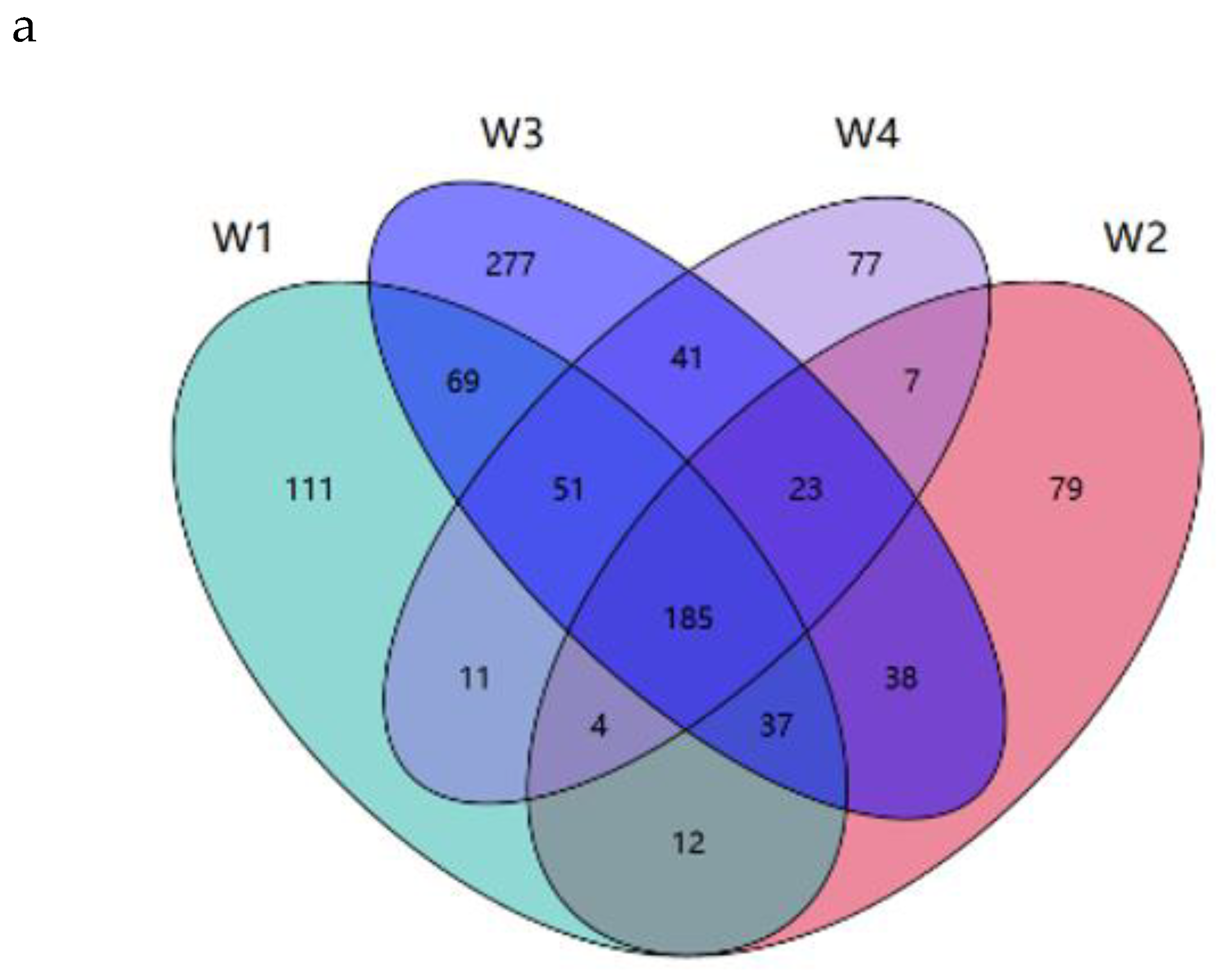
A total of 59 VOCs, including 17 esters, 14 aldehydes, 15 hydrocarbons, 6 alcohols, 9 acids and 7 others, were detected in the four differently-aged Xuanwei hams (
Table 2). To visualize the distribution of the types, quantities and contents of the compounds, stacked bar charts, Wayne diagrams and Asahi diagrams were plotted and analyzed (
Figure 3). Compound types and contents contained in differently-aged Xuanwei hams were quite different (
Figure 3a,b) . W2 had highest content of VOCs substances at 4417.35 μg/100 g, which indicated that amino acid catabolism, fatty acid metabolism, and carbohydrate decomposition were directly involved in VOCs formation [
35], followed by W4 (2944.08 μg/100 g), W1 (2763.44 μg/100 g) and W3 (1022.62 μg/100 g). Twenty-four VOCs were found in differently-aged Xuanwei hams, of which 26 were found in W1, W2 and W3 in total, 27 in W2, W3 and W4, 41 in W2 and W4, and 26 in W1 and W3 in total (
Figure 3c). Additionally, two VOCs were detected in W1 only, three in W2 only (
Figure 3d?), and three in W3 and W4 respectively(
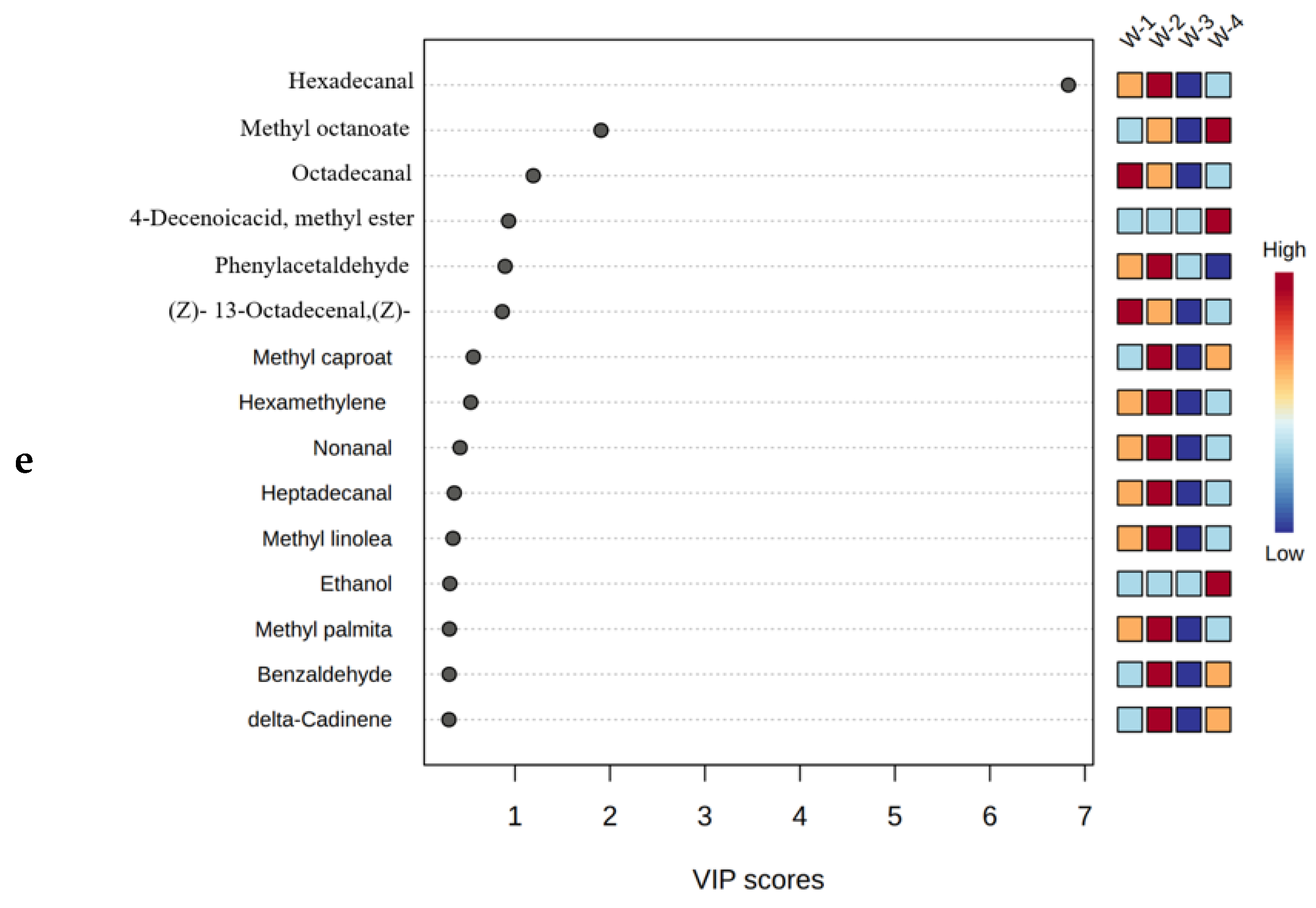
Figure 3d). The VOCs with VIP scores>1 were (Z)- 13-Octadecenal, (Z)- 2, Phenylacetaldehyde, 4-Decenoicacid, methyl ester, Octadecanal, Methyl octanoate and Hexadecanal (
Figure 3e). Hexadecanal, and the VIP scores of Methyl octanoate and Hexadecanal were>2, and the contents were higher in W4 and W2, respectively, which indicated that they had the highest contribution to Xuanwei ham flavor. Their highest content was found in W1, which contributed to its higher flavor. Hexadecanal has meat flavour and a weak aroma of flowers, and has bitter almond flavour and nutty flavour in meat as the main aromatic aldehyde [
36,
37].
W1 had a relatively short fermentation time (
Table 1), its highest VOCs were aldehydes (1729.29 μg/100 g), followed by esters (720.75 μg/100 g) and hydrocarbons (241.67 μg/100 g). Notably,hexadecanal has a weak aroma of flowers and wax, which plays an important role in improving ham flavour, and its content was higher in W2 than in all other years, especially W3 (P<0.05). There was a significant increase in flavour substances after three years of ham fermentation, which may be due to the length of time ham was associated with microbial fermentation factors. Organic acids were generated by esterification reaction with alcohol compounds, which promoted the increase of ham VOCs [
38]. Ester content in W4 was significantly higher than in the other three years (P<0.05), especially for trimethyl borate, methyl caprylate, and methyl decanoate, which played an important role in contributing to the flavour of W4.
Table 2.
VOCs content of Xuanwei ham.
Table 2.
VOCs content of Xuanwei ham.
| categories |
PK |
Library |
CAS |
RT/min |
Content /(μg/100g) |
| W1 |
W2 |
W3 |
W4 |
| Salts |
A1 |
Trimethyl borate |
000121-43-7 |
6.18 |
118.26±2.63d |
239.82±5.46a |
179.07±9.51b |
152.94±2.91c |
| A2 |
Methyl butyrate |
000623-42-7 |
8.11 |
11.86±2.13c |
21.96±1.46b |
\ |
26.94±3.03a |
| A3 |
Methyl 2-methylbutyrate |
000868-57-5 |
8.71 |
43.47±1.79a |
\ |
\ |
38.71±2.01b |
| A4 |
Methyl isovalerate |
000556-24-1 |
9.03 |
41.64±1.72b |
53.93±2.68a |
\ |
33.49±3.12c |
| A5 |
Methyl caproate |
000106-70-7 |
14.13 |
89.44±1.86b |
213.30±2.49a |
10.29±1.22c |
93.04±2.35b |
| A6 |
Methyl octanoate |
000111-11-5 |
20.30 |
70.34±2.04c |
234.75±3.22b |
\ |
364.52±3.69a |
| A7 |
Methyl n-caprate |
000110-42-9 |
25.74 |
25.50±2.00c |
51.34±2.19b |
7.57±2.07d |
261.30±1.85a |
| A8 |
Dodecanoic acid, methylester |
000111-82-0 |
30.56 |
4.36±1.83c |
12.07±1.34b |
5.41±1.94c |
18.25±2.15a |
| A9 |
methyl myrist |
000124-10-7 |
34.88 |
27.54± 0.99b |
39.53±2.25a |
18.08±2.71c |
37.64±1.67a |
| A10 |
Methyl hexadecanoate |
000112-39-0 |
38.85 |
85.20±1.93b |
141.80±1.49a |
63.71±1.50d |
75.93±3.28c |
| A11 |
1,6-Hexanediol diacrylate |
013048-33-4 |
38.92 |
62.45±2.52b |
91.83±1.44a |
13.55±2.00d |
27.63±1.25c |
| A12 |
Octadecanoic acid,methyl ester |
000112-61-8 |
42.51 |
22.16±1.53b |
31.57±2.13a |
17.43±1.81c |
21.82±1.53b |
| A13 |
Methyl oleate |
000112-62-9 |
42.86 |
55.45±1.96c |
84.05±2.43a |
57.44±2.15c |
67.26±0.84b |
| A14 |
Methyl linoleate |
000112-63-0 |
43.66 |
63.07±2.61b |
83.39±2.71a |
20.04±3.44d |
44.55±1.99c |
| A15 |
Methyln-nonanoate |
001731-84-6 |
23.16 |
\ |
\ |
\ |
13.53±1.81 |
| A16 |
Ethyl caprate |
000110-38-3 |
25.78 |
\ |
\ |
\ |
6.61±1.92 |
| A17 |
4-Decenoic acid, methylester, (4Z)- |
007367-83-1 |
27.05 |
\ |
\ |
\ |
132.56±2.37 |
| |
Subtotal |
|
|
720.75±5.59c |
1308.35±2.02b |
392.61±9.97d |
1416.70±0.64a |
| Aldehyde |
B1 |
Isovaleraldehyde |
000590-86-3 |
6.46 |
32.01±1.77c |
114.82±1.50a |
9.40±1.07d |
43.44±1.79b |
| B2 |
Hexanal |
000066-25-1 |
11.05 |
10.31±1.95b |
15.59±2.14a |
4.44±1.12c |
\ |
| B3 |
1-Nonana |
000124-19-6 |
20.50 |
59.30±1.73b |
147.36±2.16a |
11.70±1.35c |
56.38±1.00b |
| B4 |
Phenylmethana |
000100-52-7 |
24.21 |
62.51±2.16c |
157.20±2.41a |
15.51±1.27d |
74.66±0.74b |
| B5 |
Phenylacetaldehyde |
000122-78-1 |
27.02 |
76.54±0.54b |
122.30±1.75a |
28.93±1.70c |
\ |
| B6 |
trans,trans-2,4-Decadien-1-al |
025152-84-5 |
30.97 |
8.45±2.18b |
18.97±1.66a |
\ |
\ |
| B7 |
Tetradecanal |
000124-25-4 |
33.20 |
18.92±1.57b |
29.09±1.47a |
\ |
16.63±2.04b |
| B8 |
Pentadecanal |
002765-11-9 |
35.34 |
26.31±2.19b |
44.21±2.00a |
\ |
24.52±2.16b |
| B9 |
Hexadecanal |
000629-80-1 |
37.41 |
1029.03±2.34b |
1334.18±2.37a |
169.45±1.84d |
638.27±1.84c |
| B10 |
Heptadecanal |
1000376-70-0 |
39.35 |
68.20±2.04b |
72.42±1.82a |
14.37±1.25d |
46.29±2.06c |
| B11 |
Octadecanal |
000638-66-4 |
41.23 |
195.35±2.03a |
143.28±1.77b |
25.42±2.09d |
98.52±2.35c |
| B12 |
13-Octadecenal, (13Z)- |
058594-45-9 |
41.66 |
142.35±1.82a |
112.64±1.96b |
17.48±1.92d |
75.19±2.05c |
| B13 |
2-Methylbutyraldehyde |
000096-17-3 |
6.38 |
\ |
16.66±1.50a |
\ |
12.56±2.07b |
| B14 |
2-Undecenal |
002463-77-6 |
29.61 |
\ |
\ |
27.33±1.87 |
\ |
| |
Subtotal |
|
1729.29±10.07b 2328.71±3.92a |
324.03±10.79d |
1086.46±4.30c |
| Hydrocarbons |
C1 |
Valencene |
004630-07-3 |
28.07 |
8.35±2.25ab |
5.54±2.07b |
\ |
9.27±2.05a |
| C2 |
trans-Caryophyllene |
000087-44-5 |
28.90 |
21.90±1.47c |
39.36±2.15a |
7.39±1.08d |
29.25±2.01b |
| C3 |
alpha-himachalene |
003853-83-6 |
28.98 |
27.63±1.90c |
54.13±1.00a |
11.94±1.48d |
35.92±2.54b |
| C4 |
delta-Cadinene |
000483-76-1 |
29.60 |
53.59±1.80c |
124.63±0.98a |
\ |
56.91±1.74b |
| C5 |
germacrene d |
023986-74-5 |
29.70 |
13.50±1.96c |
25.53±1.97a |
6.41±0.80d |
17.25±2.21b |
| C6 |
α-curcumene |
000644-30-4 |
29.93 |
22.45±2.03c |
48.73±2.53a |
11.54±1.00d |
32.57±0.99b |
| C7 |
Cuparene |
016982-00-6 |
31.13 |
14.32±1.02b |
25.86±2.02a |
6.51±0.92c |
25.51±1.79a |
| C8 |
Calamenene |
000483-77-2 |
31.29 |
12.44±2.07c |
30.58±2.13a |
\ |
21.73±1.33b |
| C9 |
D-Limonene |
005989-27-5 |
14.37 |
\ |
22.39±2.04 |
\ |
\ |
| C10 |
Pentadecane |
000629-62-9 |
23.57 |
20.56±0.96b |
34.51±1.04a |
7.84±1.50c |
18.65±2.35b |
| C11 |
n-Hexadecane |
000544-76-3 |
26.07 |
32.38±2.12b |
49.43±1.94a |
\ |
19.26±1.02c |
| C12 |
n-Heptadecane |
000629-78-7 |
28.41 |
14.57±1.01b |
23.59±2.01a |
\ |
9.32±1.56c |
| C13 |
Hexane |
000110-54-3 |
3.69 |
\ |
45.68±1.79b |
88.89±1.66a |
23.53±1.87c |
| C14 |
Heptane |
000142-82-5 |
4.00 |
\ |
41.91±1.53a |
37.26±0.86b |
16.35±1.05c |
| C15 |
Cyclohexane |
000110-82-7 |
4.09 |
\ |
78.6±2.04a |
64.43±2.06b |
38.59±2.08c |
| |
Subtotal |
|
|
241.67±2.80c |
650.50±3.49a |
242.21±2.13c |
354.10±9.46b |
| Alcohol |
D1 |
Mushroom alcoho |
003391-86-4 |
21.96 |
7.58±1.79b |
28.03±1.48a |
\ |
10.05±1.86b |
| D2 |
Dodecyl alcohol |
000112-53-8 |
33.92 |
16.48±0.89b |
37.23±1.86a |
4.42±0.99c |
\ |
| D3 |
1-Octanol |
000111-87-5 |
24.76 |
\ |
30.31±1.03 |
\ |
\ |
| D4 |
2-Phenylethanol |
000060-12-8 |
32.90 |
\ |
14.35±0.85a |
\ |
7.39±2.18b |
| D5 |
Dodecyl alcohol |
000112-53-8 |
33.92 |
\ |
\ |
33.51±1.88a |
9.30±1.20b |
| |
Subtotal |
|
|
24.06±2.64d |
109.92±0.38a |
37.92±0.89c |
32.74±3.57b |
| Else |
E1 |
Butylated hydroxytoluene |
000128-37-0 |
32.85 |
8.10±1.68b |
11.47±0.96a |
\ |
5.56±1.93b |
| E2 |
Phenol |
000108-95-2 |
34.73 |
12.37±2.03 |
\ |
\ |
\ |
| E3 |
2,4-Di-tert-butylphenol |
000096-76-4 |
40.28 |
\ |
8.41±2.02 |
\ |
\ |
| E4 |
Methyl tridecyl ketone |
002345-28-0 |
35.18 |
7.69±1.52b |
\ |
\ |
10.19±1.40a |
| E5 |
2,6-Dimethyl pyrazine |
000108-50-9 |
18.94 |
19.52±1.95 |
\ |
\ |
\ |
| E6 |
Estragole |
000140-67-0 |
31.29 |
\ |
\ |
10.45±0.84 |
\ |
| E7 |
Dimethyl sulfoxide |
000067-68-5 |
26.07 |
\ |
\ |
15.40±2.07 |
\ |
| |
Subtotal |
|
|
47.67±7.08a |
19.88±1.07bc |
25.85±1.34b |
15.75±0.57c |
| Aggregate |
59 |
|
|
|
2763.44±3.88c |
4417.35±3.60a |
1022.62±20.76d
2944.08±9.71b |
Figure 2.
Distribution and content of VOCs in differently-aged Xuanwei ham samples. (a) Classification and quantity of VOCs; (b) Classification and content of VOCs; (c) Venn diagram analysis of VOCs; (d) Characteristic VOCs; and (e) PLS-DA diagram of Xuanwei hams.
Figure 2.
Distribution and content of VOCs in differently-aged Xuanwei ham samples. (a) Classification and quantity of VOCs; (b) Classification and content of VOCs; (c) Venn diagram analysis of VOCs; (d) Characteristic VOCs; and (e) PLS-DA diagram of Xuanwei hams.
3.3. Microbial Community Analysis
The volatile flavour characteristics of differently-aged Xuanwei ham were analysed with a Fox 4000 electronic nose equipped with 10 metal sensors, in which their corresponding response substances and category substances are shown in
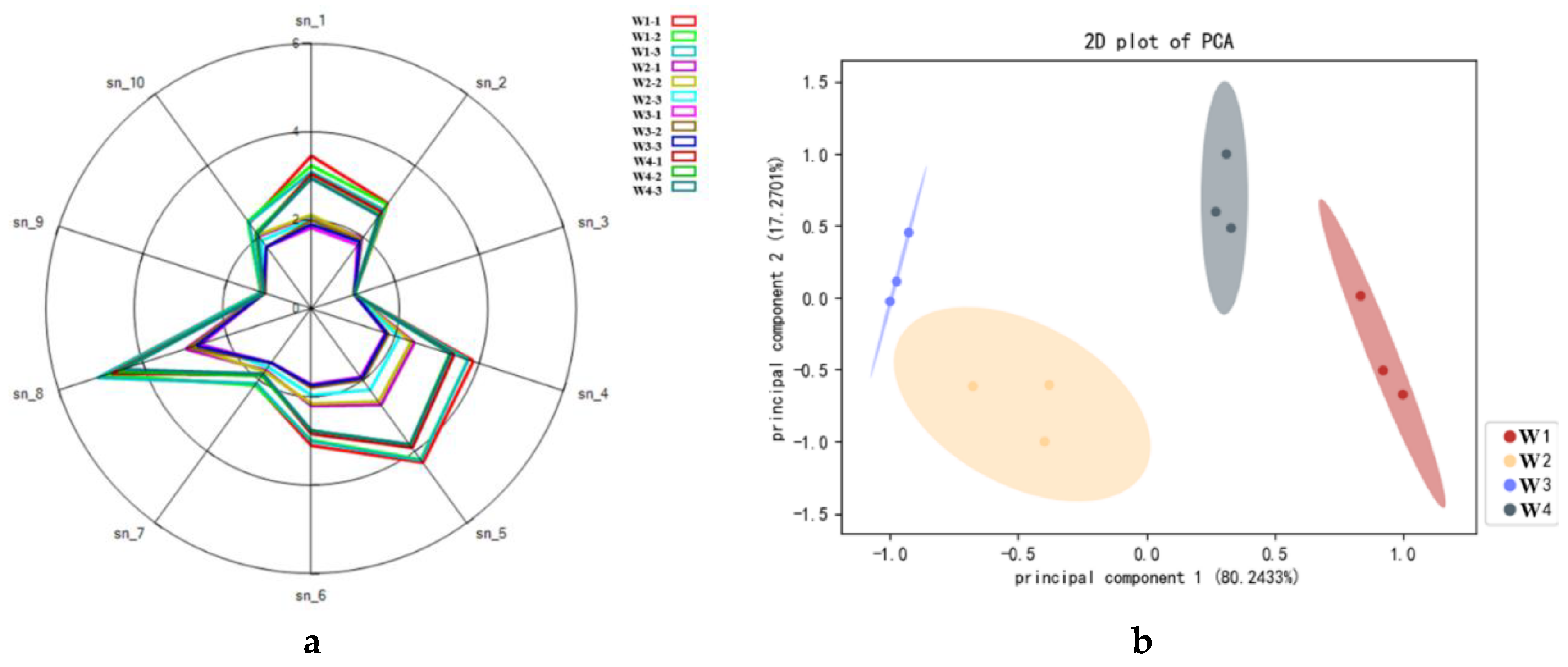
Table 3. Fig. 5 is the radar chart made by the sensor response value to Xuanwei ham odor, and the electronic nose requires that the sensor's maximum response value of the tested sample to be > 0.5, and all the experimental samples met the detection requirements. Its aroma depends not only on the flavor molecular composition, but also its concentration. Sensors sn-1, sn-2, sn-4, sn-5, sn-6, and sn-8, had higher response values for the flavour of differently-aged Xuanwei hams, of which sn-4, sn-5, sn-8 had a higher degree of differentiation (
Figure 5). Sensor sn-8 had the largest response value, and sensed signal intensity was W1 > W4 > W2 > W3, and W1 intensity was significantly higher than during the other storage periods, which coincided with the volatile flavor substances measured by GC-MS. W2 and W3 radar fingerprints almost overlapped, indicating that similar volatile components existed in the samples. Response values of sn-3 and sn-9 sensors were significantly smaller than the other sensors, indicating that they were insensitive to the response of methane, ozone and other flavor substances or the content of this type of flavor substance was lower, thus the sensor response was smaller. Overall, radargrams were effective in distinguishing between differently-aged Xuanwei ham volatile components.
PCA uses orthogonal transformations to convert a set of observations of observable correlated variables into a set of linearly uncorrelated variable values of principal components [
39]. The contribution of the 1st principal component was 80.2433%, the contribution of the 2nd was17.2701%, and the sum of the two was 97.5134%, indicating that PC1 and PC2 extracted the volatile compounds main characteristics in the storage period of Xuanwei ham [
40], which can be used to characterize the overall information of the four selected ham samples (
Figure 6). Thus, it can be used to analyze the changing law of volatile aroma components of Xuanwei ham during different storage periods. The aromas of W2 and W3 Xuanwei ham samples were closer, and W1 was farther away from the other three (
Figure 6). This may be due to the shortest ripening time, whereby ripe ham aroma could not be fully generated and released, but with the extension of ripening time, new volatile substances may be generated, so that Xuanwei ham has its unique ripe aroma.
Table 3.
Corresponding information of electronic nose sensors.
Table 3.
Corresponding information of electronic nose sensors.
| Transducers |
Responsive substance |
Category substances |
| S1 |
Alkanes, fumes |
Propane, natural gas, fumes |
| S2 |
Alcohols, aldehydes, short-chain alkanes |
Alcohol, fumes, isobutane, formaldehyde |
| S3 |
ozone (O3) |
\ |
| S4 |
sulfide |
hydrogen sulfide |
| S5 |
organic amine |
Ammonia, methylamine, ethanolamine |
| S6 |
Organic gases, benzophenones, alcohols and aldehydes, aromatic compounds |
Toluene, acetone, ethanol, hydrogen, other organic vapours |
| S7 |
Short-chain burnt hydrocarbons |
Methane, natural gas, biogas |
| S8 |
Aromatic compounds, alcohols and aldehydes |
Toluene, formaldehyde, benzene, alcohol, acetone |
| S9 |
hydrogen-containing gas |
hydrogen (gas) |
| S10 |
Flammable gases |
methane CH4 |
Figure 3.
Radar fingerprints of Xuanwei ham flavour(a) and PCA(b).
Figure 3.
Radar fingerprints of Xuanwei ham flavour(a) and PCA(b).
3.4. Figures, Tables and Schemes
3.4.1. Macrogenomic Data Overview
To determine the microbial community information present in the ham fermentation process, differently-aged Xuanwei hams were analyzed using macro-genome sequencing. The process yielded 78.75 Gbp of Raw Base (
Table 4). The sequencing errors of Clean Q30> 97% of reads of differently-aged Xuanwei hams were < 1‰, which showed the high quality of the sequencing procedure and that it met the analysis requirements. A total of 87,508,391 reads were obtained from Illumina MiSeq sequencing of Xuanwei ham, of which The Chao1 index and Shannon index in W3 were significantly higher than in W1, W2 and W4 (405.41 and 4.61, respectively), indicating that the population variability of the W3 microbial community was relatively large and community diversity was high. This study was analyzed using observed_features, and compared to Observed_otus it better described the different ways in which QlME 2 uses non-categorical features. Chao1 index results were similar to those of observed_features, further demonstrating high diversity as well as overall abundance of bacterial microbial communities in W3.
3.4.2. Bacterial microbiological composition of Xuanwei ham
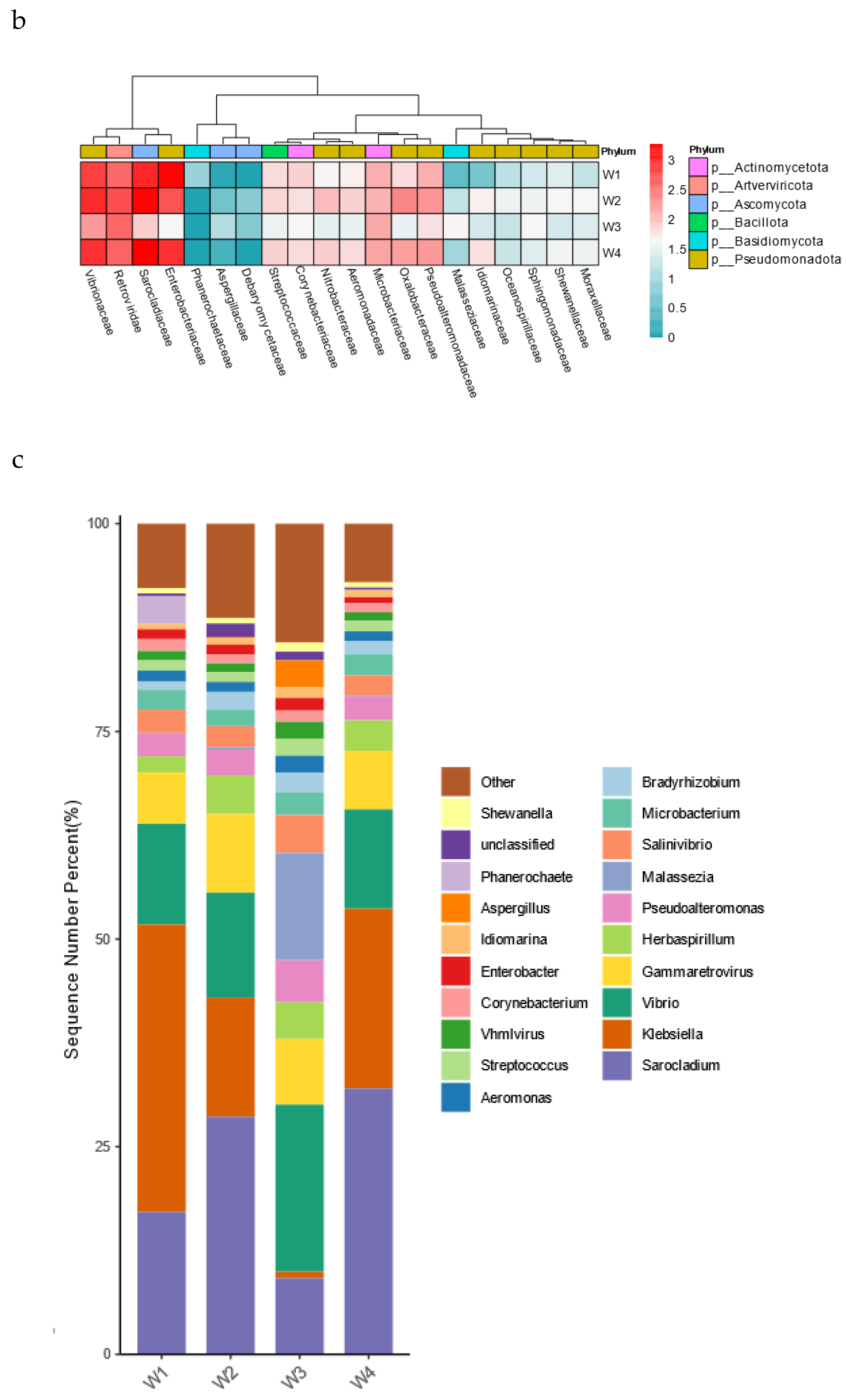
Macro-genome sequencing was performed in differently-aged Xuanwei hams (W1, W2, W3 and W4) to analyse changes in microbial composition over time. The dominant phylum of Xuanwei ham in all four years was Pseudomonadota, which concurs with Li Cong's findings [
11]. The most abundant families in W1, W2, and W4 were Enterobacteriaceae, Sarocladiaceae, Retroviridae, and Vibrionaceae, and Enterobacteriaceae relative abundance was lowest in W2, while the main families in W3 were Retroviridae and Vibrionaceae (
Figure 4a). A total of 185 microorganisms, which accounted for only 9.3% of overall microorganisms indicates that ham fermentation is an open environment, and maturation involves numerous unknown and uncultured microorganisms (
Figure 4b). To fully characterize the microorganisms, a more scientific analysis of microbial composition at the genus level is required. The most abundant genera in W1, W2 and W4 were Sarocladium, Klebsiella and Vibrio, and Klebsiella was the most abundant genus in W1, indicating that there were few characteristic microorganisms in W1, W2 and W4 at the genus level (
Figure 4c). This was consistent with the results of the characteristic microorganisms analyses where the most abundant genus in W3 was Vibrio, as was the second most abundant genus. and the second most abundant genera were Sarocladium and Gammaretrovirus.This is similar to the results of comparative analysis of microbial diversity of five dry-cured hams from Yunnan studied by Lin Junyi et al [
41].
3.4.3. Xuanwei Ham LEfSe Analysis
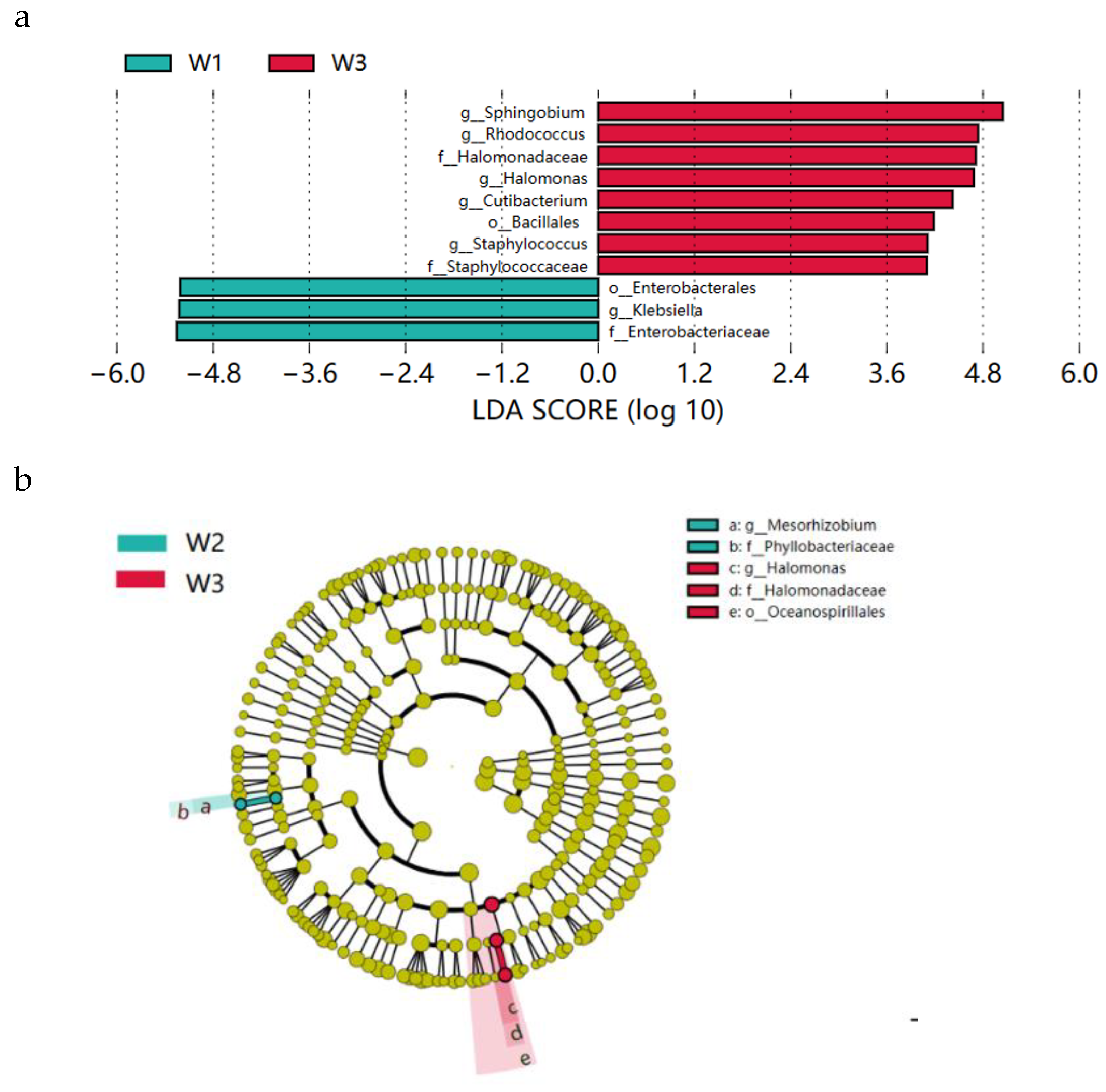
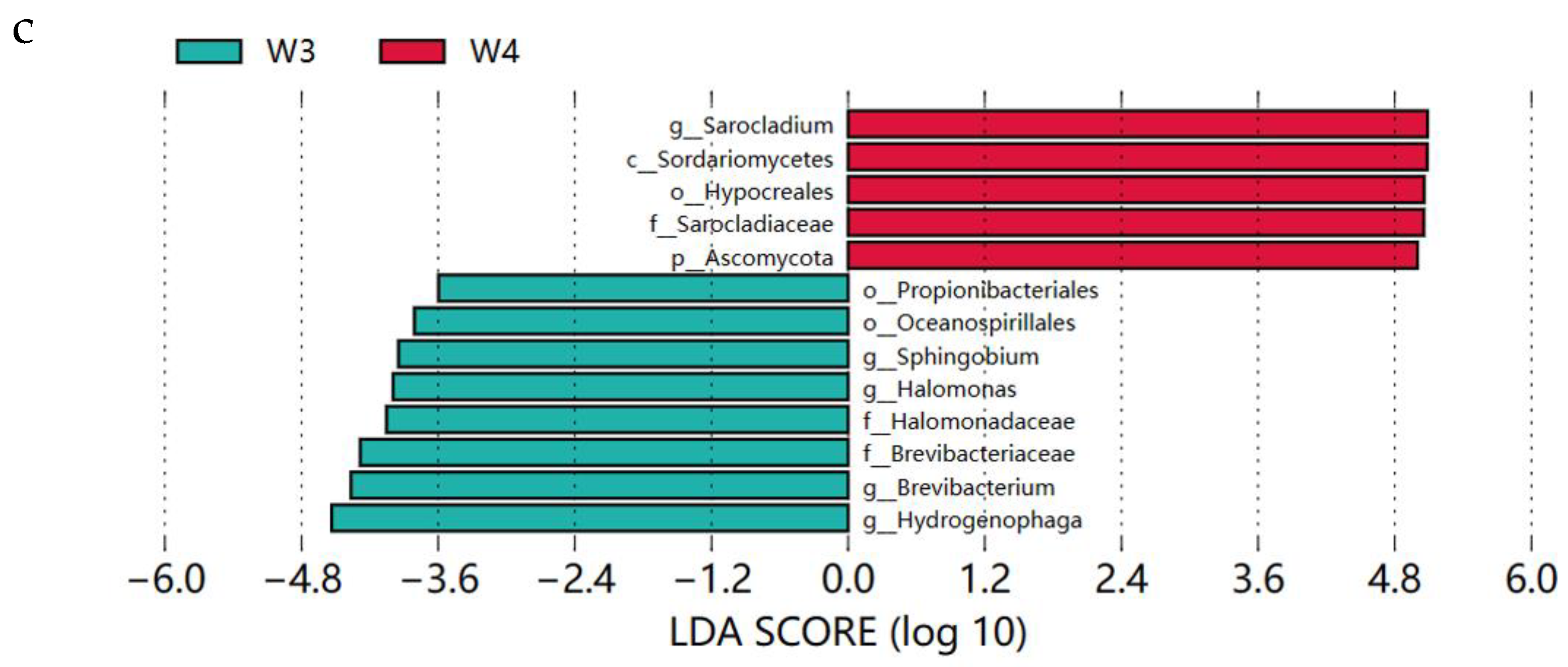
From the above analysis (Figure. 4), it is clear that W1, W2 and W4 have similar microbial abundance in phylum, family and genus, so there are few characteristic flora present among them.We compared W3 with W1, W2 and W4 respectively, to identify characteristic microorganisms present in differently-aged Xuanwei hams. Through LEfSe analysis, the first three characteristic bacteria were represented. In W1 and W3, the characteristic bacteria in W1 were Enterobacteriaceae, Klebsiella and Enterobacterales, while in W3 they were Sphingobium, Rhodococcus and Halomonadaceae (
Figure 5a). In W2 and W3, the characteristic bacteria in W2 were Phyllobacteriaceae, Mesorhizobium, while in W3 were Oceanospirillales, Halomonadaceae and Halomonas (
Figure 5b). In W3 and W4, the representative bacterial microorganisms in W3 were Hydrogenophaga, Brevibacterium and Brevibacteriaceae, while in W4 they were Sarocladium, Sordariomycetes and Hypocreales (
Figure 5c).
Figure 5.
LEfSe analyses of Xuanwei ham in (a) W1 and W3 (b) W2 and W3, and (c) W3 and W4. Prefixes k, p, c, o, f, and g of the bacterial name stand for kingdom, phylum, class, order, family, and genus, respectively.
Figure 5.
LEfSe analyses of Xuanwei ham in (a) W1 and W3 (b) W2 and W3, and (c) W3 and W4. Prefixes k, p, c, o, f, and g of the bacterial name stand for kingdom, phylum, class, order, family, and genus, respectively.
3.4.4. Correlation Analysis of Phylum and Genus Microbiota with Free Amino Acids, VOCs
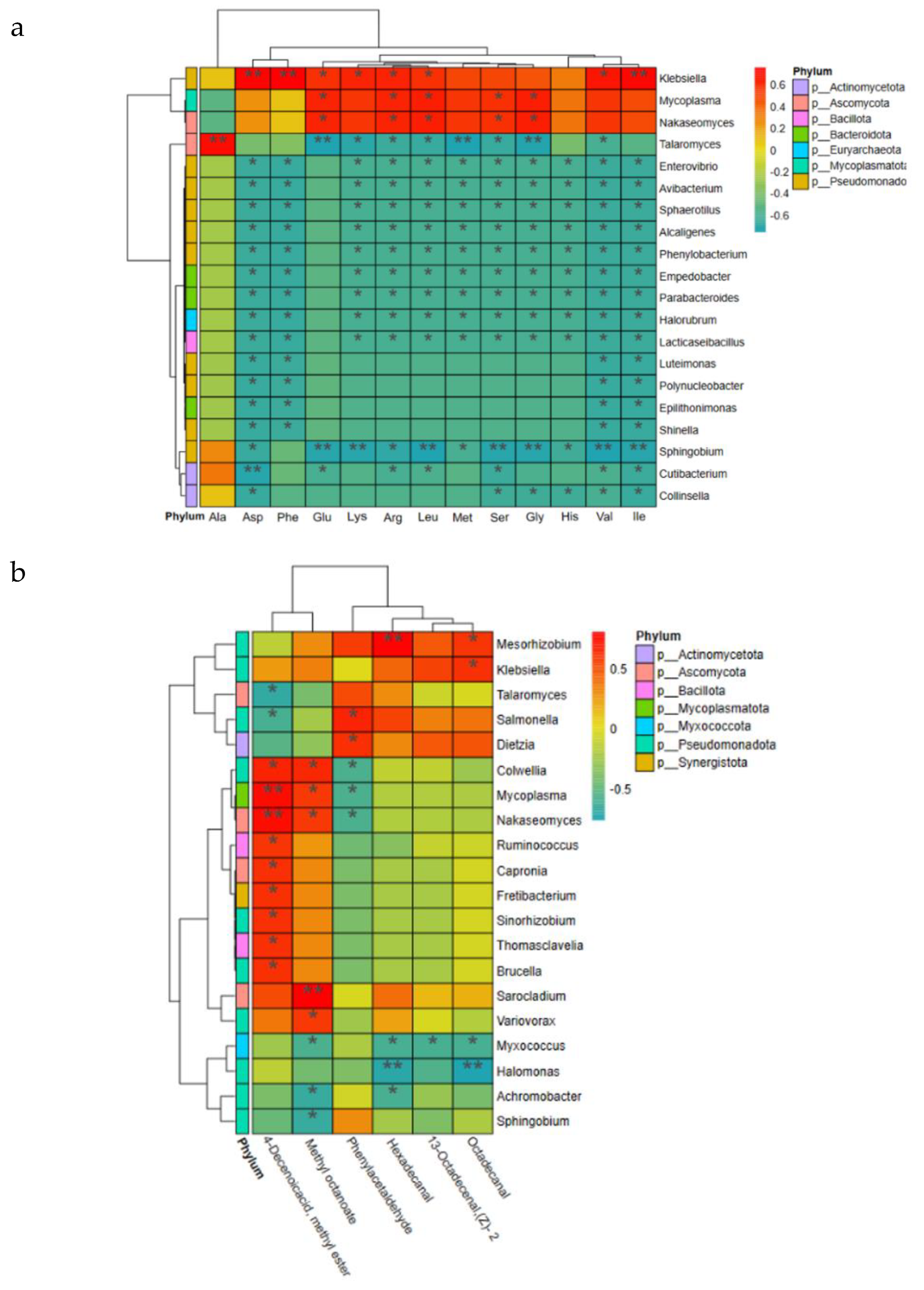
Correlation heatmap was used to study the relationship of bacterial microbial communities with free amino acids and VOCs (
Figure 6). Correlation analyses of free amino acids with microorganisms with large TAV values and high contributions were performed, and Ascomycota showed a highly significant positive correlation with Ala at the phylum level (P < 0.01), and Glu, Arg, Leu, and Ser at the genus level (
Figure 6a). Gly were positively correlated with Mycoplasma and Nakaseomyces (P<0.05), and Klebsiella showed highly significant positive correlation (P < 0.01) with Asp, Phe and Ile; indicating that Klebsiella may promote their production, and the high W4 total content of free amino acids may not be separated from the presence of Klebsiella. Correlation analysis was performed using VOCs with VIP scores > 1 and microorganisms, and at the phylum level Mycoplasmatota and Ascomycota showed highly significant positive correlation (P < 0.01) with 4-Decenoicacid, methyl ester (
Figure 6b). At the genus level Colwellia, Mycoplasma, Nakaseomyces, Ruminococcus, Capronia, Fretibacterium, Sinorhizobium, Thomasclavelia, Brucella showed significant positive correlation (P < 0.05) with 4-Decenoicacid, methyl ester and Mycoplasma. Nakaseomyces showed a highly significant positive correlation (P < 0.01) with 4-Decenoicacid, methyl ester, suggesting that many microorganisms are favourable for its production which has a papaya aroma [
42] and was present only in W4, suggesting that it can confer a unique flavour to high vintage hams. Mesorhizobium and Sarocladium showed highly significant positive correlations (P < 0.01) with Hexadecanal and Methyl octanoate, respectively; confirming the main microorganisms in hams that contribute to flavour formation.
Figure 6.
Correlation analysis of microbial community with Xuanwei ham (a) free amino acids, and (b) volatile flavour.
Figure 6.
Correlation analysis of microbial community with Xuanwei ham (a) free amino acids, and (b) volatile flavour.
4. Conclusions
During ripening, the highest total free amino acid content and significantly higher glutamic acid content, and the main fresh flavour amino acid, were found in 4-year aged Xuanwei ham (P < 0.05), and could not be separated from the presence of Klebsiella. Lin [
43] et al. found that Klebsiella could enhance the flavour of traditional Mongolian cheeses during the ripening process. Of the 59 VOCs detected, 17 were esters, and the highest ester content was found in 4-year aged Xuanwei ham.Hexadecanal VIP scores were significantly higher than those of other VOCs, and the highest content was found in 1-year aged Xuanwei ham, which contributed to its higher flavour. Bacterial Chao1 and Shannon indices(VIP scores) of 3-year aged Xuanwei ham were significantly higher than other 3-year aged Xuanwei ham.Bacterial composition of 3-year-old Xuanwei ham differed greatly from other ham samples, with Pseudomonadota being the dominant bacterial group at the phylum level, and Retroviridae and Vibrionaceae dominating at the family level. At the genus level, differently-aged Xuanwei hams were mainly dominated by Sarocladium and Vibrio.The characteristic microorganisms present between 3-year and 1-, 2- and 4-year aged Xuanwei hams were Sphingobium, Rhodococcus, Halomonadaceae, Oceanospirillales, and Halomonas. Glu, Arg, Leu, Ser, and Gly were significantly and positively correlated with Mycoplasma and Nakaseomyces. The production of Asp, Phe, and Ile was directly or indirectly correlated with the action of Klebsiella microorganisms. Mycoplasmatota, Ascomycota, Colwellia, and Mycoplasma were significantly positively correlated with 4-Decenoic acid, and methyl ester (4Decenoic acid, methyl ester), and methyl ester was significantly positively correlated (P < 0.05) and was only present in 4-year-old Xuanwei ham. This substance may only be present in high vintage Xuanwei hams. Since we studied only four years of ham samples, follow-up studies are required to further confirm this possibility. The correlation between Mycoplasmatota et al. and 4-Decenoicacid, methyl ester was only hypothesised on the basis of high-throughput sequencing, and its contribution to VOCs was not directly confirmed, so follow-up studies are required on the basis of isolation and purification of the relevant dominant bacteria and back-joining of fermentation. Our results provide a better scientific basis for Xuanwei ham flavour formation and quality control.
Author Contributions
G.L.; conceptualization, methodology, validation, investigation, and writ ing—original draft preparation. S.L.; methodology, validation, and writing review and editing. Y.W. and J.Y.; data curation and software. Z.L.; conceptualization, writing—review and editing, visualization, project administration, and funding acquisition. Y.C.; investigation and methodology. and L.L.; conceptualization, methodology, project administration, and funding ac quisition. W.H. resources. P.W.and W.W; visualization All authors have read and agreed to the published version of the manu script.
Funding
Tibet College of Agriculture and Animal Husbandry Graduate Education Innovation Programme Project(YJS2024-54), Major Science and Technology Projects of the Tibet Autonomous Region (XZ202101ZD0005N)and Graduate Teaching Reform and Construction Proiect of University-Xizang Agriculture and Animal Husbandry College (YJSJG2023-015).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated or analyzed in this study are available within the manuscript and are available from the corresponding authors upon request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Dong Yinchu. Modernisation of traditional Chinese flavour meat products[J]. Meat Research, 1998, (02):3-6.
- Wu W, Zhou Y, Wang G, et al. Changes in the physicochemical properties and volatile flavor compounds of dry-cured Chinese Laowo ham during processing[J]. Journal of Food Processing and Preservation, 2020, 44(8): e14593. [CrossRef]
- Huang A X, Ge C R, Huang Q C. The study of ingredients and processing techniques of Xuanwei style ham[J]. Journal of food processing and preservation, 2010, 34: 136-148. [CrossRef]
- YI Yongxin, WANG Xuemin, XIANG Jun, et al. Effects of curing agent and rinsing process on the flavour of Xuanwei ham[J]. Food and Fermentation Industry, 2024, 50(04): 85-92. [CrossRef]
- Li Z, Wang Y, Pan D, et al. Insight into the relationship between microorganism communities and flavor quality of Chinese dry-cured boneless ham with different quality grades[J]. Food Bioscience, 2022, 50: 102174. [CrossRef]
- Guo X, Wang Y, Lu S, et al. Changes in proteolysis, protein oxidation, flavor, color and texture of dry-cured mutton ham during storage[J]. Lwt, 2021, 149: 111860. [CrossRef]
- Zhu Y, Guo Y, Yang F, et al. Combined application of high-throughput sequencing and UHPLC-Q/TOF-MS-based metabolomics in the evaluation of microorganisms and metabolites of dry-cured ham of different origins[J]. International Journal of Food Microbiology, 2021, 359: 109422. [CrossRef]
- Wang Y, Shen Y, Wu Y, et al. Comparison of the microbial community and flavor compounds in fermented mandarin fish (Siniperca chuatsi): Three typical types of Chinese fermented mandarin fish products[J]. Food Research International, 2021, 144: 110365. [CrossRef]
- Yu, Y. R., Wang, G. Y., Sun, Y. H., Ge, C. R., & Liao, G. Z. (2020). Changes in physicochemical parameters, free fatty acid profile and water-soluble compounds of Yunnan dry-cured beef during processing. Journal of Food Processing and Preservation, 44(4), e14380. [CrossRef]
- Virgili, R., Saccani, G., Gabba, L., Tanzi, E., & Soresi Bordini, C. (2007). Changes of free amino acids and biogenic amines during extended ageing of Italian dry-cured ham. Lwt–food Science and Technology, 40(5), 871–878. [CrossRef]
- Li C, Zheng Z, Wang G, et al. Revealing the intrinsic relationship between microbial communities and physicochemical properties during ripening of Xuanwei ham[J]. Food Research International, 2024: 114377. [CrossRef]
- Ge Q, Pei H, Liu R, et al. Effects of Lactobacillus plantarum NJAU-01 from **hua ham on the quality of dry-cured fermented sausage[J]. LWT, 2019, 101: 513-518. [CrossRef]
- Wang Y, Li F, Chen J, et al. High-throughput sequencing-based characterization of the predominant microbial community associated with characteristic flavor formation in **hua Ham[J]. Food Microbiology, 2021, 94: 103643. [CrossRef]
- Wang Y, Shen Y, Wu Y, et al. Comparison of the microbial community and flavor compounds in fermented mandarin fish (Siniperca chuatsi): Three typical types of Chinese fermented mandarin fish products[J]. Food Research International, 2021, 144: 110365. [CrossRef]
- Zhong A, Chen W, Duan Y, et al. The potential correlation between microbial communities and flavors in traditional fermented sour meat[J]. LWT, 2021, 149: 111873. [CrossRef]
- Deng J, Xu H, Li X, et al. Correlation of characteristic flavor and microbial community in **hua ham during the post-ripening stage[J]. LWT, 2022, 171: 114067. [CrossRef]
- Qiu D, Duan R, Wang Y, et al. Effects of different drying temperatures on the profile and sources of flavor in semi-dried golden pompano (Trachinotus ovatus)[J]. Food chemistry, 2023, 401: 134112. [CrossRef]
- Zhu N, Wang S, Zhao B, et al. Label-free proteomic strategy to identify proteins associated with quality properties in sauced beef processing[J]. Food Bioscience, 2021, 42: 101163. [CrossRef]
- Xu X, You M, Song H, et al. Investigation of umami and kokumi taste-active components in bovine bone marrow extract produced during enzymatic hydrolysis and Maillard reaction[J]. International journal of food science & technology, 2018, 53(11): 2465-2481. [CrossRef]
- CAO Chenchen, FENG Meiqin, SUN Jian, et al. Effects of functional fermentation agents on oxidative stability and volatile flavour substances of fermented sausages[J]. Food Science,2019,40(20):106-113. [CrossRef]
- Marcel M. Cutadapt removes adapter sequences from high-throughput sequencing reads[J]. The European Molecular Biology Network, 2011, 17: 1. [CrossRef]
- Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets[J]. Bioinformatics, 2011, 27(6): 863-864. [CrossRef]
- Langmead B, Salzberg S L. Fast gapped-read alignment with Bowtie 2[J]. Nature methods, 2012, 9(4): 357-359. [CrossRef]
- Wood D E, Salzberg S L. Kraken: ultrafast metagenomic sequence classification using exact alignments[J]. Genome biology, 2014, 15: 1-12. [CrossRef]
- Lu J, Breitwieser F P, Thielen P, et al. Bracken: estimating species abundance in metagenomics data[J]. PeerJ Computer Science, 2017, 3: e104. [CrossRef]
- Mandal S, Van Treuren W, White R A, et al. Analysis of composition of microbiomes: a novel method for studying microbial composition[J]. Microbial ecology in health and disease, 2015, 26(1): 27663. [CrossRef]
- Brum J R, Ignacio-Espinoza J C, Roux S, et al. Patterns and ecological drivers of ocean viral communities[J]. Science, 2015, 348(6237): 1261498. [CrossRef]
- Gao Y, Zhang G, Jiang S, et al. Wekemo Bioincloud: A user-friendly platform for meta-omics data analyses[J]. iMeta, 2024: e175. [CrossRef]
- Li Qin. Identification of characteristic flavour substances in Agaricus bisporus soup and study on the release of flavour substances during simmering[D]. Jiangnan University,2011.
- LIU Biqin, WANG Xinrui, ZHAO Wenhua, et al. Physicochemical and flavour properties of Norden ham from different sources and years[J]. Meat Research, 2021, 35(08): 1-8. [CrossRef]
- Ur-Rehman S, Fox P F. Effect of added α-ketoglutaric acid, pyruvic acid or pyridoxal phosphate on proteolyis and quality of Cheddar cheese[J]. Food Chemistry, 2002, 76(1): 21-26. [CrossRef]
- Zhao Jingli. Study on the Melad reaction involving free amino acids during the flavour formation of Jinhua ham[D]. Henan Agricultural University, 2013.
- Michikawa K, Konosu S. Sensory identification of effective components for masking bitterness of arginine in synthetic extract of scallop[C]//Olfaction and Taste XI: Proceedings of the 11th International Symposium on Olfaction and Taste and of the 27th Japanese Symposium on Taste and Smell. Joint Meeting held at Kosei-nenkin Kaikan, Sapporo, Japan, July 12–16, 1993. Springer Japan, 1994: 278-278. [CrossRef]
- Sforza S, Pigazzani A, Motti M, et al. Oligopeptides and free amino acids in Parma hams of known cathepsin B activity[J]. Food Chemistry, 2001, 75(3): 267-273. [CrossRef]
- Li P, Bao Z, Wang Y, et al. Role of microbiota and its ecological succession on flavor formation in traditional dry-cured ham: a review[J]. Critical Reviews in Food Science and Nutrition, 2023: 1-17. [CrossRef]
- ZHANG Mingfang, WU Guofang, MA Yao, et al. Effects of lactic acid bacteria complex on growth performance, slaughter performance, meat quality and volatile flavour composition of black Tibetan sheep[J]. China Feed,2023,(21):73-80. [CrossRef]
- Chen Yanhua. Analysis of microflora of low-salt and high-moisture Sichuan bacon and the effect of pasteurisation treatment on its storage quality[D]. Sichuan Agricultural University, 2022. [CrossRef]
- Reyes-Díaz R, González-Córdova A F, del Carmen Estrada-Montoya M, et al. Volatile and sensory evaluation of Mexican Fresco cheese as affected by specific wild Lactococcus lactis strains[J]. Journal of Dairy Science, 2020, 103(1): 242-253. [CrossRef]
- Papadopoulou O S, Panagou E Z, Mohareb F R, et al. Sensory and microbiological quality assessment of beef fillets using a portable electronic nose in tandem with support vector machine analysis[J]. Food Research International, 2013, 50(1): 241-249. [CrossRef]
- Rattray N J W, Hamrang Z, Trivedi D K, et al. Taking your breath away: metabolomics breathes life in to personalized medicine[J]. Trends in biotechnology, 2014, 32(10): 538-548. [CrossRef]
- LIN Junyi, SONG Chunlian, ZHANG Ying, et al. Comparative analysis of microbial diversity of five dry-cured hams from Yunnan[J]. China Food Additives, 2023, 34(10): 233-241. [CrossRef]
- ZHANG Wei, JIANG Li, JIANG Yongxin, et al. GC-MS analysis of aroma components of white-flowered papaya fruit[J]. Southern China Fruit Tree, 2017, 46(04): 117-120. [CrossRef]
- Lin Jiawei. Correlation between microbial community structure and flavour substances in the ripening process of traditional Mongolian cheese [D]. Inner Mongolia Agricultural University, 2023. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).