Submitted:
17 May 2024
Posted:
20 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Survey and Isolation of Fusarium Species
2.2. Molecular Identification and Phylogenetic Analyses
2.3. Pathogenicity Tests
2.4. Statistical Analyses
3. Results
3.1. Fungal Isolation and Prevalence of Fusarium Species
3.2. Molecular Identification and Phylogenetic Analyses
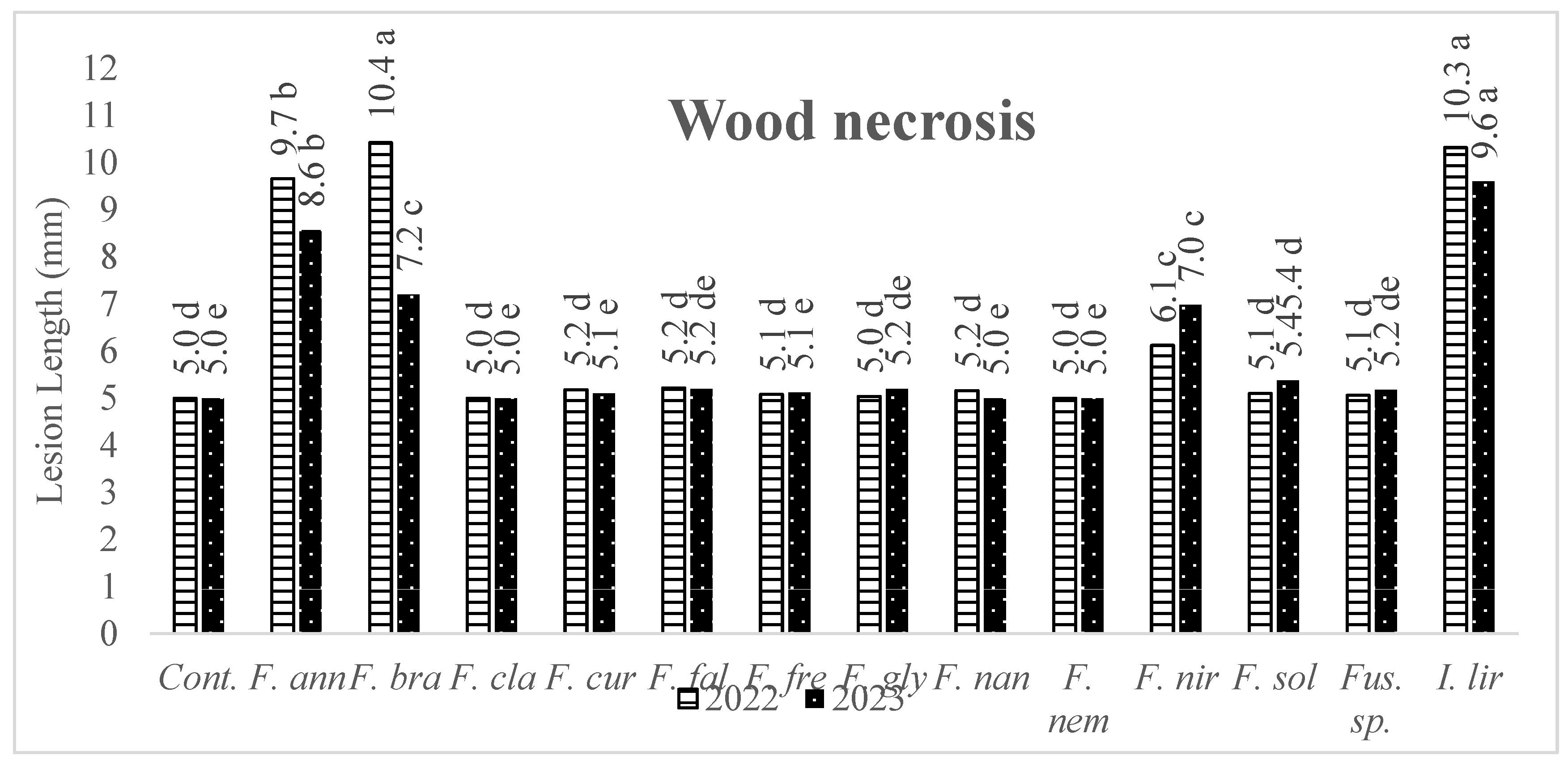
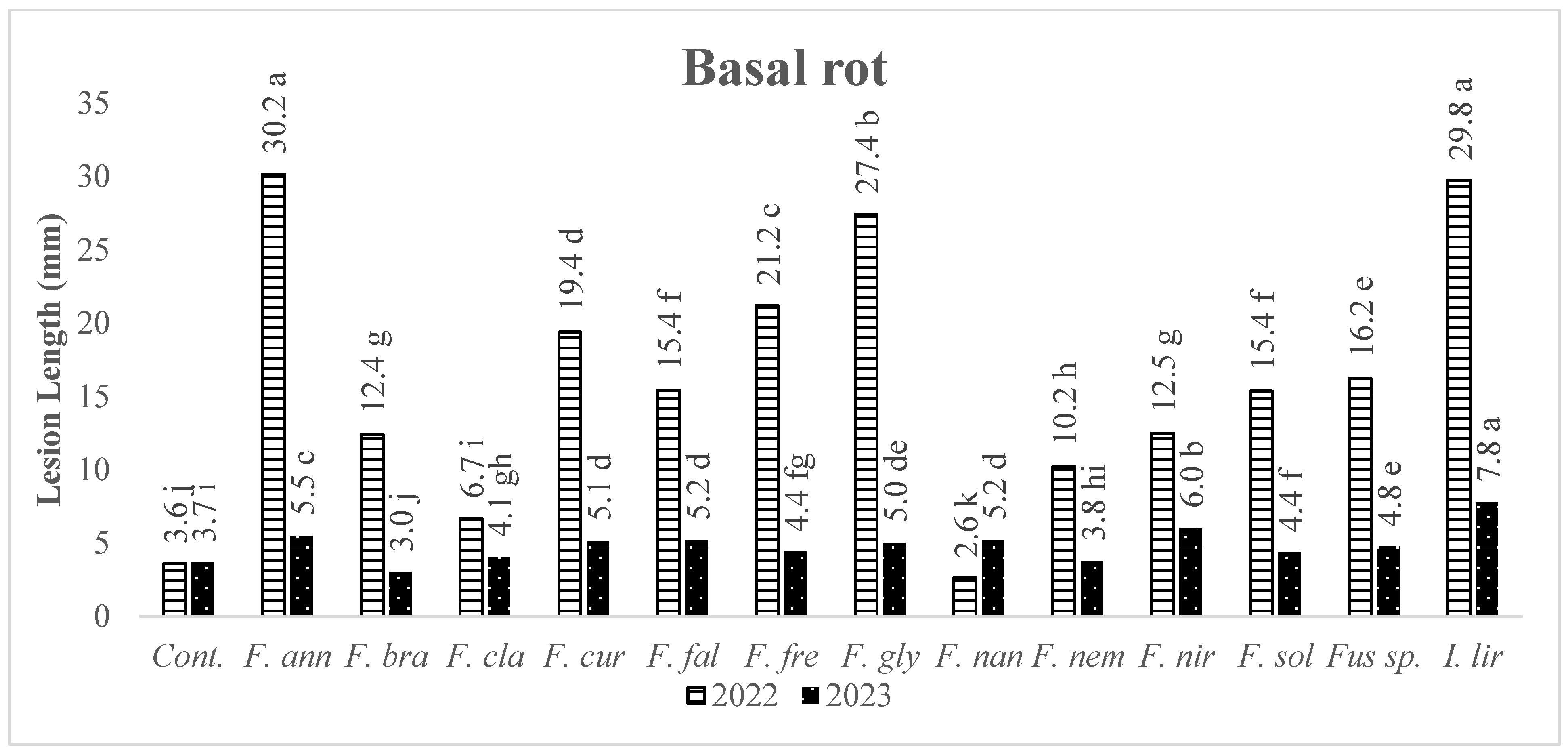
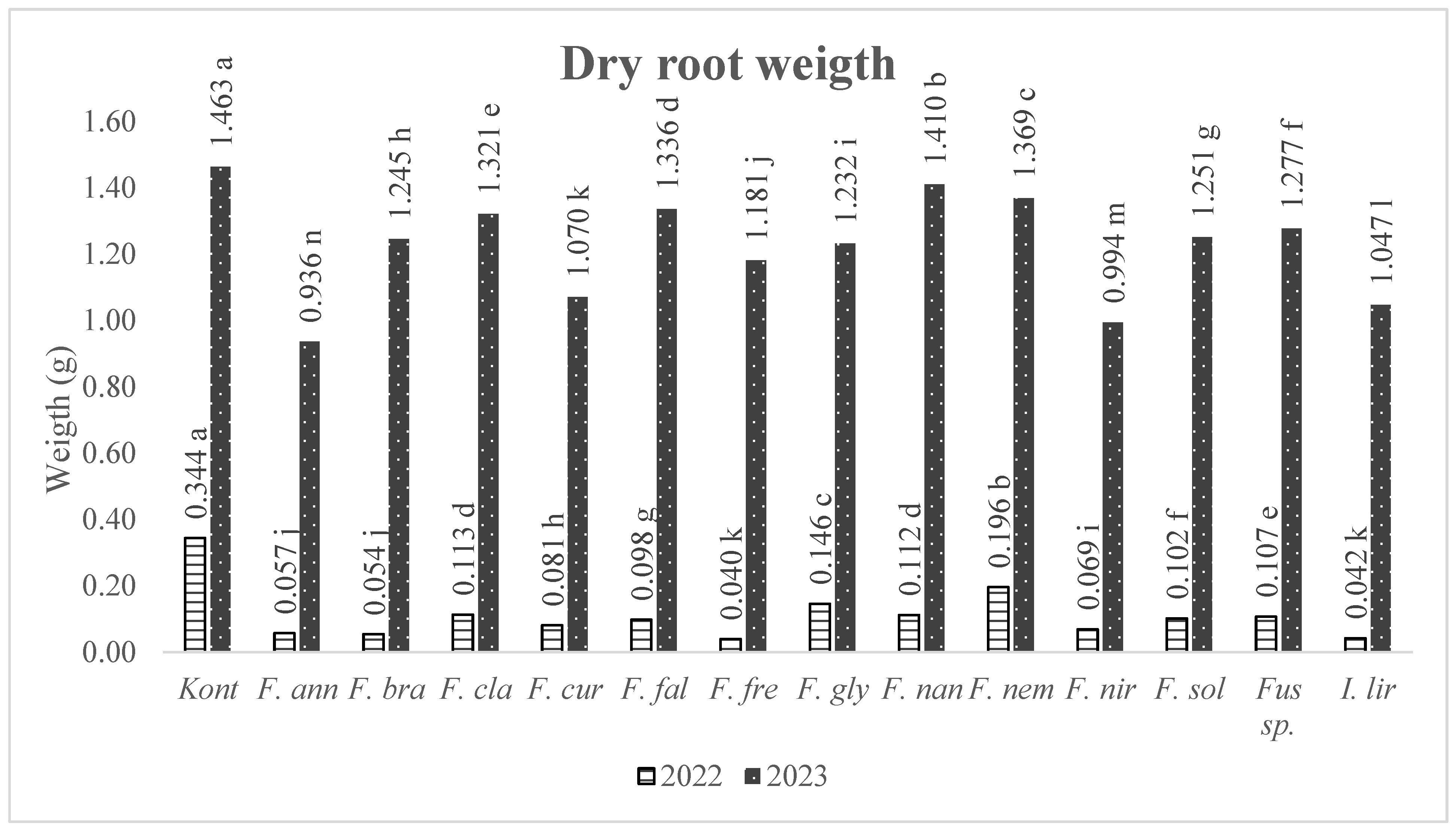
3.2. Pathogenicity of Fusarium Isolates and Species
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Anonymous. Personal communication with the official staff. General Directorate of Plant Production at Turkish Ministry of Agriculture and Forestry 2023. [Google Scholar]
- Gubler, W.D.; Baumgartner, K.; Browne, G.T.; Eskalen, A.; Rooney-Latham, S. , Petit, E.; Bayramian, L.A. Root disease of grapevines. Australas. Plant Path. 2004, 33, 157–165. [Google Scholar] [CrossRef]
- Gramaje, D.; Armengol, J. Fungal trunk pathogens in the grapevine propagation process: Potential inoculum sources, detection, identification, and management strategies. Plant Dis. 2012, 95, 1040–1055. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K; Whitaker, B. K.; Laraba, I.; Proctor, R.H.; Brown, D.W.; Broders, K.; Kim, H.S.; McCormick, S.P.; Busman, M., Aoki, T.; Torres-Cruz, T.J.; Geiser, D.M. DNA sequence-based identification of Fusarium: A work in progress. Plant Dis. 2022, 106, 1597–1609. [Google Scholar] [CrossRef] [PubMed]
- Summerell, B.A. Resolving Fusarium: current status of the genus. Annu. Rev. Phytopathol. 2019, 57, 323–339. [Google Scholar] [CrossRef]
- Halleen, F.; Crous, P.W.; Petrini, O. Fungi associated with healthy grapevine cuttings in nurseries, with special reference to pathogens involved in the decline of young vines. Australas. Plant Pathol. 2003, 32, 47–52. [Google Scholar] [CrossRef]
- Highet, A.S.; Nair, N.G. Fusarium oxysporum associated with grapevine decline in the Hunter Valley, NSW, Australia. Australian Journal of Grape and Wine Research 1995, 1, 48–50. [Google Scholar] [CrossRef]
- Reveglia, P.; Cinelli, T.; Cimmino, A.; Masi, M.; Evidente, A. The main phytotoxic metabolite produced by a strain of Fusarium oxysporum inducing grapevine plant declining in Italy. Nat. Prod. Res. 2018, 32, 2398–2407. [Google Scholar] [CrossRef]
- Vilvert, E.; Costa, M.D.; Cangahuala-Inocente, G.C.; Lovato, P.E. Root proteomic analysis of grapevine rootstocks inoculated with Rhizophagus irregularis and Fusarium oxysporum f. sp. herbemontis. Rev. Bras. Cienc. Solo. 2017, 41, e0160134. [Google Scholar] [CrossRef]
- Úrbez-Torres, J.R.; Boulé, J.; Hrycan, J.; O’Gorman, D.T. Potential role of Fusarium spp. in grapevine decline. Phytopathol. Mediterr. 2023, 62, 269–281. [Google Scholar] [CrossRef]
- Bustamente, M.I.; Todd, C.; Elfar, K.; Hamid, M.I.; Garcia, J.F.; Cantu, D.; Rolshausen, P.E.; Eskalen, A. Identification and pathogenicity of Fusarium species associated with young vine decline in California. Plant Dis. 2024, 108, 1053–1061. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, X.; Zhang, W.; Zhang, J.; Wang, H.; Peng, J.; Wang, X.; Yan, J. Belowground microbiota analysis indicates that Fusarium spp. exacerbate grapevine trunk disease. Environmental Microbiome 2023, 18, 29. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, Y.Y.; Li, X.H.; Zhang, W.; Li, Y.H.; Wang, X.D.; Yan, J.Y. First report of Fusarium commune associated with root rot of grapevine in China. Plant Dis. 2023, 107, 1238. [Google Scholar] [CrossRef]
- Akgül, D.S.; Ahioğlu, M. Fungal pathogens associated with young grapevine decline in the Southern Turkey vineyards. BIO 42nd World Congress of Vine and Wine, Web of Conferences, 2019, 15, 01027.
- Leslie, F.J.; Summerell, A.B. ; The Fusarium Laboratory Manual. Blackwell Publishing, London, UK. 2006; pp 220.
- O’Donnell, K.; Cigelnik, E.; Nirenberg, H.I. Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia 1998, 90, 465–493. [Google Scholar] [CrossRef]
- Crous, P.W.; Lombard, L.; Sandoval-Denis, M.; Seifert, K.; Schroers, H.J.; Chaverri, P.; … Thines, M. Fusarium: more than a node or a foot-shaped basal cell. Stud. Mycol. 2021, 98, 100116. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.; Sutton, D.A.; Rinaldi, M.G.; Sarver, B.A.J.; Balajee, S.A.; Schroers, H.J.; … Geiser, D.M. Internet-accessible DNA sequence database for identifying Fusaria from human and animal infections. J. Clin. Microbiol. 2010, 48, 3708–3718. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Haeseler, A.V.; Lanfear, R. IQ-TREE 2: New Models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Haeseler, A.V.; Jermiin, L.S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; Haeseler, A.V.; Minh, B.Q.; Vinh, L.S. ; UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Úrbez-Torres, J.R.; Haag, P.; Bowen, P.; O’Gorman, D.T. ; Grapevine trunk diseases in British Columbia: Incidence and characterization of the fungal pathogens associated with esca and Petri diseases of grapevine. Plant Dis. 2014, 98, 456–468. [Google Scholar] [CrossRef]
- Gomez, K.A.; Gomez, A.A. Statistical procedures for agricultural research (2nd ed.). Wiley, UK. 1984, pp 680.
- Granett, J.; Omer, A.D.; Pessereau, P.; Walker, M.A. Fungal infections of grapevine roots in phylloxera-infested vineyards. Vitis 1998, 37, 39–42. [Google Scholar] [CrossRef]
- Pintos, C.; Redondo, V.; Costas, D.; Aguin, O.; Mansilla, P. Fungi associated with grapevine trunk diseases in nursery-produced Vitis vinifera plants. Phytopathol. Mediterr. 2018, 57, 407–424. [Google Scholar] [CrossRef]
- Lombard, L.; Sandoval-Denis, M.; Lamprecht, S.C.; Crous, P.W. Epitypification of Fusarium oxysporum clearing the taxonomic chaos. Persoonia 2019, 43, 1–47. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.F.; Hwang, S.F.; Conner, R.L.; Gossen, B.D. First report of Fusarium proliferatum causing root rot in soybean (Glycine max L.) in Canada. Crop Prot. 2015, 67, 52–58. [Google Scholar] [CrossRef]
- Akbar, A.; Hussain, S.; Ullah, K.; Fahim, M.; Ali, G.S. Detection, virulence and genetic diversity of Fusarium species infecting tomato in Northern Pakistan. PloS One. [CrossRef]
- Sanna, M.; Martino, I.; Guarnaccia, V.; Mezzalama, M. Diversity and pathogenicity of Fusarium species associated with stalk and crown rot in maize in Northern Italy. Plants, 2023, 12, 3857. [Google Scholar] [CrossRef] [PubMed]
- Rajput, N.; Zaman, B.; Huo, C.; Cao, J.; Atiq, M.; Lodhi, A.M.; Syed, R.N.; Khan, B.; Iqbal, O.; Zhao, Z. First report of Fusarium equiseti causing stem rot disease of grape (Vitis vinifera L.) in Afghanistan. J. Plant Pathol. 2020, 102, 1277. [Google Scholar] [CrossRef]
- Parra, M.Á.; Gómez, J.; Aguilar, F.W.; Martínez, J.A. Fusarium annulatum causes Fusarium rot of cantaloupe melons in Spain. Phytopathol. Mediterr. 2022, 2022. 61, 269–277. [Google Scholar] [CrossRef]
- Mirghasempour, S.A.; Studholme, D.J.; Chen, W.; Zhu, W.; Mao, B. Molecular and pathogenic characterization of Fusarium species associated with corm rot disease in saffron from China. J. Fungi 2022, 8, 515. [Google Scholar] [CrossRef] [PubMed]
- Vetrova, S.; Alyokhina, K.; Engalycheva, I.; Kozar, E.; Mukhina, K.; Sletova, M.; Krivenkov, L.; Tikhonova, T.; Kameneva, A.; Frolova, S.; et al. Identification and pathogenicity of Fusarium species associated with onion basal rot in the Moscow Region of Russian Federation. J. Fungi 2024, 10, 331. [Google Scholar] [CrossRef]
- Zhang, H.; Sha, H.D.; Chen, W.L.; Mao, B.Z. First Report of Fusarium annulatum Causing Blight on Bletilla striata (Baiji) in China. Plant Dis. 2024, 108, 800. [Google Scholar] [CrossRef]
- Aiello, D.; Fiorenza, A.; Leonardi, G.R.; Vitale, A.; Polizzi, G. Fusarium nirenbergiae (Fusarium oxysporum Species Complex) causing the wilting of passion fruit in Italy. Plants 2021, 10, 2011. [Google Scholar] [CrossRef]
- Zhao, X.; Li, H.; Zhou, L.; Chen, F.; Chen, F. Wilt of Acer negundo L. caused by Fusarium nirenbergiae in China. J. For. Res. 2020, 31, 2013–2022. [Google Scholar] [CrossRef]
- López-Moral, A.; Antón-Domínguez, B.I.; Lovera, M.; Arquero, O.; Trapero, A.; Agusti-Brisach, C. Identification and pathogenicity of Fusarium species associated with wilting and crown rot in almond (Prunus dulcis). Sci. Rep. 2024, 14, 5720. [Google Scholar] [CrossRef] [PubMed]
- Mirghasempour, S.A.; Michailides, T.; Chen, W.; Mao, B. Fusarium spp. associated with Dendrobium officinale dieback disease in China. J. Fungi 2022, 8, 919. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, W.; Li, X.; Ji, S.; Chethana, K.W.T.; Hyde, K.D.; Yan, J. Fusarium species associated with cherry leaf spot in China. Plants 2022, 11, 2760. [Google Scholar] [CrossRef] [PubMed]
- Murali, M.; Naziya, B.; Ansari, M.A.; Alomary, M.N.; AlYahya, S.; Almatroudi, A.; Thriveni, M.C.; Gowtham, H.G.; Singh, S.B.; Aiyaz, M. Bioprospecting of rhizosphere-resident fungi: Their role and importance in sustainable agriculture. J. Fungi 2021, 7, 314. [Google Scholar] [CrossRef]
- Chauhan, P.; Sharma, N.; Tapwal, A.; Kumar, A.; Verma, G.S.; Meena, M.; Seth, C.S.; Swapnil, P. Soil microbiome: Diversity, benefits and interactions with plants. Sustainability 2023, 15, 14643. [Google Scholar] [CrossRef]
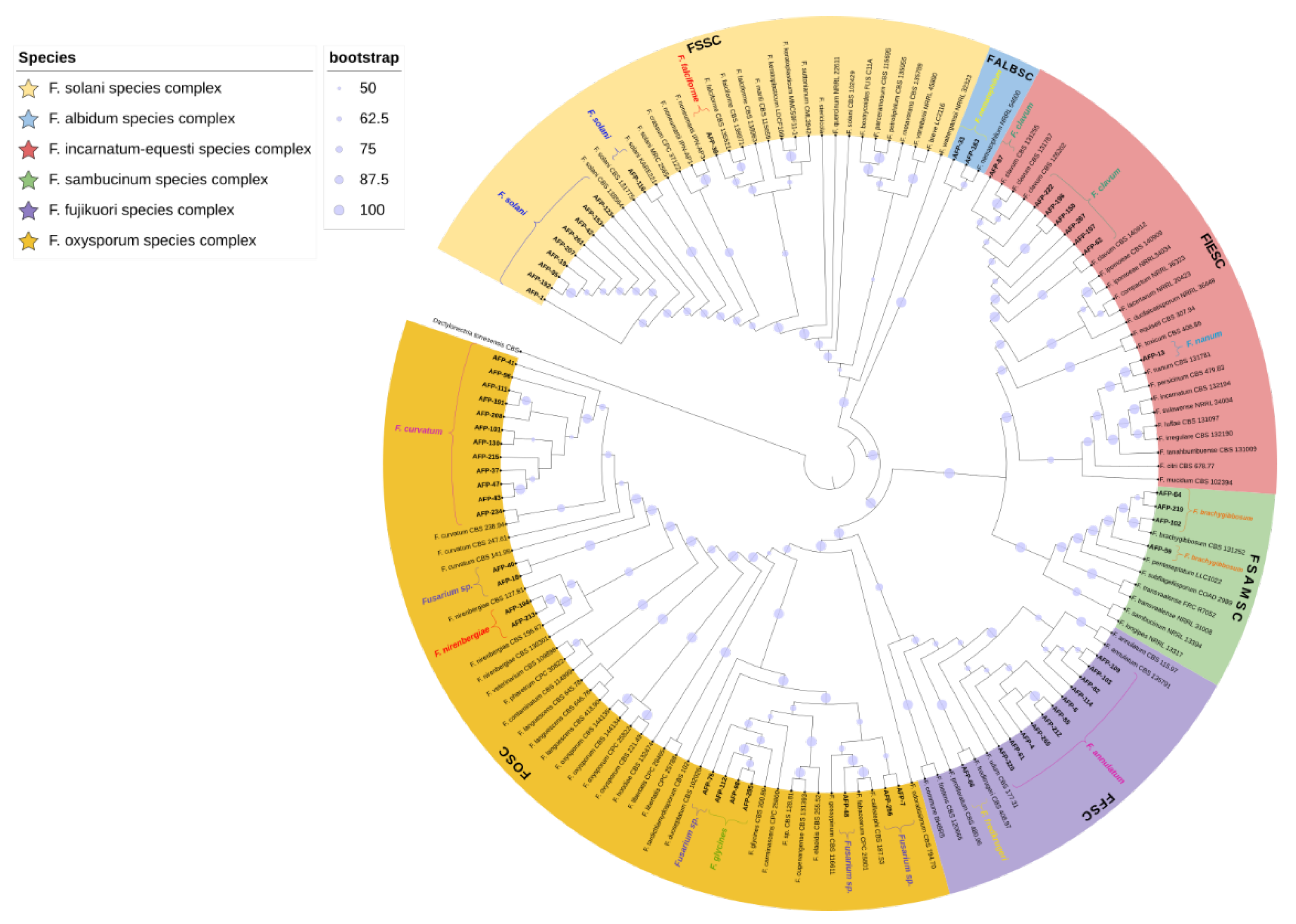


| Reference Species | Isolate | GenBank Accession Numbers | |
| TEF1-α | RPB2 | ||
| F. solani | CBS 138564 | KT272100 | KT272102 |
| " | CBS 131775 | JX118990 | JX237778 |
| " | KARE_221 | MK077042 | MK077080 |
| " | MRC_2565 | MH582420 | MH582410 |
| " | CBS 102429 | KM231936 | KM232376 |
| F. crassum | CPC_37122 | MW248760 | MW446594 |
| F. noneumartii | IPN-AP1 | OP902594 | OP902591 |
| " | IPN-AP3 | OP902596 | OP902593 |
| F. falciforme | CBS 135521 | KU711733 | KU604357 |
| " | CBS 138971 | KT716212 | KT716187 |
| " | CBS 138963 | KT716213 | KT716188 |
| F. martii | CBS 115659 | JX435156 | JX435256 |
| F. keratoplasticum | LDCF109 | OP184958 | OP186372 |
| " | MMC59F11-1 | MF069182 | MF069181 |
| F. suttonianum | CML3942 | MK158921 | MH709236 |
| F. stericicola | N/A | LR583659 | LR583888 |
| F. quercinum | NRRL:22611 | DQ246841 | EU329518 |
| F. bostrycoides | FUS C11A | PP105767 | PP125181 |
| F. parceramosum | CBS 115695 | JX435149 | JX435249 |
| F. petroliphilum | CBS 135955 | KU711768 | KU604337 |
| F. metavorans | CBS 135789 | KU711773 | KU604374 |
| F. vanettenii | NRRL 45880 | FJ240352 | JX171655 |
| F. breve | LC2116 | MW620163 | MW474688 |
| F. waltergamsii | NRRL 32323 | DQ246951 | EU329576 |
| F. nematophilum | NRRL_54600 | N/A | JX171664 |
| F. clavum | CBS 131255 | MN170460 | MN170393 |
| " | CBS 131787 | MN170461 | MN170394 |
| " | CBS 126202 | MN170456 | MN170389 |
| " | CBS 140912 | MN170462 | MN170395 |
| F. ipomoeae | CBS 140909 | MN170479 | MN170412 |
| " | NRRL 34034 | GQ505636 | GQ505814 |
| F. compactum | NRRL 36323 | GQ505648 | GQ505826 |
| F. lacertarum | NRRL 20423 | GQ505593 | GQ505771 |
| F. duofalcatisporum | NRRL 36448 | GQ505652 | GQ505830 |
| F. equiseti | CBS 307.94 | KR071777 | KU604327 |
| F. toxicum | CBS 406.86 | MN170508 | MN170441 |
| F. nanum | CBS 131781 | MN170487 | MN170420 |
| F. persicinum | CBS 479.83 | MN170495 | MN170428 |
| F. incarnatum | CBS 132194 | KF255470 | KF255542 |
| F. sulawense | NRRL 34004 | GQ505628 | GQ505806 |
| F. luffae | CBS 131097 | MN170482 | MN170415 |
| F. irregulare | CBS 132190 | MN170480 | MN170413 |
| F. tanahbumbuense | CBS 131009 | MN170506 | MN170439 |
| F. citri | CBS 678.77 | MN170453 | MN170386 |
| F. mucidum | CBS 102394 | MN170484 | MN170417 |
| F. brachygibbosum | CBS 131252 | JQ429334 | JX162526 |
| F. pentaseptatum | LLC1022 | OP487255 | OP486819 |
| F. subflagellisporum | COAD 2989 | MT774486 | MZ970426 |
| F. transvaalense | FRC R7052 | MW233161 | MW233505 |
| " | NRRL 31008 | MW233102 | MW233446 |
| F. sambucinum | NRRL 13394 | MW233064 | MW233407 |
| F. longipes | NRRL 13317 | MW233058 | MG282411 |
| F. annulatum | CBS 115.97 | MW401973 | MW402785 |
| " | CBS 135791 | MW402054 | MW402746 |
| F. udum | CBS 177.31 | MH484957 | MH484866 |
| F. fredkrugeri | CBS 408.97 | MW402126 | MW402814 |
| F. proliferatum | CBS 480.96 | MN534059 | MN534272 |
| F. foetens | CBS 120665 | MH485009 | MH484918 |
| F. commune | BHBR5 | OR900978 | OR888540 |
| F. odoratissimum | CBS 794.70 | MH484969 | MH484878 |
| F. callistephi | CBS 187.53 | MH484966 | MH484875 |
| F. fabacearum | CPC 25801 | MH485029 | MH484938 |
| F. gossypinum | CBS 116611 | MH484998 | MH484907 |
| F. elaeidis | CBS 255.52 | MH484965 | MH484874 |
| F. cugenangense | CBS 131393 | MH485019 | MH484928 |
| Fusarium sp. | CBS 128.81 | MH484975 | MH484884 |
| F. carminascens | CPC 25800 | MH485028 | MH484937 |
| F. glycines | CBS 200.89 | MH484979 | MH484888 |
| F. duoseptatum | CBS 102026 | MH484987 | MH484896 |
| F. tardichlamydosporum | CBS 102028 | MH484988 | MH484897 |
| F. libertatis | CPC 25788 | MH485024 | MH484933 |
| " | CPC 28465 | MH485035 | MH484944 |
| F. hoodiae | CBS 132474 | MH485020 | MH484929 |
| F. oxysporum | CBS 221.49 | MH484963 | MH484872 |
| " | CPC 25822 | MH485034 | MH484943 |
| " | CBS 144134 | MH485044 | MH484953 |
| " | CBS 144135 | MH485045 | MH484954 |
| F. languescens | CBS 413.90 | MH484981 | MH484890 |
| " | CBS 646.78 | MH484972 | MH484881 |
| " | CBS 645.78 | MH484971 | MH484880 |
| F. contaminatum | CBS 114899 | MH484992 | MH484901 |
| F. pharetrum | CPC 30822 | MH485042 | MH484951 |
| F. veterinarium | CBS 109898 | MH484990 | MH484899 |
| F. nirenbergiae | CBS 130301 | MH485017 | MH484926 |
| " | CBS 196.87 | MH484977 | MH484886 |
| " | CBS 127.81 | MH484974 | MH484883 |
| F. curvatum | CBS 141.95 | MH484985 | MH484894 |
| " | CBS 247.61 | MH484967 | MH484876 |
| " | CBS 238.94 | MH484984 | MH484893 |
| Dactylonectria torresensis | CBS 129086 | JF735870 | KM232347 |
| Nursery | Location | Rootstock or Cultivar | Isolation | ||
| Frequency (%) | |||||
| 1 | Bursa | 1103P-Trakya İlkeren | 32.9 | ||
| 2 | Mersin | 1103P- Victoria | 32.9 | ||
| 3 | Salihli, Manisa | Thompson Seedless | - | ||
| 4 | Salihli, Manisa | Sultana Seedless | 28.5 | ||
| 5 | Salihli, Manisa | Sultana Seedless | - | ||
| 6 | Salihli, Manisa | Sultana Seedless | 38.5 | ||
| 7 | Salihli, Manisa | 1103P / Sultana Seedless | 40.0 | ||
| 8 | Alaşehir, Manisa | Sultana Seedless | 34.3 | ||
| 9 | Alaşehir, Manisa | Sultana Seedless | 4.3 | ||
| 10 | Alaşehir, Manisa | Sultana Seedless | 24.3 | ||
| 11 | Sarıgöl, Manisa | Sultana Seedless | 58.6 | ||
| 12 | Salihli, Manisa | Sultana Seedless | 17.1 | ||
| 13 | Tekirdağ | Kober 5BB / Sultan 1 | 8.6 | ||
| 14 | Tekirdağ | Kober 5BB / Bozbey | 17.2 | ||
| 15 | Tekirdağ | 1103P-Tekirdağ Çekirdeksizi | 27.1 | ||
| 16 | Tekirdağ | 110R-Yapıncak | 2.9 | ||
| 17 | Denizli | 41B / Sultana Seedless | 20.0 | ||
| 18 | Denizli | 41B / Sultana Seedless | 40.0 | ||
| 19 | Denizli | 41B / Sultana Seedless | 65.7 | ||
| 20 | Denizli | 41B / Sultana Seedless | 50.0 | ||
| 21 | Denizli | 41B / Michele Palieri | 45.7 | ||
| 22 | Şanlıurfa | 1103P - Ergin Çekirdeksizi | 12.9 | ||
| 23 | Şanlıurfa | 110R - Horozkarası | 11.5 | ||
| 24 | Şanlıurfa | 99R - Çiloreş | 12.9 | ||
| 25 | Şanlıurfa | 1103P - Victoria | 5.7 | ||
| 26 | Manisa | 41B / Red Globe | 47.1 | ||
| 27 | Manisa | Kober 5BB / Royal | 20.0 | ||
| 28 | Manisa | 1103P - Sultana Seedless | 20.0 | ||
| 29 | Manisa | Kober 5BB - Sultana Seedless | 21.4 | ||
| 30 | Manisa | 1103P - Crimson Seedless | 17.1 | ||
| 31 | Manisa | 110R / Alicante Bouschet | 26.3 | ||
| 32 | Alaşehir, Manisa | 1103P - Thompson Seedless | 60.0 | ||
| 33 | Manisa | Kober 5BB / Ata Sarısı | 22.9 | ||
| 34 | Turgutlu, Manisa | Kober 5BB /Sultana Seedless | 28.6 | ||
| 35 | Manisa | Kober 5BB / Trakya İlkeren | 8.6 | ||
| 36 | Tokat | 1103P - Narince | 40.0 | ||
| 37 | Tokat | 1103P/Narince | 21.4 | ||
| 38 | Tokat | 1103P/Narince | 11.4 | ||
| 39 | Tokat | 1103P/Sultan7 | 12.9 | ||
| 40 | Tokat | 1103P/Narince | 20.0 | ||
| 41 | Tokat | Du Lot / Narince | 12.9 | ||
| 42 | Adıyaman | Kober 5BB / Hatun Parmağı | 17.9 | ||
| 43 | Mersin | 1103P / Victoria | 31.3 | ||
| Mean | 24.9 | ||||
| Isolate | Fusarium Species | Location | Rootstock / Cultivar | GenBank Accession Numbers | |
| TEF1-α | RPB2 | ||||
| AFP004 | Fusarium annulatum | Bursa | 1103 Paulsen | PP449277 | PP449217 |
| AFP006 | " | Bursa | 1103 Paulsen | PP449278 | PP449218 |
| AFP061 | " | Manisa | Kober 5BB | PP449279 | PP449219 |
| AFP082 | " | Tokat | 1103 Paulsen | PP449280 | PP449220 |
| AFP085 | " | Tokat | 1103 Paulsen | PP449281 | PP449221 |
| AFP103 | " | Manisa | Kober 5BB | PP449282 | PP449222 |
| AFP109 | " | Manisa | 110 Richter | PP449283 | PP449223 |
| AFP114 | " | Manisa | Kober 5BB | PP449284 | PP449224 |
| AFP212 | " | Tokat | 1103 Paulsen | PP449285 | PP449225 |
| AFP265 | " | Manisa | Sultana Seedless | PP449286 | PP449226 |
| AFP320 | " | Tekirdağ | 1103 Paulsen | PP449287 | PP449227 |
| AFP059 | Fusarium brachygibbosum | Manisa | Kober 5BB | PP449288 | PP449228 |
| AFP064 | " | Manisa | 41B | PP449289 | PP449229 |
| AFP102 | " | Manisa | Ramsey | PP449290 | PP449230 |
| AFP219 | " | Şanlıurfa | 1103 Paulsen | PP449291 | PP449231 |
| AFP062 | Fusarium clavum | Manisa | Kober 5BB | PP449292 | PP449232 |
| AFP087 | " | Tokat | 1103 Paulsen | PP449293 | PP449233 |
| AFP107 | " | Manisa | Ramsey | PP449294 | PP449234 |
| AFP150 | " | Tokat | 1103 Paulsen | PP449295 | PP449235 |
| AFP196 | " | Manisa | Ramsey | PP449296 | PP449236 |
| AFP222 | " | Tokat | 1103 Paulsen | PP449297 | PP449237 |
| AFP267 | " | Manisa | Sultana Seedless | PP449298 | PP449238 |
| AFP037 | Fusarium curvatum | Denizli | 140 Ruggeri | PP449299 | PP449239 |
| AFP041 | " | Denizli | 1103 Paulsen | PP449300 | PP449240 |
| AFP043 | " | Denizli | 140 Ruggeri | PP449301 | PP449241 |
| AFP047 | " | Denizli | 140 Ruggeri | PP449302 | PP449242 |
| AFP096 | " | Manisa | Kober 5BB | PP449303 | PP449243 |
| AFP101 | " | Manisa | Ramsey | PP449304 | PP449244 |
| AFP111 | " | Manisa | 110 Richter | PP449305 | PP449245 |
| AFP130 | " | Mersin | 1103 Paulsen | PP449306 | PP449246 |
| AFP191 | " | Tokat | 1103 Paulsen | PP449307 | PP449247 |
| AFP208 | " | Tokat | 1103 Paulsen | PP449308 | PP449248 |
| AFP215 | " | Şanlıurfa | 1103 Paulsen | PP449309 | PP449249 |
| AFP234 | " | Şanlıurfa | 1104 Paulsen | PP449310 | PP449250 |
| AFP038 | Fusarium falciforme | Denizli | 140 Ruggeri | PP449311 | PP449251 |
| AFP066 | Fusarium fredkrugeri | Manisa | Kober 5BB | PP449312 | PP449252 |
| AFP098 | Fusarium glycines | Manisa | Ramsey | PP449313 | PP449253 |
| AFP112 | " | Manisa | 110 Richter | PP449314 | PP449254 |
| AFP295 | " | Manisa | Sultana Seedless | PP449315 | PP449255 |
| AFP013 | Fusarium nanum | Mersin | 140 Ruggeri | PP449316 | PP449256 |
| AFP033 | Fusarium nematophilum | Manisa | Kober 5BB | PP449317 | PP449257 |
| AFP163 | " | Tokat | 1103 Paulsen | PP449318 | PP449258 |
| AFP194 | Fusarium nirenbergiae | Manisa | Ramsey | PP449319 | PP449259 |
| AFP213 | " | Tokat | 1103 Paulsen | PP449320 | PP449260 |
| AFP001 | Fusarium solani | Bursa | 1103 Paulsen | PP449321 | PP449261 |
| AFP019 | " | Manisa | 1103 Paulsen | PP449322 | PP449262 |
| AFP042 | " | Denizli | 140 Ruggeri | PP449323 | PP449263 |
| AFP095 | " | Manisa | Kober 5BB | PP449324 | PP449264 |
| AFP116 | " | Manisa | Kober 5BB | PP449325 | PP449265 |
| AFP123 | " | Mersin | 1103 Paulsen | PP449326 | PP449266 |
| AFP153 | " | Tokat | 1103 Paulsen | PP449327 | PP449267 |
| AFP192 | " | Manisa | Ramsey | PP449328 | PP449268 |
| AFP207 | " | Mersin | 1103 Paulsen | PP449329 | PP449269 |
| AFP261 | " | Manisa | Sultana Seedless | PP449330 | PP449270 |
| AFP007 | Fusarium sp. | Bursa | 1103 Paulsen | PP449331 | PP449271 |
| AFP018 | " | Manisa | 1103 Paulsen | PP449332 | PP449272 |
| AFP040 | " | Denizli | 140 Ruggeri | PP449333 | PP449273 |
| AFP048 | " | Denizli | 1103 Paulsen | PP449334 | PP449274 |
| AFP075 | " | Denizli | 140 Ruggeri | PP449335 | PP449275 |
| AFP256 | " | Manisa | Sultana Seedless | PP449336 | PP449276 |
| Isolates | 2022 | Lesion | Isolates | 2023 | Lesion | ||||
| Species | (mm) | Species | (mm) | ||||||
| AFP006 | F. annulatum | 12.0 | a* | AFP061 | F. annulatum | 8.9 | a* | ||
| AFP114 | F. annulatum | 11.9 | a | AFP115 | Ilyonectria liriodendri | 8.6 | a | ||
| AFP109 | F. annulatum | 11.0 | b | AFP213 | F. nirenbergiae | 8.4 | a | ||
| AFP103 | F. annulatum | 10.9 | bc | AFP103 | F. annulatum | 8.0 | b | ||
| AFP059 | F. brachygibbosum | 10.4 | bc | AFP265 | F. annulatum | 7.7 | bc | ||
| AFP115 | Ilyonectria liriodendri | 10.3 | c | AFP114 | F. annulatum | 7.5 | c | ||
| AFP265 | F. annulatum | 7.9 | d | AFP111 | F. curvatum | 7.5 | c | ||
| AFP194 | F. nirenbergiae | 7.1 | e | AFP194 | F. nirenbergiae | 7.5 | c | ||
| AFP061 | F. annulatum | 6.9 | e | AFP006 | F. annulatum | 6.3 | d | ||
| AFP004 | F. annulatum | 6.9 | e | AFP004 | F. annulatum | 5.9 | de | ||
| AFP096 | F. curvatum | 5.3 | f | AFP096 | F. curvatum | 5.2 | e | ||
| AFP213 | F. nirenbergiae | 5.3 | f | AFP109 | F. annulatum | 5.1 | e | ||
| AFP256 | Fusarium sp. | 5.3 | f | AFP059 | F. brachygibbosum | 5.1 | e | ||
| AFP043 | F. curvatum | 5.2 | f | AFP043 | F. curvatum | 5.1 | e | ||
| AFP101 | F. curvatum | 5.2 | f | AFP101 | F. curvatum | 5.0 | e | ||
| AFP038 | F. falciforme | 5.2 | f | AFP098 | F. glycines | 5.0 | e | ||
| AFP040 | Fusarium sp. | 5.2 | f | AFP019 | F. solani | 5.0 | e | ||
| AFP013 | F. nanum | 5.2 | f | AFP041 | F. curvatum | 5.0 | e | ||
| AFP130 | F. curvatum | 5.1 | f | AFP130 | F. curvatum | 5.0 | e | ||
| AFP019 | F. solani | 5.1 | f | AFP037 | F. curvatum | 5.0 | e | ||
| AFP037 | F. curvatum | 5.1 | f | AFP040 | Fusarium sp. | 5.0 | e | ||
| AFP075 | Fusarium sp. | 5.1 | f | AFP066 | F. fredkrugeri | 5.0 | e | ||
| AFP095 | F. solani | 5.1 | f | AFP123 | F. solani | 5.0 | e | ||
| AFP111 | F. curvatum | 5.1 | f | AFP256 | Fusarium sp. | 5.0 | e | ||
| AFP191 | F. curvatum | 5.1 | f | AFP191 | F. curvatum | 5.0 | e | ||
| AFP041 | F. curvatum | 5.1 | f | AFP038 | F. falciforme | 5.0 | e | ||
| AFP001 | F. solani | 5.1 | f | AFP001 | F. solani | 5.0 | e | ||
| AFP007 | Fusarium sp. | 5.1 | f | AFP018 | Fusarium sp. | 5.0 | e | ||
| AFP222 | F. clavum | 5.1 | f | AFP222 | F. clavum | 5.0 | e | ||
| AFP261 | F. solani | 5.1 | f | AFP261 | F. solani | 5.0 | e | ||
| AFP066 | F. fredkrugeri | 5.1 | f | AFP075 | Fusarium sp. | 5.0 | e | ||
| AFP033 | F. nematophilum | 5.0 | f | AFP033 | F. nematophilum | 5.0 | e | ||
| AFP048 | Fusarium sp. | 5.0 | f | AFP048 | Fusarium sp. | 5.0 | e | ||
| AFP062 | F. clavum | 5.0 | f | AFP062 | F. clavum | 5.0 | e | ||
| AFP123 | F. solani | 5.0 | f | AFP095 | F. solani | 5.0 | e | ||
| AFP196 | F. clavum | 5.0 | f | AFP196 | F. clavum | 5.0 | e | ||
| AFP098 | F. glycines | 5.0 | f | AFP013 | F. nanum | 5.0 | e | ||
| AFP018 | Fusarium sp. | 5.0 | f | AFP007 | Fusarium sp. | 5.0 | e | ||
| Non-inoculated Control | 5.0 | f | Non-inoculated Control | 5.0 | e | ||||
| LSD = 0.65 | LSD = 0.92 | ||||||||
| Isolates | 2022 | Basal | Isolates | 2023 | Basal | |||
| Fungal Species | Necrosis (mm) | Fungal Species | Necrosis (mm) | |||||
| AFP061 | F. annulatum | 37.0 | a | AFP115 | Ilyonectria liriodendri | 7.8 | a | |
| AFP103 | F. annulatum | 34.6 | ab | AFP004 | F. annulatum | 6.2 | ab | |
| AFP114 | F. annulatum | 30.8 | a-c | AFP101 | F. curvatum | 6.2 | ab | |
| AFP041 | F. curvatum | 29.8 | a-d | AFP194 | F. nirenbergiae | 6.2 | ab | |
| AFP115 | Ilyonectria liriodendri | 29.8 | a-d | AFP103 | F. annulatum | 6.0 | a-c | |
| AFP004 | F. annulatum | 28.4 | a-e | AFP111 | F. curvatum | 6.0 | a-c | |
| AFP109 | F. annulatum | 27.6 | b-f | AFP114 | F. annulatum | 5.8 | b-d | |
| AFP098 | F. glycines | 27.4 | b-f | AFP213 | F. nirenbergiae | 5.8 | b-d | |
| AFP006 | F. annulatum | 26.8 | b-g | AFP037 | F. curvatum | 5.6 | b-e | |
| AFP265 | F. annulatum | 26.4 | b-g | AFP109 | F. annulatum | 5.6 | b-e | |
| AFP019 | F. solani | 25.2 | c-h | AFP256 | Fusarium sp. | 5.4 | b-f | |
| AFP111 | F. curvatum | 23.8 | c-i | AFP006 | F. annulatum | 5.2 | b-f | |
| AFP018 | Fusarium sp. | 23.2 | c-i | AFP013 | F. nanum | 5.2 | b-f | |
| AFP256 | Fusarium sp. | 22.8 | c-i | AFP038 | F. falciforme | 5.2 | b-f | |
| AFP037 | F. curvatum | 21.2 | d-j | AFP048 | Fusarium sp. | 5.2 | b-f | |
| AFP066 | F. fredkrugeri | 21.2 | d-j | AFP061 | F. annulatum | 5.2 | b-f | |
| AFP075 | Fusarium sp. | 19.8 | d-j | AFP191 | F. curvatum | 5.2 | b-f | |
| AFP096 | F. curvatum | 19.0 | f-k | AFP040 | Fusarium sp. | 5.0 | b-f | |
| AFP123 | F. solani | 17.8 | g-l | AFP098 | F. glycines | 5.0 | b-f | |
| AFP101 | F. curvatum | 17.0 | h-m | AFP265 | F. annulatum | 5.0 | b-f | |
| AFP043 | F. curvatum | 16.0 | i-m | AFP075 | Fusarium sp. | 4.8 | b-g | |
| AFP007 | Fusarium sp. | 15.4 | i-n | AFP123 | F. solani | 4.8 | b-g | |
| AFP038 | F. falciforme | 15.4 | i-n | AFP001 | F. solani | 4.6 | b-g | |
| AFP130 | F. curvatum | 15.0 | i-n | AFP019 | F. solani | 4.6 | b-g | |
| AFP261 | F. solani | 15.0 | i-n | AFP041 | F. curvatum | 4.6 | b-g | |
| AFP191 | F. curvatum | 13.6 | j-o | AFP043 | F. curvatum | 4.6 | b-g | |
| AFP194 | F. nirenbergiae | 12.8 | j-o | AFP222 | F. clavum | 4.6 | b-g | |
| AFP059 | F. brachygibbosum | 12.4 | j-p | AFP007 | Fusarium sp. | 4.4 | b-g | |
| AFP213 | F. nirenbergiae | 12.2 | j-p | AFP066 | F. fredkrugeri | 4.4 | b-g | |
| AFP095 | F. solani | 10.4 | k-q | AFP096 | F. curvatum | 4.4 | b-g | |
| AFP033 | F. nematophilum | 10.2 | k-q | AFP130 | F. curvatum | 4.2 | c-g | |
| AFP040 | Fusarium sp. | 9.8 | l-q | AFP018 | Fusarium sp. | 4.0 | d-g | |
| AFP001 | F. solani | 8.6 | m-q | AFP095 | F. solani | 4.0 | d-g | |
| AFP196 | F. clavum | 8.4 | m-q | AFP261 | F. solani | 4.0 | d-g | |
| AFP062 | F. clavum | 6.6 | n-q | AFP033 | F. nematophilum | 3.8 | e-g | |
| AFP048 | Fusarium sp. | 6.4 | n-q | AFP062 | F. clavum | 3.8 | e-g | |
| AFP222 | F. clavum | 5.2 | o-q | AFP196 | F. clavum | 3.8 | e-g | |
| AFP013 | F. nanum | 3.6 | p-q | AFP059 | F. brachygibbosum | 3.7 | f-g | |
| Non-inoculated Control | 2.6 | q | Non-inoculated Control | 3.0 | g | |||
| LSD = 9.06 | LSD = 1.87 | |||||||
| Isolates | 2022 | Root Dry | Isolates | 2023 | Root Dry | |||
| Species | Weight (g) | Species | Weight (g) | |||||
| Non-inoculated Control | 0.344 | a* | Non-inoculated Control | 1.463 | a | |||
| AFP041 | F. curvatum | 0.224 | b | AFP222 | F. clavum | 1.436 | b | |
| AFP033 | F. nematophilum | 0.196 | c | AFP048 | Fusarium sp. | 1.425 | c | |
| AFP256 | Fusarium sp. | 0.182 | d | AFP062 | F. clavum | 1.417 | d | |
| AFP123 | F. solani | 0.150 | e | AFP013 | F. nanum | 1.410 | e | |
| AFP098 | F. glycines | 0.146 | f | AFP101 | F. curvatum | 1.399 | f | |
| AFP018 | Fusarium sp. | 0.134 | g | AFP033 | F. nematophilum | 1.369 | g | |
| AFP062 | F. clavum | 0.125 | h | AFP075 | Fusarium sp. | 1.353 | h | |
| AFP261 | F. solani | 0.125 | h | AFP261 | F. solani | 1.349 | i | |
| AFP048 | Fusarium sp. | 0.119 | i | AFP038 | F. falciforme | 1.336 | j | |
| AFP013 | F. nanum | 0.112 | j | AFP001 | F. solani | 1.327 | k | |
| AFP130 | F. curvatum | 0.111 | j | AFP095 | F. solani | 1.286 | l | |
| AFP196 | F. clavum | 0.109 | jk | AFP007 | Fusarium sp. | 1.282 | m | |
| AFP019 | F. solani | 0.108 | k | AFP019 | F. solani | 1.266 | n | |
| AFP109 | F. annulatum | 0.104 | l | AFP040 | Fusarium sp. | 1.251 | o | |
| AFP222 | F. clavum | 0.103 | l | AFP059 | F. brachygibbosum | 1.245 | p | |
| AFP038 | F. falciforme | 0.098 | m | AFP098 | F. glycines | 1.232 | q | |
| AFP004 | F. annulatum | 0.087 | n | AFP256 | Fusarium sp. | 1.231 | q | |
| AFP001 | F. solani | 0.083 | o | AFP066 | F. fredkrugeri | 1.181 | r | |
| AFP040 | Fusarium sp. | 0.082 | o | AFP191 | F. curvatum | 1.172 | s | |
| AFP007 | Fusarium sp. | 0.078 | p | AFP103 | F. annulatum | 1.160 | t | |
| AFP194 | F. nirenbergiae | 0.072 | q | AFP018 | Fusarium sp. | 1.118 | u | |
| AFP096 | F. curvatum | 0.070 | q | AFP196 | F. clavum | 1.108 | v | |
| AFP213 | F. nirenbergiae | 0.065 | r | AFP096 | F. curvatum | 1.101 | w | |
| AFP043 | F. curvatum | 0.063 | r | AFP006 | F. annulatum | 1.098 | x | |
| AFP114 | F. annulatum | 0.058 | s | AFP213 | F. nirenbergiae | 1.069 | y | |
| AFP059 | F. brachygibbosum | 0.054 | t | AFP043 | F. curvatum | 1.056 | z | |
| AFP101 | F. curvatum | 0.054 | t | AFP115 | Ilyonectria liriodendri | 1.047 | a1 | |
| AFP103 | F. annulatum | 0.051 | tu | AFP123 | F. solani | 1.026 | b1 | |
| AFP006 | F. annulatum | 0.050 | u | AFP111 | F. curvatum | 1.016 | c1 | |
| AFP075 | Fusarium sp. | 0.049 | uv | AFP109 | F. annulatum | 1.012 | d1 | |
| AFP095 | F. solani | 0.047 | vw | AFP037 | F. curvatum | 0.974 | e1 | |
| AFP037 | F. curvatum | 0.045 | wx | AFP041 | F. curvatum | 0.921 | f1 | |
| AFP111 | F. curvatum | 0.044 | x | AFP130 | F. curvatum | 0.919 | f1 | |
| AFP115 | Ilyonectria liriodendri | 0.042 | xy | AFP194 | F. nirenbergiae | 0.919 | f1 | |
| AFP066 | F. fredkrugeri | 0.040 | yz | AFP114 | F. annulatum | 0.901 | g1 | |
| AFP191 | F. curvatum | 0.038 | z | AFP004 | F. annulatum | 0.893 | h1 | |
| AFP265 | F. annulatum | 0.026 | a1 | AFP265 | F. annulatum | 0.880 | i1 | |
| AFP061 | F. annulatum | 0.022 | b1 | AFP061 | F. annulatum | 0.599 | j1 | |
| LSD = | 0.002 | LSD = | 0.003 | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





