Submitted:
05 June 2024
Posted:
06 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Lessons from the Human Cell Atlas (HCA)
2.1. Cross-Tissue Studies in Health
2.2. Cross-Tissue Studies in Health: Challenges and Potential Solutions
2.2.1. Methods for Sample Processing
2.2.2. Challenges of Variation between scSeq Platforms
2.2.3. Challenges of Data Access
2.2.4. Methods for Data Integration
2.2.4. Methods for Benchmarking of Data Integration
2.2.5. Methods for Cell Type/State Annotation
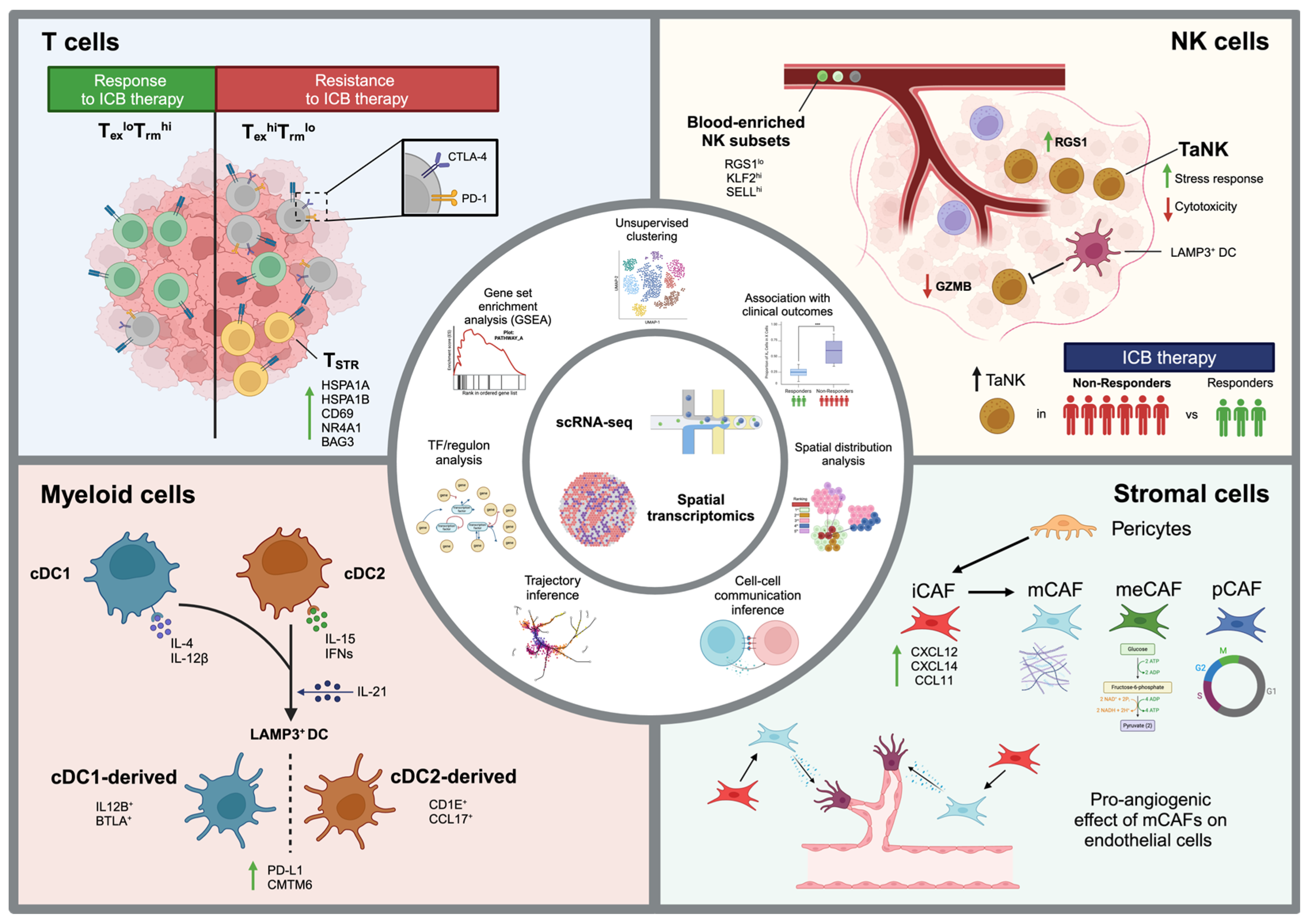
3. Learnings from Pan-Cancer Studies
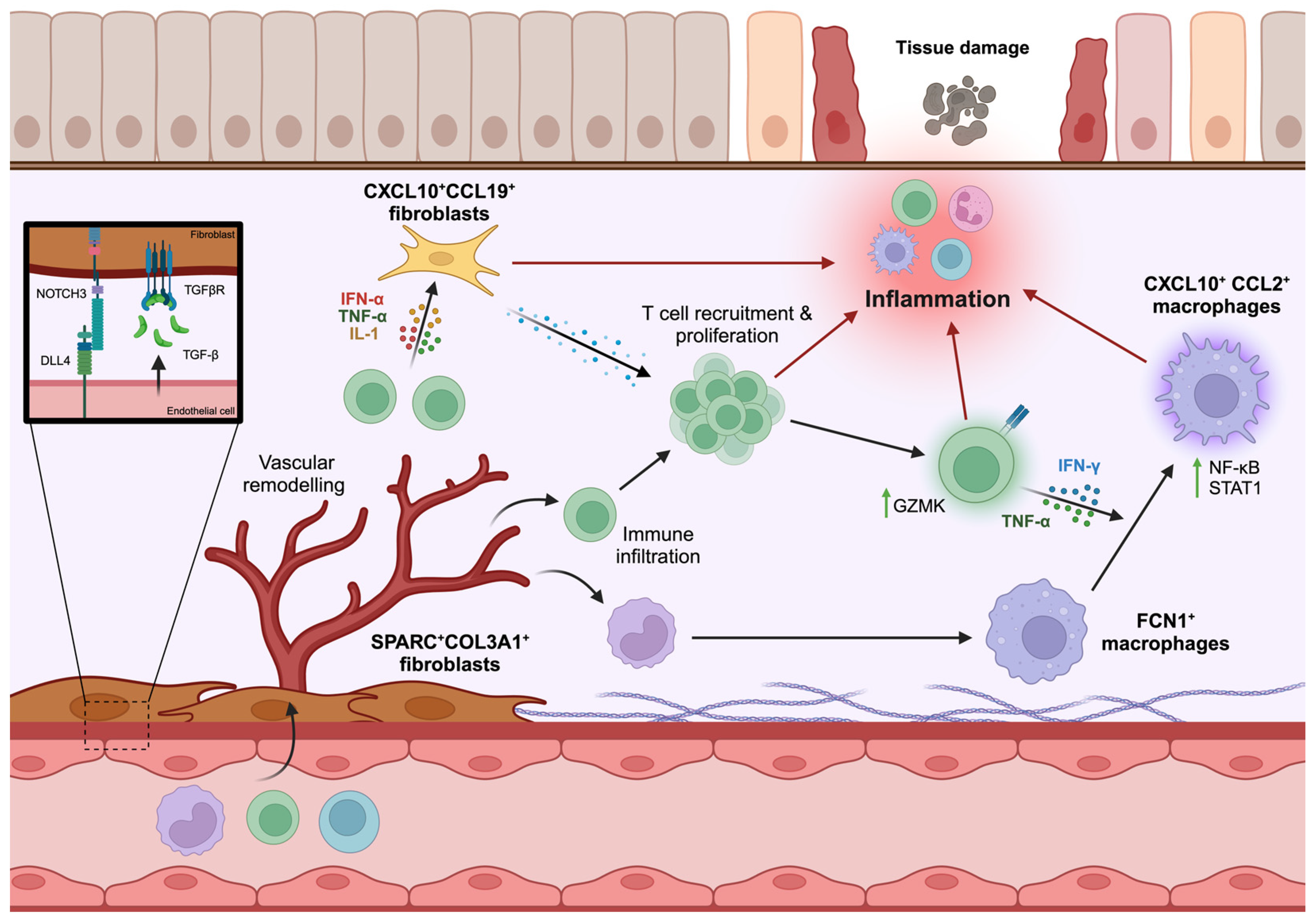
4. Cross-Tissue Studies of IMIDs
5. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Monteleone, G.; Moscardelli, A.; Colella, A.; Marafini, I.; Salvatori, S. Immune-Mediated Inflammatory Diseases: Common and Different Pathogenic and Clinical Features. Autoimmunity Reviews 2023, 22 (10), 103410. [CrossRef]
- Conrad, N.; Misra, S.; Verbakel, J. Y.; Verbeke, G.; Molenberghs, G.; Taylor, P. N.; Mason, J.; Sattar, N.; McMurray, J. J. V.; McInnes, I. B.; et al. Incidence, Prevalence, and Co-Occurrence of Autoimmune Disorders over Time and by Age, Sex, and Socioeconomic Status: A Population-Based Cohort Study of 22 Million Individuals in the UK. The Lancet 2023, 401 (10391), 1878–1890. [CrossRef]
- Monaco, C.; Nanchahal, J.; Taylor, P.; Feldmann, M. Anti-TNF Therapy: Past, Present and Future. International Immunology 2015, 27 (1), 55–62. [CrossRef]
- Landewé, R.; Braun, J.; Deodhar, A.; Dougados, M.; Maksymowych, W. P.; Mease, P. J.; Reveille, J. D.; Rudwaleit, M.; Heijde, D. van der; Stach, C.; et al. Efficacy of Certolizumab Pegol on Signs and Symptoms of Axial Spondyloarthritis Including Ankylosing Spondylitis: 24-Week Results of a Double-Blind Randomised Placebo-Controlled Phase 3 Study. Annals of the Rheumatic Diseases 2014, 73 (1), 39–47. [CrossRef]
- Torres, J.; Mehandru, S.; Colombel, J.-F.; Peyrin-Biroulet, L. Crohn’s Disease. The Lancet 2017, 389 (10080), 1741–1755. [CrossRef]
- Ungaro, R.; Mehandru, S.; Allen, P. B.; Peyrin-Biroulet, L.; Colombel, J.-F. Ulcerative Colitis. The Lancet 2017, 389 (10080), 1756–1770. [CrossRef]
- Buckley, C. D.; Chernajovsky, L.; Chernajovsky, Y.; Modis, L. K.; O’Neill, L. A.; Brown, D.; Connor, R.; Coutts, D.; Waterman, E. A.; Tak, P. P. Immune-Mediated Inflammation across Disease Boundaries: Breaking down Research Silos. Nat Immunol 2021, 22 (11), 1344–1348. [CrossRef]
- Clark, A. D.; Nair, N.; Anderson, A. E.; Thalayasingam, N.; Naamane, N.; Skelton, A. J.; Diboll, J.; Barton, A.; Eyre, S.; Isaacs, J. D.; et al. Lymphocyte DNA Methylation Mediates Genetic Risk at Shared Immune-Mediated Disease Loci. Journal of Allergy and Clinical Immunology 2020, 145 (5), 1438–1451. [CrossRef]
- Papalexi, E.; Satija, R. Single-Cell RNA Sequencing to Explore Immune Cell Heterogeneity. Nat Rev Immunol 2018, 18 (1), 35–45. [CrossRef]
- Hao, Y.; Stuart, T.; Kowalski, M. H.; Choudhary, S.; Hoffman, P.; Hartman, A.; Srivastava, A.; Molla, G.; Madad, S.; Fernandez-Granda, C.; et al. Dictionary Learning for Integrative, Multimodal and Scalable Single-Cell Analysis. Nat Biotechnol 2024, 42 (2), 293–304. [CrossRef]
- Wolf, F. A.; Angerer, P.; Theis, F. J. SCANPY: Large-Scale Single-Cell Gene Expression Data Analysis. Genome Biology 2018, 19 (1), 15. [CrossRef]
- Regev, A.; Teichmann, S. A.; Lander, E. S.; Amit, I.; Benoist, C.; Birney, E.; Bodenmiller, B.; Campbell, P.; Carninci, P.; Clatworthy, M.; et al. The Human Cell Atlas. eLife 2017, 6, e27041. [CrossRef]
- Lindeboom, R. G. H.; Regev, A.; Teichmann, S. A. Towards a Human Cell Atlas: Taking Notes from the Past. Trends in Genetics 2021, 37 (7), 625–630. [CrossRef]
- Domínguez Conde, C.; Xu, C.; Jarvis, L. B.; Rainbow, D. B.; Wells, S. B.; Gomes, T.; Howlett, S. K.; Suchanek, O.; Polanski, K.; King, H. W.; et al. Cross-Tissue Immune Cell Analysis Reveals Tissue-Specific Features in Humans. Science 2022, 376 (6594), eabl5197. [CrossRef]
- Suo, C.; Dann, E.; Goh, I.; Jardine, L.; Kleshchevnikov, V.; Park, J.-E.; Botting, R. A.; Stephenson, E.; Engelbert, J.; Tuong, Z. K.; et al. Mapping the Developing Human Immune System across Organs. Science 2022, 376 (6597), eabo0510. [CrossRef]
- Eraslan, G.; Drokhlyansky, E.; Anand, S.; Fiskin, E.; Subramanian, A.; Slyper, M.; Wang, J.; Van Wittenberghe, N.; Rouhana, J. M.; Waldman, J.; et al. Single-Nucleus Cross-Tissue Molecular Reference Maps toward Understanding Disease Gene Function. Science 2022, 376 (6594), eabl4290. [CrossRef]
- The Tabula Sapiens Consortium. The Tabula Sapiens: A Multiple-Organ, Single-Cell Transcriptomic Atlas of Humans. Science 2022, 376 (6594), eabl4896. [CrossRef]
- Donlin, L. T.; Rao, D. A.; Wei, K.; Slowikowski, K.; McGeachy, M. J.; Turner, J. D.; Meednu, N.; Mizoguchi, F.; Gutierrez-Arcelus, M.; Lieb, D. J.; et al. Methods for High-Dimensional Analysis of Cells Dissociated from Cryopreserved Synovial Tissue. Arthritis Research & Therapy 2018, 20 (1), 139. [CrossRef]
- Zheng, G. X. Y.; Terry, J. M.; Belgrader, P.; Ryvkin, P.; Bent, Z. W.; Wilson, R.; Ziraldo, S. B.; Wheeler, T. D.; McDermott, G. P.; Zhu, J.; et al. Massively Parallel Digital Transcriptional Profiling of Single Cells. Nat Commun 2017, 8 (1), 14049. [CrossRef]
- Ortolano, N. The neXt generation of single cell RNA-seq: An introduction to GEM-X technology. 10x Genomics. https://www.10xgenomics.com/blog/the-next-generation-of-single-cell-rna-seq-an-introduction-to-gem-x-technology (accessed 2024-05-13).
- Miga, K. H.; Newton, Y.; Jain, M.; Altemose, N.; Willard, H. F.; Kent, W. J. Centromere Reference Models for Human Chromosomes X and Y Satellite Arrays. Genome Res. 2014, 24 (4), 697–707. [CrossRef]
- Martin, F. J.; Amode, M. R.; Aneja, A.; Austine-Orimoloye, O.; Azov, A. G.; Barnes, I.; Becker, A.; Bennett, R.; Berry, A.; Bhai, J.; et al. Ensembl 2023. Nucleic Acids Research 2023, 51 (D1), D933–D941. [CrossRef]
- Freeberg, M. A.; Fromont, L. A.; D’Altri, T.; Romero, A. F.; Ciges, J. I.; Jene, A.; Kerry, G.; Moldes, M.; Ariosa, R.; Bahena, S.; et al. The European Genome-Phenome Archive in 2021. Nucleic Acids Research 2022, 50 (D1), D980–D987. [CrossRef]
- Barrett, T.; Wilhite, S. E.; Ledoux, P.; Evangelista, C.; Kim, I. F.; Tomashevsky, M.; Marshall, K. A.; Phillippy, K. H.; Sherman, P. M.; Holko, M.; et al. NCBI GEO: Archive for Functional Genomics Data Sets—Update. Nucleic Acids Research 2013, 41 (D1), D991–D995. [CrossRef]
- Ryu, Y.; Han, G. H.; Jung, E.; Hwang, D. Integration of Single-Cell RNA-Seq Datasets: A Review of Computational Methods. Molecules and Cells 2023, 46 (2), 106–119. [CrossRef]
- Haghverdi, L.; Lun, A. T. L.; Morgan, M. D.; Marioni, J. C. Batch Effects in Single-Cell RNA-Sequencing Data Are Corrected by Matching Mutual Nearest Neighbors. Nat Biotechnol 2018, 36 (5), 421–427. [CrossRef]
- Luecken, M. D.; Büttner, M.; Chaichoompu, K.; Danese, A.; Interlandi, M.; Mueller, M. F.; Strobl, D. C.; Zappia, L.; Dugas, M.; Colomé-Tatché, M.; et al. Benchmarking Atlas-Level Data Integration in Single-Cell Genomics. Nat Methods 2022, 19 (1), 41–50. [CrossRef]
- Butler, A.; Hoffman, P.; Smibert, P.; Papalexi, E.; Satija, R. Integrating Single-Cell Transcriptomic Data across Different Conditions, Technologies, and Species. Nat Biotechnol 2018, 36 (5), 411–420. [CrossRef]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W. M.; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177 (7), 1888-1902.e21. [CrossRef]
- Hie, B.; Bryson, B.; Berger, B. Efficient Integration of Heterogeneous Single-Cell Transcriptomes Using Scanorama. Nat Biotechnol 2019, 37 (6), 685–691. [CrossRef]
- Polański, K.; Young, M. D.; Miao, Z.; Meyer, K. B.; Teichmann, S. A.; Park, J.-E. BBKNN: Fast Batch Alignment of Single Cell Transcriptomes. Bioinformatics 2020, 36 (3), 964–965. [CrossRef]
- Barkas, N.; Petukhov, V.; Nikolaeva, D.; Lozinsky, Y.; Demharter, S.; Khodosevich, K.; Kharchenko, P. V. Joint Analysis of Heterogeneous Single-Cell RNA-Seq Dataset Collections. Nat Methods 2019, 16 (8), 695–698. [CrossRef]
- Tran, H. T. N.; Ang, K. S.; Chevrier, M.; Zhang, X.; Lee, N. Y. S.; Goh, M.; Chen, J. A Benchmark of Batch-Effect Correction Methods for Single-Cell RNA Sequencing Data. Genome Biology 2020, 21 (1), 12. [CrossRef]
- Korsunsky, I.; Millard, N.; Fan, J.; Slowikowski, K.; Zhang, F.; Wei, K.; Baglaenko, Y.; Brenner, M.; Loh, P.; Raychaudhuri, S. Fast, Sensitive and Accurate Integration of Single-Cell Data with Harmony. Nat Methods 2019, 16 (12), 1289–1296. [CrossRef]
- Welch, J. D.; Kozareva, V.; Ferreira, A.; Vanderburg, C.; Martin, C.; Macosko, E. Z. Single-Cell Multi-Omic Integration Compares and Contrasts Features of Brain Cell Identity. Cell 2019, 177 (7), 1873-1887.e17. [CrossRef]
- Lin, Y.; Ghazanfar, S.; Wang, K. Y. X.; Gagnon-Bartsch, J. A.; Lo, K. K.; Su, X.; Han, Z.-G.; Ormerod, J. T.; Speed, T. P.; Yang, P.; et al. scMerge Leverages Factor Analysis, Stable Expression, and Pseudoreplication to Merge Multiple Single-Cell RNA-Seq Datasets. Proceedings of the National Academy of Sciences 2019, 116 (20), 9775–9784. [CrossRef]
- Lopez, R.; Regier, J.; Cole, M. B.; Jordan, M. I.; Yosef, N. Deep Generative Modeling for Single-Cell Transcriptomics. Nat Methods 2018, 15 (12), 1053–1058. [CrossRef]
- Xu, C.; Lopez, R.; Mehlman, E.; Regier, J.; Jordan, M. I.; Yosef, N. Probabilistic Harmonization and Annotation of Single-cell Transcriptomics Data with Deep Generative Models. Molecular Systems Biology 2021, 17 (1), e9620. [CrossRef]
- Lotfollahi, M.; Wolf, F. A.; Theis, F. J. scGen Predicts Single-Cell Perturbation Responses. Nat Methods 2019, 16 (8), 715–721. [CrossRef]
- Lotfollahi, M.; Naghipourfar, M.; Theis, F. J.; Wolf, F. A. Conditional Out-of-Distribution Generation for Unpaired Data Using Transfer VAE. Bioinformatics 2020, 36 (Supplement_2), i610–i617. [CrossRef]
- Lütge, A.; Zyprych-Walczak, J.; Kunzmann, U. B.; Crowell, H. L.; Calini, D.; Malhotra, D.; Soneson, C.; Robinson, M. D. CellMixS: Quantifying and Visualizing Batch Effects in Single-Cell RNA-Seq Data. Life Science Alliance 2021, 4 (6). [CrossRef]
- Büttner, M.; Miao, Z.; Wolf, F. A.; Teichmann, S. A.; Theis, F. J. A Test Metric for Assessing Single-Cell RNA-Seq Batch Correction. Nat Methods 2019, 16 (1), 43–49. [CrossRef]
- Rousseeuw, P. J. Silhouettes: A Graphical Aid to the Interpretation and Validation of Cluster Analysis. Journal of Computational and Applied Mathematics 1987, 20, 53–65. [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. Journal of Machine Learning Research 2011, 12 (85), 2825–2830.
- Hubert, L.; Arabie, P. Comparing Partitions. Journal of Classification 1985, 2 (1), 193–218. [CrossRef]
- Pasquini, G.; Arias, J. E. R.; Schäfer, P.; Busskamp, V. Automated Methods for Cell Type Annotation on scRNA-Seq Data. Computational and Structural Biotechnology Journal 2021, 19, 961–969. [CrossRef]
- Xie, B.; Jiang, Q.; Mora, A.; Li, X. Automatic Cell Type Identification Methods for Single-Cell RNA Sequencing. Computational and Structural Biotechnology Journal 2021, 19, 5874–5887. [CrossRef]
- Shao, X.; Liao, J.; Lu, X.; Xue, R.; Ai, N.; Fan, X. scCATCH: Automatic Annotation on Cell Types of Clusters from Single-Cell RNA Sequencing Data. iScience 2020, 23 (3). [CrossRef]
- Cao, Y.; Wang, X.; Peng, G. SCSA: A Cell Type Annotation Tool for Single-Cell RNA-Seq Data. Front. Genet. 2020, 11. [CrossRef]
- Zhang, Z.; Luo, D.; Zhong, X.; Choi, J. H.; Ma, Y.; Wang, S.; Mahrt, E.; Guo, W.; Stawiski, E. W.; Modrusan, Z.; et al. SCINA: A Semi-Supervised Subtyping Algorithm of Single Cells and Bulk Samples. Genes 2019, 10 (7), 531. [CrossRef]
- Zhang, A. W.; O’Flanagan, C.; Chavez, E. A.; Lim, J. L. P.; Ceglia, N.; McPherson, A.; Wiens, M.; Walters, P.; Chan, T.; Hewitson, B.; et al. Probabilistic Cell-Type Assignment of Single-Cell RNA-Seq for Tumor Microenvironment Profiling. Nat Methods 2019, 16 (10), 1007–1015. [CrossRef]
- Kiselev, V. Y.; Yiu, A.; Hemberg, M. Scmap: Projection of Single-Cell RNA-Seq Data across Data Sets. Nat Methods 2018, 15 (5), 359–362. [CrossRef]
- Aran, D.; Looney, A. P.; Liu, L.; Wu, E.; Fong, V.; Hsu, A.; Chak, S.; Naikawadi, R. P.; Wolters, P. J.; Abate, A. R.; et al. Reference-Based Analysis of Lung Single-Cell Sequencing Reveals a Transitional Profibrotic Macrophage. Nat Immunol 2019, 20 (2), 163–172. [CrossRef]
- Hou, R.; Denisenko, E.; Forrest, A. R. R. scMatch: A Single-Cell Gene Expression Profile Annotation Tool Using Reference Datasets. Bioinformatics 2019, 35 (22), 4688–4695. [CrossRef]
- de Kanter, J. K.; Lijnzaad, P.; Candelli, T.; Margaritis, T.; Holstege, F. C. P. CHETAH: A Selective, Hierarchical Cell Type Identification Method for Single-Cell RNA Sequencing. Nucleic Acids Research 2019, 47 (16), e95. [CrossRef]
- Alquicira-Hernandez, J.; Sathe, A.; Ji, H. P.; Nguyen, Q.; Powell, J. E. scPred: Accurate Supervised Method for Cell-Type Classification from Single-Cell RNA-Seq Data. Genome Biology 2019, 20 (1), 264. [CrossRef]
- Tan, Y.; Cahan, P. SingleCellNet: A Computational Tool to Classify Single Cell RNA-Seq Data Across Platforms and Across Species. cels 2019, 9 (2), 207-213.e2. [CrossRef]
- Kimmel, J. C.; Kelley, D. R. Semisupervised Adversarial Neural Networks for Single-Cell Classification. Genome Res. 2021, 31 (10), 1781–1793. [CrossRef]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W. M.; Zheng, S.; Butler, A.; Lee, M. J.; Wilk, A. J.; Darby, C.; Zager, M.; et al. Integrated Analysis of Multimodal Single-Cell Data. Cell 2021, 184 (13), 3573-3587.e29. [CrossRef]
- Lotfollahi, M.; Naghipourfar, M.; Luecken, M. D.; Khajavi, M.; Büttner, M.; Wagenstetter, M.; Avsec, Ž.; Gayoso, A.; Yosef, N.; Interlandi, M.; et al. Mapping Single-Cell Data to Reference Atlases by Transfer Learning. Nat Biotechnol 2022, 40 (1), 121–130. [CrossRef]
- Kang, J. B.; Nathan, A.; Weinand, K.; Zhang, F.; Millard, N.; Rumker, L.; Moody, D. B.; Korsunsky, I.; Raychaudhuri, S. Efficient and Precise Single-Cell Reference Atlas Mapping with Symphony. Nat Commun 2021, 12 (1), 5890. [CrossRef]
- Zheng, L.; Qin, S.; Si, W.; Wang, A.; Xing, B.; Gao, R.; Ren, X.; Wang, L.; Wu, X.; Zhang, J.; et al. Pan-Cancer Single-Cell Landscape of Tumor-Infiltrating T Cells. Science 2021, 374 (6574), abe6474. [CrossRef]
- Chu, Y.; Dai, E.; Li, Y.; Han, G.; Pei, G.; Ingram, D. R.; Thakkar, K.; Qin, J.-J.; Dang, M.; Le, X.; et al. Pan-Cancer T Cell Atlas Links a Cellular Stress Response State to Immunotherapy Resistance. Nat Med 2023, 29 (6), 1550–1562. [CrossRef]
- Tang, F.; Li, J.; Qi, L.; Liu, D.; Bo, Y.; Qin, S.; Miao, Y.; Yu, K.; Hou, W.; Li, J.; et al. A Pan-Cancer Single-Cell Panorama of Human Natural Killer Cells. Cell 2023, 186 (19), 4235-4251.e20. [CrossRef]
- Efremova, M.; Vento-Tormo, M.; Teichmann, S. A.; Vento-Tormo, R. CellPhoneDB: Inferring Cell–Cell Communication from Combined Expression of Multi-Subunit Ligand–Receptor Complexes. Nat Protoc 2020, 15 (4), 1484–1506. [CrossRef]
- Cheng, S.; Li, Z.; Gao, R.; Xing, B.; Gao, Y.; Yang, Y.; Qin, S.; Zhang, L.; Ouyang, H.; Du, P.; et al. A Pan-Cancer Single-Cell Transcriptional Atlas of Tumor Infiltrating Myeloid Cells. Cell 2021, 184 (3), 792-809.e23. [CrossRef]
- Ma, C.; Yang, C.; Peng, A.; Sun, T.; Ji, X.; Mi, J.; Wei, L.; Shen, S.; Feng, Q. Pan-Cancer Spatially Resolved Single-Cell Analysis Reveals the Crosstalk between Cancer-Associated Fibroblasts and Tumor Microenvironment. Mol Cancer 2023, 22 (1), 170. [CrossRef]
- Street, K.; Risso, D.; Fletcher, R. B.; Das, D.; Ngai, J.; Yosef, N.; Purdom, E.; Dudoit, S. Slingshot: Cell Lineage and Pseudotime Inference for Single-Cell Transcriptomics. BMC Genomics 2018, 19 (1), 477. [CrossRef]
- Zhang, F.; Wei, K.; Slowikowski, K.; Fonseka, C. Y.; Rao, D. A.; Kelly, S.; Goodman, S. M.; Tabechian, D.; Hughes, L. B.; Salomon-Escoto, K.; et al. Defining Inflammatory Cell States in Rheumatoid Arthritis Joint Synovial Tissues by Integrating Single-Cell Transcriptomics and Mass Cytometry. Nat Immunol 2019, 20 (7), 928–942. [CrossRef]
- Zhang, F.; Jonsson, A. H.; Nathan, A.; Millard, N.; Curtis, M.; Xiao, Q.; Gutierrez-Arcelus, M.; Apruzzese, W.; Watts, G. F. M.; Weisenfeld, D.; et al. Deconstruction of Rheumatoid Arthritis Synovium Defines Inflammatory Subtypes. Nature 2023, 623 (7987), 616–624. [CrossRef]
- Reynolds, G.; Vegh, P.; Fletcher, J.; Poyner, E. F. M.; Stephenson, E.; Goh, I.; Botting, R. A.; Huang, N.; Olabi, B.; Dubois, A.; et al. Developmental Cell Programs Are Co-Opted in Inflammatory Skin Disease. Science 2021, 371 (6527), eaba6500. [CrossRef]
- Kong, L.; Pokatayev, V.; Lefkovith, A.; Carter, G. T.; Creasey, E. A.; Krishna, C.; Subramanian, S.; Kochar, B.; Ashenberg, O.; Lau, H.; et al. The Landscape of Immune Dysregulation in Crohn’s Disease Revealed through Single-Cell Transcriptomic Profiling in the Ileum and Colon. Immunity 2023, 56 (2), 444-458.e5. [CrossRef]
- Arazi, A.; Rao, D. A.; Berthier, C. C.; Davidson, A.; Liu, Y.; Hoover, P. J.; Chicoine, A.; Eisenhaure, T. M.; Jonsson, A. H.; Li, S.; et al. The Immune Cell Landscape in Kidneys of Patients with Lupus Nephritis. Nat Immunol 2019, 20 (7), 902–914. [CrossRef]
- Zhang, F.; Mears, J. R.; Shakib, L.; Beynor, J. I.; Shanaj, S.; Korsunsky, I.; Nathan, A.; Donlin, L. T.; Raychaudhuri, S.; Accelerating Medicines Partnership Rheumatoid Arthritis and Systemic Lupus Erythematosus (AMP RA/SLE) Consortium. IFN-γ and TNF-α Drive a CXCL10+ CCL2+ Macrophage Phenotype Expanded in Severe COVID-19 Lungs and Inflammatory Diseases with Tissue Inflammation. Genome Medicine 2021, 13 (1), 64. [CrossRef]
- Jonsson, A. H.; Zhang, F.; Dunlap, G.; Gomez-Rivas, E.; Watts, G. F. M.; Faust, H. J.; Rupani, K. V.; Mears, J. R.; Meednu, N.; Wang, R.; et al. Granzyme K+ CD8 T Cells Form a Core Population in Inflamed Human Tissue. Science Translational Medicine 2022, 14 (649), eabo0686. [CrossRef]
- Korsunsky, I.; Wei, K.; Pohin, M.; Kim, E. Y.; Barone, F.; Major, T.; Taylor, E.; Ravindran, R.; Kemble, S.; Watts, G. F. M.; et al. Cross-Tissue, Single-Cell Stromal Atlas Identifies Shared Pathological Fibroblast Phenotypes in Four Chronic Inflammatory Diseases. Med 2022, 3 (7), 481-518.e14. [CrossRef]
- Curion, F.; Rich-Griffin, C.; Agarwal, D.; Ouologuem, S.; Thomas, T.; Theis, F. J.; Dendrou, C. A. Panpipes: A Pipeline for Multiomic Single-Cell and Spatial Transcriptomic Data Analysis. bioRxiv December 18, 2023, p 2023.03.11.532085. [CrossRef]
- Rao, D. A.; Gurish, M. F.; Marshall, J. L.; Slowikowski, K.; Fonseka, C. Y.; Liu, Y.; Donlin, L. T.; Henderson, L. A.; Wei, K.; Mizoguchi, F.; et al. Pathologically Expanded Peripheral T Helper Cell Subset Drives B Cells in Rheumatoid Arthritis. Nature 2017, 542 (7639), 110–114. [CrossRef]
- Bocharnikov, A. V.; Keegan, J.; Wacleche, V. S.; Cao, Y.; Fonseka, C. Y.; Wang, G.; Muise, E. S.; Zhang, K. X.; Arazi, A.; Keras, G.; et al. PD-1hiCXCR5– T Peripheral Helper Cells Promote B Cell Responses in Lupus via MAF and IL-21. JCI Insight 2019, 4 (20). [CrossRef]
- Ekman, I.; Ihantola, E.-L.; Viisanen, T.; Rao, D. A.; Näntö-Salonen, K.; Knip, M.; Veijola, R.; Toppari, J.; Ilonen, J.; Kinnunen, T. Circulating CXCR5−PD-1hi Peripheral T Helper Cells Are Associated with Progression to Type 1 Diabetes. Diabetologia 2019, 62 (9), 1681–1688. [CrossRef]
- Yong, L.; Chunyan, W.; Yan, Y.; Wanyu, L.; Huifan, J.; Pingwei, Z.; Yanfang, J. Expanded Circulating Peripheral Helper T Cells in Primary Biliary Cholangitis: Tph Cells in PBC. Molecular Immunology 2021, 131, 44–50. [CrossRef]
- Wang, X.; Li, T.; Si, R.; Chen, J.; Qu, Z.; Jiang, Y. Increased Frequency of PD-1hiCXCR5- T Cells and B Cells in Patients with Newly Diagnosed IgA Nephropathy. Sci Rep 2020, 10 (1), 492. [CrossRef]
- Zhang, P.; Wang, M.; Chen, Y.; Li, J.; Liu, Z.; Lu, H.; Fei, Y.; Feng, R.; Zhao, Y.; Zeng, X.; et al. Expanded CD4+CXCR5-PD-1+ Peripheral T Helper like Cells and Clinical Significance in IgG4-Related Disease. Clinical Immunology 2022, 237, 108975. [CrossRef]
- EU-STANDS4PM. Harmonised Data Access Agreement (hDAA) for sharing and using controlled access data. https://www.eu-stands4pm.eu/data_access.
- Schneider, V. A.; Graves-Lindsay, T.; Howe, K.; Bouk, N.; Chen, H.-C.; Kitts, P. A.; Murphy, T. D.; Pruitt, K. D.; Thibaud-Nissen, F.; Albracht, D.; et al. Evaluation of GRCh38 and de Novo Haploid Genome Assemblies Demonstrates the Enduring Quality of the Reference Assembly. Genome Res. 2017, 27 (5), 849–864. [CrossRef]
- Sharma, A.; Mohammad, A. J.; Turesson, C. Incidence and Prevalence of Giant Cell Arteritis and Polymyalgia Rheumatica: A Systematic Literature Review. Seminars in Arthritis and Rheumatism 2020, 50 (5), 1040–1048. [CrossRef]
- Zou, X.; Huo, F.; Sun, L.; Huang, J. Peripheral Helper T Cells in Human Diseases. Journal of Autoimmunity 2024, 145, 103218. [CrossRef]
| Language | Dimension Reduction |
Similarity search level |
Output type (G/E/W) |
Notes | Reference | |
|---|---|---|---|---|---|---|
| MNN | R | - | Cell | G | [26] | |
| fastMNN | R | PCA | Cell | G | Good for simple integration tasks [27] | [26] |
| Seurat v2 (CCA) | R | CCA | Cell | E | [28] | |
| Seurat v3 | R | CCA | Cell | G | High usability [27] | [29] |
| Scanorama | Python | SVD | Cell | G/E | Good for simple integration tasks [27] | [30] |
| BBKNN | Python | PCA | Cell | W | High speed and usability [27] | [31] |
| Conos | R | PCA | Cell | W | [32] | |
| Harmony | R | PCA | Cluster | E | Good for simple integration tasks. High speed and usability [27,33] | [34] |
| LIGER | R | iNMF | Cluster | E | [35] | |
| scMerge | R | PCA | Cluster | G | [36] | |
| scVI | Python | VAE | - | E | Good for complex integration tasks. Memory efficient. [27] | [37] |
| scANVI | Python | VAE | - | E | Good for complex integration tasks. Memory efficient. Requires cell annotations. [27] | [38] |
| scGen | Python | VAE | - | G | Requires cell annotations | [39] |
| trVAE | Python | VAE | - | E | [40] |
| Metric name | Level | Notes | Reference | |
|---|---|---|---|---|
| Batch mixing | iLISI | Cell | Inverse of the sum of batch probabilities within a weighted kNN. Reflects the number of batches in a neighbourhood. Graph variant scales to large datasets | [27,34] |
| kBET | Cell type | Comparison of label composition of a k-nearest neighbourhood of a cell and the expected (global) label composition | [42] | |
| Graph connectivity | Cell type | Determines how well the kNN graph of the integrated data connects cells of the same label | [27] | |
| ASW batch | Cell | Relationship between within-batch and between batch distances of a cell. Reflects separation between batches | [43] | |
| PCR batch | Global | Correlation of batch variable with principal components weighted by variance contribution. Reflects the total variance explained by the batch variable | [42] | |
| Bio-conservation | cLISI | Cell | Inverse of the sum of cell type probabilities within a weighted kNN. Reflects the number of cell types in a neighbourhood. Graph variant scales to large datasets | [27,34] |
| ASW label | Cell type | Relationship between within-label and between-label distances of a cell. Reflects separation between cell type clusters | [43] | |
| Isolated label | Cell type | Determines how well cell type labels that are shared by few batches are separated from other cell type labels | [27] | |
| KMeans NMI | Cell type | Overlap between predicted clustering and provided cell type labels | [44] | |
| KMeans ARI | Cell type | Overlap between predicted clustering and provided cell type labels (after correcting for overlap by chance) | [45] |
| Method name | Language | Approach | Reference | |
|---|---|---|---|---|
| Marker-based | scCATCH | R | Scoring system | [48] |
| SCSA | R | Scoring system | [49] | |
| SCINA | Python | Bi-modal distribution fit to marker genes | [50] | |
| CellAssign | R | Probabilistic Bayesian model | [51] | |
| Reference-based | scmap-cell | R | Cosine similarity | [52] |
| scmap-cluster | R | Cosine similarity, Pearson/Spearman correlation | [52] | |
| SingleR | R | Spearman correlation | [53] | |
| scMatch | Python | Spearman correlation | [54] | |
| CHETAH | R | Spearman correlation | [55] | |
| CellTypist | Python | Logistic regression classifier | [14] | |
| scPred | R | SVM | [56] | |
| SingleCellNet | R | Random forest | [57] | |
| scNym | Python | Adversarial neural network | [58] | |
| Seurat (Azimuth) | R | Reference mapping + Transfer learning | [59] | |
| scArches | Python | Reference mapping + Transfer learning | [60] | |
| Symphony | R | Reference mapping + Transfer learning | [61] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





