1. Introduction
Reportedly,
Salmonella Typhimurium, a food-borne pathogen, has raised public health concerns because of its involvement in 1.35 million infections, over 26,500 hospitalizations and numerous mortality rates in the United States alone [
1,
2]. Several
Salmonella serotypes exist, but
Salmonella Typhimurium is unique among the over 2,600 serotypes because of its prevalence, stability, and frequent implication in infections over the years.
Salmonella Typhimurium poses a complex array of virulent factors contributing to its pathogenicity and facilitating infections in humans. The virulence factors aid the bacteria to invade and adhere to both the M and intestinal epithelial cells using several adhesion molecules on their surface [
3,
4]. Once the bacteria gain successful entry into the host, they manipulate and rearrange the host cellular organization, forming membrane ruffles. They may also be engulfed directly by dendritic cells from the underlying tissues. One typical virulence factor that enables
Salmonella Typhimurium to achieve these feet is its flagellum, a dynamic molecular structure that facilitates motility and environmental sensing [
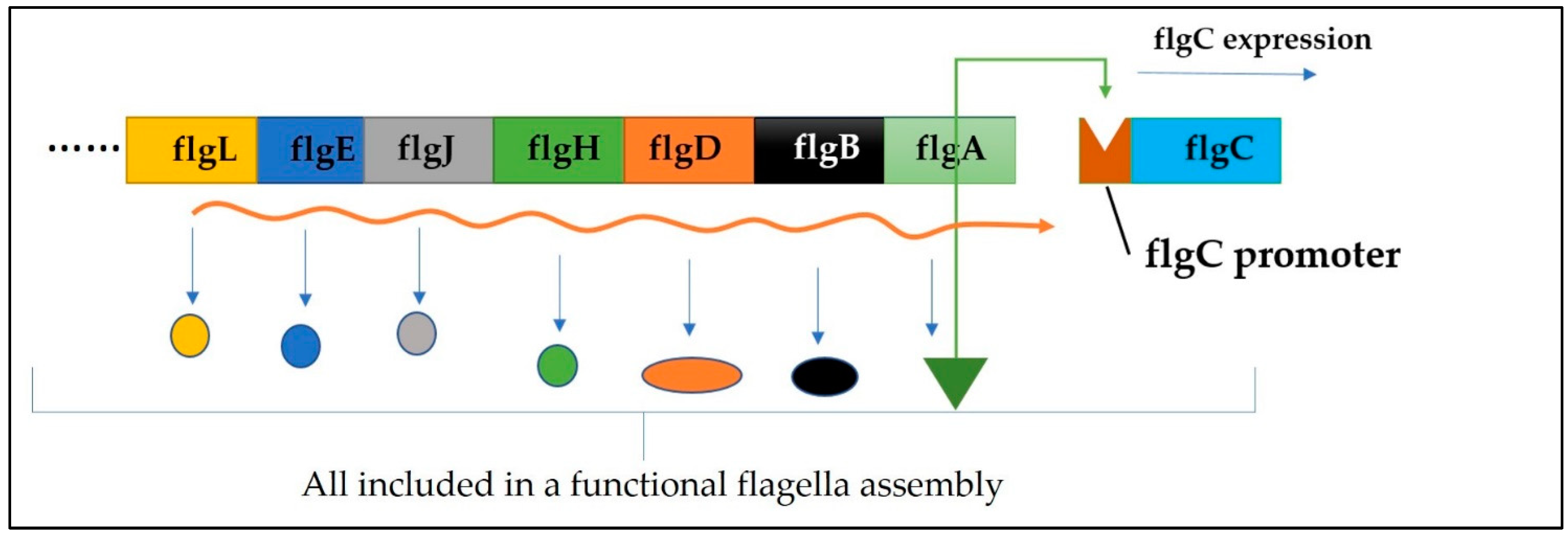
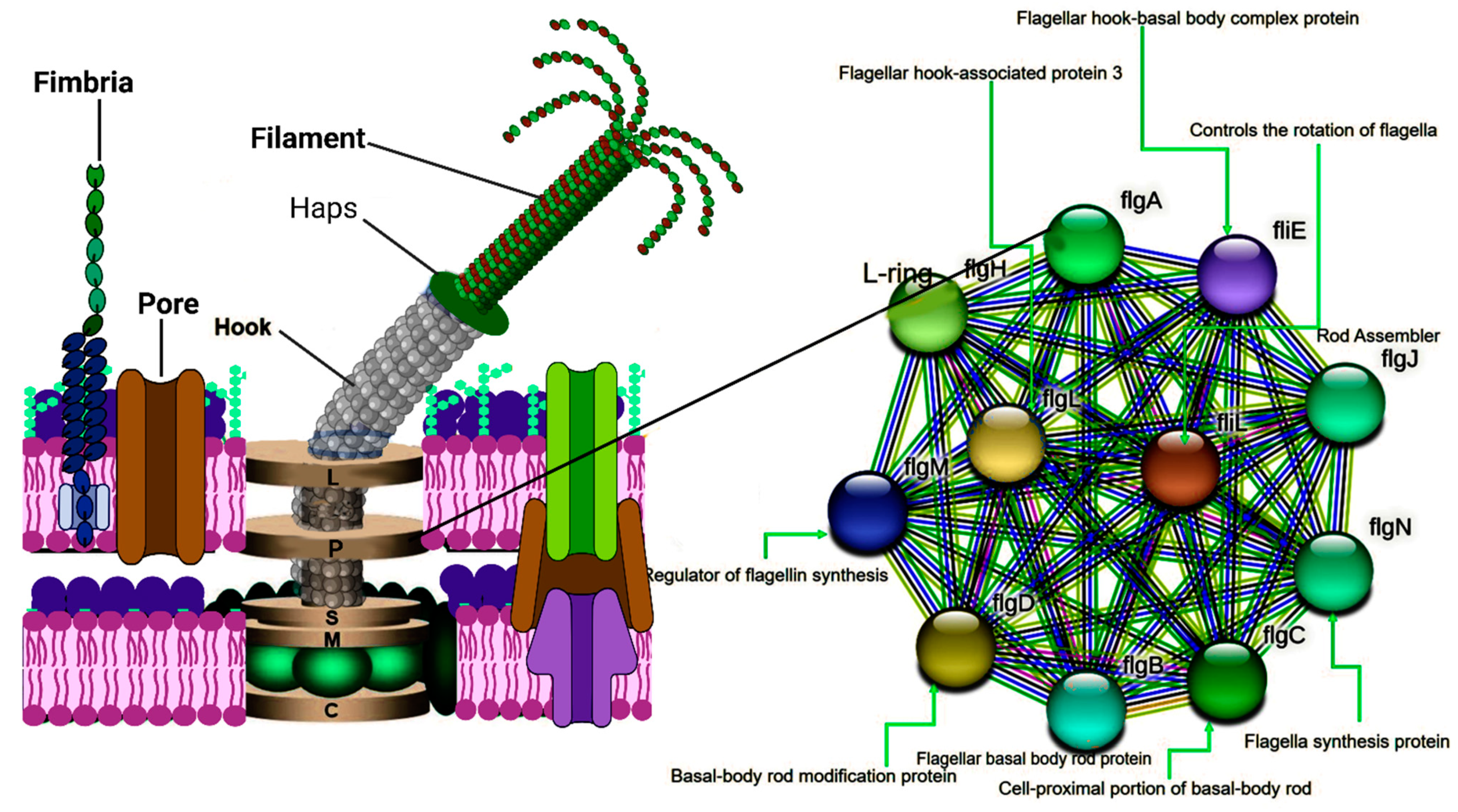
5]. Flagella are whip-like appendages and extensions on the surface of several bacteria. Bacteria can swim/move freely through a liquid medium with the aid of their flagella. A typical flagellum comprises a filament, a hook, and a basal body working in sync to propel the bacteria into motility. The basal body is responsible for the rotation of the flagellum, which drives energy through the proton motive force. The proper formation and function of the flagella are intricately tied to the presence and expression of the flgA gene responsible for encoding the flgA protein [
6]. This gene is crucial for the assembly of the P-ring in the basal body of the flagellum (
Figure 2), coupled with several other genes (
Figure 1) responsible for the flagella's dynamic role. The P-ring is critical for firmly holding the flagellum to the peptidoglycan layer of the cell wall, aiding the firm rotation of the flagella. The proper function of the P-ring relies on the presence of a functional flgA protein, underscoring the paramount role of flgA in maintaining bacterial movement, an essential physiological process for achieving its biological goals such as chemotaxis, colonization, and pathogenesis [
7].
The flagellum is powered by a motor with a speed of over 60,000 revolutions per minute, with an intricate ability to reverse its rotation direction to navigate complex environments swiftly [
8]. FlgA is the base protein responsible for the assembly of this flagellum, as well as many other proteins. FlgA is crucial for building up the P-ring, a critical component of the flagella machinery, and it is also intricately linked with the transcription of flgC (
Figure 1) [
9].
Figure 1.
Diagrammatical illustration of the crucial genes needful in the biosynthesis of a flagellum. All associated genes must be expressed, including flgA to ensure the formation of a functional flagellum. In addition, flgA has been demonstrated to serve as a transcriptional factor and is noted to be a pre-requirement for flgC expression.
Figure 1.
Diagrammatical illustration of the crucial genes needful in the biosynthesis of a flagellum. All associated genes must be expressed, including flgA to ensure the formation of a functional flagellum. In addition, flgA has been demonstrated to serve as a transcriptional factor and is noted to be a pre-requirement for flgC expression.
Figure 2.
Flagellum structure and String network of flgA.
Figure 2.
Flagellum structure and String network of flgA.
Among the dozens of phytocannabinoids, Cannabidiol (CBD), the non-psychoactive constituent, has captured the interest of several researchers globally due to its alleged pharmacological qualities, such as its anxiolytic, anti-inflammatory and analgesic properties [
10]. Contrary to delta-9-tetrahydrocannabinol (THC), CBD is free from any psychotic effects, making it suitable for treating patients with zero or minimal feelings of “high” associated with traditional cannabis use [
11]. However, most of the exploration of CBD’s potential has been focused on its impact on humans and mammals, targeting conditions such as chronic pains, anxiety, epilepsy, and multiple sclerosis [
12]. These explorations have generally centered on CBD's association with the endocannabinoid system, which is responsible for modulating several critical cognitive and physiological conditions such as mood, memory, and pain perception. Recently, researchers have shifted perspective due to the urgent need for novel compounds to target the persistent antimicrobial resistance crisis. Hence, there is a burgeoning field of research focusing on unveiling the role of CBD on microorganisms, with a particular focus on microbial physiology and genetics, as well as CBDs’ role as an antimicrobial. This shift in perspective has significantly expanded the scope of phytocannabinoid research, embracing micro and cellular biology in its emblem. Several early-stage studies have demonstrated CBDs’ ability to curtail Gram-Negative and Gram-Positive bacteria from growing, highlighting CBDs' ability to function as an antibacterial therapy [
13]. Several other studies have compared CBDs’ efficacy as antibacterial to other popular antibiotics, such as Kanamycin and polymyxin B, with CBD reportedly functioning more effectively than the latter [
14]. Moreover, exploring the impact of CBD on microbial gene expression is an exciting area to endeavor. Our laboratory previously explored the mechanisms
Salmonella Typhimurium uses to develop resistance to CBD after we induced the bacteria to develop resistance to CBD [
15]. CBD may influence various cellular responses in microbes by altering gene expression patterns. The objectives of this study were focused on the effects of prolonged exposure of
Salmonella Typhimurium to CBD and its impact on the expression of the flgA gene. We hypothesize that CBD treatment may lead to an upregulation of the flgA gene, potentially influencing the bacterium's ability to withstand environmental stresses.
2. Materials and Methods
2.1. Media, Chemicals, Bacterial Strains, and Other Reagents
All chemicals (Hexane, methanol –LC-MS (≥99.9%), water, and ethanol absolute proof (≥99.5%) employed in this research were of HPLC grade. Our laboratory has elucidated that
Salmonella Typhimurium is susceptible to CBD at micromolar concentrations [
13]. The two PCR ribo-types of
Salmonella Typhimurium used in this study were called CBD-resistant
Salmonella Typhimurium and CBD-Susceptible
Salmonella Typhimurium that were created from the lab-coded BV4012 strains. The BV4012 is
Salmonella Typhimurium LT2 strain MS1868 (a gift from Dr. Anthony R. Poteete, University of Massachusetts). CBD-resistant strains were previously created in our laboratory through prolonged exposure of
Salmonella Typhimurium to CBD doses [
13]. The resistant colonies were picked and plated on LB agar premixed with CBD at 10 μg/ml final concentration, with continuous re-culturing in higher CBD doses. Henceforth, all cultures of CBD-resistant strains received 10 μg/ml CBD concentration or more in their growth media. For the maintenance of the bacteria, Luria Broth (BD, Difco, Franklin Lakes, NJ, USA) and Luria Agar (BD, Difco, Franklin Lakes, NJ, USA) were the media used. Sustainable CBD LLC conducted the CBD extraction and purification process. (Salem, AL, USA) and has been described elsewhere [
14].
The protocol provided with the RNA extraction kit, Rneasy kit (Qiagen Sciences, Maryland, USA), was used to extract RNA from both CBD-susceptible and CBD-resistant strains of Salmonella. Extracted RNA samples were treated with DNase I (Thermo Scientific™, Vilnius, Lithuania) to remove contaminating genomic DNA. Following DNase-I treatment, the concentration and purity of the RNA were determined by Nanodrop (1000 spectrophotometer, ThermoFisher Scientific, Madison, WI, USA).
Prepare RNA Sample: The RNA samples were diluted to 1 μg /mL, for which 1 μg of the total RNA was used for each reverse transcription reaction. The reaction mix for each sample was as follows. 1 μg of RNA was mixed with 1 μL dNTP mix (10 mM each), 1 μL of 50 μM Random oligo(dT) primers and Nuclease-free water was then added to a final volume of 13 μL. The RNA and primer mix was incubated at 65°C for 5 minutes and then placed on ice for 1 minute. 4 μL 5X SuperScript IV reaction buffer was added to the denatured RNA-primer mix, followed by the addition of 1 μL 100 mM DTT, 1 μL RNaseOUT (40 U/μL), 1 μL of 200 U/μL SuperScript IV Reverse Transcriptase (lot# 00916637, Invitrogen, ThermoFisher Scientific, Vilnius, Lithuania). The mixture was incubated at 50°C for 10 minutes, then the reaction inactivated by heating at 80°C for 10 minutes. The resulting cDNA was stored at -20°C for later use.
To investigate the relative expression of the flgA gene, the total RNA of both CBD-resistant and CBD-susceptible
Salmonella Typhimurium strain (n = 3) was extracted using the Rneasy kit (Qiagen Sciences, Maryland, USA) after an overnight culture at pH 7 LB broth. After successful extraction, the RNA was dissolved in RNase-free water, and the concentration was determined by Nanodrop (1000 spectrophotometer, ThermoFisher Scientific, Madison, WI, USA). Following the RNA extraction and reverse transcription into cDNA using the SSIV Cell Direct cDNA Synthesis kit, a qPCR reaction was performed on the samples. The qPCR was performed using primers specific for the flgA gene: Forward: 5'-TTCATCGCCCAATCTGCTAC -3'; Reverse: 5'- GACGGTTCCACTCACGATTT -3', using CFX96TM real-time PCR (Bio-Rad Laboratories, Hercules, CA, USA) following similar procedures as in our previous work [
14], and the DNA gyrase B (gyrB) gene served as the housekeeping gene. The primers for gyrB used for the study were Fw-gyrB GTTGGTGAAGGTTTCGTGGC and Rv-gyrB ATATCGGCGACACGGATGAC.
The motility of
Salmonella Typhimurium strains was assessed using motility agar (0.3% agar) prepared at pH levels 3, 7, and 12, as described by [
15]. The pH adjustment was achieved by dropping 1N HCL for acidic targets and 1N NaOH for essential targets while stirring. Bacterial cultures were inoculated at the center of the agar plates and incubated at 37°C for specified time points. The migration of bacteria from the inoculation point was documented through photographs.
The growth kinetics of Salmonella Typhimurium were evaluated in LB broth adjusted to pH levels 3, 5, 7, and 12. The pH adjustment was achieved by dropping 1N HCL for acidic treatments and 1N NaOH for basic treatments targets while stirring. Overnight cultures of Salmonella Typhimurium were inoculated into the broth and incubated at 37°C with shaking. Aliquots of 200 μl bacterial cells were seeded in a 96-well microtiter plate (Fisherbrand™, Fisher Scientific, Fair Lawn, NJ, United States) at an OD of 0.3 to mimic the early-log phase. Growth was monitored by measuring the optical density at 600 nm (OD600) at regular intervals using SpectraMax plate reader (Molecular Devices SpectraMax®, ABS Plus) (Molecular Devices LLC., San Jose, CA, United States). Growth curves were plotted, and significant inhibition at extreme pH levels was noted.
To investigate the influence of nutrient starvation, minimal media without nutrients such as carbon, nitrogen and phosphorus were prepared and supplemented with CBD at 5 µg/mL concentrations, 50 µg/mL, and 100 µg/mL, following similar techniques employed by [
16]. Both CBD-resistant and CBD-susceptible strains were inoculated into the media and incubated at 37°C under shaking. Growth was monitored by measuring the optical density at 600 nm (OD600) at regular intervals using a SpectraMax plate reader (Molecular Devices SpectraMax
® ABS Plus) (Molecular Devices LLC., San Jose, CA, United States). Growth curves were plotted, and significant inhibition at extreme pH levels was recorded.
Bacterial cultures were grown in a 96-well plate under both CBD treatment and untreated control conditions, incubated at 37°C for 48 h. Biofilms were stained with crystal violet, washed, and solubilized in ethanol. Absorbance at 412 nm was measured to quantify biofilm formation.
CBD-resistant and CBD-susceptible Salmonella Typhimurium strains were grown in 6-well plates (Fisherbrand™, Fisher Scientific, Fair Lawn, NJ, USA) supplemented with LB broth to induce biofilm formation. Salmonella Typhimurium biofilm samples were treated with 5 μg/ml and 100 μg/ml of CBD. Deionized water and hydrogen peroxide were used as the control groups. We used a scanning electron microscope (SEM) to view the microstructure and morphology of the biofilm formed. The prepared specimens were vacuum-dried and sprayed with gold using an EMS Quorum (EMS 150R ES) ion-sputtering instrument. Using an analytical SEM (JEOL JSM-6010LA, Tokyo, Japan) coupled with IntouchScopesoftware (JSM-IT200 InTouchScope™ SEM Series, JEOL Solutions, Tokyo, Japan). We fixed the specimens using 10% formaldehyde solution at room temperature for 10 minutes, then washed them with PDS thrice. We dehydrated the washed specimens serially using 50%, 70%, and 90% absolute ethanol. The specimens were finally dried in a vacuum, sprayed with gold using an EMS quorum ion-sputtering instrument, and observed using an analytical SEM (JEOL JSM-6010LA, Tokyo, Japan) coupled with IntouchScopesoftware (JSM-IT200 InTouchScope™ SEM Series, JEOL Solutions, Tokyo, Japan).
Two different Salmonella Typhimurium strains (CBD-resistant and CBD-susceptible strains) were used for these experiments, and the experiments were conducted in triplicates. The results are presented as means ± SEM, and means ± STD, p-values lower or equal to 0.05 were considered significant using student paired t-test. All statistical analyses were conducted, and graphs were plotted on Microsoft Excel (Microsoft 2010), microscopic images were processed using ImageJ (an opensource NIH software).
3. Results
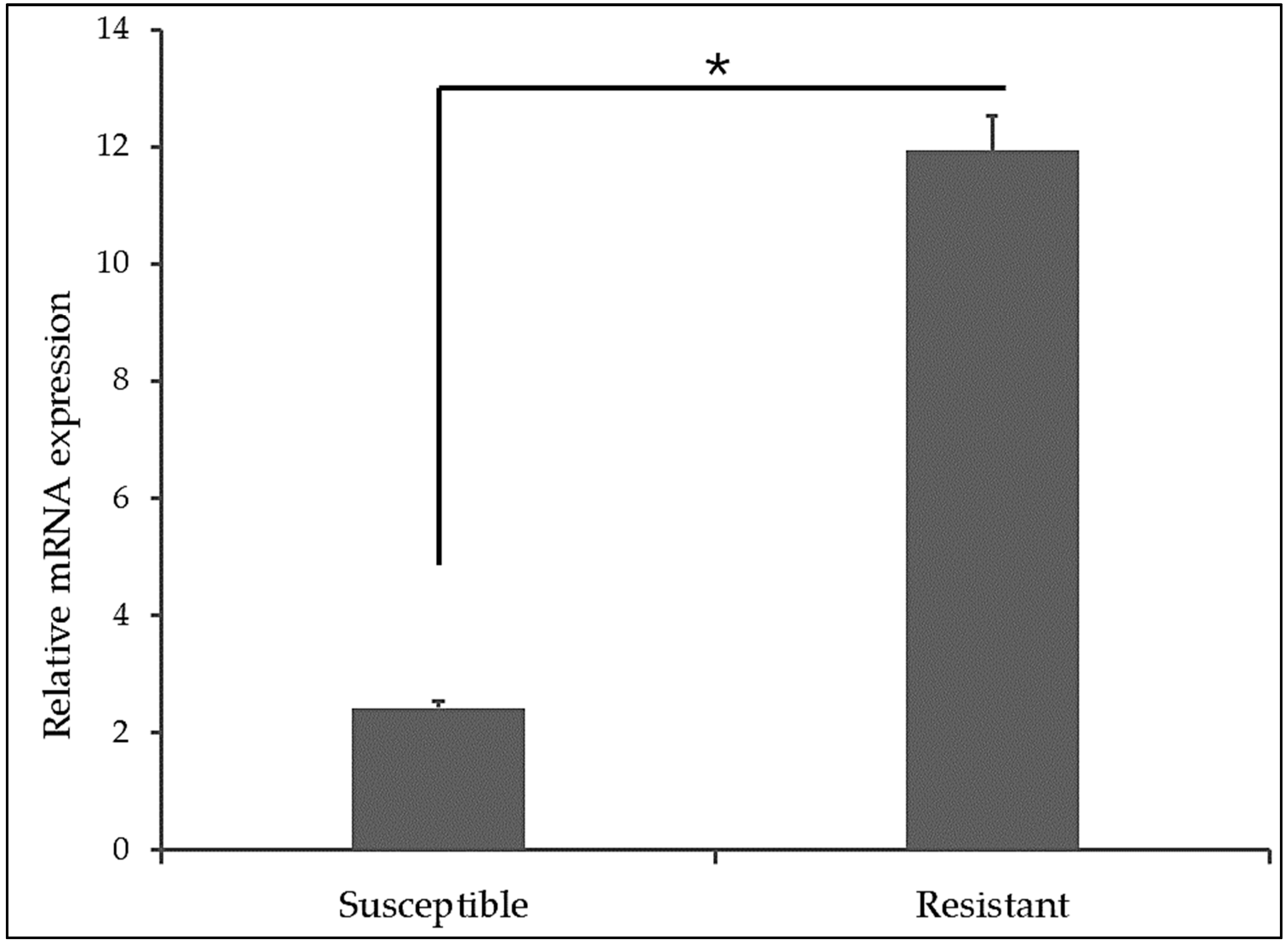
A comparative analysis of the flgA gene expression between the CBD-resistant and CBD-susceptible strains revealed that, the CBD-resistant strains have higher expression of the flgA gene (almost six times higher expression of relative mRNA) than the CBD-susceptible strain of
Salmonella Typhimurium as shown in
Figure 3.
The motility assay, as influenced by the expression of the flgA gene, showed significant differences between CBD-resistant and CBD-susceptible
Salmonella Typhimurium strains.
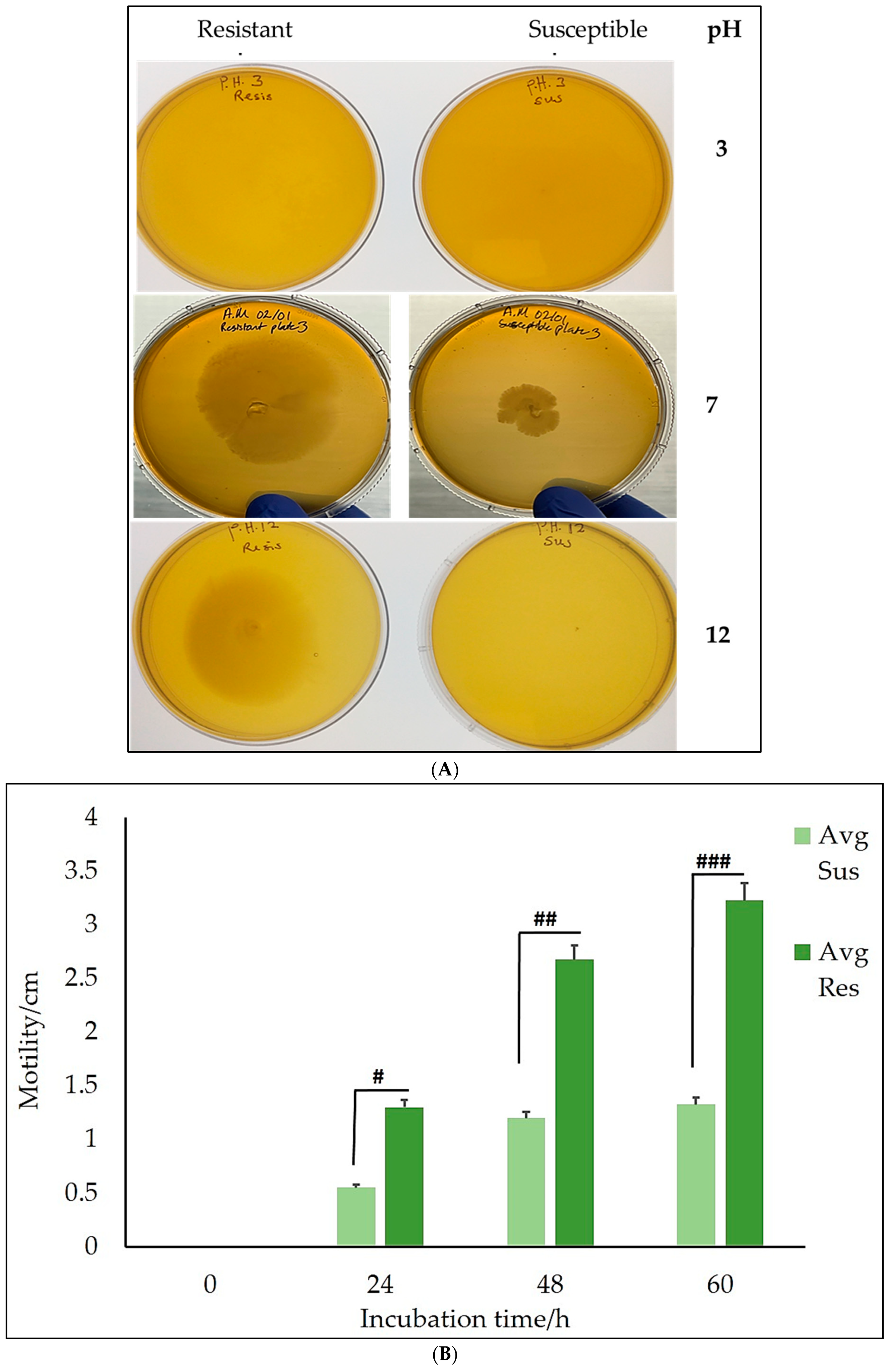
Figure 4A shows that
Salmonella Typhimurium motility was significantly minimized at higher pH levels. Additionally, at pH 3, there was no growth for both the CBD-resistant and CBD-susceptible strains of
Salmonella Typhimurium. At pH 7, growth was recorded for both strains, with the CBD-resistant strains exhibiting higher growth than the CBD-susceptible strains. At pH 12, there was no growth for the CBD-susceptible strains, but growth was recorded for the CBD-resistant strains.
After 48 h of incubation, both strains' average radius in cm traveled was 0.0 cm, while the CBD-resistant strains traveled an average radius of 1.6 ± 2.26 cm at pH 12. This suggests that extreme pH (pH 3) deleteriously impacts the strains. Additionally, the CBD-susceptible strains did not survive at pH 12, occupying an average radius of 0.0 cm, as indicated in
Table 1.
The CBD-resistant strains exhibited improved mobility at pH 7 compared to the CBD-susceptible strains, as indicated in
Figure 4B, at all the different time points. This implies that CBD resistance may be linked to increased bacterial mobility, possibly contributing to
Salmonella Typhimurium ability to invade host tissues efficiently and grow. Furthermore, the resistant strains maintained higher motility than the susceptible strains, as seen across all the pH levels tested (
Figure 4A). This suggests a possible resilience and adaptive advantage of the CBD-resistant strains in different environmental (pH) situations, encouraging their survival and growth.
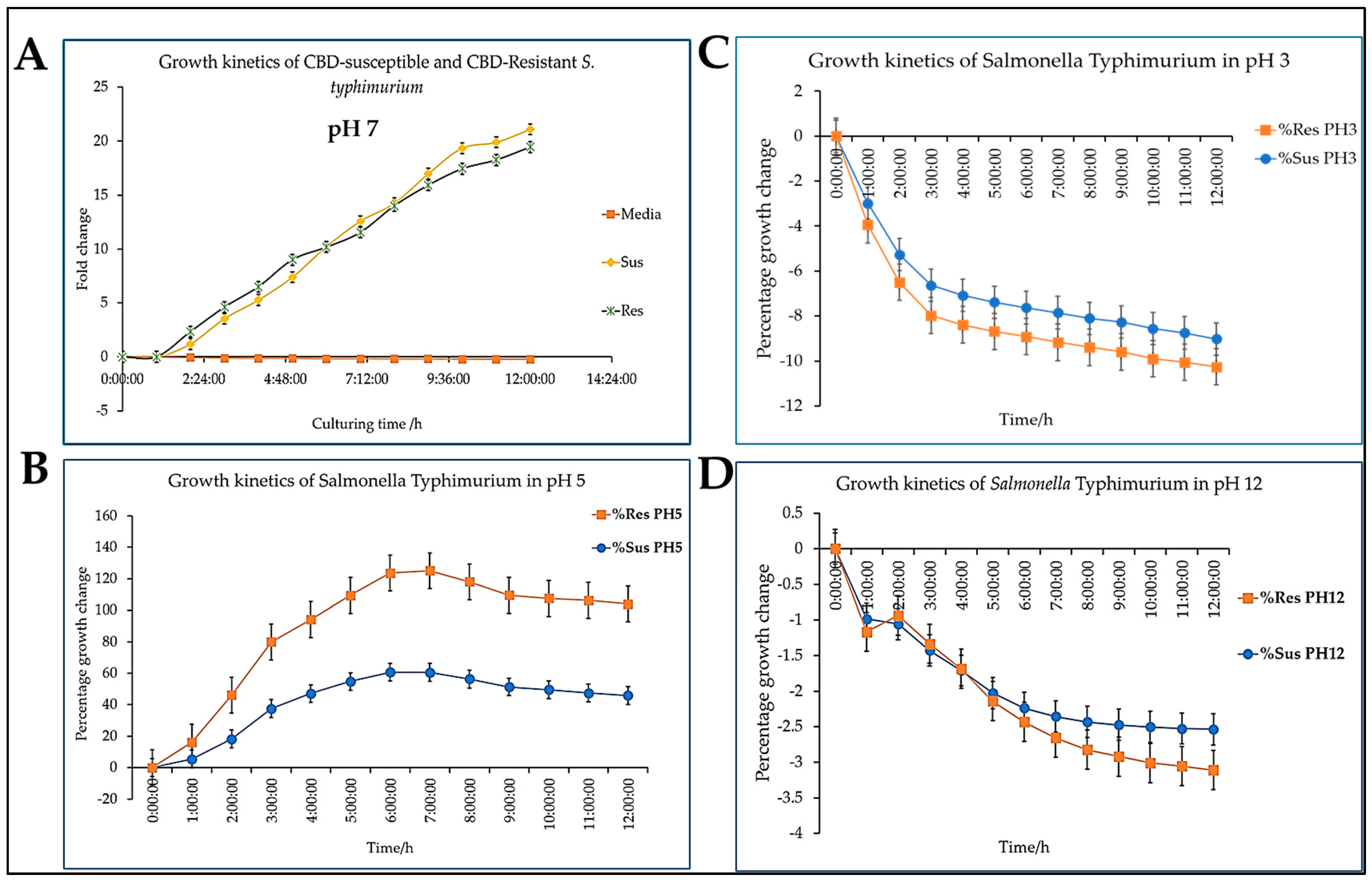
Different pH levels have different impact on the survival and growth of both CBD-resistant and CBD-susceptible strains of
Salmonella Typhimurium, as evidenced in
Figure 5. pH 7 favored the proliferation of both the CBD-susceptible and CBD-resistant strains, as shown in
Figure 5A. Compared to the control group (media), pH 7 did not negatively impact the survival of both strains. This observation is valid also for strains at pH 5, as shown in
Figure 5C. However, regarding percentage proliferation, the CBD-resistant strain withstood pH 5 stress better than the CBD-susceptible strain. Both strains nevertheless follow a similar trend. Thus, the growth of both strains peaked between the fifth and sixth-time points and declined at the eighth-hour time point, as shown in
Figure 5C. pH 3 and pH 12 have detrimental impacts on both strains as extreme killing was observed for all the time points (
Figure 5B and 5D). At extreme pH (both acidic and basic), the CBD-resistant strains declined slightly higher than the susceptible strains at the end of the 12 h period.
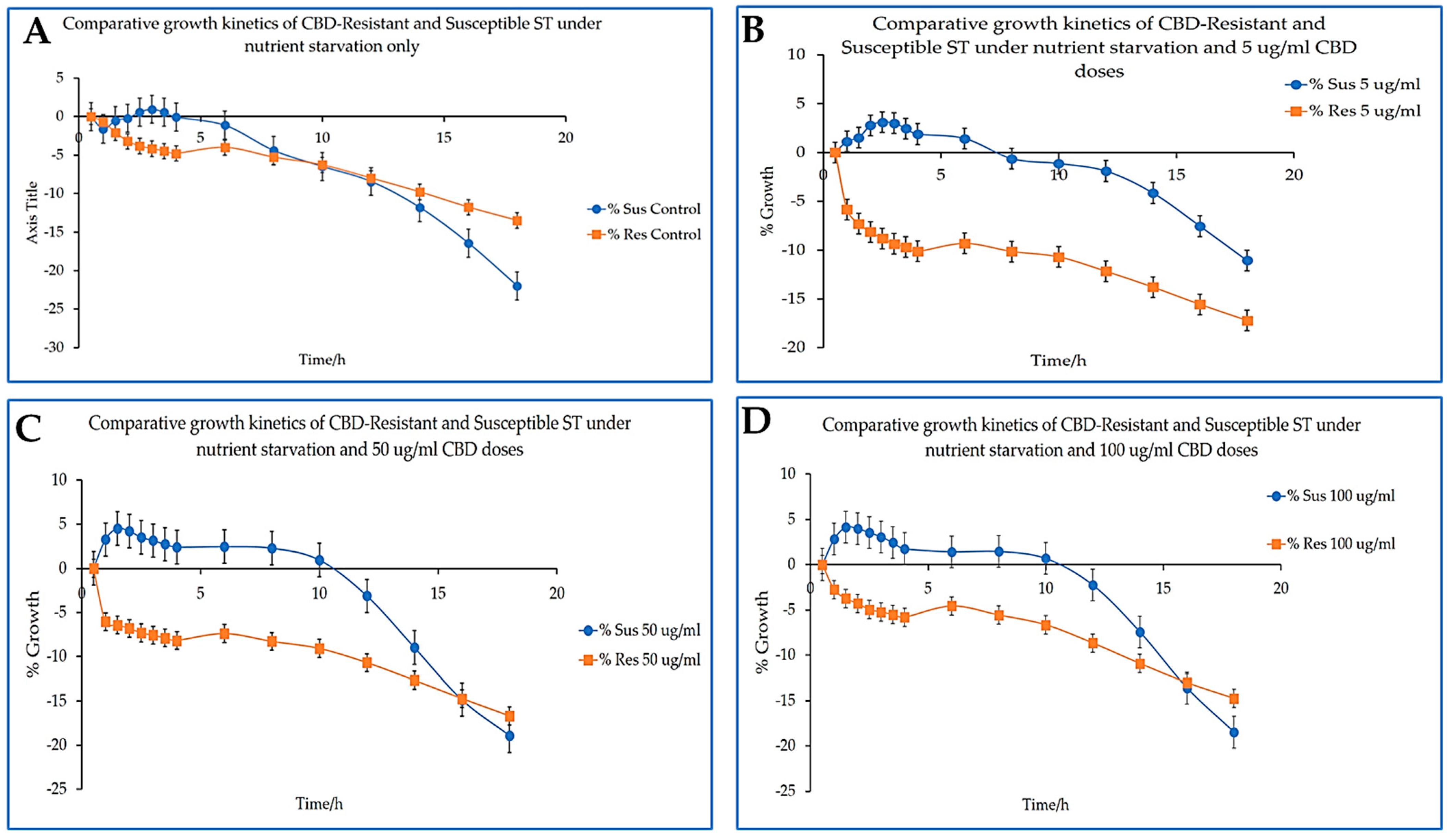
Under nutrient starvation, both CBD-resistant and CBD-susceptible strains were severely impacted negatively under all the doses of the CBD treatments, as shown in
Figure 4A-D. Interestingly, the CBD-susceptible strains demonstrated a level of resistance to nutrient starvation. However, their resistance was short-lived. The CBD-susceptible strains try to grow a little at all CBD doses by achieving a growth rate of nearly 5%. However, the CBD-resistant strains could not withstand nutrient starvation in the least, as shown in
Figure 4A-4D.
Figure 5A and 5B represent the separate growth kinetics of CBD-susceptible and CBD-resistant
Salmonella Typhimurium strains under different CBD doses. As stated above in
Figure 4, CBD-susceptible strains demonstrated tolerance to nutrient starvation better than CBD-resistant strains. Though the CBD-susceptible strain's growth was insignificant, they were able to maintain a minimal steady growth until the 10th hour before actually starting to die, as shown in
Figure 5B. The CBD-resistant strain could not demonstrate this physiology as they could not grow to the least and start dying immediately under nutrient starvation.
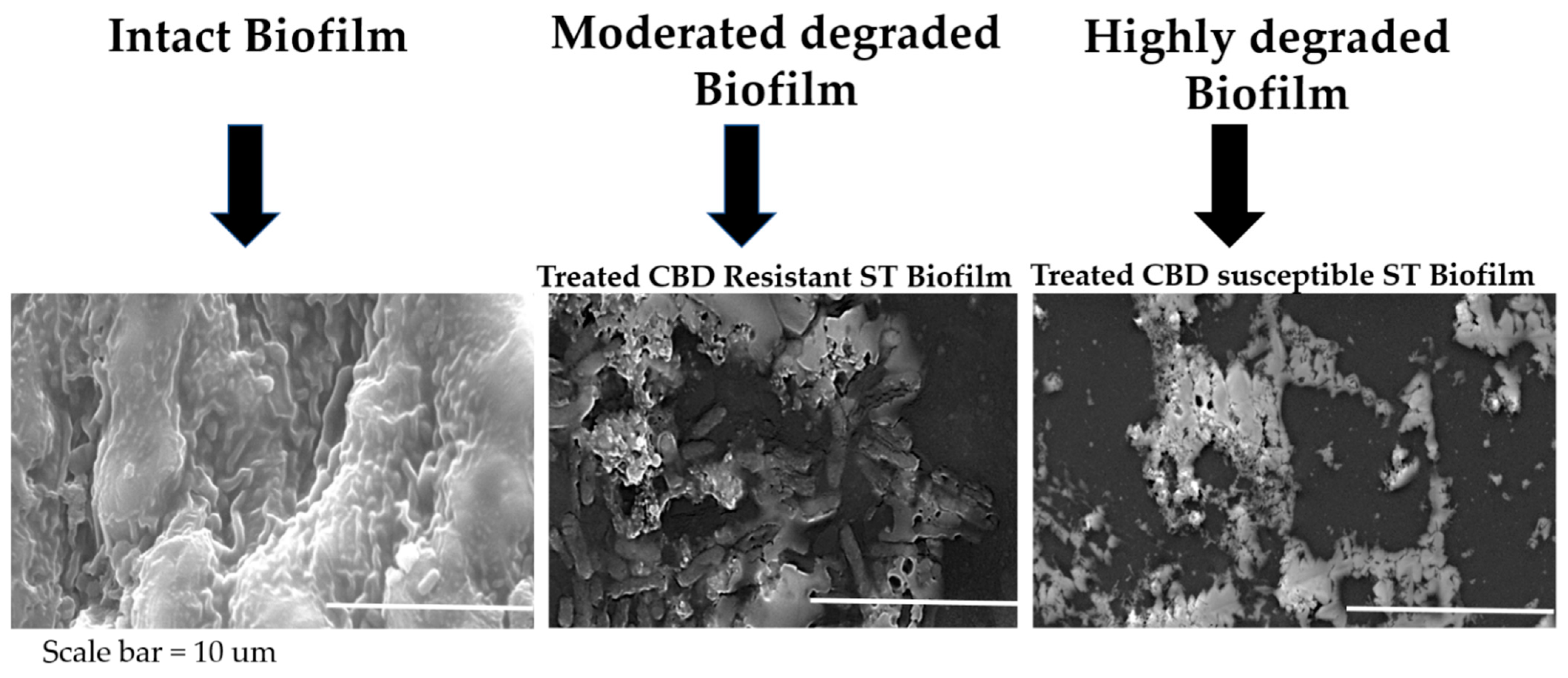
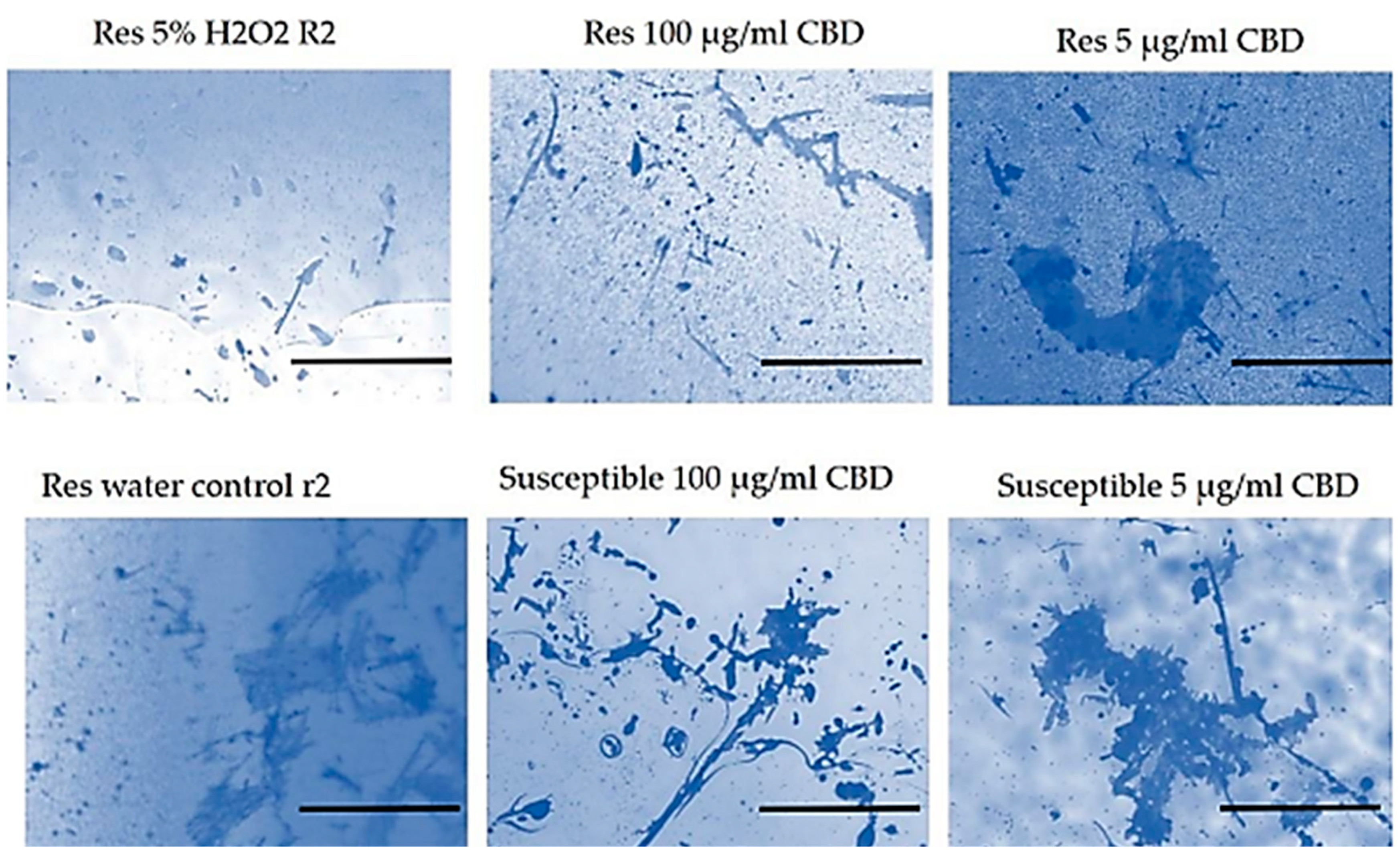
Scanning electron microscopy revealed that CBD-resistant strains formed more robust biofilms than CBD-susceptible strains. Under CBD treatment, the resistant strains showed less biofilm degradation as compared to the intact biofilm, suggesting that CBD resistance may enhance the bacteria's ability to form and maintain biofilms, a critical factor in bacterial survival and infection persistence, as indicated in
Figure 6.
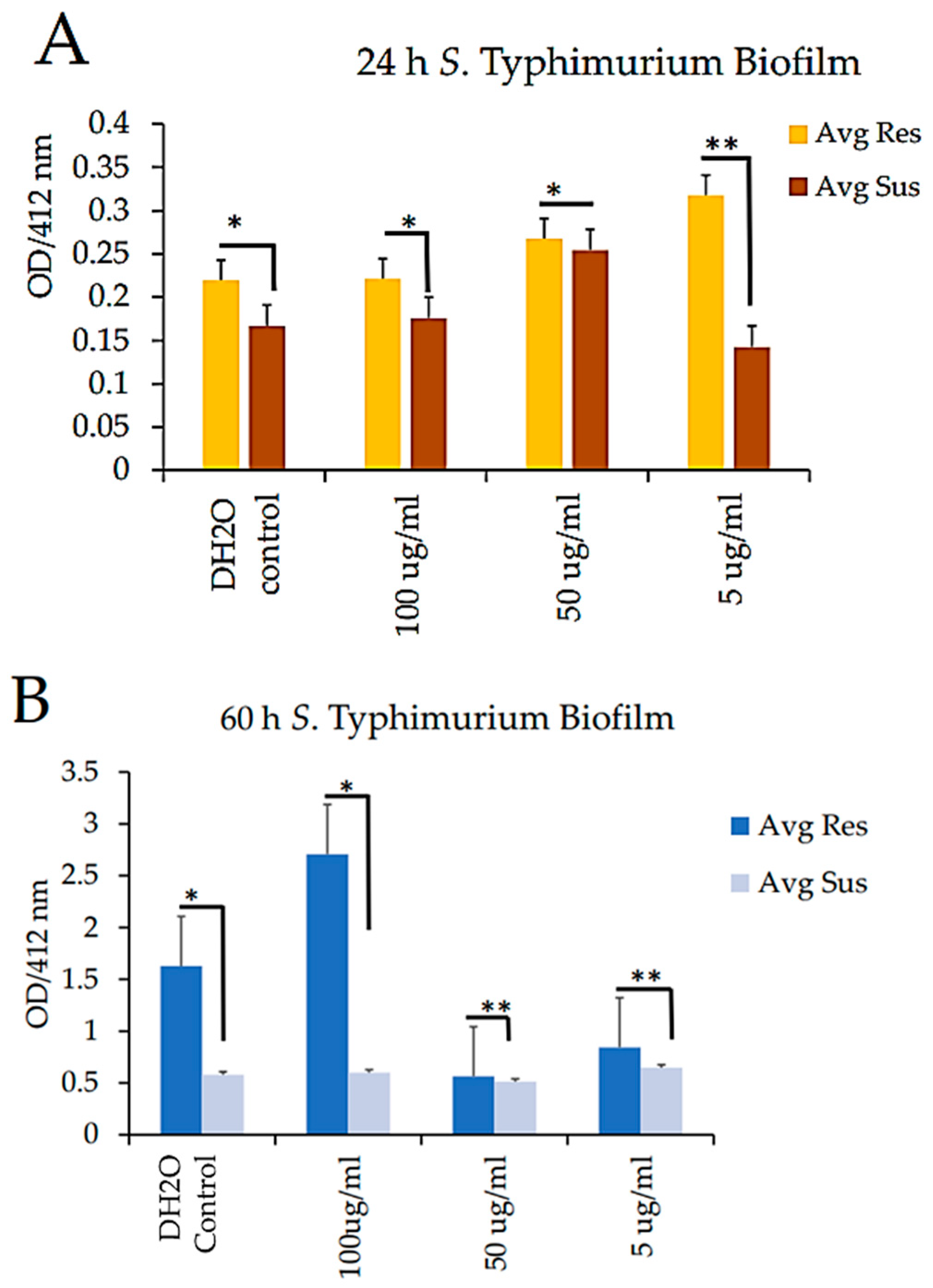
As shown in
Figure 6, we incubated both strains for 24 h and 60 h for the quantitative assessment of biofilm formation. Following incubation, the CBD-resistant strains exhibited significant growth for 24 h and 60 h for the different CBD concentrations tested. However, there were no significant differences in the growth rates at 60 h for the 50 µg/ml and 5 µg/ml for both the 24h and 60 h incubation, as shown in
Figure 7A and B.
Microphotographs of crystal violet stained biofilms indicated that the CBD-resistant strains had less damaged (slightly intact) biofilms, with the 5 µg/ml CDB concentration recording more robust biofilm formation than the 100 µg/ml concentrations. The CBD-susceptible strains recorded damaged/degraded biofilm with the 5 µg/ml CBD-concentration degraded more than the 100 µg/ml concentrations compared to the control group where deionized water was used, as shown in
Figure 8. This suggests that the CBD concentrations used in the biofilm analysis had resulted in degrading biofilm formed by the CBD-susceptible strains better than the CBD-resistant strains.
Figure 8.
SEM Analysis of Salmonella Typhimurium biofilm under CBD treatment and untreated control.
Figure 8.
SEM Analysis of Salmonella Typhimurium biofilm under CBD treatment and untreated control.
Figure 9.
Quantitative analysis of biofilm formation in both resistant and susceptible S. Typhimurium after A. 24 h incubation and B. 60 h incubation. * = Not significant (
Figure 9A), ** p-value < 0.05 ((
Figure 9A), * p-value < 0.01 (
Figure 9B), ** = Not significant (
Figure 9B).
Figure 9.
Quantitative analysis of biofilm formation in both resistant and susceptible S. Typhimurium after A. 24 h incubation and B. 60 h incubation. * = Not significant (
Figure 9A), ** p-value < 0.05 ((
Figure 9A), * p-value < 0.01 (
Figure 9B), ** = Not significant (
Figure 9B).
Figure 10.
Microphotographs of crystal violet stained biofilms of S. Typhimurium formed under different treatments. Scale bar = 100 µm.
Figure 10.
Microphotographs of crystal violet stained biofilms of S. Typhimurium formed under different treatments. Scale bar = 100 µm.
4. Discussion
The flag gene plays an essential role in the assembly and function of bacteria flagella, which is crucial for bacterial motility. This gene encodes the flgA protein, which is critical for assembling the P-ring of the flagellar basal body in
Salmonella [
17]. The P-ring functions in the stabilization of the flagellum by holding it firm to the bacteria cell wall. FlgA is crucial for adequate flagellar formation. The P-ring cannot be formed without flgA, leading to improper flagellar assembly and impaired motility [
18]. The flgA gene is located within the flagellar operon, with its expression being coordinated with other flagellar genes to enhance proper flagellar synthesis. Bacteria motility is paramount to allowing bacteria to move towards nutrient-rich environments (chemotaxis), which is vital for bacteria proliferation. Bacteria evade hostile environments such as antibiotics, toxins, and unfavorable conditions with motility [
19]. This increases their chances of survival. Motility is especially critical for pathogenic bacteria, as their colonization of host tissues and their establishment of infections is by maneuvering through the mucus and tissue barriers.
Additionally, flagella enhance bacteria's ability to aggregate and form biofilm, enabling them to withstand hostile environments and contributing to bacteria resistance development [
20]. It also supports the exchange of genes through conjugation, spreading traits such as resistance genes. Finally, it enhances bacterial survival, adaptation, and growth, enabling their ecological and evolutionary success [
21].
In this study, our primary aim was to assess how prolonged exposure of Salmonella Typhimurium to CBD influences flgA gene expression. Motility assays, pH induction, nutrient starvation, biofilm assays and mRNA expression were employed to achieve this aim.
Our analysis demonstrates that CBD-resistant strains of
Salmonella Typhimurium exhibited significantly higher levels of flgA mRNA expression than the CBD-susceptible strains, as shown in
Figure 3. This elevated expression suggests active transcription of the flgA gene in CBD-resistant strains, potentially playing a crucial role in the bacteria's survival and growth. The upregulation of the flgA gene may be associated with enhanced motility, adherence, and overall bacterial fitness, which are vital for Salmonella's survival, growth, and virulence. In a study conducted by Smith et al. [
22] to elucidate gene expression profiles in antibiotic-resistant
Salmonella Typhimurium, they recorded a 3-fold increase in flgA gene expression in CBD-resistant
Salmonella strains compared to the susceptible strains. Their findings suggest that flgA gene plays a paramount role in resistance development, possibly through increased motility and biofilm formation synonymous to our observations, where robust biofilm formation was associated with the CBD-resistant strains as compared to the highly degraded biofilms associated with the CBD-susceptible strains indicated in
Figure 8. Another study was carried out by Johnson et al. [
23] to stipulate the essence of the flgA gene in
Salmonella pathogenicity. In similar fashion to our reports, they found higher expression of flgA genes, which enhances rapid colonization and evasion of the host immune system. These findings suggest a strong correlation between the flagella gene and bacteria growth and survival. A 2.5-fold increase in gene expression was reported in a transcriptomic analysis study involving resistant and susceptible strains of
Salmonella by Lee et al. [
24]. Their findings demonstrated striking similarities to our observation, emphasizing the flgA gene's dynamic role in enhancing
Salmonella's adaptability to antibiotic stress. Furthermore, Jones et al. [
25] and Davis and Kumar [
26] explored several genetic routes that enhance resistance development. The latter studies focused on the significance of motility genes, reporting a 4-fold increase in flgA gene expression in the resistant strains, while the former focused on the regulatory mechanisms of flagella gene expression in resistant strains of
Salmonella, recording a 3.5-fold increase in flgA gene expression. Jones et al., [
25] attributed the increased expression to enhanced biofilm formation and survival in harsh environments, a finding that is similar to our observation when we carried a comparative biofilm analysis of the CBD-resistant and CBD-susceptible strains of
Salmonella Typhimurium as shown in
Figure 8. Davis and Kumar [
27] attributed their findings to flgA gene upregulation and concluded that this gene may be responsible for
Salmonella pathogenicity and virulence. Our study demonstrates a consistent pattern of the CBD-resistant strains exhibiting higher motility in all the time points as compared to the CBD-susceptible strains, as shown in
Figure 4B, suggesting that there is more transcription of flgA in the CBD-resistant strain than the in the CBD-susceptible strain. This indicates that the flgA gene plays an enormous role in resistance development mechanisms because of the gene involvement in
Salmonella Typhimurium survival, growth, and pathogenicity under duress. Thompson et al. analyzed the significance of flgA in
Salmonella pathogenicity [
28]. They found a higher expression of flgA genes in the resistant strains than in the susceptible strains, enhancing their better evasion and colonization of the host immune system. Similar findings were reported by Chen et al. [
29], Kim et al. [
30], and Nguyen et al. [
31].
pH plays a multifaceted role in the survival, proliferation, virulence, and the ability of pathogenic bacteria to cause infections. Bacteria exhibit a range of tolerance to pH for their growth and survival. Deviation from this optimal range (primarily pH 6-7) harms the survival and growth of most pathogens [
32]. However, several pathogenic bacteria have evolved and developed strategies to withstand an increase in basic and acidic environments, reminiscent of the stomach's acidity, and express virulence factors in that respect. Bacterial colonization of host tissues and the ability to form biofilm, both critical for establishing chronic infections, are partly influenced by pH. Understanding pH-orchestrated associations is essential for understanding the mode of operandi of bacterial pathogenesis and developing therapeutic strategies [
33].
The observations in
Figure 4A underscore the role of pH levels and the development of resistance in both CBD-resistant and CBD-susceptible
Salmonella Typhimurium. At pH 3, there was no growth for both CBD-resistant and CBD-susceptible strains. This observation may insinuate that extreme pH negatively influences bacterial survival and growth. These findings are similar to those of Smith et al. [
22], who reported that extreme pH levels (both basic and acidic) significantly inhibited bacterial growth and antibiotic resistance.
Additionally, Johnson et al. [
23] reported in their study that
Salmonella strains exhibited different resistance levels based on the pH of the medium, showing that the resistant strains exhibited minimal inhibition at pH 7. Their findings are consistent with those in our observation, where both CBD-resistant and CBD-susceptible strains were not significantly inhibited at pH 7, as shown in
Figure 3A, suggesting that neutral pH conditions are favorable for the proliferation of the CBD-resistant and CBD-susceptible strains. Lee et al. [
24] reported in their inhibition study that their observation of clear zones indicated areas that prevent bacterial growth due to antibiotics or other unfavorable conditions. Our findings align with this, where pH 12 significantly inhibited the growth of both strains of
Salmonella Typhimurium (
Figure 4A and
Figure 4D). David and Kumar [
34] reported that certain antibiotics' effectiveness depends on the host system's pH and that certain antibiotics were especially less effective at extreme pH, as we have observed at pH 3 in our study (
Figure 5A and
Figure 5C).
Furthermore, a comprehensive analysis of bacterial growth under varied environmental conditions was undertaken by Martin et al. [
35]. The varied environmental conditions considered in their study were pH and the presence of antibiotics. Similar to our findings, bacterial growth varied with pH, indicating the role of pH in regulating antibiotic resistance. Studies conducted in 2018 by Miller et al. [
36] showed that extreme pH levels can inhibit bacterial growth significantly, as we have observed with the trends in
Figure 5A-D, where extreme pH (3 and 12) significantly reduced the growth of both CBD-resistant and CBD-susceptible strains of
Salmonella Typhimurium [
37]. Similar studies show that resistance development can impact proliferation dynamics, explaining the variations seen in
Figure 5A and
Figure 5C between the CBD-resistant and CBD-susceptible strains [
38]. CBD-resistance development has potentially affected the growth rates of the susceptible strains differently than the resistant strains.
Salmonella Typhimurium shows consistent and robust growth in neutral and near neutral pH (
Figure 5A and
Figure 5C), where both CBD-resistant and CBD-susceptible strains are experiencing massive proliferation. However, though the CBD-susceptible strains edged slightly over the CBD-resistant strain at the neutral pH, this observation differed from pH 5, where the CBD-resistant strains outperformed the CBD-susceptible strains. This observation implies that the CBD resistance acquired enhances
Salmonella Typhimurium's ability to withstand a slightly hostile environment compared to the susceptible strain. Our findings further suggest that extreme pHs (both acidic and basic) have a deleterious impact on the survival of both the resistant and the susceptible strains of Salmonella Typhimurium, as shown in
Figure 5C and
Figure 5D. Notably, in
Figure 5C and
Figure 5D, it is observed that the CBD-resistant strain curves have declined more than the CBD-susceptible strains. This signifies the sensitivity of
Salmonella Typhimurium to extreme pH, which impacts its growth kinetics [
39]. While several studies reported that resistant bacterial strains often thrive better in hostile environments, our findings contradict this notion, as we observed a more significant decline in growth in the CBD-resistant strain compared to the CBD-susceptible strains. However, the growth of both strains was severely impacted at pH 3 and pH 12 (
Figure 5B and
Figure 5D).
Bacteria metabolize nutrients via glycolysis and the TCA cycle, generating ATP that fuels cellular activities such as flagellar motility. Nutrients play several crucial roles, including the synthesis of flagella, by utilizing the components (lipids, sugars, and proteins) needed to form flagella and its functionality [
40] properly. Additionally, the sustenance and upkeep of cellular machinery require nutrients to influence motility, such as membrane transporters and motor proteins, to maintain cellular integrity and functionality. Nutrients are paramount to chemotaxis, acting as a compass for bacteria movement in nutrient-rich environments, enhancing movement towards suitable conditions [
41]. Furthermore, nutrients are intricately linked to motility, providing the energy and resources required for flagellar rotation and motility. Another critical factor influenced by nutrients is biofilm formation [
42].
In this study, we have comprehensively analyzed the growth kinetics of both CBD-resistant and CBD-susceptible strains of
Salmonella Typhimurium under nutrient starvation and varying CBD concentrations as presented in
Figure 6A-D over a 20 h period. This is to measure the influence of nutrient starvation and CBD concentration on the growth of
Salmonella Typhimurium.
Figure 6A represents the growth kinetics of CBD-resistant and CBD-susceptible strains under nutrient starvation without CBD. Both strains recorded a decline in growth, with the resistant strains showing a slow decline over time. This aligns with the findings of Miller et al. [
36] that nutrient starvation hampers bacteria growth kinetics. While nutrient starvation showed a deleterious impact on the growth and survival of both strains in all treatment conditions, the CBD-susceptible strains generally exhibited a level of tolerance to nutrient starvation better than the CBD-resistant strains, as seen in
Figure 6A-D. Except for the control (
Figure 6A), where both bacteria growth declined at almost the same rate, the growth decline for the CBD-susceptible strains was different when CBD in varied doses was applied to the cells. Thus, in the presence of CBD, the susceptible strains could metabolize and use CBD for growth, even though their growth was short-lived.
On the contrary, while we expected CBD-resistant strains to have withstood nutrient starvation, they declined rapidly compared to susceptible strains. This observation was not expected but aligns with scientific reports. In
Figure 3, we showed that the flgA gene was expressed in the CBD-resistant strains almost six times that of the CBS-susceptible strains. The flgA gene is responsible for assembling the basal body of the flagella, and the process requires a substantial amount of energy. The energy requirement of flagella in bacteria is influenced by the proton motive force (PMF) generated by the electron transport chain (ETC) [
43]. As protons fall back into the cell via the motor protein complex in the flagellar basal body, rotational energy drives flagellar movement. Furthermore, ATP hydrolysis contributes to flagellar movement, especially in twitching or gliding motility. However, nutrient starvation can hinder flagellar movement by limiting the availability of ATP and proton motive force, thereby impairing bacterial motility and navigation towards nutrients. Since flagella synthesis is an energy-intensive process [
43], the CBD-resistant strains possibly could not operate in the least without any form of energy. In addition, while the CBD-susceptible strains might have utilized CBD as an energy source, the CBD-resistant strains did not have any room for CBD utility. This is because they have already developed resistance to CBD and hence might have used its efflux pumps that pump out CBD automatically. In our previous study, where we analyzed the mechanisms of
Salmonella Typhimurium resistance to CBD, we noticed an upregulation of the ompC (outer membrane protein C) gene, which may be responsible for depriving the CBD-resistant strains the temporal benefit of utilizing CBD for energy as enjoyed by the CBD-susceptible strains [
15].
In the presence of 5 µg/mL CBD under nutrient starvation, both strains exhibited a significant decrease in growth, with the susceptible strains experiencing a more moderate decline (thus, the susceptible strains tolerated the nutrient starvation and grew to almost 5% during the first 6 h of incubation)
Figure 6B. The CBD-resistant strains demonstrate instant decline. This death trajectory was observed in all the different CBD concentrations employed. This unexpected observation suggests that CBD-susceptible strains are more adaptive to nutrient starvation than their CBD-resistant counterparts. This observation sharply contrasts Thompson et al. who reported that resistant strains often perform better under nutrient starvation due to enhanced survival mechanisms under hostile situations [
44].
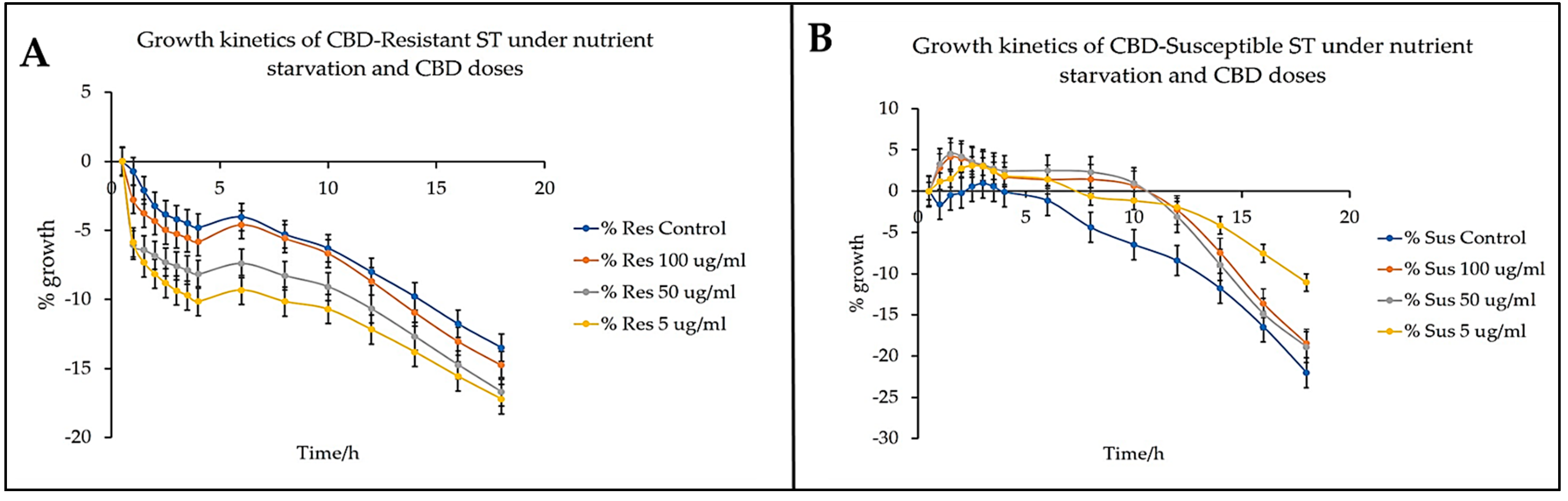
Similar to
Figure 6,
Figure 7 provides a comprehensive analysis of the growth kinetics of CBD-resistant and CBD-susceptible strains of
Salmonella Typhimurium under nutrient starvation and varying doses of CBD. This investigation aimed to understand the growth behavior of the strains under nutrient starvation and varied CBD concentrations. This knowledge is paramount in the development of targeted antimicrobial strategies. The findings suggest the essence of delving into the resistance pathways and adaptive mechanisms to understand the intricacies of bacterial infection and methods to develop a robust intervention against them [
45,
46]. Contrary to the results shown in
Figure 7B, the untreated control for the susceptible strains produced the sharpest decline in growth, dipping into the negative quadrant in the first three and half hours, and continued to die exponentially. The CBD-susceptible strains' 5 µg/mL treatment group staggered in the positive quadrant for close to seven hours before dipping into the negative quadrant. The 50 µg/mL and 100 µg/mL treatment groups produced resilient growth curves. Both stayed in the positive quadrant until after the tenth hour before dipping into the negative quadrant. The inference obtained from these observations is that treatment of the bacteria to CBD upon nutrient starvation substantially affected the resistant strains due to several reasons, among which might include the following: the resistant strains demonstrated a 6-fold higher expression of flgA gene, indicating a 6-fold utilization of cellular energetics for transcriptional and translational mechanisms targeting the biosynthesis of flagella. In nutrient-deficient conditions, such processes are unsustainable, leading to the death of the bacteria. The CBD-susceptible strains, however, seem to develop the ability to utilize CBD as a nutrient source. This observation is supported by the growth curves recorded in
Figure 6 and 7 as treatment to CBD extended the growth of CBD-susceptible strains longer than the untreated control.
Additionally, the CBD-resistant strains showed higher motility compared to the CBD-susceptible strains. The mechanized movement of the flagellum requires a considerable expenditure of cellular ATPs. Nutrient starvation provides a breakdown wherein there are little to no nutrients available. Thus, the strains have no energy source to replenish any ATP used. This situation leads to higher deaths from bacteria. Furthermore, CBD-resistant strains have been demonstrated elsewhere to develop efflux pumps that continuously pump CBD out of the intracellular compartments of the bacteria in its environment. This mechanism requires ATP utilization as well as the complete exclusion of the adoption of CBD as a nutrient source; this, therefore, pushes more CBD-resistant strains to die rapidly.