1. Introduction
In December 2019, the emergence of a novel coronavirus, COVID-19, originated in Wuhan, China [
1]. Within a short span, the virus spread rapidly, traversing borders to infect populations in over 100 countries [
2]. As of August 22nd, 2020, the global count of confirmed COVID-19 cases surged past 23,036,919, with the death toll surpassing 800,945 [
3]. Employing smart technologies holds promise for early detection of potential COVID-19 cases [
4]. The realms of artificial intelligence (AI) and machine learning have witnessed extensive utilization across diverse sectors, particularly in recent years [
5]. While AI techniques have found widespread application in healthcare [
3], the advent of COVID-19 underscores the urgent need to leverage these technologies for identifying, predicting, and containing its spread. It is envisioned that AI techniques will revolutionize the healthcare landscape, necessitating their integration into the ongoing battle against the COVID-19 pandemic [
6]. Enhancing the accuracy of COVID-19 diagnosis is imperative for promptly confirming positive cases, thereby curbing further transmission and ensuring timely intervention.
To identify research gaps, this study conducted a bibliometric analysis of existing literature on the application of machine learning algorithms to COVID-19 using the VOS viewer tool. Based on data from the Web of Science database in August 2020, the analysis revealed two primary clusters of research themes. The first cluster predominantly focused on predicting COVID-19 infections using weather datasets, while the second cluster emphasized the utilization of deep learning algorithms for analyzing Chest CT and X-Ray images. Despite the acknowledged high sensitivity of COVID-19 diagnosis through CT and X-Ray imaging [
7], widespread adoption of these diagnostic modalities for screening poses challenges due to factors such as radiation exposure, cost constraints, and equipment availability. Consequently, accurately distinguishing between positive and negative COVID-19 cases remains a pressing challenge, underscoring the need for innovative solutions to mitigate the pandemic [
8].
Health constitutes the cornerstone of society, yet the world currently grapples with an unprecedented health crisis precipitated by the COVID-19 pandemic. Globally, lives are being lost, and the far-reaching impact of the virus is palpable across various spheres. Infection and mortality rates continue to escalate, exacerbating economic downturns worldwide. Existing medical infrastructure strains under the burden of COVID-19 cases, while efforts to develop a vaccine are underway. Nations worldwide deploy diverse strategies to stem the virus's spread, with early detection emerging as a crucial imperative. Physicians recommend COVID testing for individuals exhibiting symptoms [
9], highlighting the critical need for swift detection to preempt transmission. Various technologies and strategies are deployed to expedite patient detection, with ongoing research focused on advanced techniques for COVID patient detection. As the virus spreads rapidly, there is an escalating demand for rapid and effective decision-making in the medical domain. Machine learning techniques offer promising avenues for real-time tracking and diagnosis of COVID patients [
10]. By analyzing symptom patterns, machine learning algorithms can classify individuals at risk of infection [
11], [
12], [
13].
Biomedical data classification using machine learning has emerged as a significant research domain in recent years. Various algorithms have been employed to enhance the accuracy of data classification methods, with deep learning techniques exhibiting particular promise. Recent literature highlights the utilization of machine learning for analyzing CT scan lung images of COVID patients [
9]. In this study, a deep neural network classifier is employed to classify COVID data based on symptom profiles, facilitating self-assessment and aiding in decision-making [
1], [
2]. This approach also assists in identifying individuals requiring testing, thereby contributing to infection control efforts.
The remainder of this paper is organized as follows:
Section 2 discusses prior research on machine learning-based COVID-19 analysis,
Section 3 provides a detailed description of the methodology employed,
Section 4 presents the classification results and compares them with other methods, and finally,
Section 5 concludes the study, outlining future avenues for research.
2. Literature Review
Machine learning and data mining techniques play a critical role in biomedical data analysis, enabling faster and more accurate detection and diagnosis of various diseases. Researchers have explored numerous algorithms to classify diseases and patients, utilizing machine learning-based automatic disease diagnosis systems trained on diverse biomedical signals, radiological images, pathological reports, and medical health records. Despite the global health crisis posed by COVID-19, significant efforts have been made in patient detection and classification using machine learning approaches [
9].
Deep learning, an advanced machine learning technique, has emerged as a powerful tool in biomedical data analysis due to its ability to process large volumes of images without extensive preprocessing. For instance, chest CT scans are crucial for diagnosing COVID-19 patients, and researchers have developed convolutional neural network-based segmentation methods to automate the identification of infected regions [
8]. Additionally, deep learning approaches have been utilized for patient classification, leveraging both 2D and 3D techniques.
Beyond radiological image analysis, machine learning has been applied to various aspects of COVID-19 analysis, including drug detection, patient screening, and epidemic prediction. Mathematical models, driven by machine learning algorithms, offer insights into the global spread of the virus and aid in designing effective containment strategies. Innovations such as smartphone-based patient detection systems demonstrate the potential for cost-effective and accessible healthcare solutions. These technologies, combined with AI-driven predictive models, contribute to early detection and intervention efforts, mitigating the impact of the pandemic. While significant progress has been made, there is still room for improvement, particularly in leveraging deep learning techniques for comprehensive COVID-19 data analysis. Future research should explore additional factors beyond symptom analysis and leverage ensemble learning approaches to enhance prediction accuracy. In summary, machine learning and data mining techniques hold promise in addressing the challenges posed by the COVID-19 pandemic. However, their effectiveness depends on collaborative efforts between researchers, healthcare professionals, and technology developers [
7].
A study retrospectively analyzed 114 subjects from the Taizhou hospital of Zhejiang Province in China between January 17, 2020, and February 1, 2020. Informed consent was obtained from all patients, with 59.6% males and 40.4% females participating. Real-time polymerase chain reaction (real-time PCR) was used for COVID-19 diagnosis, resulting in 32 positive and 82 negative cases. Clinical symptoms of infected patients included high fever, cough, mucus sputum, headache, fatigue, pharyngalgia (pharynx pain), and chest tightness [
7]. Positive COVID-19 cases had a higher mean age and male prevalence compared to negative cases. Random selection methods were employed to address gender imbalances, and statistical analysis showed no significant difference in infection rates between genders.
3. Methodology
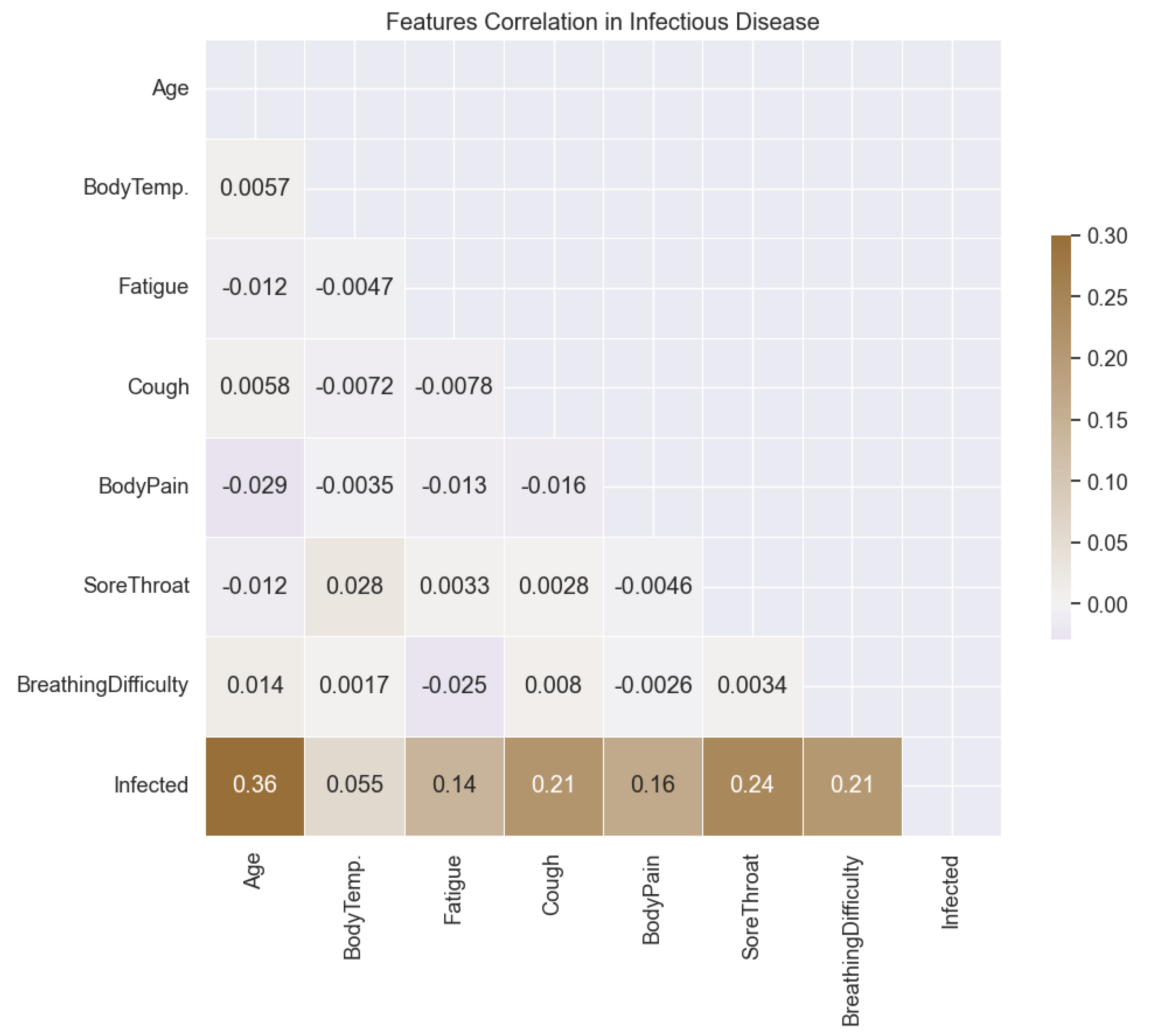
In this study, we begin by conducting an exploratory analysis of the COVID-19 dataset to understand the relationship between different features and the target variable, infectious probability. To visualize this relationship, we employ a correlation plot (
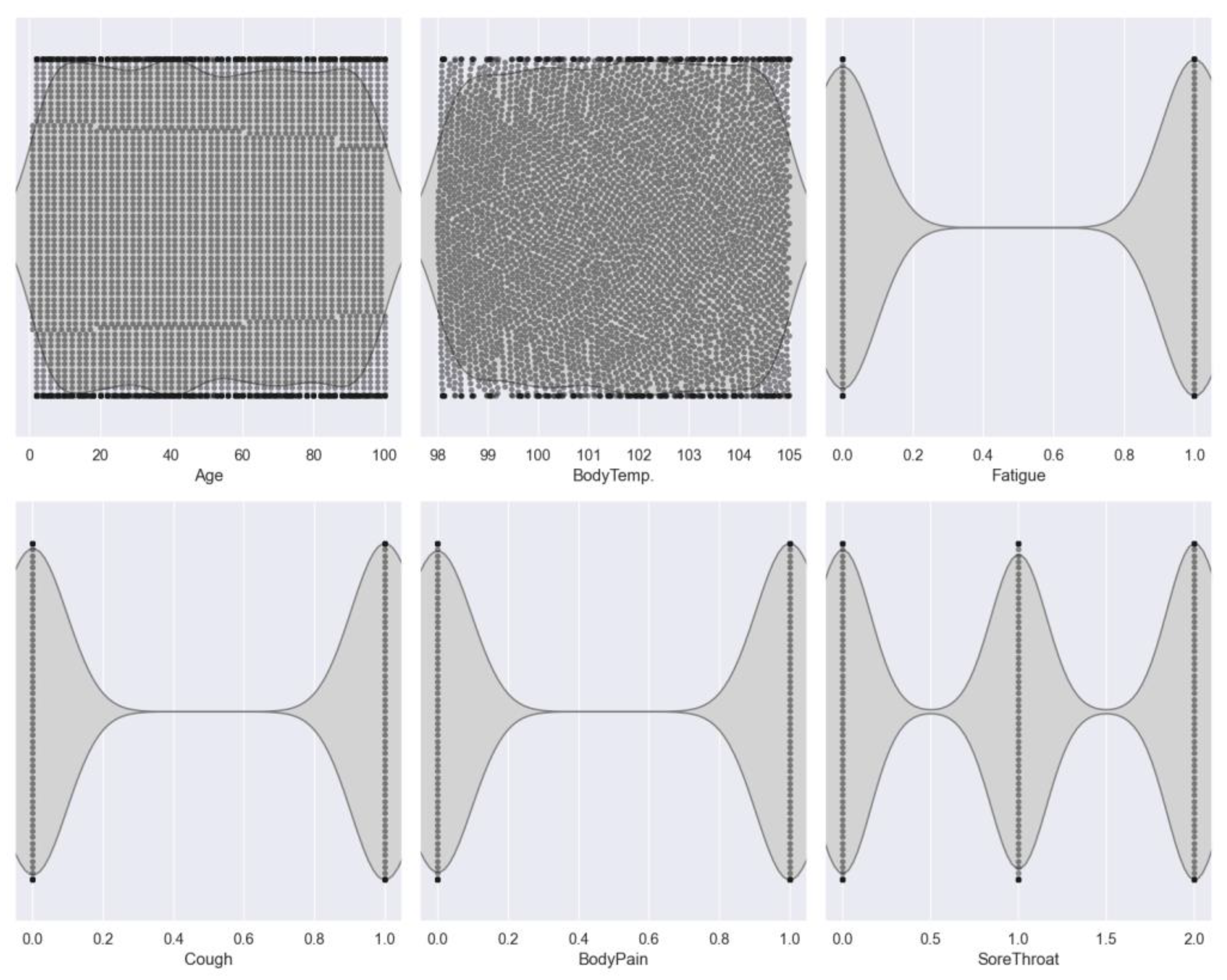
Figure 1), which reveals the correlations among features and their correlation with the target variable. Notably, the analysis highlights a significant correlation, particularly between age and infectious probability, with a correlation coefficient of 0.36. Subsequently, we utilize violin plots (
Figure 2) to illustrate the distribution of features across different classes.
The dataset comprises various features related to COVID-19 symptoms, including 'Fatigue', 'Cough', 'Body pain', 'Sore throat', and 'Breathing difficulty', along with the target variable 'Infected'. The values for 'Fatigue', 'Cough', and 'Body pain' are encoded as 0 for 'No' and 1 for 'Yes', indicating the presence or absence of these symptoms. Conversely, 'Sore throat' and 'Breathing difficulty' are encoded on a scale of 0 to 2, representing the severity of symptoms. It's essential to emphasize that the dataset used for this study is a randomly generated sample, and in real-world scenarios, governments can collect data from a more extensive population, ranging from 50,000 to 60,000 individuals worldwide.
Moving forward, we train and test various machine learning models on the collected data. The features considered for training the models include 'age' and 'symptoms', while the target variable is 'probability of a patient getting infected'. This approach enables us to develop a predictive model that can estimate the likelihood of a patient being affected by COVID-19 based on their age and reported symptoms. For instance, when a new patient provides information such as age, body temperature, and symptoms (e.g., fatigue, cough, body pain, sore throat, and breathing difficulty), the trained machine learning model can predict the probability of the patient being infected. As an example, if a patient aged 42 reports symptoms with specific severity levels, the model may output a probability estimate, such as 46% (note: this is an illustrative example and not derived from the model's prediction).
In this study, we evaluate the performance of several machine learning models, including 'DNN', 'Voting', 'Bagging', 'SVC_rbf', 'SVC_linear', 'SVC_polynomial', and 'SVC_sigmoid'. These models are assessed based on their ability to accurately predict the probability of COVID-19 infection, considering the input features of age and symptoms.
3.2. Dataset Splitting
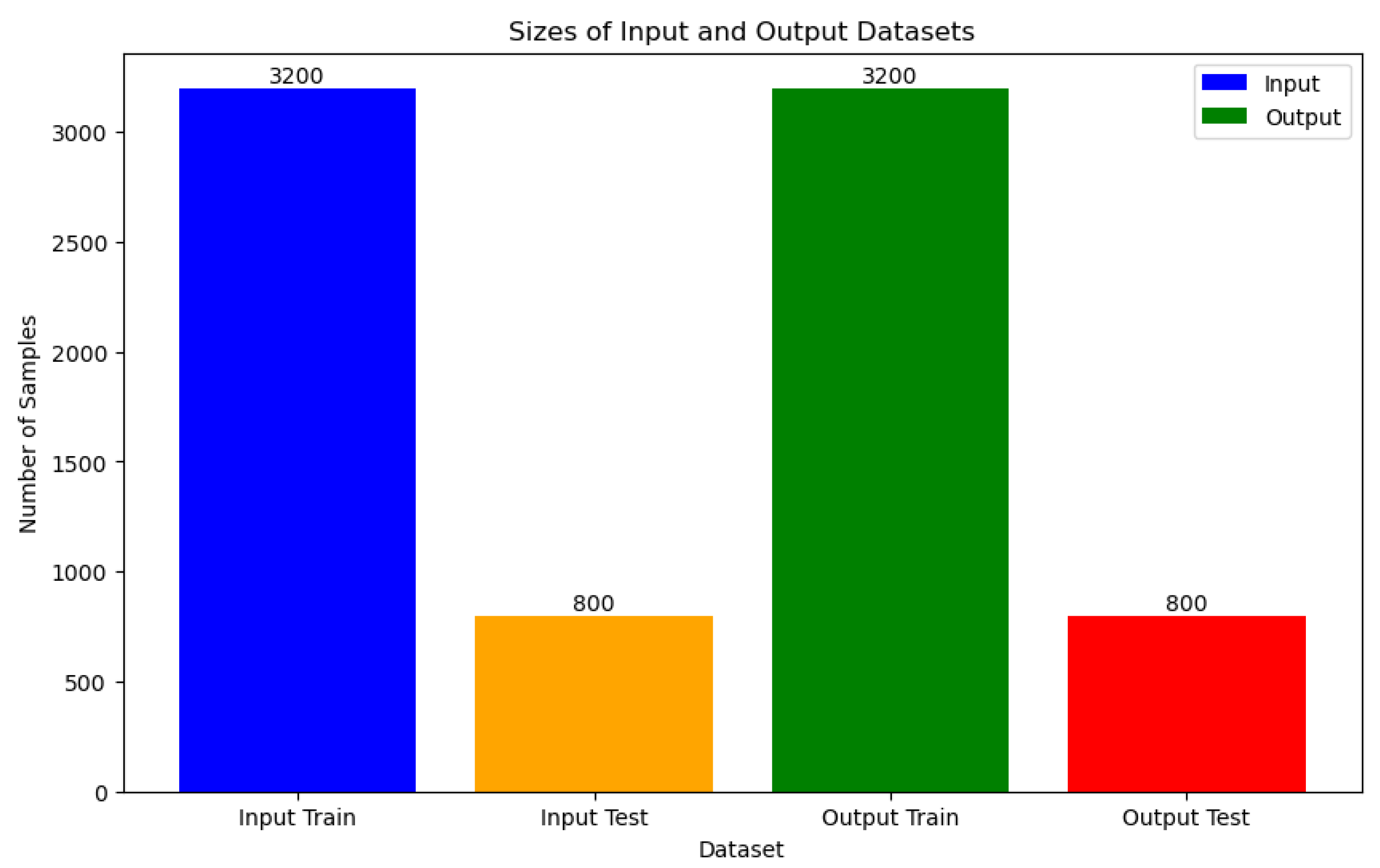
The COVID-19 dataset, consisting of 4000 instances, undergoes a structured data splitting process to facilitate model training and evaluation. Specifically, 3200 instances are designated for training purposes, while the remaining 800 instances serve as a separate test set to assess model performance. To mitigate the risk of overfitting, a k-fold cross-validation technique with k=5 is applied. This approach partitions the dataset into five subsets, allowing for robust training and assessment of machine learning models. It's noteworthy that the dataset is curated from various published works and the World Health Organization (WHO), ensuring its diverse and pertinent representation of the COVID-19 pandemic landscape.
3.3. ML Models
3.3.1. Linear Classification Algorithm
A classification algorithm, known as a classifier, determines its classifications based on a linear predictor function that combines a set of weights with the feature vector, as shown in the equation below:
Here, represents the output, is the activation function, denotes the weights, is the feature vector, and represents the dot product of weights and features.
Given the widespread use of linear machine learning (ML) algorithms, this study focuses on employing two well-known models: logistic regression and support vector machine (SVM) with a linear kernel, also known as Support Vector Classification (SVC).
Support Vector Machine (SVM)
Linear Function Kernel
Support Vector Machines (SVMs) are a type of machine learning algorithm that predicts by identifying a hyperplane separating data points of different categories [
14]. This hyperplane is positioned to maximize the margin, or distance, between the nearest data points and the decision boundary. In the case of classification with a linear kernel, the decision function of SVM can be expressed mathematically as follows:
Where,
represents the decision function that predicts the class label of an input sample
,
is the weight vector that determines the orientation of the hyperplane,
is the input feature vector and
is the bias term.
3.3.2. Non-Linear Classification Algorithm
However, in scenarios where classes cannot be separated by a linear boundary, non-linear classifiers become essential in using machine learning models. These classifiers are adept at handling intricate classification challenges by capturing complex patterns and relationships within data. Unlike linear models, non-linear classifiers offer enhanced performance when faced with complex datasets. Below, we present the most common models fitted to our four classes of melt pool shape, encompassing tree-based algorithms, Neural Networks (NNs) algorithms, providing insight into the capabilities of non-linear algorithms.
Bagging (Bootstrap Aggregating)
Bagging, short for Bootstrap Aggregating, is an ensemble learning method that aims to improve the stability and accuracy of machine learning models by reducing variance and overfitting. It works by training multiple instances of the same base learning algorithm on different subsets of the training data and then combining their predictions through a process called aggregation, the aggregation process can be represented as follows:
Where, represents the aggregated prediction for the instance , obtained by averaging the predictions from each individual model , where is the total number of models.
Voting
In our study, we utilize voting as a technique to amalgamate predictions from multiple individual models, specifically Random Forest and Gradient Boosting. By combining these two methods, each of which demonstrates strong classification capabilities independently, we aim to achieve a comprehensive assessment of their collective performance, the aggregation process in soft voting can be represented as:
Where, represents the final prediction for the instance x, obtained by averaging the predicted probabilities from each individual model , and is the total number of models.
Neural Networks (NNs)
Neural Networks (NNs) are versatile machine learning algorithms suitable for both regression and classification tasks [
15]. Within these networks, individual neurons perform linear and nonlinear transformations on input data, producing outputs that are adjusted through the iterative process of backpropagation, wherein weights and biases are updated to optimize model performance.
Multilayer Perceptrons (MLPs)
Multilayer Perceptrons (MLPs) represent a class of neural networks distinguished by their layered architecture comprising interconnected neurons. These networks typically consist of an input layer, one or more hidden layers, and an output layer. Within the network, each neuron processes input data through weighted connections and applies activation functions to generate output. MLPs find extensive applications in diverse machine learning tasks such as classification, regression, and pattern recognition due to their capability to capture intricate data relationships. The general equation governing the behavior of a node in an MLP is as follows:
Where, is the output of the -th node in the layer, is the activation function applied element-wise, is the weight connecting the -th input to the -th node, is the -th input to the node, is the -th bias term for the node, and is the number of inputs to the node.
Support Vector Machine (SVM)
Radial Basis Function (RBF) Kernel
Support Vector Classification (SVC) with Radial Basis Function (RBF) Kernel is a variant of Support Vector Machines (SVM), which is a powerful supervised learning algorithm used for classification tasks [
16]. The RBF kernel is particularly effective in handling non-linear relationships between features in the dataset. It works by transforming the input space into a higher-dimensional space where the classes can be more easily separated by a hyperplane. This transformation is achieved using a Gaussian radial basis function. The RBF kernel has two main parameters: gamma (γ) and regularization parameter (C), which control the flexibility of the decision boundary and the trade-off between maximizing the margin and minimizing classification errors, respectively.
Polynomial Function Kernel
In SVC, Polynomial Function Kernel as another type of SVM, data points are mapped to a higher-dimensional space using polynomial transformations, allowing for the creation of non-linear decision boundaries. The polynomial kernel function computes the dot product between pairs of data points in the transformed space, enabling effective classification in cases where the relationship between features and classes is non-linear. The polynomial kernel function is defined as:
where are input feature vectors, is a constant term, and is the degree of the polynomial. This kernel function allows SVC to capture complex patterns and achieve high accuracy in classification tasks.
Sigmoid Function Kernel
Support Vector Classifier (SVC) with Sigmoid Function Kernel is another variant of the support vector machine (SVM) algorithm, which utilizes a sigmoid function as its kernel. The sigmoid kernel function is defined as:
Where, are input feature vectors, and are constants, and is the hyperbolic tangent function.
The sigmoid kernel function allows SVC to create non-linear decision boundaries by transforming the input space into a higher-dimensional space. It is particularly useful for classification tasks where the relationship between features and classes is non-linear. However, it may be more sensitive to noise and less robust compared to other kernel functions like the polynomial or radial basis function (RBF) kernels.
4. Results and discussion
The utilization of machine learning (ML) models in healthcare presents a significant advancement in saving both time and cost compared to traditional clinical and practical investigations. By leveraging ML algorithms, healthcare professionals can analyze vast amounts of data efficiently, leading to quicker diagnoses and treatment decisions. However, one crucial aspect to consider when employing ML models is the risk of overfitting, where the model learns the training data too well, resulting in poor generalization to unseen data. Therefore, monitoring and mitigating overfitting is essential to ensure the reliability and effectiveness of ML models in healthcare applications.
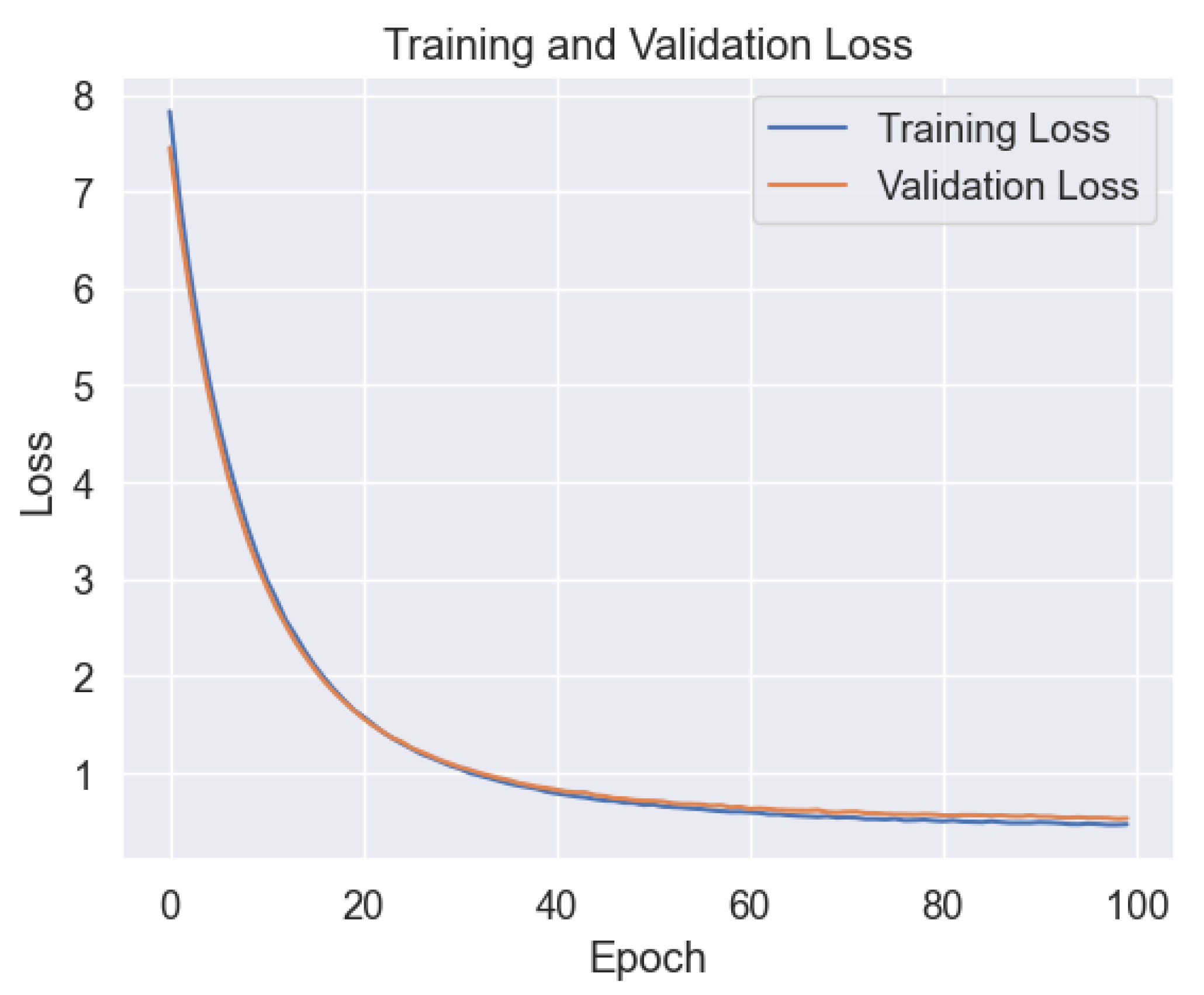
In our study, we first applied a deep neural network (DNN) to predict the probability of COVID-19 infection.
Figure 4 illustrates the training and validation performance of the DNN model across epochs. Interestingly, the plot reveals minimal signs of overfitting, indicating that the model efficiently learns from the training data without excessively memorizing it.
Subsequently, we evaluated the performance of other ML models, including Voting and Bagging, in mitigating overfitting.
Figure 5 demonstrates the impact of adjusting hyperparameters such as learning rate and minimum child weight in the Voting model. Despite variations in hyperparameters, the model shows a consistent performance with negligible overfitting. Similarly,
Figure 6 illustrates the stability of the Bagging model across different parameter ranges, indicating robustness against overfitting.
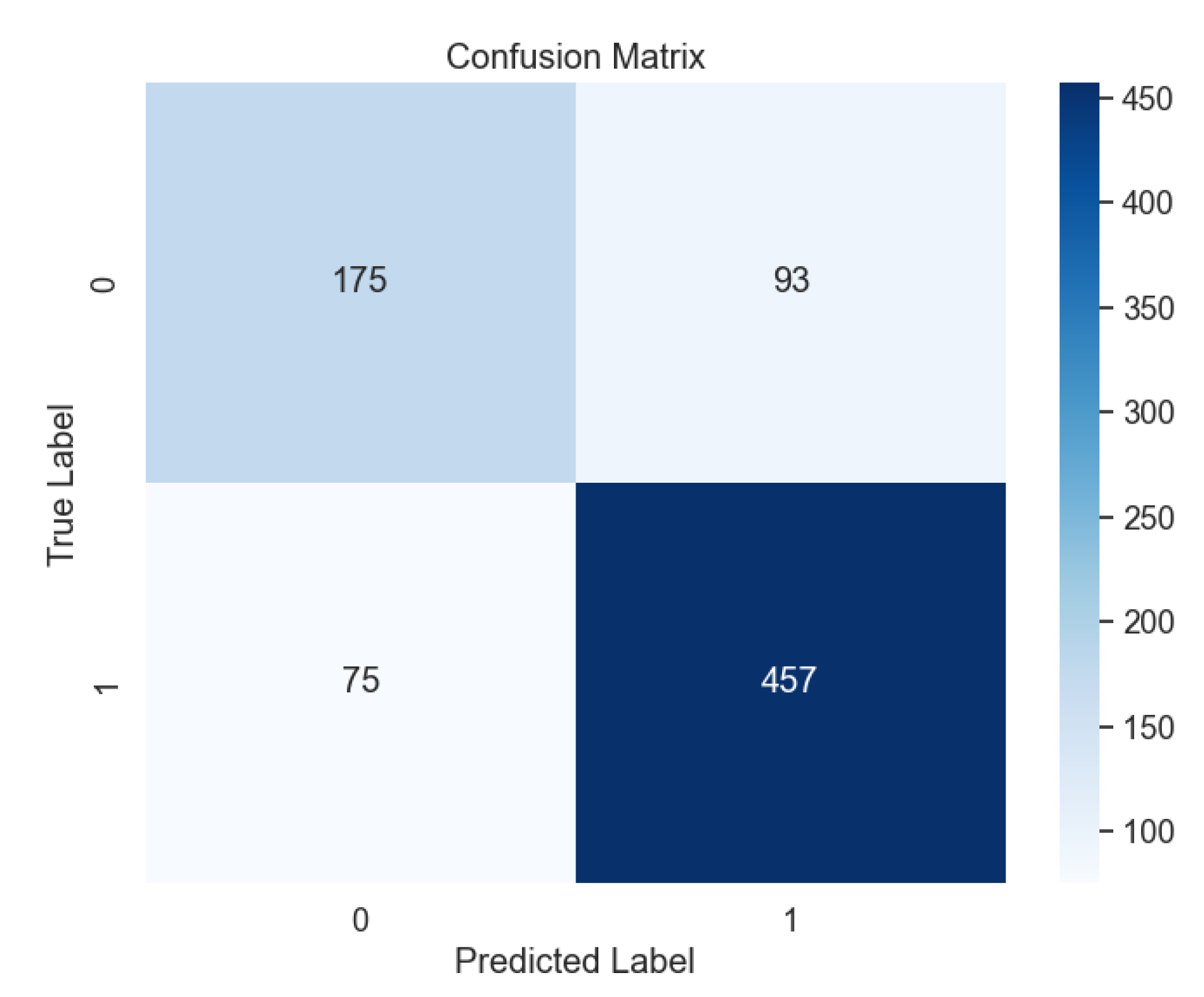
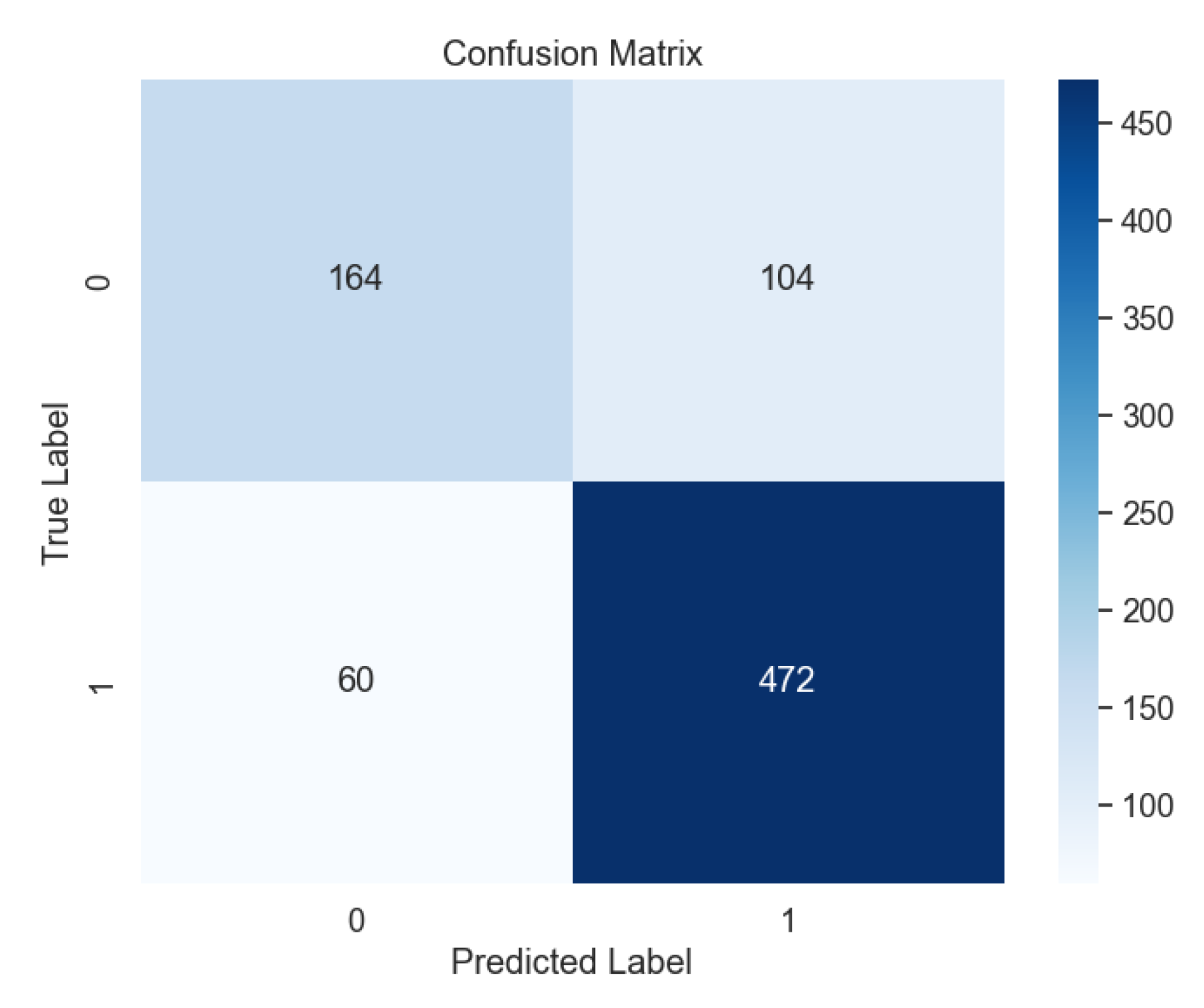
In contrast, the performance of Support Vector Machine (SVM) models varied based on kernel types.
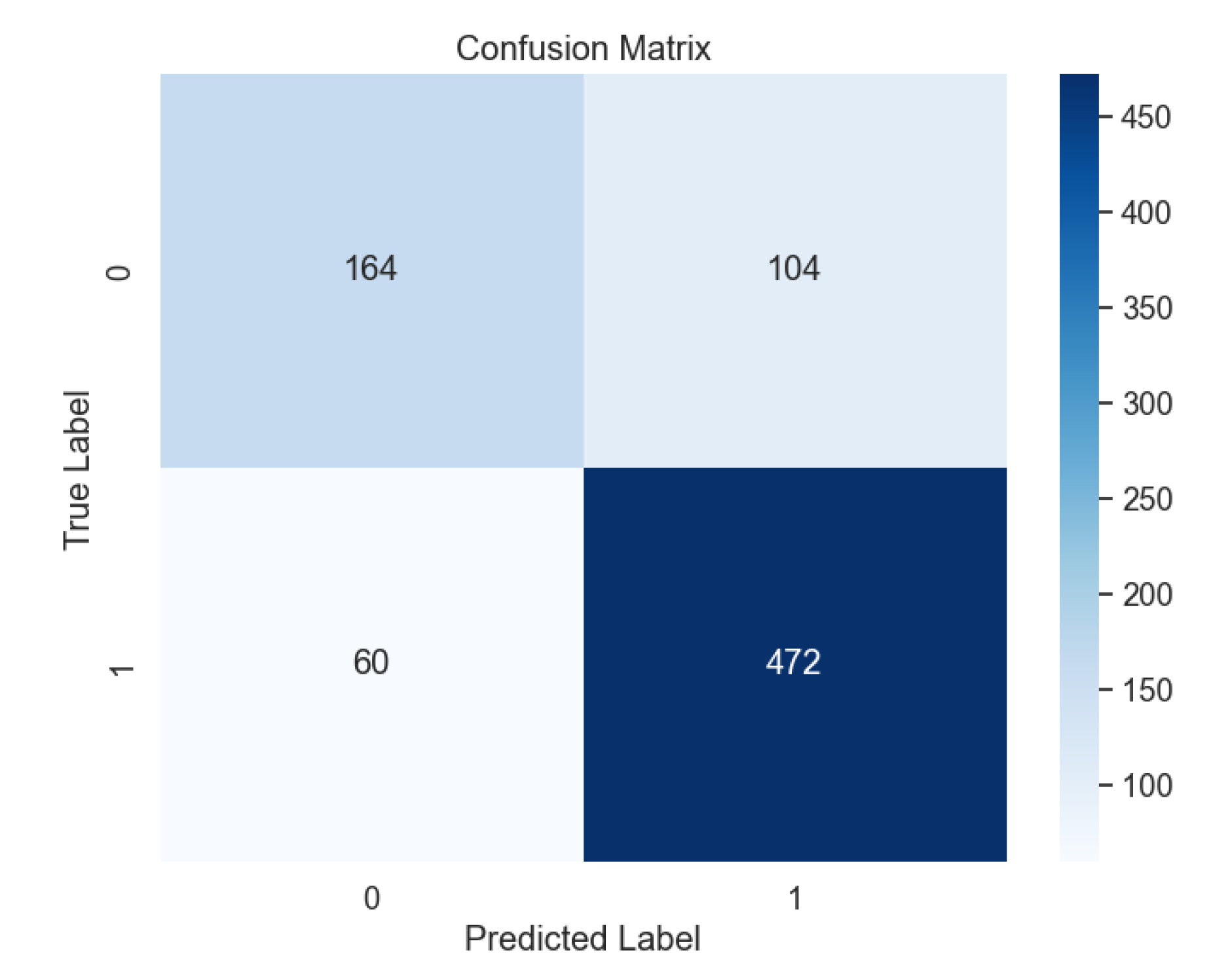
Figure 7 shows the confusion matrix for the SVM model with the radial basis function (RBF) kernel, outperforming other kernel types such as Linear (
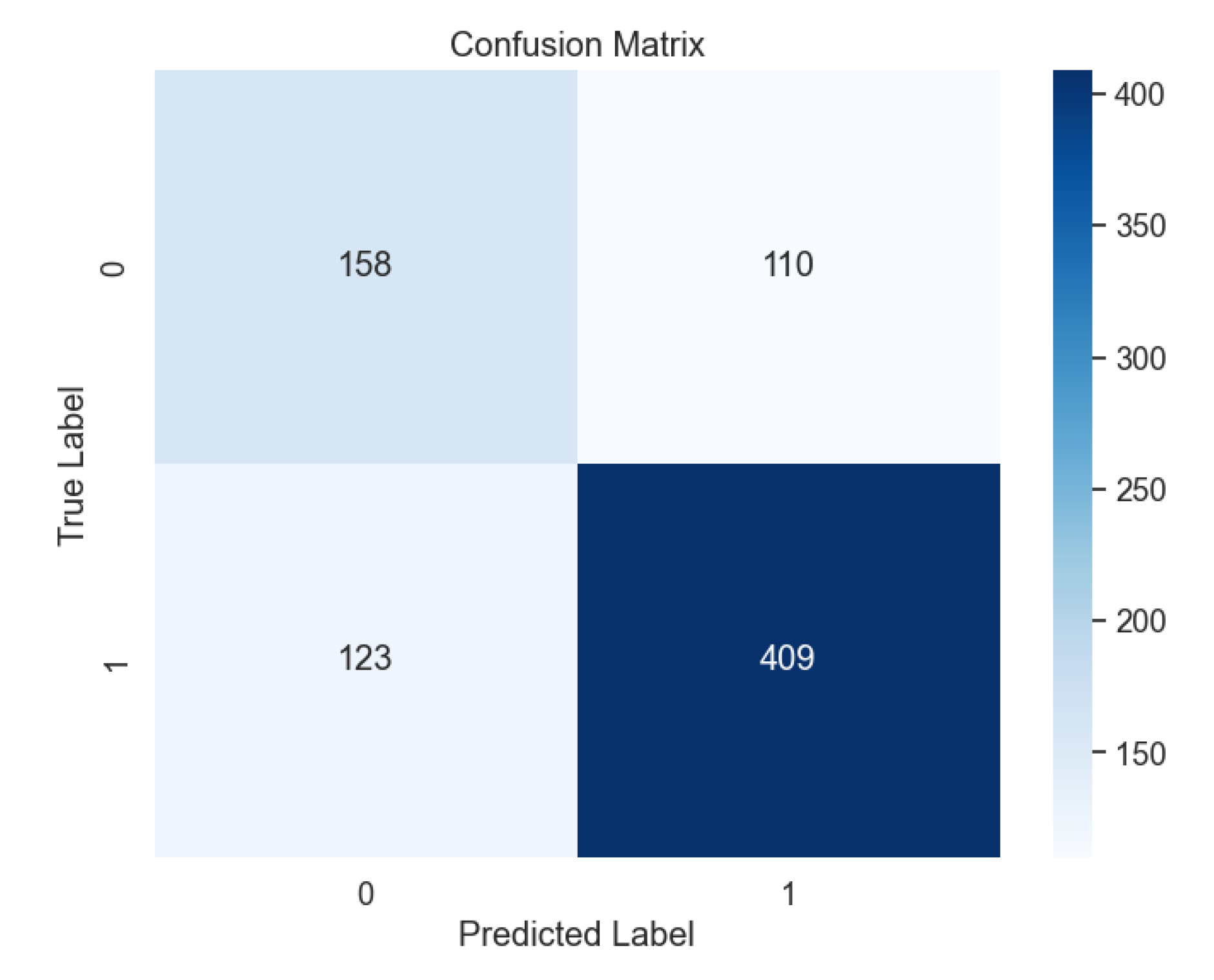
Figure 8), Polynomial (
Figure 9), and Sigmoid (
Figure 10).
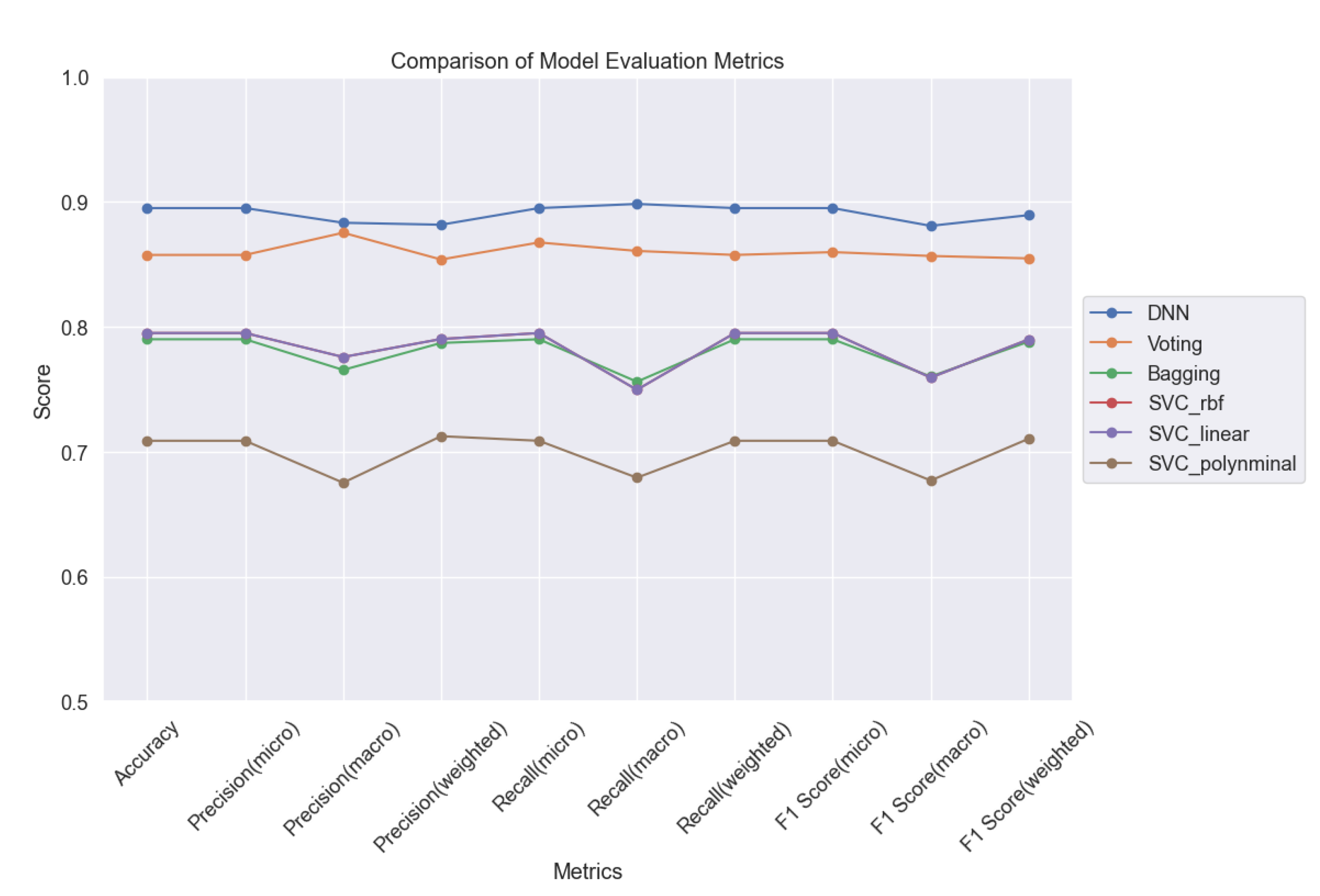
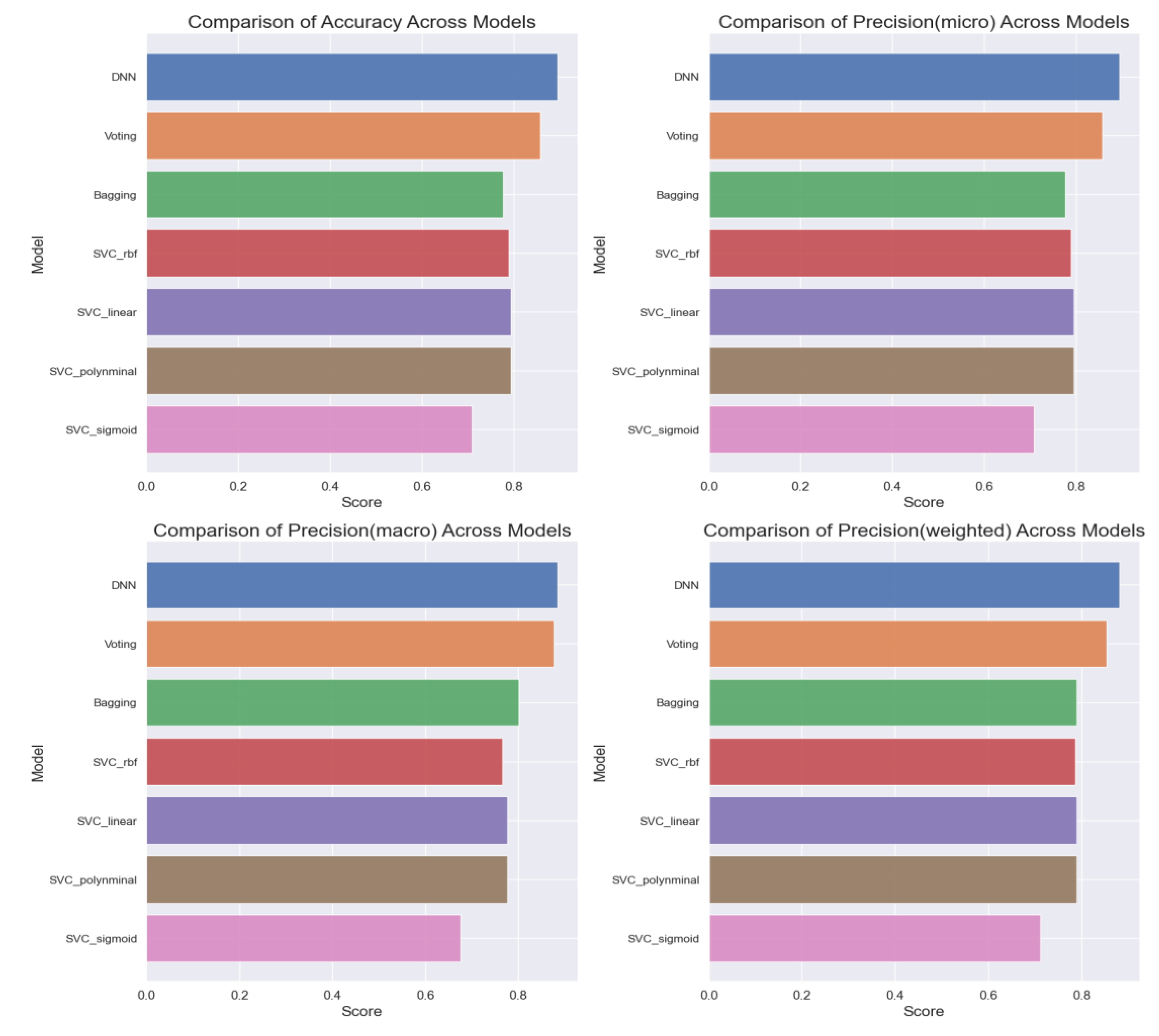
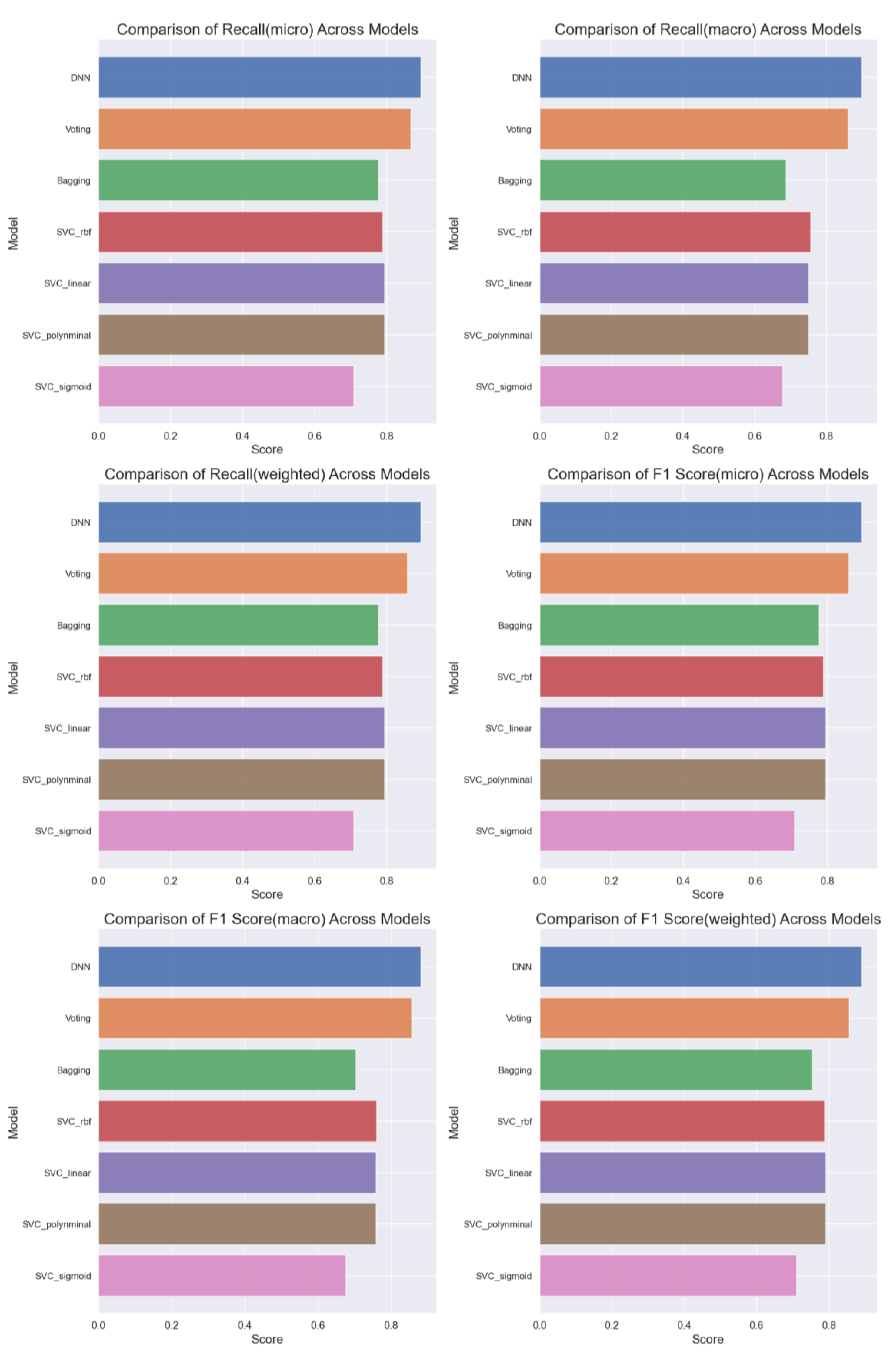
Overall, a comprehensive comparison of all ML models (
Figure 11) based on accuracy, precision, recall, and F1 score highlights the superior performance of the DNN model, achieving an accuracy of 89%. Further analysis (
Figure 12 and
Figure 13) delves into the intricate details of each model's performance, particularly focusing on the challenges in predicting uninfected patients. While the DNN model demonstrates exceptional accuracy in detecting infected patients, its performance in identifying uninfected patients is affected by various factors such as regional differences, individual characteristics, and the complexity of symptoms associated with different diseases.
In conclusion, our study underscores the importance of ML models in healthcare, particularly in the context of COVID-19 detection. While the DNN model emerges as the top performer, further research is warranted to address the challenges associated with predicting uninfected patients accurately. Additionally, ongoing efforts to optimize ML algorithms and refine feature selection techniques are essential for enhancing the overall performance and reliability of healthcare-focused ML models.
5. Conclusions
In conclusion, our study underscores the significant role of machine learning (ML) models in healthcare, particularly in the context of COVID-19 detection. By leveraging ML algorithms, healthcare professionals can analyze extensive datasets efficiently, leading to quicker diagnoses and treatment decisions. Throughout our investigation, we have demonstrated the effectiveness of various ML models, including deep neural networks (DNN), Voting, Bagging, and Support Vector Machine (SVM), in predicting the probability of COVID-19 infection.
Our findings highlight the superior performance of the DNN model, which achieved an accuracy of 89% in predicting COVID-19 infection probability. Despite minimal signs of overfitting, our analysis emphasizes the importance of monitoring and mitigating overfitting to ensure the reliability and generalization capability of ML models.
Furthermore, our study sheds light on the challenges associated with predicting uninfected patients accurately, which are influenced by regional differences, individual characteristics, and the complexity of symptoms associated with various diseases. Addressing these challenges requires ongoing research efforts to optimize ML algorithms and refine feature selection techniques.
Overall, the results of our study underscore the potential of ML models to revolutionize healthcare by providing efficient and accurate tools for disease detection and diagnosis. As we continue to advance in this field, collaborative efforts between researchers, healthcare professionals, and technology developers will be essential in harnessing the full potential of ML in improving healthcare outcomes and mitigating the impact of pandemics such as COVID-19.
References
- W. De Ceukelaire and C. Bodini, “We Need Strong Public Health Care to Contain the Global Corona Pandemic,” International Journal of Health Services, vol. 50, no. 3, p. 276, Jul. 2020, doi: 10.1177/0020731420916725. [CrossRef]
- J. Bian and F. Modave, “The rapid growth of intelligent systems in health and health care,” https://doi.org/10.1177/1460458219896899, vol. 26, no. 1, pp. 5–7, Jan. 2020, doi: 10.1177/1460458219896899. [CrossRef]
- B. McCall, “COVID-19 and artificial intelligence: protecting health-care workers and curbing the spread,” Lancet Digit Health, vol. 2, no. 4, pp. e166–e167, Apr. 2020, doi: 10.1016/s2589-7500(20)30054-6. [CrossRef]
- K. Asada et al., “Application of Artificial Intelligence in COVID-19 Diagnosis and Therapeutics,” J Pers Med, vol. 11, no. 9, Sep. 2021, doi: 10.3390/JPM11090886. [CrossRef]
- S. R. Srinivasa Rao and J. A. Vazquez, “Identification of COVID-19 can be quicker through artificial intelligence framework using a mobile phone–based survey when cities and towns are under quarantine,” Infect Control Hosp Epidemiol, vol. 41, no. 7, pp. 826–830, Jul. 2020, doi: 10.1017/ICE.2020.61. [CrossRef]
- “COVID - Coronavirus Statistics - Worldometer.” Accessed: Mar. 09, 2024. [Online]. Available: https://www.worldometers.info/coronavirus/.
- Rehman, M. A. Iqbal, H. Xing, and I. Ahmed, “COVID-19 Detection Empowered with Machine Learning and Deep Learning Techniques: A Systematic Review,” Applied Sciences 2021, Vol. 11, Page 3414, vol. 11, no. 8, p. 3414, Apr. 2021, doi: 10.3390/APP11083414. [CrossRef]
- D. Orso, N. Federici, R. Copetti, L. Vetrugno, and T. Bove, “Infodemic and the spread of fake news in the COVID-19-era,” Eur J Emerg Med, vol. 27, no. 5, pp. 327–328, Oct. 2020, doi: 10.1097/MEJ.0000000000000713. [CrossRef]
- L. Chen et al., “[Analysis of clinical features of 29 patients with 2019 novel coronavirus pneumonia],” Zhonghua Jie He He Hu Xi Za Zhi, vol. 43, no. 0, p. E005, Feb. 2020, doi: 10.3760/CMA.J.ISSN.1001-0939.2020.0005. [CrossRef]
- B. Kadam and S. R. Atre, “Social media panic and COVID-19 in India,” J Travel Med, vol. 27, no. 3, Apr. 2020, doi: 10.1093/JTM/TAAA057. [CrossRef]
- M. A. Ilani and M. Khoshnevisan, “Study of surfactant effects on intermolecular forces (IMF) in powder-mixed electrical discharge machining (EDM) of Ti-6Al-4V,” International Journal of Advanced Manufacturing Technology, vol. 116, no. 5–6, pp. 1763–1782, Sep. 2021, doi: 10.1007/S00170-021-07569-3/FIGURES/30. [CrossRef]
- M. A. Ilani and M. Khoshnevisan, “An evaluation of the surface integrity and corrosion behavior of Ti-6Al-4 V processed thermodynamically by PM-EDM criteria,” International Journal of Advanced Manufacturing Technology, vol. 120, no. 7–8, pp. 5117–5129, Jun. 2022, doi: 10.1007/S00170-022-09093-4/FIGURES/18. [CrossRef]
- R. Hosseini Rad, S. Baniasadi, P. Yousefi, H. Morabbi Heravi, M. Shaban Al-Ani, and M. Asghari Ilani, “Presented a Framework of Computational Modeling to Identify the Patient Admission Scheduling Problem in the Healthcare System,” J Healthc Eng, vol. 2022, 2022, doi: 10.1155/2022/1938719. [CrossRef]
- D. Zhang and W. Sui, “The application of AR model and SVM in rolling bearings condition monitoring,” Communications in Computer and Information Science, vol. 152 CCIS, no. PART 1, pp. 326–331, 2011, doi: 10.1007/978-3-642-21402-8_53/COVER. [CrossRef]
- Y. Lecun, Y. Bengio, and G. Hinton, “Deep learning,” Nature 2015 521:7553, vol. 521, no. 7553, pp. 436–444, May 2015, doi: 10.1038/nature14539. [CrossRef]
- J. Li et al., “Defects Recognition in Selective Laser Melting with Acoustic Signals by SVM Based on Feature Reduction,” IOP Conf Ser Mater Sci Eng, vol. 436, no. 1, p. 012020, Oct. 2018, doi: 10.1088/1757-899X/436/1/012020. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).