Submitted:
28 May 2024
Posted:
29 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
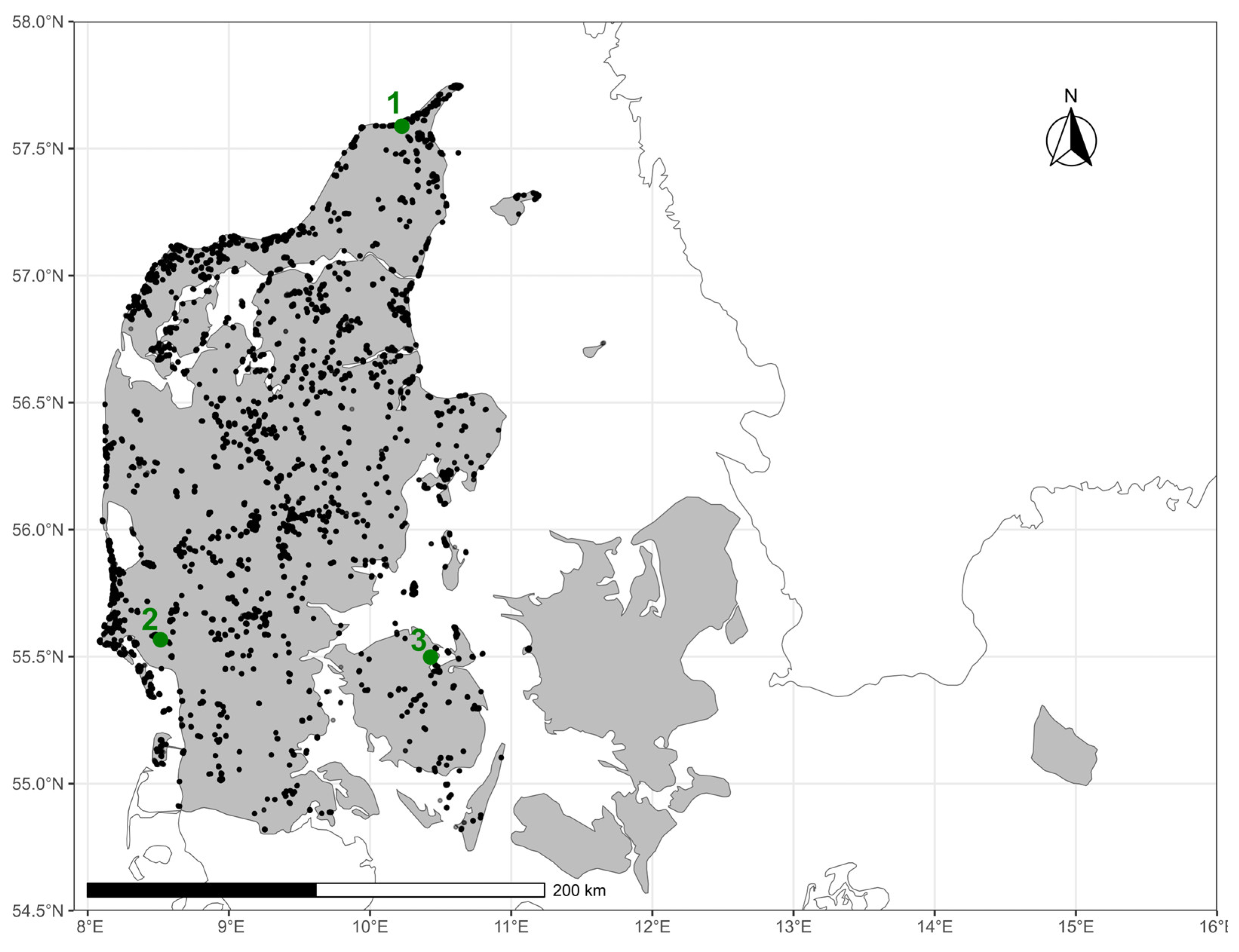
Vegetation Data
Remote Sensing Data
Preprocessing
Statistical Analysis
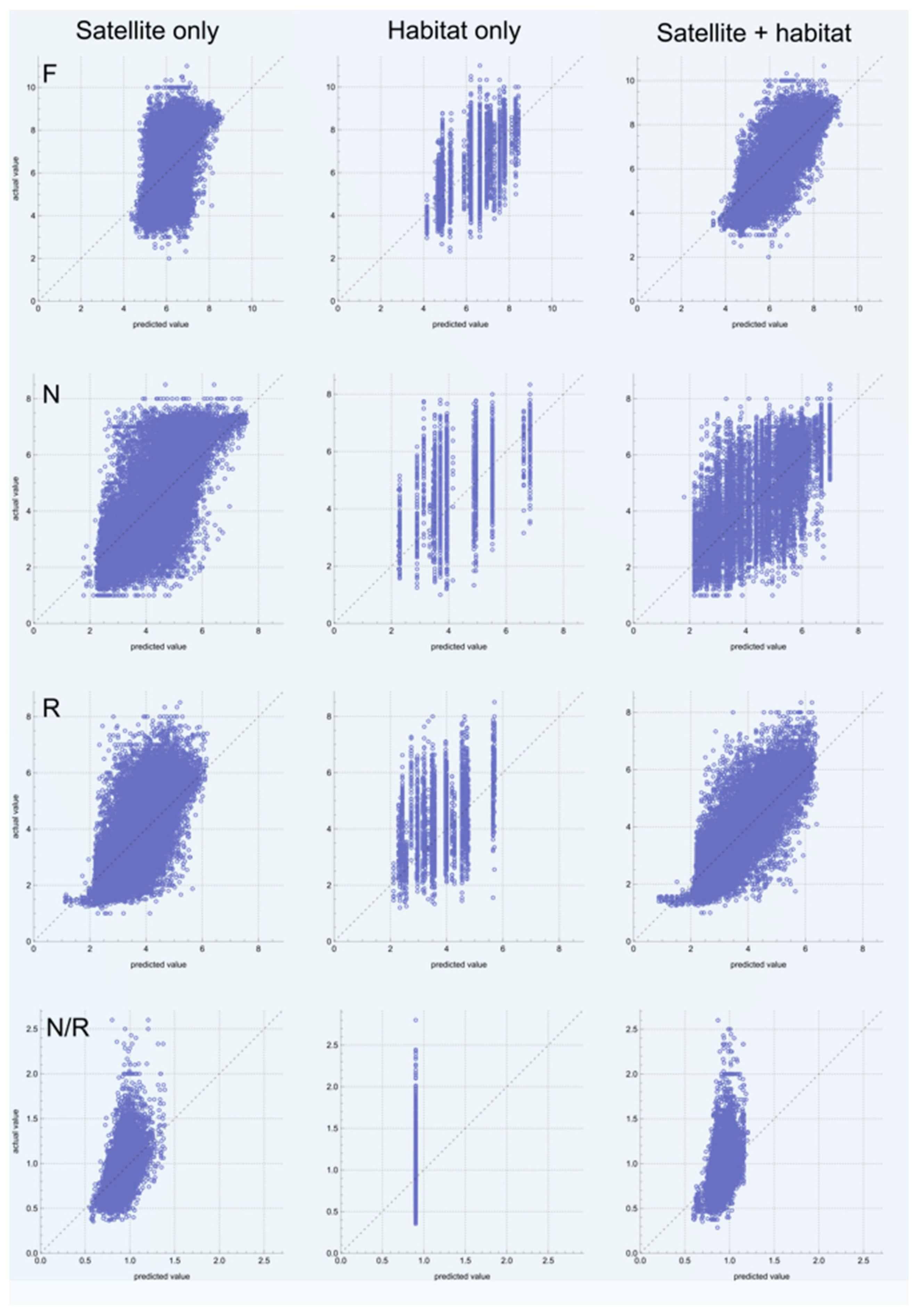
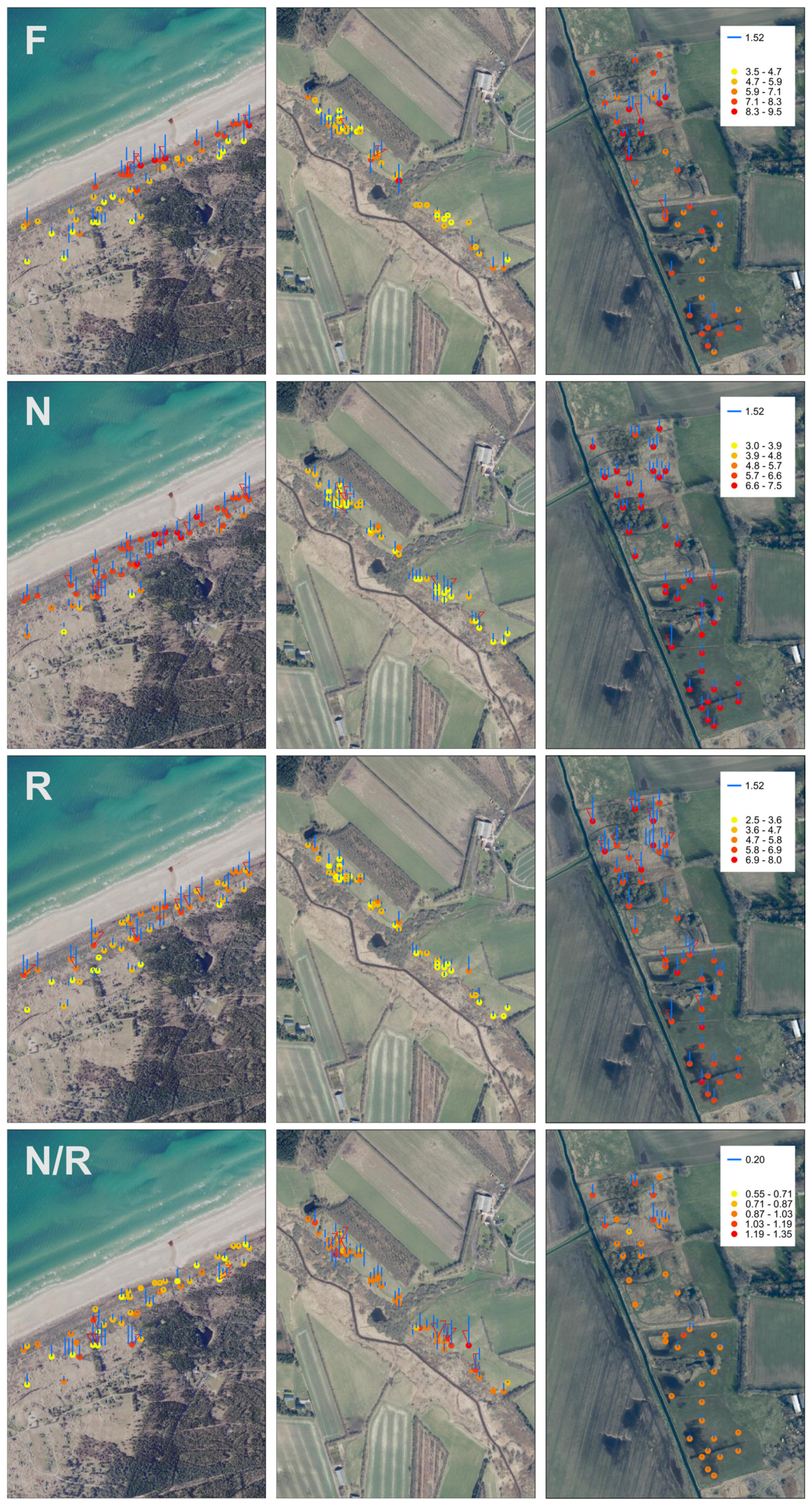
3. Results
4. Discussion
Soil Fertility and pH
Soil Moisture
Limitations and Uncertainties
Perspectives
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Phiri, D.; Simwanda, M.; Salekin, S.; Nyirenda, V.R.; Murayama, Y.; Ranagalage, M. Sentinel-2 Data for Land Cover/Use Mapping: A Review. Remote Sensing 2020, 12, 2291. [Google Scholar] [CrossRef]
- Ejrnæs, R.; Bruun, H.H. Gradient analysis of dry grassland vegetation in Denmark. Journal of Vegetation Science 2000, 11, 573–584. [Google Scholar] [CrossRef]
- Brunbjerg, A.K.; Bruun, H.H.; Brøndum, L.; Classen, A.T.; Fog, K.; Frøslev, T.G.; Goldberg, I.; Hansen, M.D.D.; Høye, T.T.; Læssøe, T.; et al. A systematic survey of regional multitaxon biodiversity: evaluating strategies and coverage. BMC Ecology 2019, 19, 43. [Google Scholar] [CrossRef] [PubMed]
- Moeslund, J.E.; Clausen, K.K.; Dalby, L.; Fløjgaard, C.; Pärtel, M.; Pfeifer, N.; Hollaus, M.; Brunbjerg, A.K. Using airborne lidar to characterize North European terrestrial high-dark-diversity habitats. Remote Sensing in Ecology and Conservation 2023, 9, 354–369. [Google Scholar] [CrossRef]
- Chytrý, M.; Hennekens, S.M.; Jiménez-Alfaro, B.; Knollová, I.; Dengler, J.; Jansen, F.; Landucci, F.; Schaminée, J.H.J.; Aćić, S.; Agrillo, E.; et al. European Vegetation Archive (EVA): an integrated database of European vegetation plots. Applied Vegetation Science 2016, 19, 173–180. [Google Scholar] [CrossRef]
- Kambach, S.; Sabatini, F.M.; Attorre, F.; Biurrun, I.; Boenisch, G.; Bonari, G.; Čarni, A.; Carranza, M.L.; Chiarucci, A.; Chytrý, M.; et al. Climate-trait relationships exhibit strong habitat specificity in plant communities across Europe. Nature Communications 2023, 14, 712. [Google Scholar] [CrossRef] [PubMed]
- Midolo, G.; Herben, T.; Axmanová, I.; Marcenò, C.; Pätsch, R.; Bruelheide, H.; Karger, D.N.; Aćić, S.; Bergamini, A.; Bergmeier, E.; et al. Disturbance indicator values for European plants. Global Ecology and Biogeography 2023, 32, 24–34. [Google Scholar] [CrossRef]
- Diekmann, M. Species indicator values as an important tool in applied plant ecology – a review. Basic and Applied Ecology 2003, 4, 493–506. [Google Scholar] [CrossRef]
- Nielsen, K.E.; Bak, J.L.; Bruus, M.; Damgaard, C.; Ejrnæs, R.; Fredshavn, J.R.; Nygaard, B.; Skov, F.; Strandberg, B.; Strandberg, M. NATURDATA.DK – Danish monitoring program of vegetation and chemical plant and soil data from non-forested terrestrial habitat types. In Biodiversity and Ecology. Special Volume: Vegetation databases for the 21st century, Dengler, J., Oldeland, J., Jansen, F., Chytrý, M., Ewald, J., Finckh, M., Glöckler, F., Lopez-Gonzalez, G., Peet, R.K., Schaminée, J.H.J., Eds.; 2012; Volume 4, pp. 375–375.
- Fredshavn, J.R.; Ejrnæs, R.; Nygaard, B. Teknisk anvisning for kortlægning af terrestriske naturtyper. TA-N3, Version 1.04. Fagdatacenter for Biodiversitet og Terrestriske Naturdata, Danmarks Miljøundersøgelser; 2010; p. 18 pp.
- Fredshavn, J.R.; Nielsen, K.E.; Ejrnæs, R.; Nygaard, B. Overvågning af terrestriske naturtyper. Version 4.1. 2018, 26 pp.
- Dronova, I.; Taddeo, S. Remote sensing of phenology: Towards the comprehensive indicators of plant community dynamics from species to regional scales. Journal of Ecology 2022, 110, 1460–1484. [Google Scholar] [CrossRef]
- Rossi, C.; Gholizadeh, H. Uncovering the hidden: Leveraging sub-pixel spectral diversity to estimate plant diversity from space. Remote Sensing of Environment 2023, 296, 113734. [Google Scholar] [CrossRef]
- Hauser, L.T.; Timmermans, J.; van der Windt, N.; Sil, Â.F.; César de Sá, N.; Soudzilovskaia, N.A.; van Bodegom, P.M. Explaining discrepancies between spectral and in-situ plant diversity in multispectral satellite earth observation. Remote Sensing of Environment 2021, 265, 112684. [Google Scholar] [CrossRef]
- Wang, Z.; Townsend, P.A.; Schweiger, A.K.; Couture, J.J.; Singh, A.; Hobbie, S.E.; Cavender-Bares, J. Mapping foliar functional traits and their uncertainties across three years in a grassland experiment. Remote Sensing of Environment 2019, 221, 405–416. [Google Scholar] [CrossRef]
- Roe, N.A.; Ducey, M.J.; Lee, T.D.; Fraser, O.L.; Colter, R.A.; Hallett, R.A. Soil chemical variables improve models of understorey plant species distributions. Journal of Biogeography 2022, 49, 753–766. [Google Scholar] [CrossRef]
- Arrouays, D.; Grundy, M.G.; Hartemink, A.E.; Hempel, J.W.; Heuvelink, G.B.M.; Hong, S.Y.; Lagacherie, P.; Lelyk, G.; McBratney, A.B.; McKenzie, N.J.; et al. Chapter Three - GlobalSoilMap: Toward a Fine-Resolution Global Grid of Soil Properties. In Advances in Agronomy, Sparks, D.L., Ed.; Academic Press: 2014; Volume 125, pp. 93–134.
- Bartels, S.F.; Caners, R.T.; Ogilvie, J.; White, B.; Macdonald, S.E. Relating bryophyte assemblages to a remotely sensed depth-to-water index in boreal forests. Frontiers in Plant Science 2018, 9, Article–858. [Google Scholar] [CrossRef] [PubMed]
- Asner, G.P.; Martin, R.E.; Knapp, D.E.; Tupayachi, R.; Anderson, C.; Carranza, L.; Martinez, P.; Houcheime, M.; Sinca, F.; Weiss, P. Spectroscopy of canopy chemicals in humid tropical forests. Remote Sensing of Environment 2011, 115, 3587–3598. [Google Scholar] [CrossRef]
- Ellenberg, H.; Weber, H.E.; Düll, R.; Wirth, V.; Werner, W. Zeigerwerte von planzen in Mitteleuropa, 3rd ed.; Erich Goltze GmbH & Co KG: Göttingen, 2001; Volume 18, p. 262. [Google Scholar]
- Schmidtlein, S. Imaging spectroscopy as a tool for mapping Ellenberg indicator values. Journal of Applied Ecology 2005, 42, 966–974. [Google Scholar] [CrossRef]
- Möckel, T.; Löfgren, O.; Prentice, H.C.; Eklundh, L.; Hall, K. Airborne hyperspectral data predict Ellenberg indicator values for nutrient and moisture availability in dry grazed grasslands within a local agricultural landscape. Ecological Indicators 2016, 66, 503–516. [Google Scholar] [CrossRef]
- Pang, H.; Zhang, A.; Yin, S.; Zhang, J.; Dong, G.; He, N.; Qin, W.; Wei, D. Estimating Carbon, Nitrogen, and Phosphorus Contents of West–East Grassland Transect in Inner Mongolia Based on Sentinel-2 and Meteorological Data. Remote Sensing 2022, 14, 242. [Google Scholar] [CrossRef]
- Council of the European Community. Council Directive 92/43/EEC of 21 May 1992 on the conservation of natural habitats and of wild fauna and flora; 1992; pp. 7–50.
- Andersen, D.K.; Nygaard, B.; Fredshavn, J.R.; Ejrnæs, R. Cost-effective assessment of conservation status of fens. Applied Vegetation Science 2013, 16, 491–501. [Google Scholar] [CrossRef]
- 26. European Space Agency. Sentinel-2 User Handbook, 2015.
- Hagolle, O.; Huc, M.; Desjardins, C.; Auer, S.; Richter, R. MAJA ATBD - Algorithm theoretical basis document; Centre National d’Études Spatiales: 2017; p. 40 pp.
- Hagolle, O.; Huc, M.; Villa Pascual, D.; Dedieu, G. A Multi-Temporal and Multi-Spectral Method to Estimate Aerosol Optical Thickness over Land, for the Atmospheric Correction of FormoSat-2, LandSat, VENμS and Sentinel-2 Images. Remote Sensing 2015, 7, 2668–2691. [Google Scholar] [CrossRef]
- Doxani, G.; Vermote, E.; Roger, J.-C.; Gascon, F.; Adriaensen, S.; Frantz, D.; Hagolle, O.; Hollstein, A.; Kirches, G.; Li, F.; et al. Atmospheric Correction Inter-Comparison Exercise. Remote Sensing 2018, 10, 352. [Google Scholar] [CrossRef] [PubMed]
- Colin, J.; Hagolle, O.; Landier, L.; Coustance, S.; Kettig, P.; Meygret, A.; Osman, J.; Vermote, E. Assessment of the Performance of the Atmospheric Correction Algorithm MAJA for Sentinel-2 Surface Reflectance Estimates. Remote Sensing 2023, 15, 2665. [Google Scholar] [CrossRef]
- Löfgren, O.; Prentice, H.C.; Moeckel, T.; Schmid, B.C.; Hall, K. Landscape history confounds the ability of the NDVI to detect fine-scale variation in grassland communities. Methods in Ecology and Evolution 2018, 9, 2009–2018. [Google Scholar] [CrossRef]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using lme4. Journal of Statistical Software 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Weber, D.; Schaepman-Strub, G.; Ecker, K. Predicting habitat quality of protected dry grasslands using Landsat NDVI phenology. Ecological Indicators 2018, 91, 447–460. [Google Scholar] [CrossRef]
- Moeslund, J.E.; Zlinszky, A.; Ejrnæs, R.; Brunbjerg, A.K.; Bøcher, P.K.; Svenning, J.-C.; Normand, S. Light detection and ranging explains diversity of plants, fungi, lichens, and bryophytes across multiple habitats and large geographic extent. Ecological Applications 2019, 29, e01907. [Google Scholar] [CrossRef] [PubMed]
- Wamelink, G.W.W.; Joosten, V.; van Dobben, H.F.; Berendse, F. Validity of Ellenberg indicator values judged from physico-chemical field measurements. Journal of Vegetation Science 2002, 13, 269–278. [Google Scholar] [CrossRef]
- Wamelink, G.W.W.; Goedhart, P.W.; Dobben, H.F.v. Measurement Errors and Regression to the Mean Cannot Explain Bias in Average Ellenberg Indicator Values. Journal of Vegetation Science 2004, 15, 847–851. [Google Scholar] [CrossRef]
- Smart, S.M.; Scott, W.A. Bias in Ellenberg indicator values ? problems with detection of the effect of vegetation type. Journal of Vegetation Science 2004, 15, 843–846. [Google Scholar] [CrossRef]
- Amani, M.; Foroughnia, F.; Moghimi, A.; Mahdavi, S.; Jin, S. Three-Dimensional Mapping of Habitats Using Remote-Sensing Data and Machine-Learning Algorithms. Remote Sensing 2023, 15, 4135. [Google Scholar] [CrossRef]
- Gaffney, R.; Augustine, D.J.; Kearney, S.P.; Porensky, L.M. Using Hyperspectral Imagery to Characterize Rangeland Vegetation Composition at Process-Relevant Scales. Remote Sensing 2021, 13, 4603. [Google Scholar] [CrossRef]
- Gillan, J.K.; Karl, J.W.; van Leeuwen, W.J.D. Integrating drone imagery with existing rangeland monitoring programs. Environmental Monitoring and Assessment 2020, 192, 269. [Google Scholar] [CrossRef] [PubMed]
- Kattenborn, T.; Eichel, J.; Wiser, S.; Burrows, L.; Fassnacht, F.E.; Schmidtlein, S. Convolutional Neural Networks accurately predict cover fractions of plant species and communities in Unmanned Aerial Vehicle imagery. Remote Sensing in Ecology and Conservation 2020, 6, 472–486. [Google Scholar] [CrossRef]
- The Danish Environmental Protection Agency. New technology can map nature areas. Available online: https://mst.dk/nyheder/2022/marts/ny-teknologi-kan-kortlaegge-naturomraader (accessed on 19 April 2024).
- Marcinkowska-Ochtyra, A.; Ochtyra, A.; Raczko, E.; Kopeć, D. Natura 2000 Grassland Habitats Mapping Based on Spectro-Temporal Dimension of Sentinel-2 Images with Machine Learning. Remote Sensing 2023, 15, 1388. [Google Scholar] [CrossRef]
- Pérez-Carabaza, S.; Boydell, O.; O’Connell, J. Habitat classification using convolutional neural networks and multitemporal multispectral aerial imagery. Journal of Applied Remote Sensing 2021, 15, 042406. [Google Scholar] [CrossRef]
| F | N | R | N/R | |||||||||
| Std. | R2 | Model | Std. | R2 | Model | Std. | R2 | Model | Std. | R2 | Model | |
| Satellite | 1.34 | 0.26 | GBT | 1.09 | 0.59 | RF | 0.93 | 0.54 | RF | 0.16 | 0.29 | RF |
| Habitat | 1.23 | 0.36 | GBT | 1.40 | 0.05 | DT | 1.29 | - | NN | 0.18 | - | LR |
| Satellite +habitat | 0.81 | 0.73 | RF | 0.92 | 0.70 | DT | 0.71 | 0.73 | RF | 0.16 | 0.23 | GBT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





