Submitted:
30 May 2024
Posted:
30 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Experimental Animal Design
2.3. Astaxanthin-Rich Dietary Supplement
2.4. Growth Performance
2.5. Tissue Sample Collection
2.6. Total RNA extraction and cDNA preparation
2.7. Bioinformatics: Genome Assembly and Gene Primer Design
2.8. Quantitative Real-Time RT-PCR (qPCR)
2.9. Gene Ontology
2.10. Tissue Histology Using Hematoxylin and Eosin Staining
2.11. Statistical Analysis
3. Results
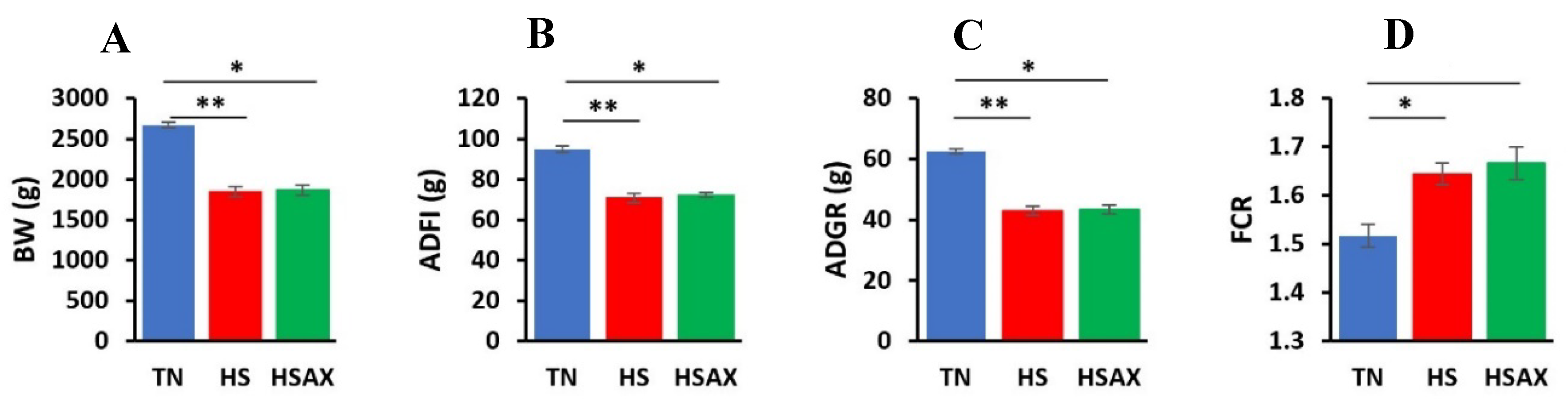
3.1. Growth Performance
3.2. Quantitative Real-time RT-PCR (qPCR) Gene Expression
3.2.1. Statistically Significant Genes Identified in Differential Expression Analysis
| Kruskal-Wallis | Dunn/Bonferroni p-value | |||
| Gene | p-value | TN vs HS | TN vs HSAX | HS vs HSAX |
| HSF1 | 0.1495 | ns | ns | ns |
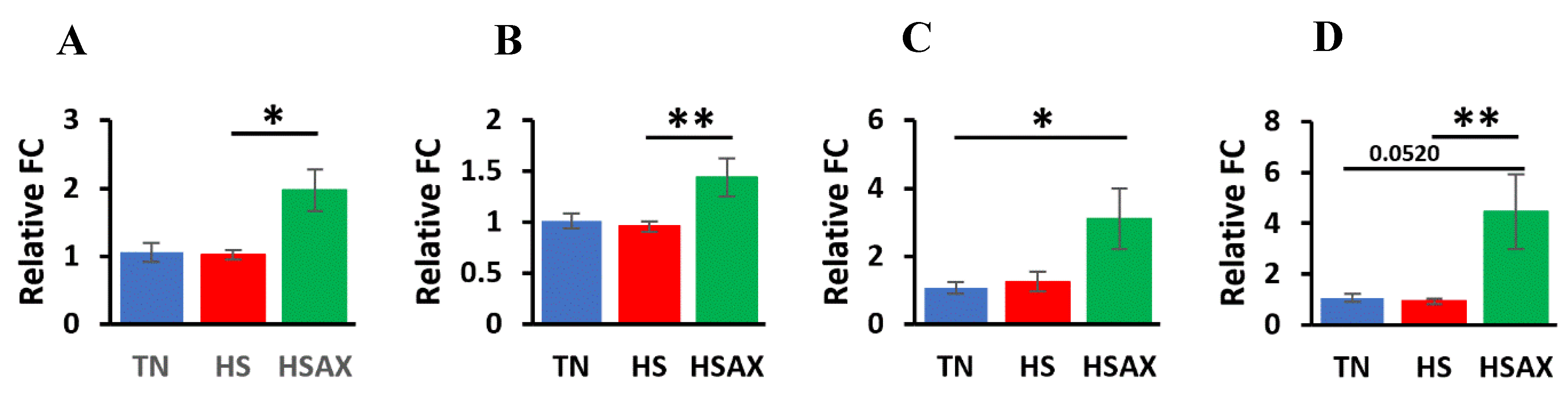
| HSF2 | 0.0222 | ns | ns | 0.0174 |
| HSF3 | 0.2199 | ns | ns | ns |
| HSP90 | 0.1299 | ns | ns | ns |
| HIFIA | 0.8948 | ns | ns | ns |
| SOD1 | 0.1990 | ns | ns | ns |
| SOD2 | 0.0030 | 0.0916 | ns | 0.0024 |
| ROS-1 | 0.3722 | ns | ns | ns |
| GPX1 | 0.0745 | ns | ns | ns |
| GPX3 | 0.0274 | ns | 0.0331 | ns |
| HMOX1 | 0.8490 | ns | ns | ns |
| CAT | 0.8342 | ns | ns | ns |
| CASPAS3 | 0.1299 | ns | ns | ns |
| GSTTI | 0.5225 | ns | ns | ns |
| TXN | 0.0018 | ns | 0.0520 | 0.0016 |
| TXN2 | 0.1034 | ns | ns | ns |
| PRDX1 | 0.0515 | ns | ns | ns |
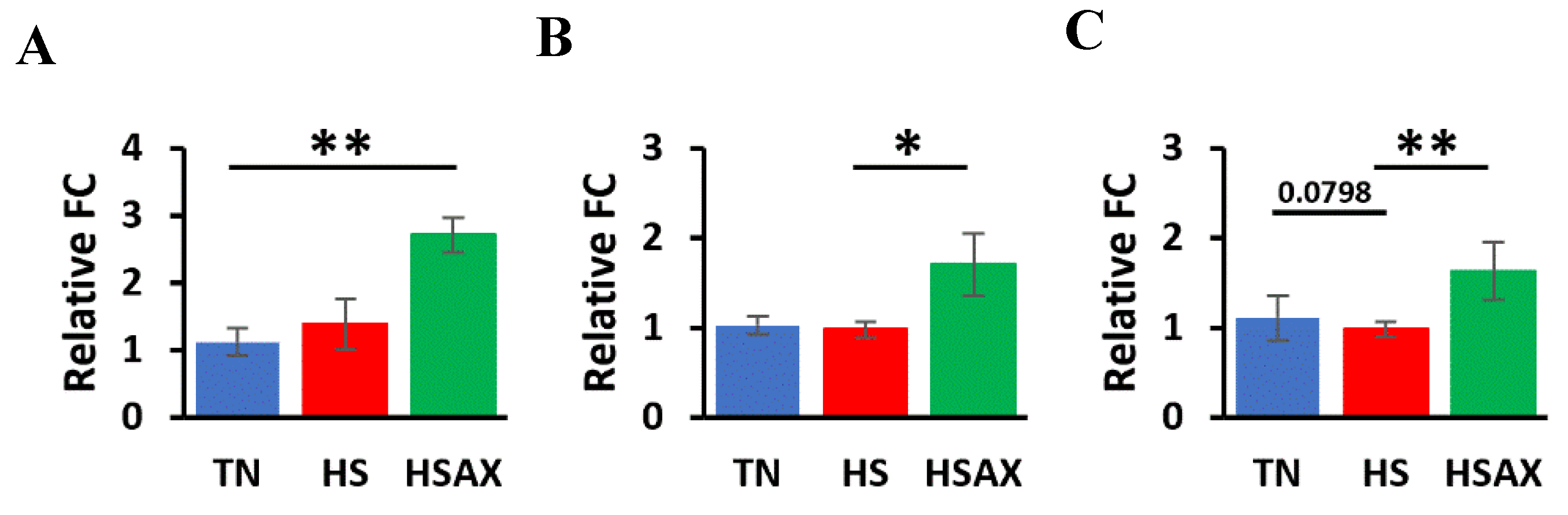
| LOX | 0.0131 | ns | 0.0105 | ns |
| MYB | 0.0589 | ns | ns | ns |
| IL-4 | 0.1169 | ns | ns | ns |
| CLDN1 | 0.0214 | ns | ns | 0.0174 |
| OCLN | 0.3679 | ns | ns | ns |
| MUC2 | 0.0035 | 0.0798 | ns | 0.0029 |
| CDH1 | 0.5671 | ns | ns | ns |
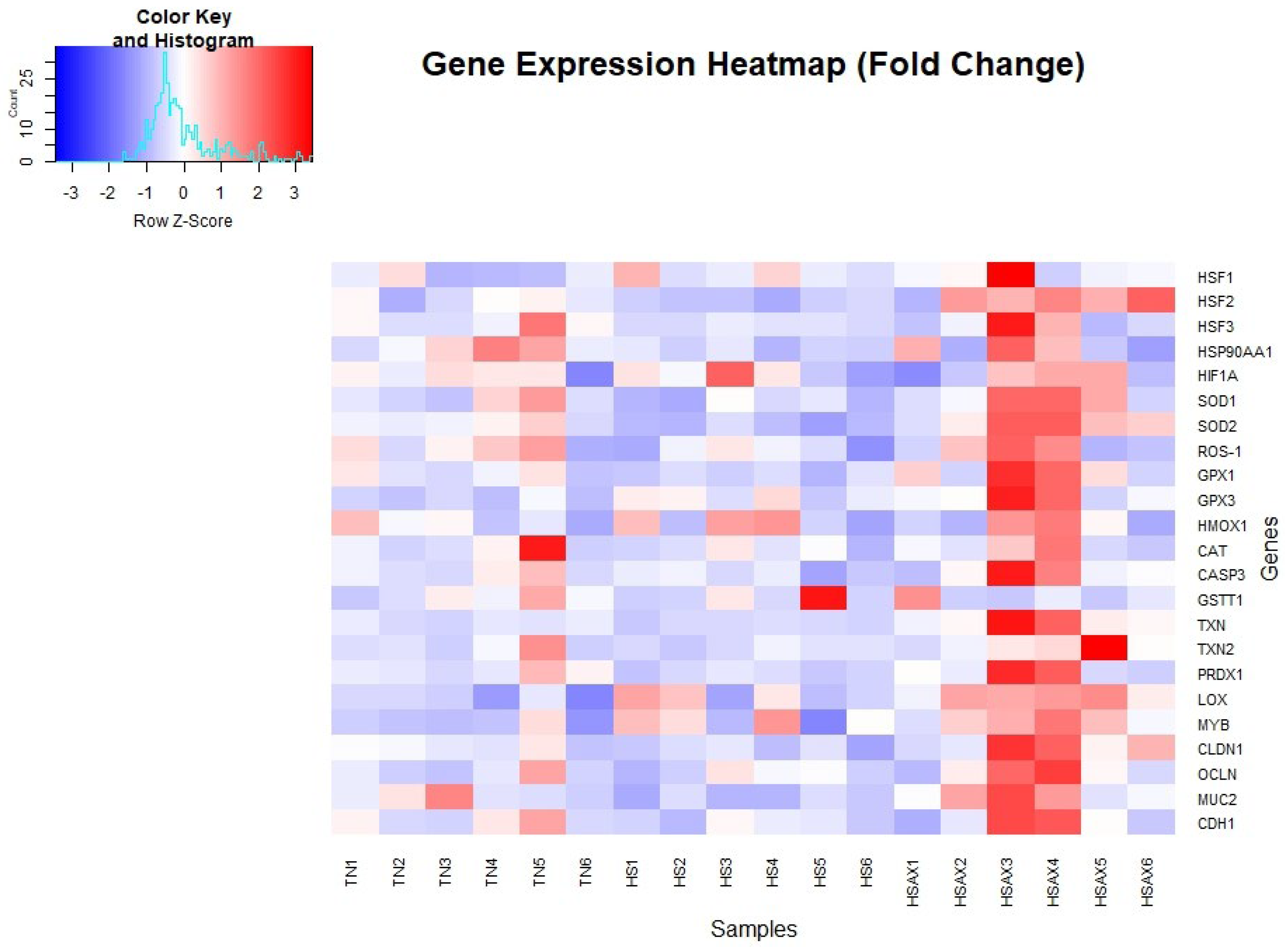
3.2.2. Cytoprotective Capacity Genes
3.2.3. Epithelial Integrity Genes
3.2.4. Gene Ontology (GO) Gene Enrichment
| Gene | Cellular component | Molecular function |
Biological process |
Transcript IDs |
| HSF2 (heat shock transcription factor 2) |
nucleus, cytoplasm | DNA-binding transcription factor activity, DNA-binding transcription activator activity, RNA polymerase II-specific, RNA polymerase II cis-regulatory region sequence-specific DNA binding | cellular response to heat, positive regulation of transcription from RNA polymerase II promoter in response to heat stress, regulation of transcription, DNA-templated | ENST00000368455 ENST00000452194 |
| SOD2 (superoxide dismutase 2, mitochondrial) | mitochondrion | superoxide dismutase activity, oxidoreductase activity, manganese ion binding, metal ion binding, identical protein binding | response to oxidative stress, oxidation-reduction process, negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway, response to hydrogen peroxide, removal of superoxide radicals | ENSGALT00000019062 |
| GPX3 (glutathione peroxidase 3) | extracellular space | glutathione peroxidase activity, selenium binding, identical protein binding | response to oxidative stress, hydrogen peroxide catabolic process | ENSGALG00010016480 |
| TXN (thioredoxin) | nucleus, cytosol, extracellular region | protein disulfide oxidoreductase activity | oxidation-reduction process, cell redox homeostasis, negative regulation of hydrogen peroxide-induced cell death, positive regulation of peptidyl-serine phosphorylation, positive regulation of peptidyl-cysteine S-nitrosylation, negative regulation of transcription by RNA polymerase II, positive regulation of DNA binding | ENSGALT00000025326 |
| LOX (lysyl oxidase) |
extracellular space | protein-lysine 6-oxidase activity, copper ion binding, collagen binding | peptidyl-lysine oxidation, collagen fibril organization | https://www.ncbi.nlm.nih. gov/gene/396474 |
| CLDN1 (claudin 1) | cell junction, bicellular tight junction, integral component of membrane, cytoplasm | structural molecule activity | bicellular tight junction assembly, cell adhesion, regulation of ion transport | ENSGALT00000077095 ENSGALT00000030190 |
| MUC2 (mucin 2) |
extracellular matrix, intracellular membrane-bounded organelle, supramolecular fiber | protein binding, virion binding, antigen binding | cholesterol homeostasis, intestinal cholesterol absorption, maintenance of gastrointestinal epithelium, negative regulation of cell growth, macrophage activation involved in immune response | https://www.ncbi.nlm.nih. gov/gene/414878 |
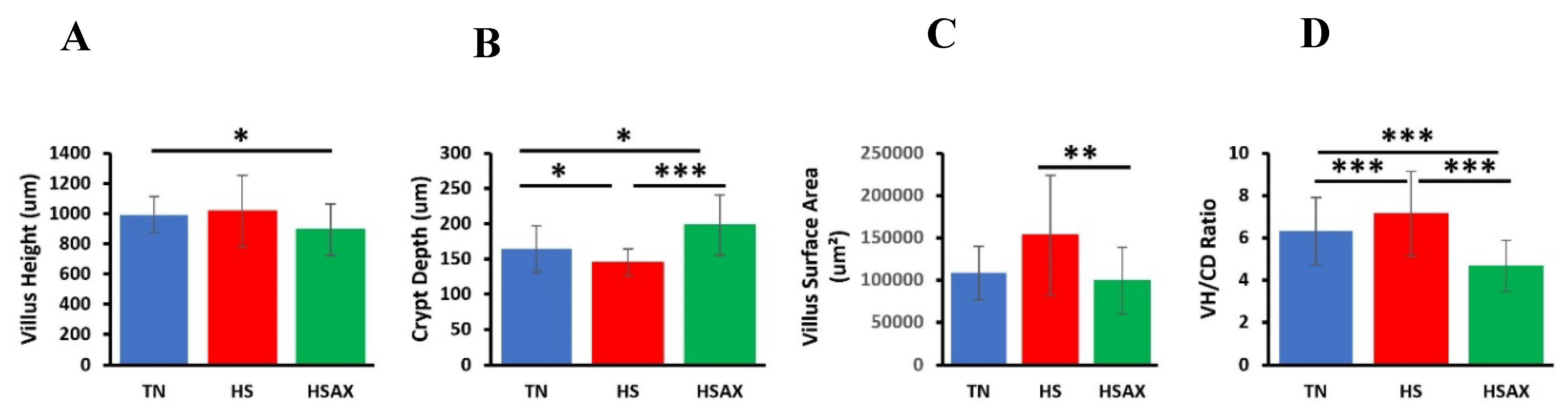
3.3. Ileum Histomorphology
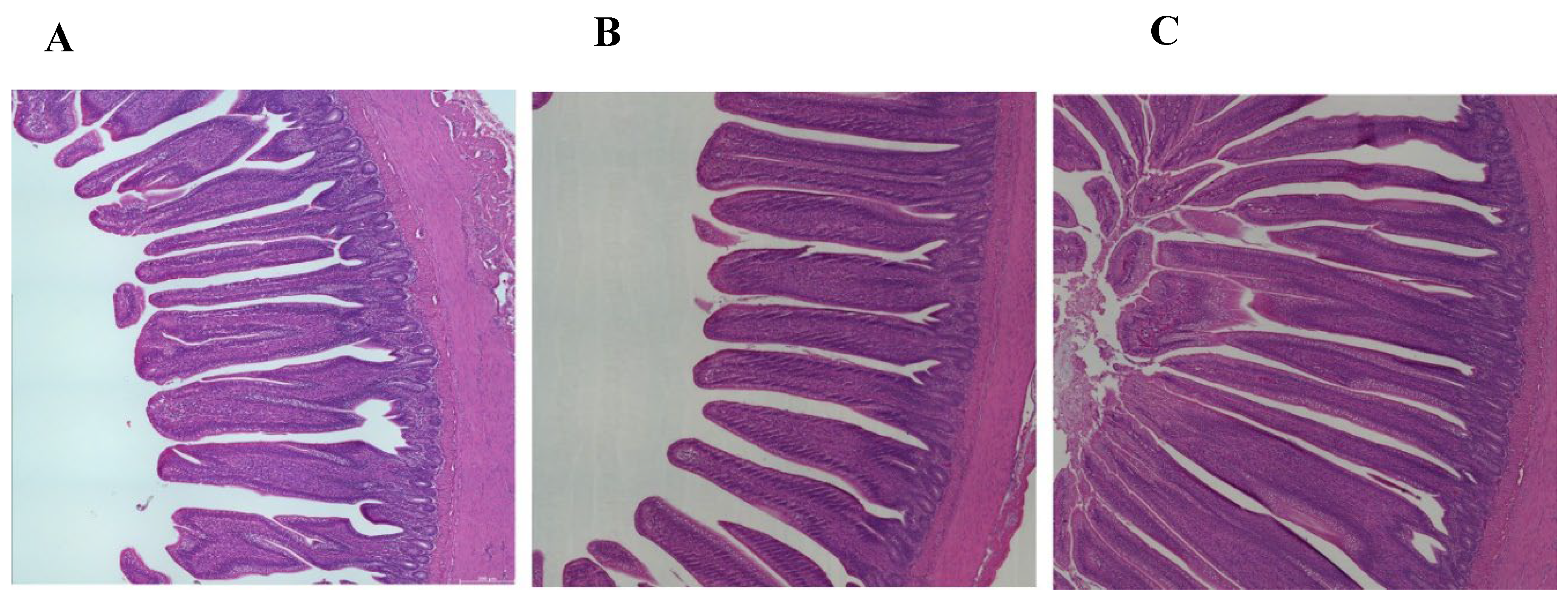
| Kruskal-Wallis |
Dunn/Bonferroni p-value |
||||||
| Measurements | TN | HS | HSAX | p-value | TN vs HS | TN vs HSAX | HS vs HSAX |
| VH (μm) | 992.79 ± 118.97 |
1061.89 ± 235.73 | 893.08 ± 170.00 | 0.0471 | 1 | 0.0552 | 0.2046 |
| CD (μm) |
164.28 ± 32.67 |
145.08 ± 19.09 |
197.87 ± 42.86 |
2.05e-06 | 3.77e-02 | 2.63e-02 | 9.30e-07 |
| VSA (μm) |
108602.30 ± 31508.84 |
152946.60 ± 70507.57 |
99497.84 ± 39296.00 |
0.0050 | 0.0995 | 0.8640 | 0.0042 |
| VH:CD (μm2) |
6.31 ± 1.59 |
7.13 ± 2.00 |
4.67 ± 1.21 |
2.39e-07 | 3.59e-01 | 4.17e-04 | 2.41e-07 |
4. Discussion
4.1. Growth Performance
4.2. Gene Ontology Enrichment and Expression Analysis
4.2.1. Cytoprotective Capacity Genes
4.2.2. Epithelial Integrity Genes
4.3. Ileum Histomorphology
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Ricke, S. C. Improving gut health in poultry. 1st Edition 2019. Burleigh Dodds Science Publishing. University of Arkansas, USA. [CrossRef]
- Bailey, Richard A. “Gut Health in Poultry: The World within - Update | The ...” Gut Health in Poultry:the World within-Update, 1 Oct. 2019, 8:30am, thepoultrysite.com/articles/gut-health-in-poultry-the-world-within-1.
- Glendinning, L., Stewart, R. D., Pallen, M. J., Watson, K.A. “Assembly of Hundreds of Novel Bacterial Genomes from the Chicken Caecum.” Genome Biology, 2019. [CrossRef]
- Kers, Jannigje G.; et al. Host and Environmental Factors Affecting the Intestinal Microbiota in Chickens. Frontiers in Microbiology, vol. 9, 2018. [CrossRef]
- Carrasco, Juan M. Diaz; et al. Microbiota, Gut Health and Chicken Productivity: What Is the Connection? Microorganisms, vol. 7, no. 10, 2019, p. 374.. [CrossRef]
- Surai, P.F., and V.I. Fisinin. Vitagenes in Poultry Production: Part 3. Vitagene Concept Development. World’s Poultry Science Journal, vol. 72, no. 4, 1 Dec. 2016, pp. 793–804. [CrossRef]
- Farag, M. R., Alagawany, M. Physiological Alterations of Poultry to the High Environmental Temperature. Journal of Thermal Biology, vol. 76, 2018, pp. 101–106.. [CrossRef]
- Lara, L., Rostagno, M. Impact of Heat Stress on Poultry Production. Animals, vol. 3, no. 2, 24 Apr. 2013, pp. 356–369, www.ncbi.nlm.nih.gov/pmc/articles/PMC4494392/. [CrossRef]
- Collier, R. J., & Gebremedhin, K. G. Thermal biology of domestic animals. Annual Review of Animal Biosciences, 2015, 3, 513–532. [CrossRef]
- Sies, H. Oxidative stress: Oxidants and antioxidants. Experimental Physiology, 1997, 82, 291–295.
- Belhadj Slimen, I., Najar, T., Ghram, A., & Abdrrabba, M. Heat stress effects on livestock: Molecular, cellular and metabolic aspects, a review. Journal of Animal Physiology and Animal Nutrition, 2016, 100(3), 401–412. [CrossRef]
- Cano-Europa, E. et al. Regulation of the redox environment. Intech, 2015. https://www.intechopen.com/books/basic-principles-and-clinical-significance-of-oxidative-stress/regulation-of-the-redox-environment.
- Montzouris, K.C., Paraskeuas, V.V., Fegeros, K. Priming of intestinal cytoprotective genes and antioxidant capacity by dietary phytogenic inclusion in broilers. Anim Nutr. 2020 Sep;6(3):305-312. [CrossRef]
- Paraskeuas, V., Griela, E., Bouziotis, D., Fegeros, K., Antonissen, G. et al., Effects of Deoxynivalenol and Fumonisins on Broiler Gut Cytoprotective Capacity. Toxins (Basel). 2021 Oct 16;13(10):729. .pimd:34679022; PMCID: PMC8538483. [CrossRef]
- Tang, D., Wu, J., Jiao, H., Wang, X., Zhao, J. et al., The development of antioxidant system in the intestinal tract of broiler chickens. Poult Sci. 2019 Feb 1:98(2):664-678. . PMID: 30289502. [CrossRef]
- Surai F, Peter. “Antioxidant Systems in Poultry Biology: Superoxide Dismutase.” Journal of Animal Research and Nutrition, vol. 01, no. 01, 2016. [CrossRef]
- Surai, P.F., and V.I. Fisinin. Vitagenes in Poultry Production: Part 1. Technological and Environmental Stresses. World’s Poultry Science Journal, vol. 72, no. 4, 1 Dec. 2016, pp. 721–734. [CrossRef]
- Surai, P. F., & Fisinin, V. I. Vitagenes in poultry production: Part 3. Vitagene concept development. World’s Poultry Science Journal, 2016, 72(4), 793–804. [CrossRef]
- 19. Liu, Guanhui; et al. Effect of Chronic Cyclic Heat Stress on the Intestinal Morphology, Oxidative Status and Cecal Bacterial Communities in Broilers. Journal of Thermal Biology, vol. 91, July 2020, p. 102619. [CrossRef]
- Morimoto, R.I., Tissieres A., and Georgopoulos, C. The biology of heat shock proteins and molecular chaperones. Cold Spring Harbor, 1994: Cold Spring Harbor Laboratory Press.
- Pirkkala, Lila; et al. Roles of the Heat Shock Transcription Factors in Regulation of the Heat Shock Response and Beyond. The FASEB Journal, vol. 15, no. 7, May 2001, pp. 1118–1131. [CrossRef]
- Vabulas, R. M. et al. Protein folding in the cytoplasm and the heat shock response. Cold Spring Harbor Perspectives in Biology, 2010, 1-19.
- Javid, M. A., Abbas, G., Waqas, M. Y., Basit, M. A., Asif, M., Akhtar, M. S., Masood, S., Saleem, M. U., Qamar, S. H., & Kiani, F. A. Evaluation of comparative effect of feed additive of allium sativum and zingeber officinale on bird growth and histomorphometric characteristics of small intestine in broilers. Revista Brasileira de Ciencia Avicola, 2019, 21(2). [CrossRef]
- Awad, W. A., Ghareeb, K., Abdel-Raheem, S., & Böhm, J. Effects of dietary inclusion of probiotic and synbiotic on growth performance, organ weights, and intestinal histomorphology of broiler chickens. Poultry Science, 2009, 88(1), 49–55. [CrossRef]
- Hsiu Chung Ou; et al. Galectin-3 Aggravates Ox-LDL-Induced Endothelial Dysfunction through LOX-1 Mediated Signaling Pathway. Environmental Toxicology, vol. 34, no. 7, 9 Apr. 2019, pp. 825–835. [CrossRef]
- Laczko, Rozalia, and Katalin Csiszar. Lysyl Oxidase (LOX): Functional Contributions to Signaling Pathways. Biomolecules, vol. 10, no. 8, 22 July 2020, p. 1093. Accessed 22 Aug. 2020. [CrossRef]
- Santos, R. R., Awati, A., Roubos-van den Hil, P. J., van Kempen, T. A. T. G., Tersteeg-Zijderveld, M. H. G. et al. Effects of feed additive blend on broilers challenged with heat stress. Avian Pathology, 2019, 48:6, 582-601. [CrossRef]
- Apalowo, O.O., Minor, R.C., Adetunji, A.O., Ekunseitan, D.A., Fasina, Y.O. Effect of Ginger Root Extract on Intestinal Oxidative Status and Mucosal Morphometrics in Broiler Chickens. Animals 2024, 14, 1084. [CrossRef]
- Akagi, R. Role of Heme Oxygenase in Gastrointestinal. Epithelial Cells. Antioxidants, 2022. [CrossRef]
- Britton, G., Liaaen-Jensen, S., & Pfander, H. Carotenoids. Isolation and analysis. Basel: Birkhäuser Verlag, 1995.
- Shahidi, F., & Barrow, C. J. Marine nutraceuticals and functional foods. Boca Raton: CRC Press, 2008.
- Astaxanthin. (n.d.). Retrieved from https://pubchem.ncbi.nlm.nih.gov/compound/Astaxanthin. Accessed 02 Apr 2024.
- Heaney, R. P. Factors Influencing the Measurement of Bioavailability, Taking Calcium as a Model. The Journal of Nutrition, 2001,131(4). [CrossRef]
- Mularczyk, M., Michalak, I., Marycz, A. Astaxanthin and other Nutrients from Haematococcus pluvialis-Multifunctional Applications. Mar Drugs. 2020 Sep 7;18(9):459. PMID: 32906619; PMCID: PMC7551667. [CrossRef]
- Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods, 2001, 25: 402-408.
- Ensembl.org. https://uswest.ensembl.org/Gallus_gallus/Info/Index Accessed 04 Apr 2024.
- Iji, P.A., Saki, A., Tivey, D.R. Body and intestinal growth of broiler chicks on a commercial starter diet. 1. Intestinal weight and mucosal development. Br Poult Sci. 2001 Sep;42(4):505-13. PMID: 11572627. [CrossRef]
- Schindelin, J., Arganda-Carreras, I., Frise, E.; et al. Fiji an open-source platform for biological-image analysis. Nature Methods, 2019, 9(7): 676-682. PMID 22743772. [CrossRef]
- R Core Team (2023) https://www.R-project.org/ Accessed 04 Apr 2024.
- Akbarian, A.; et al. Association between Heat Stress and Oxidative Stress in Poultry; Mitochondrial Dysfunction and Dietary Interventions with Phytochemicals. Journal of Animal Science and Biotechnology, vol. 7, no. 1, 28 June 2016. [CrossRef]
| Ingredients (%) | Starter | Finisher |
| Corn | 54.86 | 63.14 |
| Soybean Meal | 39.99 | 30.09 |
| Soybean oil | 2.00 | 4.50 |
| Limestone | 1.27 | 0.85 |
| Monocalcium phosphate | 0.75 | 0.50 |
| Lysine (%) | 0.23 | 0.18 |
| Methionine (%) | 0.14 | 0.12 |
| Threonine (%) | 0.20 | 0.16 |
| Sodium chloride (%) | 0.43 | 0.35 |
| Sodium bicarbonate | 0.12 | 0.10 |
| Vitamin-mineral premix | 0.50 | 0.50 |
| Astaxanthin supplement¹ | 0.01 | 0.01 |
| Total | ||
| Calculated analysis | ||
| ME (kcal/kg) | 2909 | 3203 |
| CP (%) | 22.09 | 18.07 |
| Calcium (%) | 0.75 | 0.52 |
| Total Phosphorus (%) | 0.57 | 0.47 |
| digPhosphorous (%) | 0.30 | 0.23 |
| Lysine (%) | 1.39 | 1.10 |
| dig Lysine (%) | 1.25 | 0.99 |
| Methionine (%) | 0.48 | 0.41 |
| dig Methionine (%) | 0.45 | 0.39 |
| Cysteine (%) | 0.43 | 0.38 |
| Threonine (%) | 1.03 | 0.85 |
| dig Threonine (%) | 0.85 | 0.69 |
| Tryptophan (%) | 0.33 | 0.26 |
| Methionine + cysteine (%) | 0.91 | 0.80 |
| Arginine (%) | 1.61 | 1.31 |
| Valine (%) | 1.22 | 1.03 |
| Isoleucine (%) | 0.93 | 0.76 |
| Leucine (%) | 1.89 | 1.63 |
| Neutral detergent fiber | 9.13 | 8.78 |
| Crude fiber | 3.97 | 3.46 |
| Sodium | 0.22 | 0.18 |
| Chloride | 0.30 | 0.25 |
| Choline (mg/kg) | 1419 | 1200 |
| Astaxanthin (mg/kg) | --- | 1.33 |
| ¹Astaxanthin was mixed with the soybean oil and supplemented in the diet during feed mixing. | ||
| Gene | NCBI Accession No. | Primer set (5’-3’) |
|---|---|---|
| GAPDH | X00182 | F:5' -CGCAAGGGCTAGGACGG |
| R:3' -GCGCTCTTGCGGGTACC | ||
| β-ACTIN | NM_205518.1 | F:5' -GAGAAATTGTGCGTGACATCA |
| R:3' -CCTGAACCTCTCATTGCCA | ||
| TBP | NM_205103.1 | F:5' -TAGCCCGATGATGCCGTAT |
| R:3' -GTTCCCTGTGTCGCTTGC | ||
| HSF1 | NM_001305256.1 | F:5’-AAGGAGGTGCTCCCAAAGTA |
| R:3’-TTCTTTATGCTGGACACGCTG | ||
| HSF2 | NM_001167764.2 | F:5’ -TCTTTTTACAAGCTCCGTGC |
| R:3’ -TCCCTTTGTCTCCATTTTGGT | ||
| HSF3 | NM_001305041.1 | F:5’ -TTCAGCGATGTGTTTAACCCT |
| R:3’ -GGAGGTCTTTTGGATCCTCT | ||
| HSP90AA1 | NM_001109785.1 | F:5’ -GATAACGGTGAACCTTTGGG |
| R:3’ -GGGTAGCCAATGAACTGAGA | ||
| HIF1A | XM_015287264.4 | F:5’ - GTCACCGACAAGAAGAGGAT |
| R:3’ -GTCTCTAGCTCACCAGCATC | ||
| SOD1 | NM_205064.1 | F:5' -CAACACAAATGGGTGTACCA |
| R:3' -CTCCCTTTGCAGTCACATTG | ||
| SOD2 | NM_204211.1 | F:5' -CCTTCGCAAACTTCAAGGAG |
| R:3' -AGCAATGGAATGAGACCTGT | ||
| ROS-1 | NM_205257.2 | F:5' -AAACTACAGCTGGTGTTCCC |
| R:3' -CTAAGTTCTCGGCCTTCCAT | ||
| GPX1 | NM_001277853.2 | F:5' -AATTCGGGCACCAGGAGAA |
| R:3' -CTCGAACATGGTGAAGTTGG | ||
| GPX3 | NM_001163232.3 | F:5' -AATTCGGGCACCAGGAGAA |
| R:3' -CTCGAACATGGTGAAGTTGG | ||
| HMOX1 | NM_205344.2 | F:5' -AATCGCATGAAAACAGTCCA |
| R:3' -CACATGGCAAATAAGCCCAC | ||
| CAT | NM_001031215.2 | F:5' -TGGCCAATTATCAGAGGGAC |
| R:3' -CTCGCACCTGAGACACATTA | ||
| CASP3 | NM_204725.1 | F:5' -GGTGGAGGTGGAGGAGC |
| R:3' -TGAGCGTGGTCCATCTTTTA | ||
| GSTT1 | NM_205365.1 | F:5' -AACATCCCGTTCGAGTTCAA |
| R:3' -CACTATTTGATGGCCCTGTG | ||
| TXN | NM_205453.1 | F:5' -GGCAATCTGGCTGATTTTGA |
| R:3' -ACCATGTGGCAGAGAAATCA | ||
| TXN2 | NM_001031410.1 | F:5' -CGATTGAGTACGAGGTGTCA |
| R:3' -CAGAAGAAAACCCCACAAACTT | ||
| PRDX1 | NM_001271932.1 | F:5' -GGTATTGCATACAGGGGTCT |
| R:3' -AGGGTCTCATCAACAGAACG | ||
| LOX | NM_205481.2 | F:5' -TACTTCCAGTACGGTCTGCC |
| R:3' -CTCTAACATCCGCCCGATAA | ||
| MYB | NM_205306.1 | F:5' -AGCATATACAGCAGCGATGA |
| R:3' -TTTCTCATCCTCTTCACGGG | ||
| CLDN1 | NM_001013611.2 | F:5' -CATCACTTCTCCTTCGTCAGC |
| R:3' -GCACAAAGATCTCCCAGGTC | ||
| OCLN | NM_205128.1 | F:5' -CTACAAGCAGGAGTTCGACA |
| R:3' -CTCTGCCACATCCTGGTATT | ||
| MUC2 | NM_001318434.1 | F:5' -TACAGGGAGTTCTCTGTCCA |
| R:3' -TAGGGTGTCTTGACAATCCG | ||
| CDH1 | NM_001039258.2 | F:5' -GAACTTCATCGACGAGAACC |
| R:3' -CGTTGAGGTAGTCGTAGTCC |
| Kruskal-Wallis |
Dunn/Bonferroni p-value |
||||||
| Measurements | TN | HS | HSAX | p-value | TN vs HS | TN vs HSAX | HS vs HSAX |
| BW (g) | 2673.68 ± 35.71 | 1848.85 ± 61.56 | 1867.83 ± 60.82 | 0.0047 | 0.0095 | 0.0278 | ns |
| ADFI (g) | 94.98 ± 1.45 |
70.68 ± 72.24 |
72.24 ± 1.14 |
0.0038 | 0.0047 | 0.0500 | ns |
| ADGR (g) | 62.65 ± 0.86 |
42.99 ± 1.46 |
43.45 ± 1.44 |
0.0047 | 0.0095 | 0.0278 | ns |
| FCR | 1.52 ± 0.02 |
1.64 ± 0.02 |
1.67 ± 0.03 |
0.0049 | 0.0226 | 0.0119 | ns |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





