Submitted:
02 June 2024
Posted:
03 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
1.1. Structural Domains and Classification of WRKY Transcription Factors
| Gene name | Species | Total number | Reference |
|---|---|---|---|
| AtWRKYs | Arabidopsis thaliana | 74 | [29] |
| OsWRKYs | Oryza sativa | 100+ | [30] |
| GmWRKYs | Glycine max | 197 | [31] |
| HvWRKYs | Hordeum vulgare | 45 | [32] |
| CsWRKYs | Cucumis sativus | 55 | [33] |
| SlWRKYs | Solanum lycopersicum | 81 | [34] |
| PgWRKYs | Panax ginseng | 118 | [35] |
| VuWRKYs | Vigna unguiculata | 92 | [36] |
| HvWRKYs | Hordeum vulgare | 86 | [37] |
| IbWRKYs | Ipomoea batatas | 84 | [38] |
| PhWRKYs | Petunia hybrida | 79 | [39] |
| TkWRKYs | Taraxacum kok-saghyz | 72 | [40] |
| SbWRKYs | Scutellaria baicalensis | 72 | [41] |
| HuWRKYs | Hylocereus undulatus | 70 | [42] |
| DcWRKYs | Daucus carota | 67 | [43] |
| XsWRKYs | Xanthoceras sorbifolium | 65 | [44] |
| KoWRKYs | Kandelia obovata | 64 | [45] |
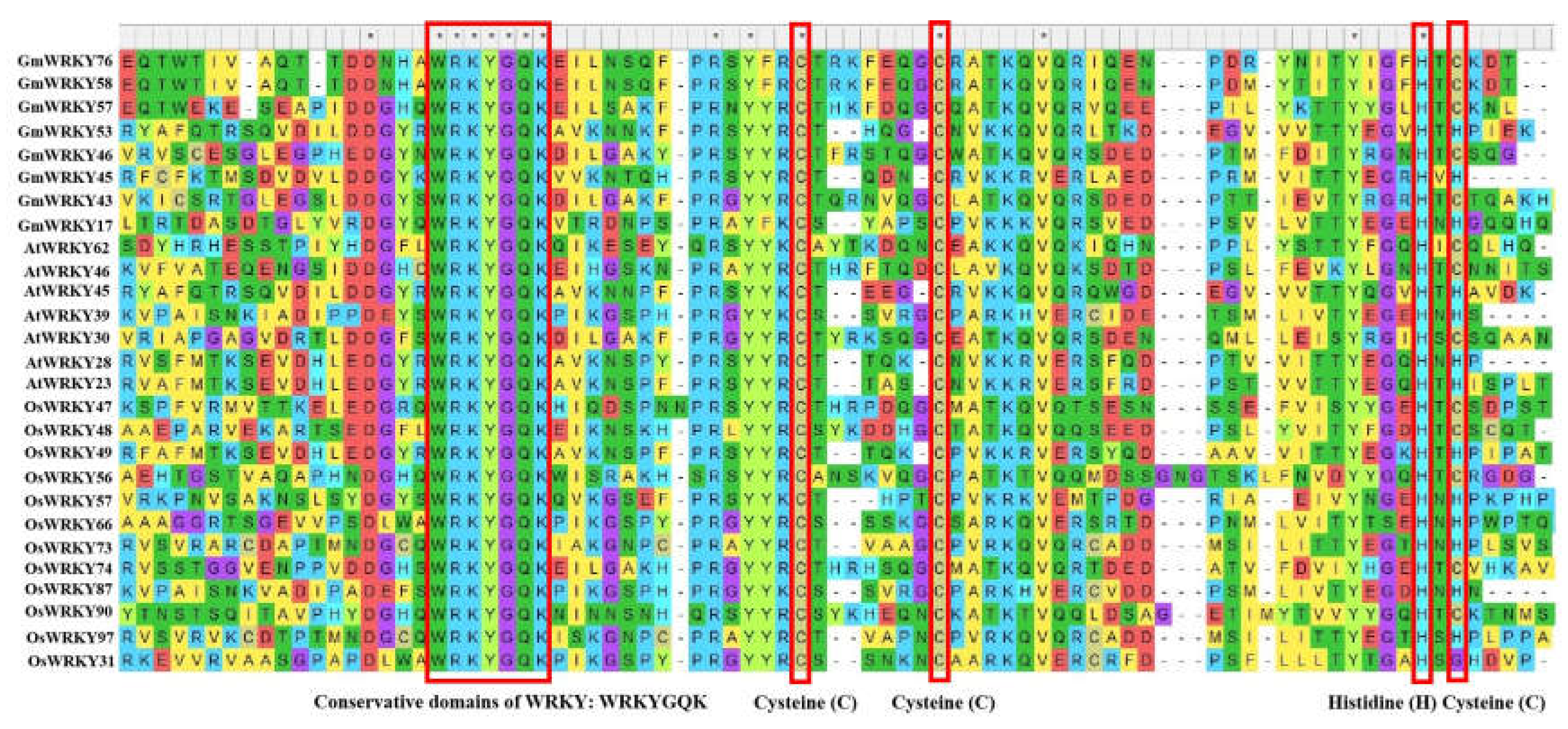
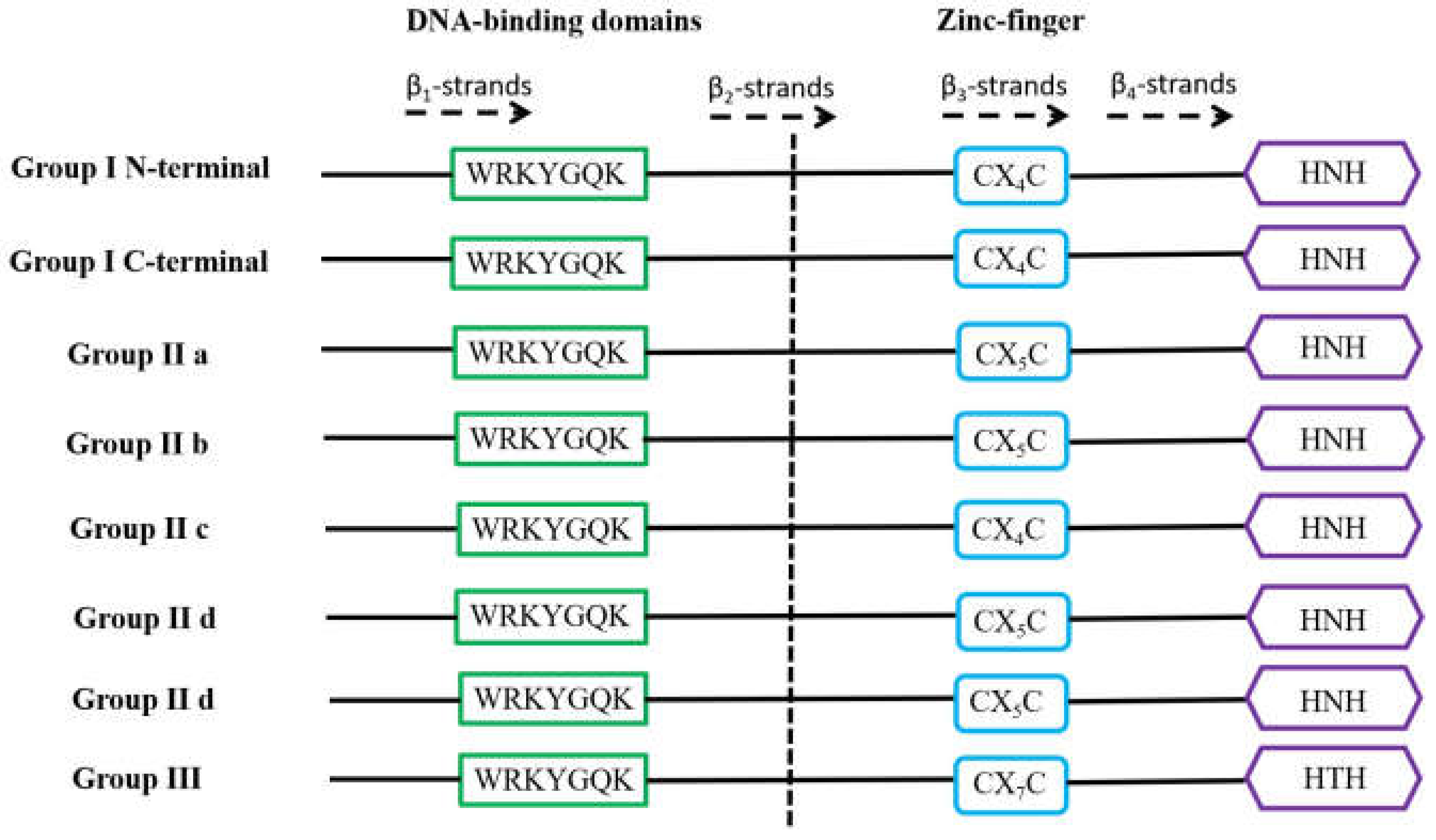
1.2. W-Box cis-Element in the Promoter Region of WRKY Downstream Genes
2. Response and Tolerance of WRKY Transcription Factor Family to Abiotic Stresses in Plant
2.1. Molecular Mechanisms of WRKY Transcription Factors Associated with Dought Stress
2.2. WRKY Transcription Factors Involved in Response to Temperature Stress
2.2.1. WRKY Transcription Factors and High Temperature Stress
2.2.1. WRKY Transcription Factors and Low Temperature Stress
2.3. WRKY Transcription Factors in Response to Salt Stress
2.4. Role of WRKY Transcription Factors in Plant Response to Heavy Metals Stress
2.5. WRKY Transcription Factors Involved in Plant Response to Nutritional Element Stress
2.6. WRKY Transcription Factors and Oxidative Stress
3. Conclusion and Prospects

| Abiotic Stress Type | WRKY transcription factors | Species | Expression patternBREAK | Reference |
|---|---|---|---|---|
| Heat | AtWRKY39 | Arabidopsis thaliana L. | Increase | [118] |
| Boron | AtWRKY47 | Arabidopsis thaliana L. | Decrease | [119] |
| Cadmium | AtWRKY13 | Arabidopsis thaliana L. | Increase | [120] |
| Salt | AtWRKY28 | Arabidopsis thaliana L. | Increase | [121] |
| Salt | AtWRKY33 | Arabidopsis thaliana L. | Increase | [122] |
| Salt | AtWRKY46 | Arabidopsis thaliana L. | Increase | [123] |
| Salt, drought | GmWRKY12 | Glycine max L. | Increase | [124] |
| Salt, drought | GmWRKY16 | Glycine max L. | Increase | [125] |
| Salt | GmWRKY20 | Glycine max L. | Increase | [126] |
| Salt, drought | GmWRKY27 | Glycine max L. | Increase | [127] |
| Salt, drought | GmWRKY54 | Glycine max L. | Increase | [128] |
| Salt | GmWRKY144, 165 | Glycine max L. | Increase | [129] |
| Salt | ZmWRKY17 | Zea mays L. | Decrease | [130] |
| Salt | ZmWRKY86 | Zea mays L. | Decrease | [79] |
| Drought | ZmWRKY104 | Zea mays L. | Increase | [131] |
| Salt | OsWRKY50 | Oryza sativa | Increase | [77] |
| Cold | OsWRKY63 | Oryza sativa | Decrease | [18] |
| Cold | OsWRKY76 | Oryza sativa | Increase | [132] |
| Salt | OsWRKY42 | Oryza sativa | Increase | [133] |
| Drought | OsWRKY55 | Oryza sativa | Decrease | [134] |
| Drought | OsWRKY5 | Oryza sativa | Decrease | [135] |
| Drought | OsWRKY97 | Oryza sativa | Increase | [136] |
| Aluminum | OsWRKY22 | Oryza sativa | Decrease | [137] |
| Salt, drought | OsWRKY87 | Oryza sativa | Increase | [138] |
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bailey-Serres, J.; Parker, J.E.; Ainsworth, E.A.; Oldroyd, G.E.D.; Schroeder, J.I. Genetic strategies for improving crop yields. Nature 2019, 575, 109–118. [CrossRef]
- Xiao, J.; Liu, B.; Yao, Y.; Guo, Z.; Jia, H.; Kong, L.; Zhang, A.; Ma, W.; Ni, Z.; Xu, S.; et al. Wheat genomic study for genetic improvement of traits in China. Sci. China Life Sci. 2022, 65, 1718–1775. [CrossRef]
- Valliyodan, B.; Ye, H.; Song, L.; Murphy, M.; Shannon, J.G.; Nguyen, H.T. Genetic diversity and genomic strategies for improving drought and waterlogging tolerance in soybeans. J. Exp. Bot. 2016, 68, 1835–1849. [CrossRef]
- Thudi, M.; Palakurthi, R.; Schnable, J.C.; Chitikineni, A.; Dreisigacker, S.; Mace, E.; Srivastava, R.K.; Satyavathi, C.T.; Odeny, D.; Tiwari, V.K.; et al. Genomic resources in plant breeding for sustainable agriculture. J. Plant Physiol. 2020, 257, 153351. [CrossRef]
- Ma, Z.; Jin, Y.-M.; Wu, T.; Hu, L.; Zhang, Y.; Jiang, W.; Du, X. OsDREB2B, an AP2/ERF transcription factor, negatively regulates plant height by conferring GA metabolism in rice. Front. Plant Sci. 2022, 13, 1007811. [CrossRef]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.K. Abiotic stress responses in plants. Nat Rev Genet. 2022, 23, 104-119.
- Ma, Z.; Wu, T.; Huang, K.; Jin, Y.-M.; Li, Z.; Chen, M.; Yun, S.; Zhang, H.; Yang, X.; Chen, H.; et al. A Novel AP2/ERF Transcription Factor, OsRPH1, Negatively Regulates Plant Height in Rice. Front. Plant Sci. 2020, 11, 709. [CrossRef]
- Zhang, H.; Zhao, Y.; Zhu, J.-K. Thriving under Stress: How Plants Balance Growth and the Stress Response. Dev. Cell 2020, 55, 529–543. [CrossRef]
- Huang, S.; Ma, Z.; Hu, L.; Huang, K.; Zhang, M.; Zhang, S.; Jiang, W.; Wu, T.; Du, X. Involvement of rice transcription factor OsERF19 in response to ABA and salt stress responses. Plant Physiol. Biochem. 2021, 167, 22–30. [CrossRef]
- Ma, Z.; Hu, L. MicroRNA: A Dynamic Player from Signalling to Abiotic Tolerance in Plants. Int. J. Mol. Sci. 2023, 24, 11364. [CrossRef]
- Gupta, A.; Rico-Medina, A.; Caño-Delgado, A.I. The physiology of plant responses to drought. Science 2020, 368, 266–269. [CrossRef]
- Ma, Z.; Hu, L.; Jiang, W. Understanding AP2/ERF Transcription Factor Responses and Tolerance to Various Abiotic Stresses in Plants: A Comprehensive Review. Int J Mol Sci. 2024, 25, 893.
- Yamada, Y.; Sato, F. Transcription factors in alkaloid biosynthesis. Int Rev Cell Mol Biol. 2013, 305, 339-82.
- Wang, H.; Chen, W.; Xu, Z.; Chen, M.; Yu, D. Functions of WRKYs in plant growth and development. Trends Plant Sci. 2023, 28, 630–645. [CrossRef]
- Guo, X.; Ullah, A.; Siuta, D.; Kukfisz, B.; Iqbal, S. Role of WRKY Transcription Factors in Regulation of Abiotic Stress Responses in Cotton. Life 2022, 12, 1410. [CrossRef]
- Su, M.; Zuo, W.; Wang, Y.; Liu, W.; Zhang, Z.; Wang, N.; Chen, X. The WKRY transcription factor MdWRKY75 regulates anthocyanins accumulation in apples (Malus domestica). Funct Plant Biol. 2022, 49, 799-809.
- Yin, Y.; Fu, H.; Mi, F.; Yang, Y.; Wang, Y.; Li, Z.; He, Y.; Yue, Z. Genomic characterization of WRKY transcription factors related to secoiridoid biosynthesis in Gentiana macrophylla. BMC Plant Biol. 2024, 24, 1–20. [CrossRef]
- Zhang, M.; Zhao, R.; Huang, K.; Huang, S.; Wang, H.; Wei, Z.; Li, Z.; Bian, M.; Jiang, W.; Wu, T.; et al. The OsWRKY63–OsWRKY76–OsDREB1B module regulates chilling tolerance in rice. Plant J. 2022, 112, 383–398. [CrossRef]
- Ma, J.; Li, C.; Sun, L.; Ma, X.; Qiao, H.; Zhao, W.; Yang, R.; Song, S.; Wang, S.; Huang, H. The SlWRKY57-SlVQ21/SlVQ16 module regulates salt stress in tomato. J Integr Plant Biol. 2023, 65, 2437-2455.
- Devaiah, B.N.; Karthikeyan, A.S.; Raghothama, K.G. WRKY75 Transcription Factor Is a Modulator of Phosphate Acquisition and Root Development in Arabidopsis. Plant Physiol. 2007, 143, 1789–1801. [CrossRef]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [CrossRef]
- Ishihama, N.; Yoshioka, H. Post-translational regulation of WRKY transcription factors in plant immunity. Curr. Opin. Plant Biol. 2012, 15, 431–437. [CrossRef]
- Lee, S.-W.; Han, S.-W.; Sririyanum, M.; Park, C.-J.; Seo, Y.-S.; Ronald, P.C. A Type I–Secreted, Sulfated Peptide Triggers XA21-Mediated Innate Immunity. Science 2009, 326, 850–853. [CrossRef]
- Chen, L.; Song, Y.; Li, S.; Zhang, L.; Zou, C.; Yu, D. The role of WRKY transcription factors in plant abiotic stresses. Biochim. et Biophys. Acta (BBA) - Gene Regul. Mech. 2012, 1819, 120–128. [CrossRef]
- Grzechowiak, M.; Ruszkowska, A.; Sliwiak, J.; Urbanowicz, A.; Jaskolski, M.; Ruszkowski, M. New aspects of DNA recognition by group II WRKY transcription factor revealed by structural and functional study of AtWRKY18 DNA binding domain. Int. J. Biol. Macromol. 2022, 213, 589–601. [CrossRef]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247-58.
- Wani, S.H.; Anand, S.; Singh, B.; Bohra, A.; Joshi, R. WRKY transcription factors and plant defense responses: latest discoveries and future prospects. Plant Cell Rep. 2021, 40, 1071–1085. [CrossRef]
- Li, W.; Pang, S.; Lu, Z.; Jin, B. Function and Mechanism of WRKY Transcription Factors in Abiotic Stress Responses of Plants. Plants 2020, 9, 1515. [CrossRef]
- Ülker, B.; Somssich, I.E. WRKY transcription factors: from DNA binding towards biological function. Curr. Opin. Plant Biol. 2004, 7, 491–498. [CrossRef]
- Wu, K.-L.; Guo, Z.-J.; Wang, H.-H.; Li, J. The WRKY Family of Transcription Factors in Rice and Arabidopsis and Their Origins. DNA Res. 2005, 12, 9–26. [CrossRef]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [CrossRef]
- Mangelsen, E.; Kilian, J.; Berendzen, K.W.; Kolukisaoglu, .H.; Harter, K.; Jansson, C.; Wanke, D. Phylogenetic and comparative gene expression analysis of barley (Hordeum vulgare) WRKY transcription factor family reveals putatively retained functions between monocots and dicots. BMC Genom. 2008, 9, 194–194. [CrossRef]
- Ling, J.; Jiang, W.; Zhang, Y.; Yu, H.; Mao, Z.; Gu, X.; Huang, S.; Xie, B. Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genom. 2011, 12, 1–20. [CrossRef]
- Huang, S.; Gao, Y.; Liu, J.; Peng, X.; Niu, X.; Fei, Z.; Cao, S.; Liu, Y. Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol. Genet. Genom. 2012, 287, 495–513. [CrossRef]
- Liu, T.; Yu, E.; Hou, L.; Hua, P.; Zhao, M.; Wang, Y.; Hu, J.; Zhang, M.; Wang, K.; Wang, Y. Transcriptome-Based Identification, Characterization, Evolutionary Analysis, and Expression Pattern Analysis of the WRKY Gene Family and Salt Stress Response in Panax ginseng. Horticulturae. 2022, 8, 756.
- Matos, M.K.d.S.; Benko-Iseppon, A.M.; Bezerra-Neto, J.P.; Ferreira-Neto, J.R.C.; Wang, Y.; Liu, H.; Pandolfi, V.; Amorim, L.L.B.; Willadino, L.; Amorim, T.C.D.V.; et al. The WRKY transcription factor family in cowpea: Genomic characterization and transcriptomic profiling under root dehydration. Gene 2022, 823, 146377. [CrossRef]
- Zheng, J.; Zhang, Z.; Tong, T.; Fang, Y.; Zhang, X.; Niu, C.; Li, J.; Wu, Y.; Xue, D.; Zhang, X. Genome-Wide Identification of WRKY Gene Family and Expression Analysis under Abiotic Stress in Barley. Agronomy 2021, 11, 521. [CrossRef]
- Liu, S.; Zhang, C.; Guo, F.; Sun, Q.; Yu, J.; Dong, T.; Wang, X.; Song, W.; Li, Z.; Meng, X.; et al. A systematical genome-wide analysis and screening of WRKY transcription factor family engaged in abiotic stress response in sweetpotato. BMC Plant Biol. 2022, 22, 1–19. [CrossRef]
- Yao, H.; Yang, T.; Qian, J.; Deng, X.; Dong, L. Genome-Wide Analysis and Exploration of WRKY Transcription Factor Family Involved in the Regulation of Shoot Branching in Petunia. Genes 2022, 13, 855. [CrossRef]
- Cheng, Y.; Luo, J.; Li, H.; Wei, F.; Zhang, Y.; Jiang, H.; Peng, X. Identification of the WRKY Gene Family and Characterization of Stress-Responsive Genes in Taraxacum kok-saghyz Rodin. Int. J. Mol. Sci. 2022, 23, 10270. [CrossRef]
- Zhang, C.; Wang, W.; Wang, D.; Hu, S.; Zhang, Q.; Wang, Z.; Cui, L. Genome-Wide Identification and Characterization of the WRKY Gene Family in Scutellaria baicalensis Georgi under Diverse Abiotic Stress. Int. J. Mol. Sci. 2022, 23, 4225. [CrossRef]
- Chen, C.; Xie, F.; Shah, K.; Hua, Q.; Chen, J.; Zhang, Z.; Zhao, J.; Hu, G.; Qin, Y. Genome-Wide Identification of WRKY Gene Family in Pitaya Reveals the Involvement of HmoWRKY42 in Betalain Biosynthesis. Int. J. Mol. Sci. 2022, 23, 10568. [CrossRef]
- Nan, H.; Gao, L.-Z. Genome-Wide Analysis of WRKY Genes and Their Response to Hormone and Mechanic Stresses in Carrot. Front. Genet. 2019, 10, 363. [CrossRef]
- Liu, Z., Saiyinduleng.; Chang, Q.; Cheng, C.; Zheng, Z.; Yu, S. Identification of yellowhorn (Xanthoceras sorbifolium) WRKY transcription factor family and analysis of abiotic stress response model. Journal of Forestry Research. 2021, 32, 987-1004.
- Du, Z.; You, S.; Zhao, X.; Xiong, L.; Li, J. Genome-Wide Identification of WRKY Genes and Their Responses to Chilling Stress in Kandelia obovata. Front. Genet. 2022, 13, 875316. [CrossRef]
- Ciolkowski, I.; Wanke, D.; Birkenbihl, R.P.; Somssich, I.E. Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol. Biol. 2008, 68, 81–92. [CrossRef]
- Maeo, K.; Hayashi, S.; Kojima-Suzuki, H.; Morikami, A.; Nakamura, K. Role of Conserved Residues of the WRKY Domain in the DNA-binding of Tobacco WRKY Family Proteins. Biosci. Biotechnol. Biochem. 2001, 65, 2428–2436. [CrossRef]
- Hrmova, M.; Hussain, S.S. Plant Transcription Factors Involved in Drought and Associated Stresses. Int. J. Mol. Sci. 2021, 22, 5662. [CrossRef]
- Khoso, M.A.; Hussain, A.; Ritonga, F.N.; Ali, Q.; Channa, M.M.; Alshegaihi, R.M.; Meng, Q.; Ali, M.; Zaman, W.; Brohi, R.D.; et al. WRKY transcription factors (TFs): Molecular switches to regulate drought, temperature, and salinity stresses in plants. Front. Plant Sci. 2022, 13, 1039329. [CrossRef]
- Wang, H.; Cheng, X.; Yin, D.; Chen, D.; Luo, C.; Liu, H.; Huang, C. Advances in the Research on Plant WRKY Transcription Factors Responsive to External Stresses. Curr. Issues Mol. Biol. 2023, 45, 2861–2880. [CrossRef]
- Krupinska, K.; Desel, C.; Frank, S.; Hensel, G. WHIRLIES Are Multifunctional DNA-Binding Proteins With Impact on Plant Development and Stress Resistance. Front. Plant Sci. 2022, 13, 880423. [CrossRef]
- Qin, Y.; Yu, H.; Cheng, S.; Liu, Z.; Yu, C.; Zhang, X.; Su, X.; Huang, J.; Shi, S.; Zou, Y.; et al. Genome-Wide Analysis of the WRKY Gene Family in Malus domestica and the Role of MdWRKY70L in Response to Drought and Salt Stresses. Genes 2022, 13, 1068. [CrossRef]
- Liu, Y.; Cao, Y. GmWRKY17-mediated transcriptional regulation of GmDREB1D and GmABA2 controls drought tolerance in soybean. Plant Mol. Biol. 2023, 113, 157–170. [CrossRef]
- Duan, D.; Yi, R.; Ma, Y.; Dong, Q.; Mao, K.; Ma, F. Apple WRKY transcription factor MdWRKY56 positively modulates drought stress tolerance. Environ. Exp. Bot. 2023, 212. [CrossRef]
- Zhang, P.; Chao, R.; Qiu, L.; Ge, W.; Liang, J.; Wen, P. ChaWRKY40 Enhances Drought Tolerance of ‘Dawei’ Hazelnuts by Positively Regulating Proline Synthesis. Forests. 2024, 15, 407.
- Wang, D.; Chen, Q.; Chen, W.; Liu, X.; Xia, Y.; Guo, Q.; Jing, D.; Liang, G. A WRKY Transcription Factor, EjWRKY17, from Eriobotrya japonica Enhances Drought Tolerance in Transgenic Arabidopsis. Int J Mol Sci. 2021, 22, 5593.
- Huang, Z.; Wang, J.; Li, Y.; Song, L.; Chen, D.; Liu, L.; Jiang, C.Z. A WRKY Protein, MfWRKY40, of Resurrection Plant Myrothamnus flabellifolia Plays a Positive Role in Regulating Tolerance to Drought and Salinity Stresses of Arabidopsis. Int J Mol Sci. 2022, 23, 8145.
- Hussain, A.; Noman, A.; Khan, M.I.; Zaynab, M.; Aqeel, M.; Anwar, M.; Ashraf, M.F.; Liu, Z.; Raza, A.; Mahpara, S.; et al. Molecular regulation of pepper innate immunity and stress tolerance: An overview of WRKY TFs. Microb. Pathog. 2019, 135, 103610. [CrossRef]
- Cheng, Z.; Luan, Y.; Meng, J.; Sun, J.; Tao, J.; Zhao, D. WRKY Transcription Factor Response to High-Temperature Stress. Plants 2021, 10, 2211. [CrossRef]
- Yoon, Y.; Seo, D.H.; Shin, H.; Kim, H.J.; Kim, C.M.; Jang, G. The Role of Stress-Responsive Transcription Factors in Modulating Abiotic Stress Tolerance in Plants. Agronomy 2020, 10, 788. [CrossRef]
- Li, S.; Khoso, M.A.; Wu, J.; Yu, B.; Wagan, S.; Liu, L. Exploring the mechanisms of WRKY transcription factors and regulated pathways in response to abiotic stress. Plant Stress 2024, 12. [CrossRef]
- Manna, M.; Thakur, T.; Chirom, O.; Mandlik, R.; Deshmukh, R.; Salvi, P. Transcription factors as key molecular target to strengthen the drought stress tolerance in plants. Physiol. Plant. 2021, 172, 847–868. [CrossRef]
- He, G.-H.; Xu, J.-Y.; Wang, Y.-X.; Liu, J.-M.; Li, P.-S.; Chen, M.; Ma, Y.-Z.; Xu, Z.-S. Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis. BMC Plant Biol. 2016, 16, 116. [CrossRef]
- Wang, Y.; Gai, W.; Yuan, L.; Shang, L.; Li, F.; Gong, Z.; Ge, P.; Wang, Y.; Tao, J.; Zhang, X.; et al. Heat-inducible SlWRKY3 confers thermotolerance by activating the SlGRXS1 gene cluster in tomato. Hortic. Plant J. 2024, 10, 515–531. [CrossRef]
- Wu, Z.; Li, T.; Cao, X.; Zhang, D.; Teng, N. Lily WRKY factor LlWRKY22 promotes thermotolerance through autoactivation and activation of LlDREB2B. Hortic. Res. 2022, 9, uhac186. [CrossRef]
- Balfagón, D.; Pascual, L.S.; Sengupta, S.; Halliday, K.J.; Gómez-Cadenas, A.; Peláez-Vico, M..; Sinha, R.; Mittler, R.; Zandalinas, S.I. WRKY48 negatively regulates plant acclimation to a combination of high light and heat stress. Plant J. 2024, 117, 1642–1655. [CrossRef]
- Ritonga, F.N.; Ngatia, J.N.; Wang, Y.; Khoso, M.A.; Farooq, U.; Chen, S. AP2/ERF, an important cold stress-related transcription factor family in plants: A review. Physiol. Mol. Biol. Plants 2021, 27, 1953–1968. [CrossRef]
- Xu, G.; Li, L.; Zhou, J.; Lyu, D.; Zhao, D.; Qin, S. Comparison of transcriptome and metabolome analysis revealed differences in cold resistant metabolic pathways in different apple cultivars under low temperature stress. Hortic. Plant J. 2023, 9, 183–198. [CrossRef]
- Mi, X.; Tang, M.; Zhu, J.; Shu, M.; Wen, H.; Zhu, J.; Wei, C. Alternative splicing of CsWRKY21 positively regulates cold response in tea plant. Plant Physiol. Biochem. 2024, 208, 108473. [CrossRef]
- Yu, H.; Li, J.; Chang, X.; Dong, N.; Chen, B.; Wang, J.; Zha, L.; Gui, S. Genome-wide identification and expression profiling of the WRKY gene family reveals abiotic stress response mechanisms in Platycodon grandiflorus. Int. J. Biol. Macromol. 2024, 257, 128617. [CrossRef]
- Wang, Y.; Dong, B.; Wang, N.; Zheng, Z.; Yang, L.; Zhong, S.; Fang, Q.; Xiao, Z.; Zhao, H. A WRKY Transcription Factor PmWRKY57 from Prunus mume Improves Cold Tolerance in Arabidopsis thaliana. Mol Biotechnol. 2023, 65, 1359-1368.
- Liu, W.; Liang, X.; Cai, W.; Wang, H.; Liu, X.; Cheng, L.; Song, P.; Luo, G.; Han, D. Isolation and Functional Analysis of VvWRKY28, a Vitis vinifera WRKY Transcription Factor Gene, with Functions in Tolerance to Cold and Salt Stress in Transgenic Arabidopsis thaliana. Int J Mol Sci. 2022, 23, 13418.
- Wang, L.; Chen, H.; Chen, G.; Luo, G.; Shen, X.; Ouyang, B.; Bie, Z. Transcription factor SlWRKY50 enhances cold tolerance in tomato by activating the jasmonic acid signaling. Plant Physiol. 2023, 194, 1075–1090. [CrossRef]
- Zhao, S.; Zhang, Q.; Liu, M.; Zhou, H.; Ma, C.; Wang, P. Regulation of Plant Responses to Salt Stress. Int. J. Mol. Sci. 2021, 22, 4609. [CrossRef]
- van Zelm, E.; Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [CrossRef]
- Gupta, A.; Mishra, R.; Rai, S.; Bano, A.; Pathak, N.; Fujita, M.; Kumar, M.; Hasanuzzaman, M. Mechanistic Insights of Plant Growth Promoting Bacteria Mediated Drought and Salt Stress Tolerance in Plants for Sustainable Agriculture. Int. J. Mol. Sci. 2022, 23, 3741. [CrossRef]
- Huang, S.; Hu, L.; Zhang, S.; Zhang, M.; Jiang, W.; Wu, T.; Du, X. Rice OsWRKY50 Mediates ABA-Dependent Seed Germination and Seedling Growth, and ABA-Independent Salt Stress Tolerance. Int. J. Mol. Sci. 2021, 22, 8625. [CrossRef]
- Huang, J.; Liu, F.; Chao, D.; Xin, B.; Liu, K.; Cao, S.; Chen, X.; Peng, L.; Zhang, B.; Fu, S.; et al. The WRKY Transcription Factor OsWRKY54 Is Involved in Salt Tolerance in Rice. Int. J. Mol. Sci. 2022, 23, 11999. [CrossRef]
- Fang, X.; Li, W.; Yuan, H.; Chen, H.; Bo, C.; Ma, Q.; Cai, R. Mutation of ZmWRKY86 confers enhanced salt stress tolerance in maize. Plant Physiol. Biochem. 2021, 167, 840–850. [CrossRef]
- Ma J.; Li C.; Sun L.; Ma X.; Qiao H.; Zhao W.; Yang R.; Song S.; Wang S.; Huang H. The SlWRKY57-SlVQ21/SlVQ16 module regulates salt stress in tomato. J Integr Plant Biol. 2023 Nov;65(11):2437-2455.
- Yu, Y.; Wu, Y.; He, L. A wheat WRKY transcription factor TaWRKY17 enhances tolerance to salt stress in transgenic Arabidopsis and wheat plant. Plant Mol. Biol. 2023, 113, 171–191. [CrossRef]
- Mirza, Z.; Haque, M.M.; Gupta, M. WRKY transcription factors: a promising way to deal with arsenic stress in rice. Mol. Biol. Rep. 2022, 49, 10895–10904. [CrossRef]
- Khan, I.; Asaf, S.; Jan, R.; Bilal, S.; Lubna; Khan, A.L.; Kim, K.-M.; Al-Harrasi, A. Genome-wide annotation and expression analysis of WRKY and bHLH transcriptional factor families reveal their involvement under cadmium stress in tomato (Solanum lycopersicum L.). Front. Plant Sci. 2023, 14, 1100895. [CrossRef]
- Abdullah.; Wani, K.I.; Naeem, M.; Jha, P.K.; Jha, U.C.; Aftab, T.; Prasad, P.V.V. Systems biology of chromium-plant interaction: insights from omics approaches. Front Plant Sci. 2024, 14, 1305179.
- Shen, C.; Yang, Y.-M.; Sun, Y.-F.; Zhang, M.; Chen, X.-J.; Huang, Y.-Y. The regulatory role of abscisic acid on cadmium uptake, accumulation and translocation in plants. Front. Plant Sci. 2022, 13, 953717. [CrossRef]
- Gu, L.; Hou, Y.; Sun, Y.; Chen, X.; Wang, G.; Wang, H.; Zhu, B.; Du, X. The maize WRKY transcription factor ZmWRKY64 confers cadmium tolerance in Arabidopsis and maize (Zea mays L.). Plant Cell Rep. 2024, 43, 1–16. [CrossRef]
- Jia, Z.; Li, M.; Wang, H.; Zhu, B.; Gu, L.; Du, X.; Ren, M. TaWRKY70 positively regulates TaCAT5 enhanced Cd tolerance in transgenic Arabidopsis. Environ. Exp. Bot. 2021, 190, 104591. [CrossRef]
- Xian, P.; Yang, Y.; Xiong, C.; Guo, Z.; Alam, I.; He, Z.; Zhang, Y.; Cai, Z.; Nian, H. Overexpression of GmWRKY172 enhances cadmium tolerance in plants and reduces cadmium accumulation in soybean seeds. Front. Plant Sci. 2023, 14, 1133892. [CrossRef]
- He, G.; Saleem, M.; Deng, T.; Zhong, Z.; He, T.; Wu, J. Unraveling the Mechanism of StWRKY6 in Potato (Solanum tuberosum)’s Cadmium Tolerance for Ensuring Food Safety. Foods 2023, 12, 2303. [CrossRef]
- Cai, Z.; Xian, P.; Wang, H.; Lin, R.; Lian, T.; Cheng, Y.; Ma, Q.; Nian, H. Transcription Factor GmWRKY142 Confers Cadmium Resistance by Up-Regulating the Cadmium Tolerance 1-Like Genes. Front. Plant Sci. 2020, 11, 724. [CrossRef]
- Chen, Y.; Kong, X.; Yang, L.; Fu, M.; Zhang, S. Genome-Wide Identification of WRKY Family Genes and the Expression Profiles in Response to Nitrogen Deficiency in Poplar. Genes 2022, 13, 2324. [CrossRef]
- Zhang, T.; Zhang, C.; Zhang, X.; Liang, Z.; Xia, P. Multi-algorithm cooperation research of WRKY genes under nitrogen stress in Panax notoginseng. Protoplasma 2022, 260, 1081–1096. [CrossRef]
- Poll, A.A.; Lee, J.; Sanderson, R.A.; Byrne, E.; Gatehouse, J.A.; Sadanandom, A.; Gatehouse, A.M.R.; Edwards, M.G. Septoria Leaf Blotch and Reduced Nitrogen Availability Alter WRKY Transcription Factor Expression in a Codependent Manner. Int. J. Mol. Sci. 2020, 21, 4165. [CrossRef]
- Javed, T.; Zhou, J.-R.; Li, J.; Hu, Z.-T.; Wang, Q.-N.; Gao, S.-J. Identification and Expression Profiling of WRKY Family Genes in Sugarcane in Response to Bacterial Pathogen Infection and Nitrogen Implantation Dosage. Front. Plant Sci. 2022, 13, 917953. [CrossRef]
- Sun, Y.; Zhang, T.; Xu, X.; Yang, Y.; Tong, H.; Mur, L.A.J.; Yuan, H. Transcriptomic Characterization of Nitrate-Enhanced Stevioside Glycoside Synthesis in Stevia (Stevia rebaudiana) Bertoni. Int. J. Mol. Sci. 2021, 22, 8549. [CrossRef]
- Zhang, L.; Chen, C.; Xie, F.; Hua, Q.; Zhang, Z.; Zhang, R.; Chen, J.; Zhao, J.; Hu, G.; Qin, Y. A Novel WRKY Transcription Factor HmoWRKY40 Associated with Betalain Biosynthesis in Pitaya (Hylocereus monacanthus) through Regulating HmoCYP76AD1. Int. J. Mol. Sci. 2021, 22, 2171. [CrossRef]
- Tang, W.; Wang, F.; Chu, H.; You, M.; Lv, Q.; Ji, W.; Deng, X.; Zhou, B.; Peng, D. WRKY transcription factors regulate phosphate uptake in plants. Environ. Exp. Bot. 2023, 208. [CrossRef]
- Zhang, Y.; Chen, H.; Liang, Y.; Lu, T.; Liu, Z.; Jin, X.; Hou, L.; Xu, J.; Zhao, H.; Shi, Y.; et al. Comparative transcriptomic and metabolomic analyses reveal the protective effects of silicon against low phosphorus stress in tomato plants. Plant Physiol. Biochem. 2021, 166, 78–87. [CrossRef]
- Nilsson, L.; Müller, R.; Nielsen, T.H. Dissecting the plant transcriptome and the regulatory responses to phosphate deprivation. Physiol. Plant. 2010, 139, 129–143. [CrossRef]
- Shukla, D.; Waigel, S.; Rouchka, E.C.; Sandhu, G.; Trivedi, P.K.; Sahi, S.V. Genome-wide expression analysis reveals contrasting regulation of phosphate starvation response (PSR) in root and shoot of Arabidopsis and its association with biotic stress. Environ. Exp. Bot. 2021, 188, 104483. [CrossRef]
- Wang, S.; Zhang, J.; Gu, M.; Xu, G. OsWRKY108 is an integrative regulator of phosphorus homeostasis and leaf inclination in rice. Plant Signal. Behav. 2021, 16. [CrossRef]
- Wang, S.; Lv, B.; Wang, A.; Hu, J.; Wu, Q.; Li, C. Cloning and functional characterization of FtWRKY29, a low phosphorus stress-inducible transcription factor in Tartary buckwheat. Plant Physiol. Biochem. 2023, 203, 107997. [CrossRef]
- Liu, X.; Yang, Y.; Wang, R.; Cui, R.; Xu, H.; Sun, C.; Wang, J.; Zhang, H.; Chen, H.; Zhang, D. GmWRKY46, a WRKY transcription factor, negatively regulates phosphorus tolerance primarily through modifying root morphology in soybean. Plant Sci. 2022, 315, 111148. [CrossRef]
- Zhang, J.; Gu, M.; Liang, R.; Shi, X.; Chen, L.; Hu, X.; Wang, S.; Dai, X.; Qu, H.; Li, H.; et al. OsWRKY21 and OsWRKY108 function redundantly to promote phosphate accumulation through maintaining the constitutive expression of OsPHT1;1 under phosphate-replete conditions. New Phytol. 2020, 229, 1598–1614. [CrossRef]
- Wang, S.; Xu, T.; Chen, M.; Geng, L.; Huang, Z.; Dai, X.; Qu, H.; Zhang, J.; Li, H.; Gu, M.; Xu, G. The transcription factor OsWRKY10 inhibits phosphate uptake via suppressing OsPHT1;2 expression under phosphate-replete conditions in rice. J Exp Bot. 2023, 74, 1074-1089.
- León, J.; Gayubas, B.; Castillo, M.-C. Valine-Glutamine Proteins in Plant Responses to Oxygen and Nitric Oxide. Front. Plant Sci. 2021, 11. [CrossRef]
- Tolosa, L.N.; Zhang, Z. The Role of Major Transcription Factors in Solanaceous Food Crops under Different Stress Conditions: Current and Future Perspectives. Plants 2020, 9, 56. [CrossRef]
- Rai, G.K.; Mishra, S.; Chouhan, R.; Mushtaq, M.; Chowdhary, A.A.; Rai, P.K.; Kumar, R.R.; Kumar, P.; Perez-Alfocea, F.; Colla, G.; et al. Plant salinity stress, sensing, and its mitigation through WRKY. Front. Plant Sci. 2023, 14, 1238507. [CrossRef]
- Khedia, J.; Agarwal, P.; Agarwal, P.K. Deciphering hydrogen peroxide-induced signalling towards stress tolerance in plants. 3 Biotech. 2019, 9, 395.
- Jia, H.; Wang, C.; Wang, F.; Liu, S.; Li, G.; Guo, X. GhWRKY68 Reduces Resistance to Salt and Drought in Transgenic Nicotiana benthamiana. PLOS ONE 2015, 10, e0120646–e0120646. [CrossRef]
- Sun, Y.; Yu, D. Activated expression of AtWRKY53 negatively regulates drought tolerance by mediating stomatal movement. Plant Cell Rep. 2015, 34, 1295–1306. [CrossRef]
- Song, H.; Cao, Y.; Zhao, L.; Zhang, J.; Li, S. Review: WRKY transcription factors: Understanding the functional divergence. Plant Sci. 2023, 334, 111770. [CrossRef]
- Bai, Y.; Sunarti, S.; Kissoudis, C.; Visser, R.G.F.; van der Linden, C.G. The Role of Tomato WRKY Genes in Plant Responses to Combined Abiotic and Biotic Stresses. Front. Plant Sci. 2018, 9, 801. [CrossRef]
- Schluttenhofer, C.; Yuan, L. Regulation of Specialized Metabolism by WRKY Transcription Factors. Plant Physiol. 2014, 167, 295–306. [CrossRef]
- Jiang, J.; Ma, S.; Ye, N.; Jiang, M.; Cao, J.; Zhang, J. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [CrossRef]
- Samad, A.F.A.; Sajad, M.; Nazaruddin, N.; Fauzi, I.A.; Murad, A.M.A.; Zainal, Z.; Ismail, I. MicroRNA and Transcription Factor: Key Players in Plant Regulatory Network. Front. Plant Sci. 2017, 8, 565. [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [CrossRef]
- Finatto, T.; Viana, V.E.; Woyann, L.G.; Busanello, C.; Maia, L.C.D.; Oliveira, A.C. Can WRKY transcription factors help plants to overcome environmental challenges? Genet Mol Biol. 2018, 41, 533-544.
- Feng, Y.; Cui, R.; Huang, Y.; Shi, L.; Wang, S.; Xu, F. Repression of transcription factor AtWRKY47 confers tolerance to boron toxicity in Arabidopsis thaliana. Ecotoxicol. Environ. Saf. 2021, 220, 112406. [CrossRef]
- Sheng, Y.; Yan, X.; Huang, Y.; Han, Y.; Zhang, C.; Ren, Y.; Fan, T.; Xiao, F.; Liu, Y.; Cao, S. The WRKY transcription factor, WRKY13, activates PDR8 expression to positively regulate cadmium tolerance in Arabidopsis. Plant Cell Environ. 2019, 42, 891-903.
- Babitha, K.C.; Ramu, S.V.; Pruthvi, V.; Mahesh, P.; Nataraja, K.N.; Udayakumar, M. Co-expression of AtbHLH17 and AtWRKY28 confers resistance to abiotic stress in Arabidopsis. Transgenic Res. 2012, 22, 327–341. [CrossRef]
- Krishnamurthy, P.; Vishal, B.; Ho, W.J.; Lok, F.C.J.; Lee, F.S.M.; Kumar, P.P. Regulation of a Cytochrome P450 Gene CYP94B1 by WRKY33 Transcription Factor Controls Apoplastic Barrier Formation in Roots to Confer Salt Tolerance. Plant Physiol. 2020, 184, 2199–2215. [CrossRef]
- Ding, Z.J.; Yan, J.Y.; Li, C.X.; Li, G.X.; Wu, Y.R.; Zheng, S.J. Transcription factor WRKY46 modulates the development of Arabidopsis lateral roots in osmotic/salt stress conditions via regulation of ABA signaling and auxin homeostasis. Plant J. 2015, 84, 56–69. [CrossRef]
- Shi, W.-Y.; Du, Y.-T.; Ma, J.; Min, D.-H.; Jin, L.-G.; Chen, J.; Chen, M.; Zhou, Y.-B.; Ma, Y.-Z.; Xu, Z.-S.; et al. The WRKY Transcription Factor GmWRKY12 Confers Drought and Salt Tolerance in Soybean. Int. J. Mol. Sci. 2018, 19, 4087. [CrossRef]
- Ma, Q.; Xia, Z.; Cai, Z.; Li, L.; Cheng, Y.; Liu, J.; Nian, H. GmWRKY16 Enhances Drought and Salt Tolerance Through an ABA-Mediated Pathway in Arabidopsis thaliana. Front. Plant Sci. 2019, 9, 1979. [CrossRef]
- Luo, X.; Bai, X.; Sun, X.; Zhu, D.; Liu, B.; Ji, W.; Cai, H.; Cao, L.; Wu, J.; Hu, M.; et al. Expression of wild soybean WRKY20 in Arabidopsis enhances drought tolerance and regulates ABA signalling. J. Exp. Bot. 2013, 64, 2155–2169. [CrossRef]
- Wang, F.; Chen, H.W.; Li, Q.T.; Wei, W.; Li, W.; Zhang, W.K.; Ma, B.; Bi, Y.D.; Lai, Y.C.; Liu, X.L.; Man, W.Q.; Zhang, J.S.; Chen, S.Y. GmWRKY27 interacts with GmMYB174 to reduce expression of GmNAC29 for stress tolerance in soybean plants. Plant J. 2015, 83, 224-36.
- Wei, W.; Liang, D.; Bian, X.; Shen, M.; Xiao, J.; Zhang, W.; Ma, B.; Lin, Q.; Lv, J.; Chen, X.; et al. GmWRKY54 improves drought tolerance through activating genes in abscisic acid and Ca2+ signaling pathways in transgenic soybean. Plant J. 2019, 100, 384–398. [CrossRef]
- Song, H.; Wang, P.; Hou, L.; Zhao, S.; Zhao, C.; Xia, H.; Li, P.; Zhang, Y.; Bian, X.; Wang, X. Global Analysis of WRKY Genes and Their Response to Dehydration and Salt Stress in Soybean. Front. Plant Sci. 2016, 7, 9. [CrossRef]
- Cai, R.; Dai, W.; Zhang, C.; Wang, Y.; Wu, M.; Zhao, Y.; Ma, Q.; Xiang, Y.; Cheng, B. The maize WRKY transcription factor ZmWRKY17 negatively regulates salt stress tolerance in transgenic Arabidopsis plants. Planta 2017, 246, 1215–1231. [CrossRef]
- Zhao, L.; Yan, J.; Xiang, Y.; Sun, Y.; Zhang, A. ZmWRKY104 Transcription Factor Phosphorylated by ZmMPK6 Functioning in ABA-Induced Antioxidant Defense and Enhance Drought Tolerance in Maize. Biology 2021, 10, 893. [CrossRef]
- Yokotani, N.; Sato, Y.; Tanabe, S.; Chujo, T.; Shimizu, T.; Okada, K.; Yamane, H.; Shimono, M.; Sugano, S.; Takatsuji, H.; et al. WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J. Exp. Bot. 2013, 64, 5085–5097. [CrossRef]
- Pillai, S.E.; Kumar, C.; Patel, H.K.; Sonti, R.V. Overexpression of a cell wall damage induced transcription factor, OsWRKY42, leads to enhanced callose deposition and tolerance to salt stress but does not enhance tolerance to bacterial infection. BMC Plant Biol. 2018, 18, 1–15. [CrossRef]
- Huang, K.; Wu, T.; Ma, Z.; Li, Z.; Chen, H.; Zhang, M.; Bian, M.; Bai, H.; Jiang, W.; Du, X. Rice Transcription Factor OsWRKY55 Is Involved in the Drought Response and Regulation of Plant Growth. Int. J. Mol. Sci. 2021, 22, 4337. [CrossRef]
- Lim, C.; Kang, K.; Shim, Y.; Yoo, S.C.; Paek, N.C. Inactivating transcription factor OsWRKY5 enhances drought tolerance through abscisic acid signaling pathways. Plant Physiol. 2022, 188, 1900-1916.
- Lv, M.; Hou, D.; Wan, J.; Ye, T.; Zhang, L.; Fan, J.; Li, C.; Dong, Y.; Chen, W.; Rong, S.; et al. OsWRKY97, an Abiotic Stress-Induced Gene of Rice, Plays a Key Role in Drought Tolerance. Plants 2023, 12, 3338. [CrossRef]
- Li, G.Z.; Wang, Z.Q.; Yokosho, K.; Ding, B.; Fan, W.; Gong, Q.Q.; Li, G.X.; Wu, Y.R.; Yang, J.L.; Ma, J.F.; Zheng, S.J. Transcription factor WRKY22 promotes aluminum tolerance via activation of OsFRDL4 expression and enhancement of citrate secretion in rice (Oryza sativa). New Phytol. 2018, 219, 149-162.
- Yan, L.; Baoxiang, W.; Jingfang, L.; Zhiguang, S.; Ming, C.; Yungao, X.; Bo, X.; Bo, Y.; Jian, L.; Jinbo, L.; et al. A novel SAPK10-WRKY87-ABF1 biological pathway synergistically enhance abiotic stress tolerance in transgenic rice (Oryza sativa). Plant Physiol. Biochem. 2021, 168, 252–262. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





