Submitted:
24 May 2024
Posted:
27 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Genomic DNA Extraction
2.3. Next-Generation Sequencing (NGS)
2.4. Sanger Sequencing
2.5. Genetic Variant Classification
3. Results
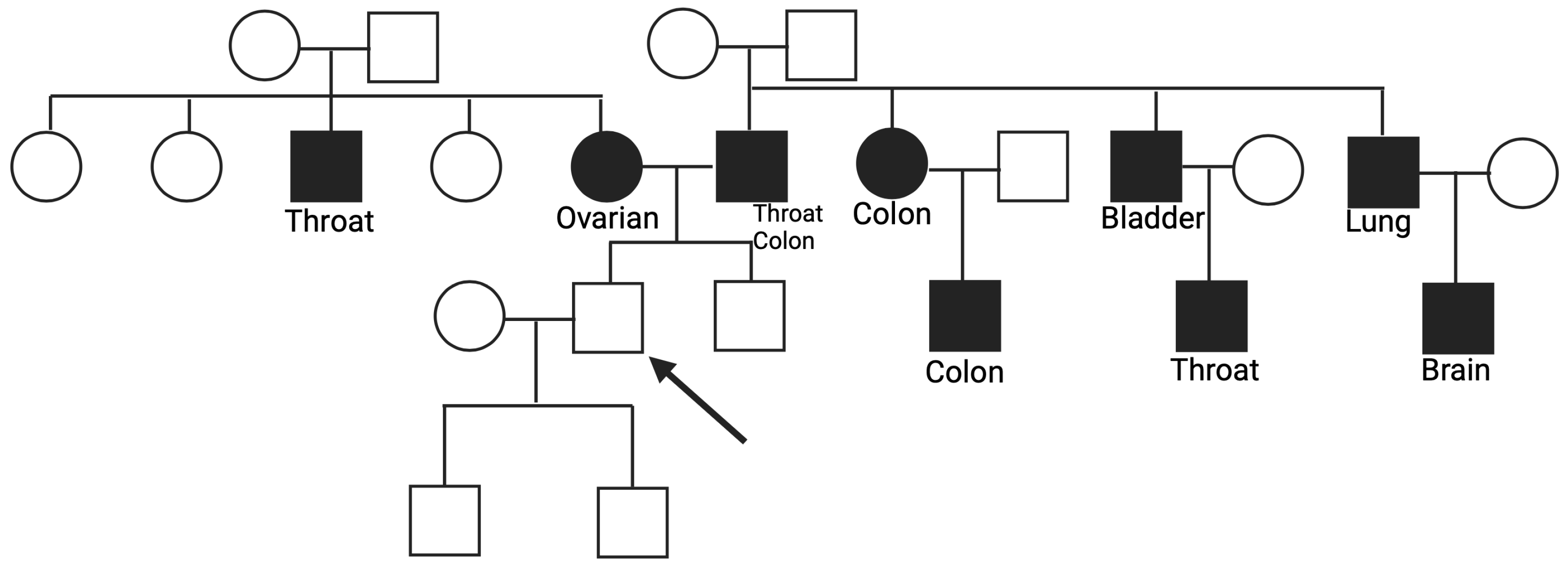
3.1. Genes Variants linked to the Homologous Recombination (HR) and related family history
3.2. Genes Variants linked to the base excision repair (BER) and related family history
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Antonucci I., Provenzano M., Sorino L., Rodrigues M., Palka G., Stuppia L. A new case of “de novo” BRCA1 mutation in a patient with early-onset breast cancer. Clin. Case Rep. 2017;5:238–240. Author 1, A.; Author 2, B. Title of the chapter. In Book Title, 2nd ed.; Editor 1, A., Editor 2, B., Eds.; Publisher: Publisher Location, Country, 2007; Volume 3, pp. 154–196. [CrossRef]
- NCCN. Detection, Prevention, and Risk Reduction. Available online: https://www.nccn.org/guidelines/category_2 (accessed on 27 February 2024).
- Knerr S, Guo B, Mittendorf KF, Feigelson HS, Gilmore MJ, Jarvik GP, Kauffman TL, Keast E, Lynch FL, Muessig KR, Okuyama S, Veenstra DL, Zepp JM, Goddard KAB, Devine B. Risk-reducing surgery in unaffected individuals receiving cancer genetic testing in an integrated health care system. Cancer. 2022 Aug 15;128(16):3090-3098. Epub 2022 Jun 9. [CrossRef] [PubMed] [PubMed Central]
- Kamps, R.; Brandão, R.D.; van den Bosch, B.J.; Paulussen, A.D.; Xanthoulea, S.; Blok, M.J.; Romano, A. Next-Generation Sequencing in Oncology: Genetic Diagnosis, Risk Prediction and Cancer Classification. Int. J. Mol. Sci. 2017, 18, 308. Author 1, A.B.; Author 2, C.D.; Author 3, E.F. Title of Presentation. In Proceedings of the Name of the Conference, Location of Conference, Country, Date of Conference (Day Month Year). [CrossRef] [PubMed]
- DQ Cancer Genetics Editorial Board. Cancer Genetics Risk Assessment and Counseling (PDQ®): Health Professional Version. 2024 Mar 6. In: PDQ Cancer Information Summaries.
- https://www.cancer.org/cancer/risk-prevention/genetics/genetic-testing-for-cancer-risk/understanding-genetic-testing-for-cancer.html.
- Di Rado S, Giansante R, Cicirelli M, Pilenzi L, Dell'Elice A, Anaclerio F, Rimoldi M, Grassadonia A, Grossi S, Canale N, Ballerini P, Stuppia L, Antonucci I. Detection of Germline Mutations in a Cohort of 250 Relatives of Mutation Carriers in Multigene Panel: Impact of Pathogenic Variants in Other Genes beyond BRCA1/2. Cancers (Basel). 2023 Dec 6;15(24):5730. [CrossRef] [PubMed] [PubMed Central]
- Petrucelli N, Daly MB, Pal T. BRCA1- and BRCA2-Associated Hereditary Breast and Ovarian Cancer. 1998 Sep 4 [Updated 2023 Sep 21]. In: Adam MP, Feldman J, Mirzaa GM, et al., editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993-2024. Available from: https://www.ncbi.nlm.nih.gov/books/NBK1247/.
- Forbes C, Fayter D, de Kock S, Quek RG. A systematic review of international guidelines and recommendations for the genetic screening, diagnosis, genetic counseling, and treatment of BRCA-mutated breast cancer. Cancer Manag Res. 2019 Mar 22;11:2321-2337. [CrossRef] [PubMed] [PubMed Central]
- Weitzel JN, Lagos VI, Cullinane CA, Gambol PJ, Culver JO, Blazer KR, Palomares MR, Lowstuter KJ, MacDonald DJ. Limited family structure and BRCA gene mutation status in single cases of breast cancer. JAMA. 2007 Jun 20;297(23):2587-95. [CrossRef] [PubMed]
- E:Bramanti SM, Trumello C, Lombardi L, Cavallo A, Stuppia L, Antonucci I, Babore A. Uncertainty following an inconclusive result from the BRCA1/2 genetic test: A review about psychological outcomes. World J Psychiatry. 2021 May 19;11(5):189-200. [CrossRef] [PubMed] [PubMed Central]
- Trottier M, Lunn J, Butler R, Curling D, Turnquest T, Francis W, Halliday D, Royer R, Zhang S, Li S, Thompson I, Donenberg T, Hurley J, Akbari MR, Narod SA. Prevalence of founder mutations in the BRCA1 and BRCA2 genes among unaffected women from the Bahamas. Clin Genet. 2016 Mar;89(3):328-31. Epub 2015 May 31. [CrossRef] [PubMed]
- Bernstein-Molho R, Singer A, Laitman Y, Netzer I, Zalmanoviz S, Friedman E. Multigene panel testing in unselected Israeli breast cancer cases: mutational spectrum and use of BRCA1/2 mutation prediction algorithms. Breast Cancer Res Treat. 2019 Jul;176(1):165-170. Epub 2019 Apr 12. [CrossRef] [PubMed]
- Ece Solmaz A, Yeniay L, Gökmen E, Zekioğlu O, Haydaroğlu A, Bilgen I, Özkınay F, Onay H. Clinical Contribution of Next-Generation Sequencing Multigene Panel Testing for BRCA Negative High-Risk Patients With Breast Cancer. Clin Breast Cancer. 2021 Dec;21(6):e647-e653. Epub 2021 Apr 12. [CrossRef] [PubMed]
- Anaclerio F, Pilenzi L, Dell'Elice A, Ferrante R, Grossi S, Ferlito LM, Marinelli C, Gildetti S, Calabrese G, Stuppia L, Antonucci I. Clinical usefulness of NGS multi-gene panel testing in hereditary cancer analysis. Front Genet. 2023 Feb 1;14:1060504. [CrossRef] [PubMed] [PubMed Central]
- Desrichard A, Bidet Y, Uhrhammer N, Bignon YJ. CHEK2 contribution to hereditary breast cancer in non-BRCA families. Breast Cancer Res. 2011;13(6):R119. Epub 2011 Nov 24. [CrossRef] [PubMed] [PubMed Central]
- Oros KK, Ghadirian P, Greenwood CM, Perret C, Shen Z, Paredes Y, Arcand SL, Mes-Masson AM, Narod SA, Foulkes WD, Provencher D, Tonin PN. Significant proportion of breast and/or ovarian cancer families of French Canadian descent harbor 1 of 5 BRCA1 and BRCA2 mutations. Int J Cancer. 2004 Nov 10;112(3):411-9. [CrossRef] [PubMed]
- Thiffault I, Saunders C, Jenkins J, Raje N, Canty K, Sharma M, Grote L, Welsh HI, Farrow E, Twist G, Miller N, Zwick D, Zellmer L, Kingsmore SF, Safina NP. A patient with polymerase E1 deficiency (POLE1): clinical features and overlap with DNA breakage/instability syndromes. BMC Med Genet. 2015 May 7;16:31. [CrossRef] [PubMed] [PubMed Central]
- Nielsen, M., Jones, N., Vogt, S., Carli, M., Vasen, H. F. A., Sampson, J. R., et al. (2009). Analysis of MUTYH genotypes and colorectal phenotypes in patients with MUTYH-associated polyposis. Gastroenterology 136 (2), 471–476. [CrossRef]
- Imyanitov EN, Kuligina ES, Sokolenko AP, Suspitsin EN, Yanus GA, Iyevleva AG, Ivantsov AO, Aleksakhina SN. Hereditary cancer syndromes. World J Clin Oncol. 2023 Feb 24;14(2):40-68. [CrossRef] [PubMed] [PubMed Central]
- Antonucci I., Provenzano M., Sorino L., Balsamo M., Aceto G.M., Battista P., Euhus D., Cianchetti E., Ballerini P., Natoli C., et al. Comparison between CaGene 5.1 and 6.0 for BRCA1/2 mutation prediction: A retrospective study of 150 BRCA1/2 genetic tests in 517 families with breast/ovarian cancer. J. Hum. Genet. 2017;62:379–387. [CrossRef]
- AIOM (Associazione Italiana Oncologia Medica) Linee guida Carcinoma Mammario in stadio precoce -Edizione 2023 (Aggiornata al 20/11/2023)23.
- Bramanti SM, Trumello C, Lombardi L, Cavallo A, Stuppia L, Antonucci I, Babore A. Uncertainty following an inconclusive result from the BRCA1/2 genetic test: A review about psychological outcomes. World J Psychiatry. 2021 May 19;11(5):189-200.
- Lombardi L, Bramanti SM, Babore A, Stuppia L, Trumello C, Antonucci I, Cavallo A. Psychological aspects, risk and protective factors related to BRCA genetic testing: a review of the literature. Support Care Cancer. 2019 Jun 15.
- Babore A, Bramanti SM, Lombardi L, Stuppia L, Trumello C, Antonucci I, Cavallo A. The role of depression and emotion regulation on parenting stress in a sample of mothers with cancer. Support Care Cancer. 2019 Dec 19.
- Rossi C, Cicalini I, Cufaro MC, et al. Breast cancer in the era of integrating "Omics" approaches. Oncogenesis. 2022;11(1):17. Published 2022 Apr 14. [CrossRef]
- Frey MK, Ahsan MD, Webster E, Levi SR, Brewer JT, Lin J, Blank SV, Krinsky H, Nchako C, Wolfe I, Thomas C, Christos P, Cantillo E, Chapman-Davis E, Holcomb K, Sharaf RN. Web-based tool for cancer family history collection: A prospective randomized controlled trial. Gynecol Oncol. 2023 Jun;173:22-30.

| Multi-gene panel | ||
|---|---|---|
| APC | ATM | BARD1 |
| BRCA1 | BRCA2 | BRIP1 |
| CDK4 | CDK12 | CDKN2A |
| CDH1 | CHEK2 | EPCAM |
| MLH1 | MSH2 | MSH6 |
| MUTYH | NBN | NF1 |
| PALB2 | POLE | POLE |
| POLD1 | PTEN | RAD51C |
| RAD51D | SMAD4 | TP53 |
| NAME | EXON | SEQUENCE |
|---|---|---|
| BRCA2_EX11F | Exon 11 | attgagatcacagctgcccc |
| BRCA2_EX11R | Exon 11 | tgaagtctgactcacagaagttt |
| CHEK2_EX13F | Exon 13 | atgtggatgtgagtcagccag |
| CHEK2_EX13R | Exon 13 | atcagctccttaagcccagacta |
| BRCA1_EX10F | Exon 10 | ttggtcagctttctgtaatcg |
| BRCA1_EX10R | Exon 10 | ccataccacgacatttgaca |
| POLE_EX8F | Exon 8 | gtcgctgctcacatgaattt |
| POLE_EX8R | Exon 8 | atttgggggaaaagcagcaa |
| MUTYH_13F | Exon 13 | agggcagtggcatgagtaac |
| MUTYH_13R | Exon 13 | gggtcaaggggttcaaatag |
| CASE ID GENE | OMIM REFSEQ | CODING PROTEIN |
|---|---|---|
| Subject 1 BRCA2 | 164757 NM_000059.3 | c.4914dupA V1639fs |
| Subject 2 CHEK2 | 604373 NM_007194.4 | c.1427C>T T476M |
| Subject 3 BRCA1 | 113705 NM_007294.4 | c.1953dup K652fs |
| Subject 4 POLE | 174762 NM_006231.3 | c.778C>T R260* |
| Subject 5 MUTYH | 608456 NM_001128425.2 | c.1187G>A G396D |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





