Submitted:
02 June 2024
Posted:
03 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
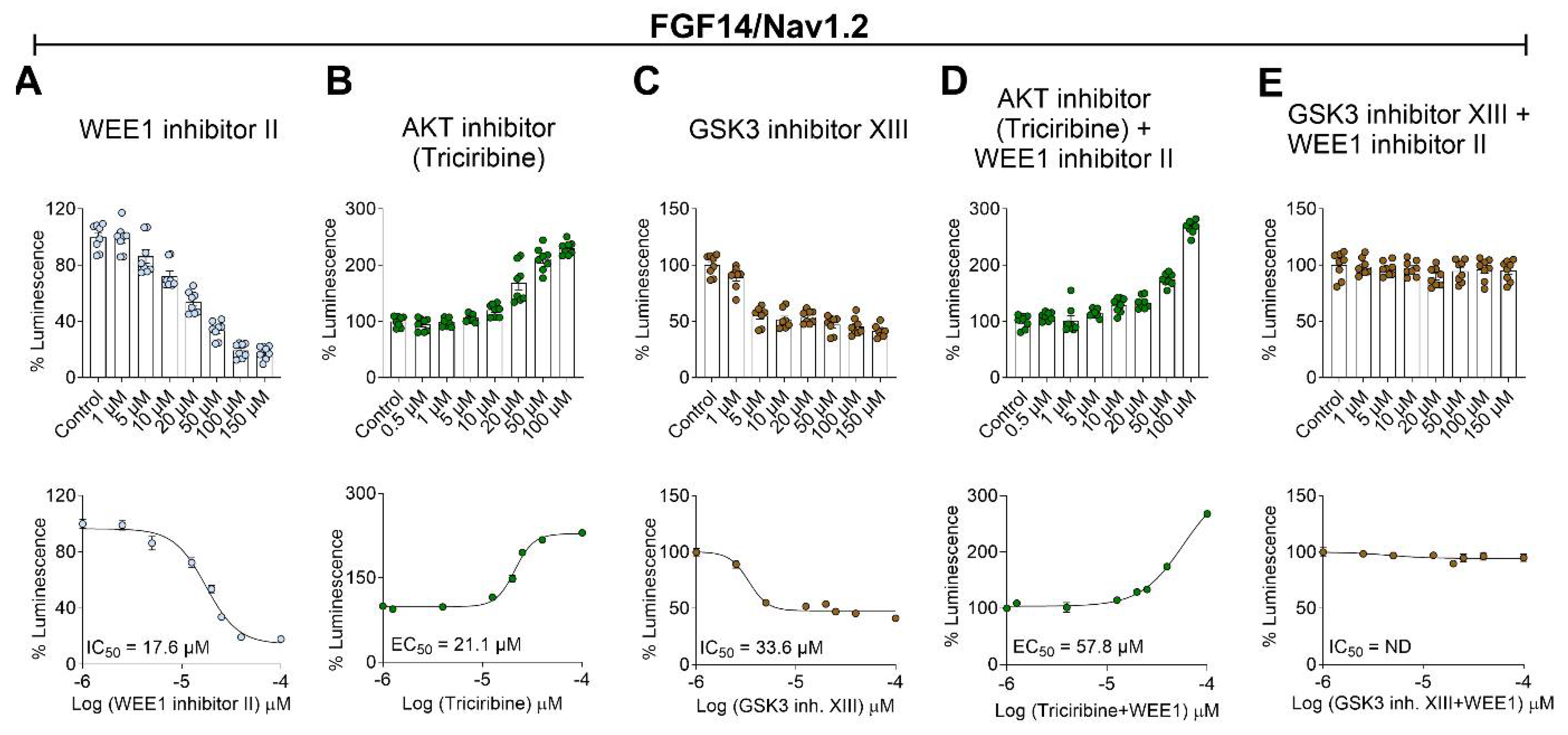
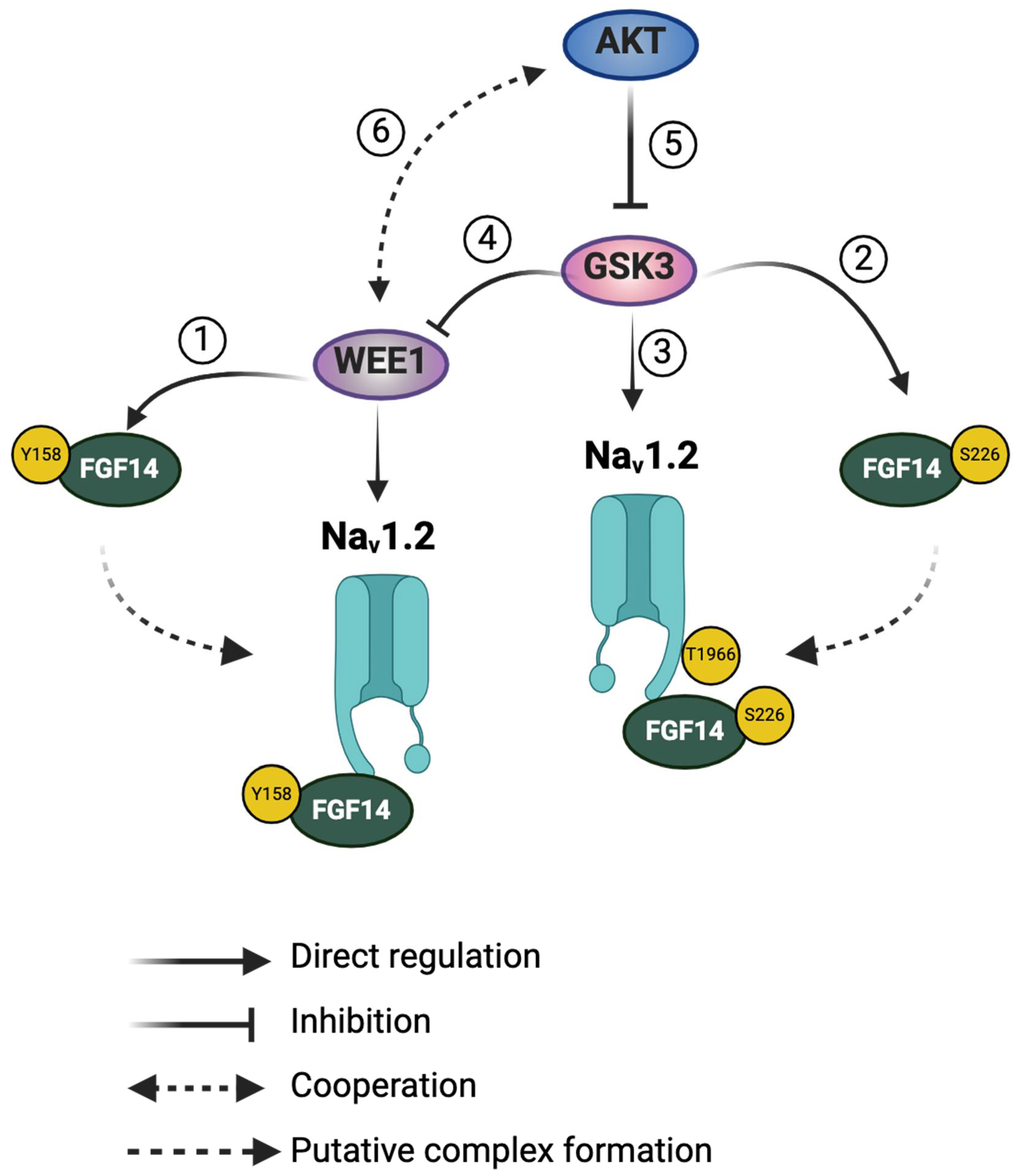
2.1. Pharmacological Interrogation of WEE1, AKT, and GSK3 Inhibitors on FGF14/Nav1.2 Complex Assembly
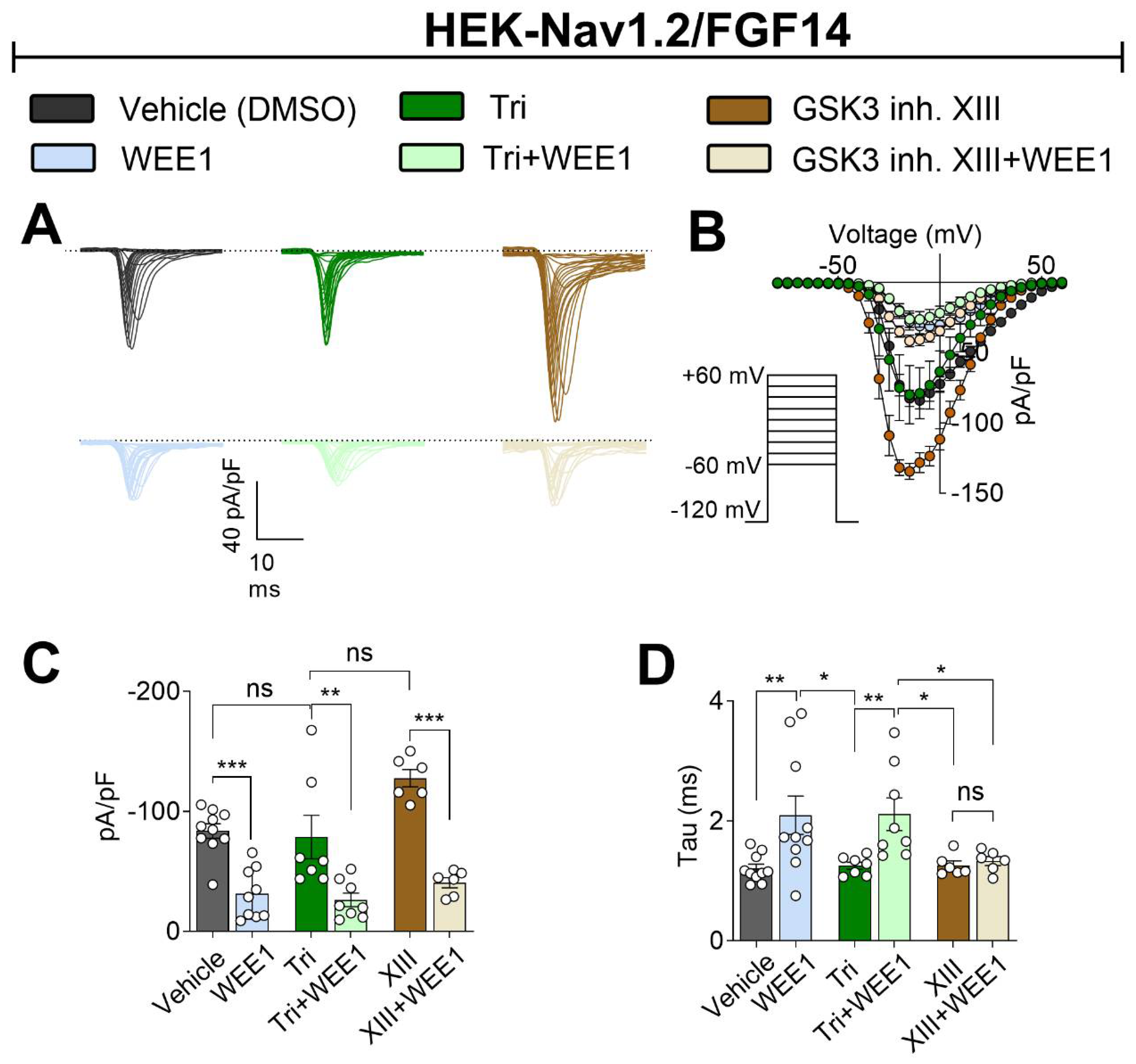
2.2. Functional Regulation of Nav1.2 -Mediated Currents through WEE1, AKT, and GSK3 Kinase Crosstalk
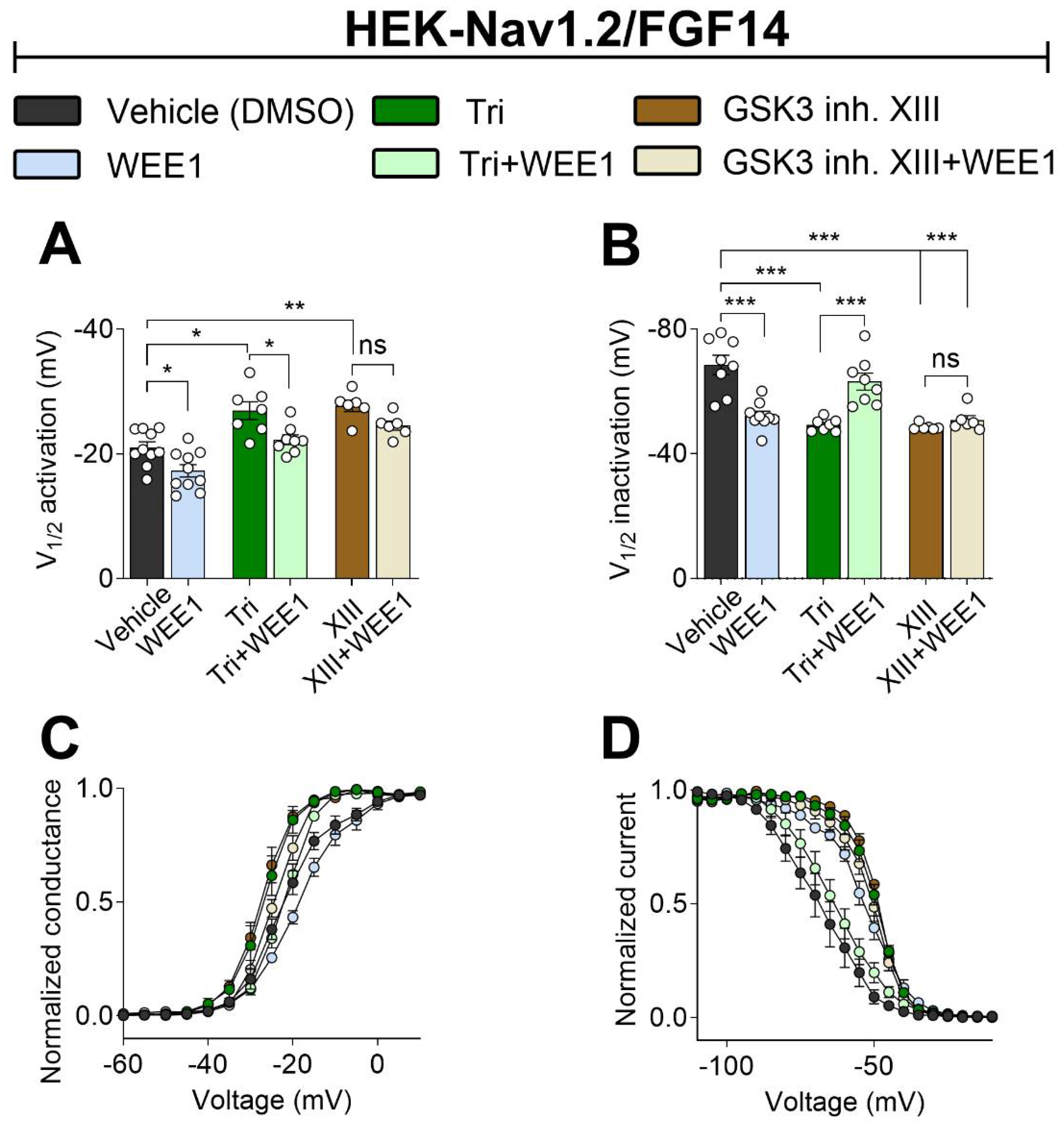
2.3. Nav1.2 Voltage Sensitivity Is Modulated by WEE1 Kinase, AKT, and GSK3
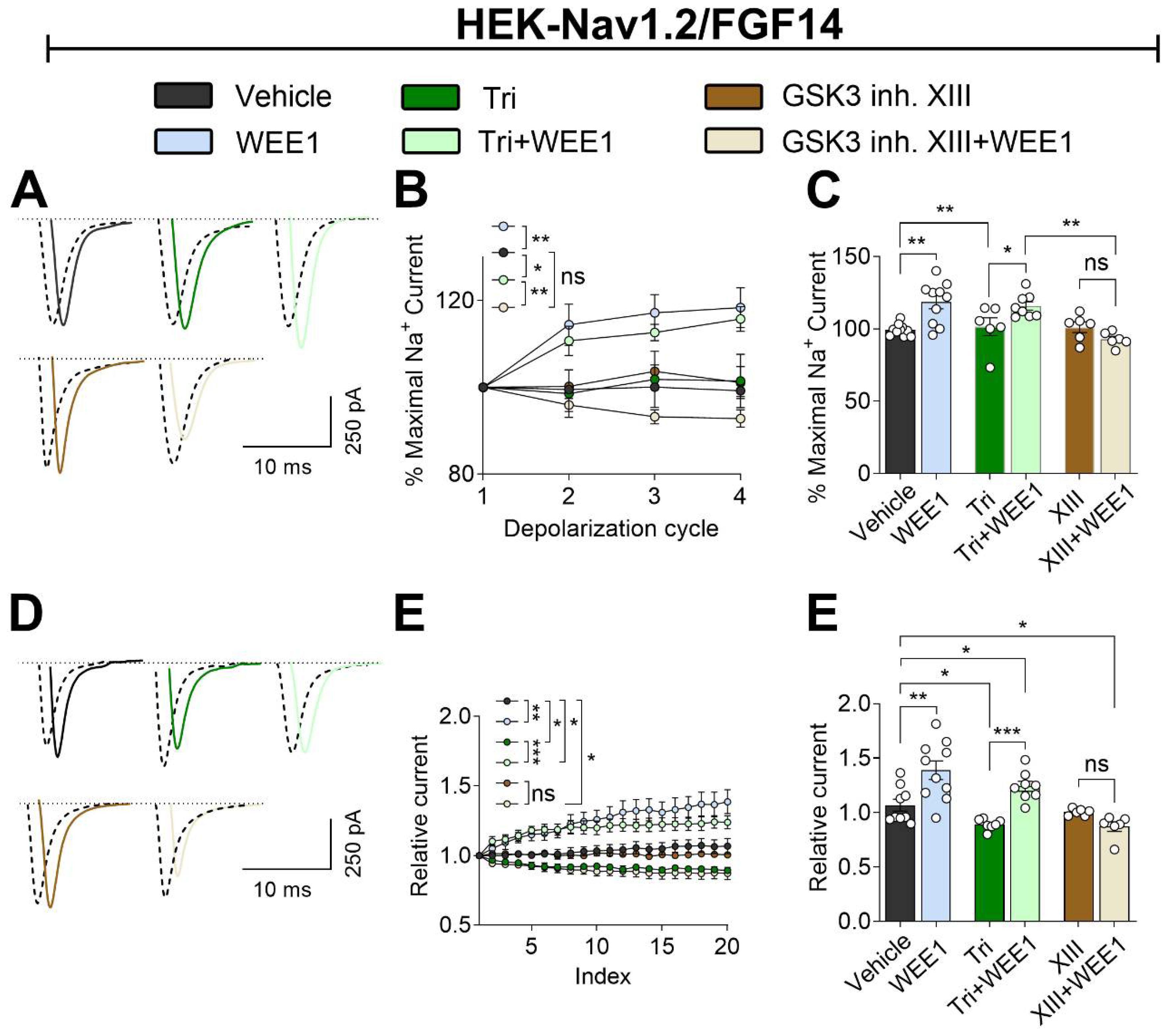
2.4. WEE1 Kinase, AKT, and GSK3 Modulate Long-Term Inactivation and Use-Dependency of the Nav1.2 Channel
3. Discussion
4. Materials and Methods
4.1. DNA Constructs
4.2. HEK293 Cell Culture
4.3. Split-Luciferase Complementation Assay
4.4. Whole-Cell Patch Clamp Electrophysiology
4.5. Statistics
Author Contributions
Acknowledgments
References
- Catterall, W.A. Voltage gated sodium and calcium channels: Discovery, structure, function, and Pharmacology. Channels 2023, 17, 2281714. [Google Scholar] [CrossRef] [PubMed]
- Leterrier, C.; Brachet, A.; Fache, M.-P.; Dargent, B. Voltage-gated sodium channel organization in neurons: Protein interactions and trafficking pathways. Neurosci. Lett. 2010, 486, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Pitt, G.S.; Lee, S. Current view on regulation of voltage-gated sodium channels by calcium and auxiliary proteins. Protein Sci. 2016, 25, 1573–1584. [Google Scholar] [CrossRef] [PubMed]
- Dvorak, N.M.; Wadsworth, P.A.; Wang, P.; Zhou, J.; Laezza, F. Development of Allosteric Modulators of Voltage-Gated Na+ Channels: A Novel Approach for an Old Target. Curr. Top. Med. Chem. 2021, 21, 841–848. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.R.; Singh, A.K.; Laezza, F. Identification of Amino Acid Residues in Fibroblast Growth Factor 14 (FGF14) Required for Structure-Function Interactions with Voltage-gated Sodium Channel Nav1.6. J. Biol. Chem. 2016, 291, 11268–11284. [Google Scholar] [CrossRef] [PubMed]
- Di Re, J.; Wadsworth, P.A.; Laezza, F. Intracellular Fibroblast Growth Factor 14: Emerging Risk Factor for Brain Disorders. Front. Cell. Neurosci. 2017, 11, 103. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.R.; Liu, Z.; Nenov, M.N.; Folorunso, O.; Singh, A.; Scala, F.; Chen, H.; James, T.F.; Alshammari, M.; Panova-Elektronova, N.I.; et al. Functional Modulation of Voltage-Gated Sodium Channels by a FGF14-Based Peptidomimetic. ACS Chem. Neurosci. 2018, 9, 976–987. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Fazel Darbandi, S.; Pochareddy, S.; Gulden, F.O.; Gilson, M.C.; Sheppard, B.K.; Sahagun, A.; An, J.Y.; Werling, D.M.; Rubenstein, J.L.R.; et al. Developmental dynamics of voltage-gated sodium channel isoform expression in the human and mouse brain. Genome Med 2021, 13, 135. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Tian, C.; Li, T.; Yang, M.; Hou, H.; Shu, Y. Distinct contributions of Nav1.6 and Nav1.2 in action potential initiation and backpropagation. Nat. Neurosci. 2009, 12, 996–1002. [Google Scholar] [CrossRef]
- Laezza, F.; Lampert, A.; Kozel, M.A.; Gerber, B.R.; Rush, A.M.; Nerbonne, J.M.; Waxman, S.G.; Dib-Hajj, S.D.; Ornitz, D.M. FGF14 N-terminal splice variants differentially modulate Nav1.2 and Nav1.6-encoded sodium channels. Mol. Cell. Neurosci. 2009, 42, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Scala, F.; Nenov, M.N.; Crofton, E.J.; Singh, A.K.; Folorunso, O.; Zhang, Y.; Chesson, B.C.; Wildburger, N.C.; James, T.F.; Alshammari, M.A.; et al. Environmental Enrichment and Social Isolation Mediate Neuroplasticity of Medium Spiny Neurons through the GSK3 Pathway. Cell Rep. 2018, 23, 555–567. [Google Scholar] [CrossRef] [PubMed]
- Hsu, W.-C.; Nenov, M.N.; Shavkunov, A.; Panova, N.; Zhan, M.; Laezza, F. Identifying a Kinase Network Regulating FGF14:Nav1.6 Complex Assembly Using Split-Luciferase Complementation. PLOS ONE 2015, 10, e0117246. [Google Scholar] [CrossRef] [PubMed]
- Laezza, F.; Shavkunov, A.; Buzhdygan, T.; Nenov, M. The FGF14:Nav channel complex is a new target of the Akt/GSK3 signaling pathway. Neuropsychopharmacol. Off. Publ. Am. Coll. Neuropsychopharmacol. 2011, 36, S89–S90. [Google Scholar]
- Shavkunov, A.S.; Wildburger, N.C.; Nenov, M.N.; James, T.F.; Buzhdygan, T.P.; Panova-Elektronova, N.I.; Green, T.A.; Veselenak, R.L.; Bourne, N.; Laezza, F. The Fibroblast Growth Factor 14·Voltage-gated Sodium Channel Complex Is a New Target of Glycogen Synthase Kinase 3 (GSK3). J. Biol. Chem. 2013, 288, 19370–19385. [Google Scholar] [CrossRef] [PubMed]
- Hsu, W.-C.J.; Wildburger, N.C.; Haidacher, S.J.; Nenov, M.N.; Folorunso, O.; Singh, A.K.; Chesson, B.C.; Franklin, W.F.; Cortez, I.; Sadygov, R.G.; et al. PPARgamma agonists rescue increased phosphorylation of FGF14 at S226 in the Tg2576 mouse model of Alzheimer's disease. Exp. Neurol. 2017, 295, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Marosi, M.; Nenov, M.N.; Di Re, J.; Dvorak, N.M.; Alshammari, M.; Laezza, F. Inhibition of the Akt/PKB Kinase Increases Nav1.6-Mediated Currents and Neuronal Excitability in CA1 Hippocampal Pyramidal Neurons. Int. J. Mol. Sci. 2022, 23, 1700. [Google Scholar] [CrossRef] [PubMed]
- Scala, F.; Nenov, M.N.; Wildburger, N.C.; Elferink, H.; Singh, A.K.; Chesson, C.B.; Buzhdygan, T.; Sohail, M.; Shavkunov, A.S.; Panova, N.I.; et al. CK2 activity is required for the interaction of FGF14 with voltage-gated sodium channels and neuronal excitability. FASEB J. 2016, 30, 2171–2186. [Google Scholar] [CrossRef] [PubMed]
- Dvorak, N.M.; Domingo, N.D.; Tapia, C.M.; Wadsworth, P.A.; Marosi, M.; Avchalumov, Y.; Fongsaran, C.; Koff, L.; Di Re, J.; Sampson, C.M.; et al. TNFR1 signaling converging on FGF14 controls neuronal hyperactivity and sickness behavior in experimental cerebral malaria. J. Neuroinflammation 2023, 20, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Wadsworth, P.A.; Singh, A.K.; Nguyen, N.; Dvorak, N.M.; Tapia, C.M.; Russell, W.K.; Stephan, C.; Laezza, F. JAK2 regulates Nav1.6 channel function via FGF14(Y158) phosphorylation. Biochim Biophys Acta Mol Cell Res 2020, 1867, 118786. [Google Scholar] [CrossRef] [PubMed]
- Dvorak, N.M.; Tapia, C.M.; Baumgartner, T.J.; Singh, J.; Laezza, F.; Singh, A.K. Pharmacological Inhibition of Wee1 Kinase Selectively Modulates the Voltage-Gated Na(+) Channel 1.2 Macromolecular Complex. Cells 2021, 10, 3103. [Google Scholar] [CrossRef] [PubMed]
- Madoux, F.; Simanski, S.; Chase, P.; Mishra, J.K.; Roush, W.R.; Ayad, N.G.; Hodder, P. An Ultra-High Throughput Cell-Based Screen for Wee1 Degradation Inhibitors. SLAS Discov. Adv. Sci. Drug Discov. 2010, 15, 907–917. [Google Scholar] [CrossRef] [PubMed]
- Milletti, G.; Colicchia, V.; Cecconi, F. Cyclers’ kinases in cell division: From molecules to cancer therapy. Cell Death Differ. 2023, 30, 2035–2052. [Google Scholar] [CrossRef] [PubMed]
- Kuzu, O.F.; Gowda, R.; Sharma, A.; Noory, M.A.; Kardos, G.; Madhunapantula, S.V.; Drabick, J.J.; Robertson, G.P. Identification of WEE1 as a target to make AKT inhibition more effective in melanoma. Cancer Biol. Ther. 2017, 19, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Penas, C.; Mishra, J.K.; Wood, S.D.; Schürer, S.C.; Roush, W.R.; Ayad, N.G. GSK3 inhibitors stabilize Wee1 and reduce cerebellar granule cell progenitor proliferation. Cell Cycle 2015, 14, 417–424. [Google Scholar] [CrossRef]
- Owens, L.; Simanski, S.; Squire, C.; Smith, A.; Cartzendafner, J.; Cavett, V.; Busby, J.C.; Sato, T.; Ayad, N.G. Activation Domain-dependent Degradation of Somatic Wee1 Kinase. J. Biol. Chem. 2010, 285, 6761–6769. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yang, S.-G.; Zhou, F.-Q. Glycogen synthase kinase 3 signaling in neural regeneration in vivo. J. Mol. Cell Biol. 2023, 15. [Google Scholar] [CrossRef]
- Marosi, M.; Arman, P.; Aceto, G.; D’ascenzo, M.; Laezza, F. Glycogen Synthase Kinase 3: Ion Channels, Plasticity, and Diseases. Int. J. Mol. Sci. 2022, 23, 4413. [Google Scholar] [CrossRef]
- Spratt, P.W.; Ben-Shalom, R.; Keeshen, C.M.; Burke, K.J., Jr.; Clarkson, R.L.; Sanders, S.J.; Bender, K.J. The Autism-Associated Gene Scn2a Contributes to Dendritic Excitability and Synaptic Function in the Prefrontal Cortex. Neuron 2019, 103, 673–685. [Google Scholar] [CrossRef] [PubMed]
- Thompson, C.H.; Ben-Shalom, R.; Bender, K.J.; George, A.L. Alternative splicing potentiates dysfunction of early-onset epileptic encephalopathy SCN2A variants. J. Gen. Physiol. 2020, 152. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-G.; Bavley, C.C.; Li, A.; Jones, R.M.; Hackett, J.E.; Bayleyen, Y.; Lee, F.S.; Rajadhyaksha, A.M.; Pitt, G.S. Scn2a severe hypomorphic mutation decreases excitatory synaptic input and causes autism-associated behaviors. J. Clin. Investig. 2021, 6. [Google Scholar] [CrossRef] [PubMed]
- Wolff, M.; Johannesen, K.M.; Hedrich, U.B.S.; Masnada, S.; Rubboli, G.; Gardella, E.; Lesca, G.; Ville, D.; Milh, M.; Villard, L.; et al. Genetic and phenotypic heterogeneity suggest therapeutic implications in SCN2A-related disorders. Brain 2017, 140, 1316–1336. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, X.; Eaton, M.; Wu, J.; Ma, Z.; Lai, S.; Park, A.; Ahmad, T.S.; Que, Z.; Lee, J.H.; et al. Severe deficiency of the voltage-gated sodium channel Na(V)1.2 elevates neuronal excitability in adult mice. Cell reports 2021, 36, 109495. [Google Scholar] [CrossRef] [PubMed]
- Asadollahi, R.; Delvendahl, I.; Muff, R.; Tan, G.; Rodríguez, D.G.; Turan, S.; Russo, M.; Oneda, B.; Joset, P.; Boonsawat, P.; et al. Pathogenic SCN2A variants cause early-stage dysfunction in patient-derived neurons. Hum Mol Genet 2023, 32, 2192–2204. [Google Scholar] [CrossRef] [PubMed]
- Mangano, G.D.; Fontana, A.; Antona, V.; Salpietro, V.; Mangano, G.R.; Giuffre, M.; Nardello, R. Commonalities and distinctions between two neurodevelopmental disorder subtypes associated with SCN2A and SCN8A variants and literature review. Mol. Genet. Genom. Med. 2022, 10, e1911. [Google Scholar] [CrossRef] [PubMed]
- Meisler, M.H.; Hill, S.F.; Yu, W. Sodium channelopathies in neurodevelopmental disorders. Nature reviews. Neuroscience 2021, 22, 152–166. [Google Scholar] [CrossRef] [PubMed]
- Goetz, R.; Dover, K.; Laezza, F.; Shtraizent, N.; Huang, X.; Tchetchik, D.; Eliseenkova, A.V.; Xu, C.-F.; Neubert, T.A.; Ornitz, D.M.; et al. Crystal Structure of a Fibroblast Growth Factor Homologous Factor (FHF) Defines a Conserved Surface on FHFs for Binding and Modulation of Voltage-gated Sodium Channels. J. Biol. Chem. 2009, 284, 17883–17896. [Google Scholar] [CrossRef] [PubMed]
- Dover, K.; Solinas, S.; D’angelo, E.; Goldfarb, M. Long-term inactivation particle for voltage-gated sodium channels. J. Physiol. 2010, 588, 3695–3711. [Google Scholar] [CrossRef] [PubMed]
- Mahling, R.; Rahlf, C.R.; Hansen, S.C.; Hayden, M.R.; Shea, M.A. Ca2+-saturated calmodulin binds tightly to the N-terminal domain of A-type fibroblast growth factor homologous factors. J. Biol. Chem. 2020, 296, 100458. [Google Scholar] [CrossRef]
- Martinez-Espinosa, P.L.; Neely, A.; Ding, J.; Lingle, C.J. Fast inactivation of Nav current in rat adrenal chromaffin cells involves two independent inactivation pathways. J. Gen. Physiol. 2021, 153. [Google Scholar] [CrossRef] [PubMed]
- James, T.F.; Nenov, M.N.; Wildburger, N.C.; Lichti, C.F.; Luisi, J.; Vergara, F.; Panova-Electronova, N.I.; Nilsson, C.L.; Rudra, J.S.; Green, T.A.; et al. The Nav1.2 channel is regulated by GSK3. Biochimica et biophysica acta 2015, 1850, 832–844. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Derderian, K.D.; Hamada, E.; Zhou, X.; Nelson, A.D.; Kyoung, H.; Ahituv, N.; Bouvier, G.; Bender, K.J. Impaired cerebellar plasticity hypersensitizes sensory reflexes in SCN2A-associated ASD. Neuron 2024, 112, 1444–1455. [Google Scholar] [CrossRef] [PubMed]
- Stertz, L.; Di Re, J.; Pei, G.; Fries, G.R.; Mendez, E.; Li, S.; Smith-Callahan, L.; Raventos, H.; Tipo, J.; Cherukuru, R.; et al. Convergent genomic and pharmacological evidence of PI3K/GSK3 signaling alterations in neurons from schizophrenia patients. Neuropsychopharmacology 2021, 46, 673–682. [Google Scholar] [CrossRef] [PubMed]
- Sowers, M.L.; Di Re, J.; Wadsworth, P.A.; Shavkunov, A.S.; Lichti, C.; Zhang, K.; Laezza, F. Sex-Specific Proteomic Changes Induced by Genetic Deletion of Fibroblast Growth Factor 14 (FGF14), a Regulator of Neuronal Ion Channels. Proteomes 2019, 7, 5. [Google Scholar] [CrossRef] [PubMed]
- Alshammari, M.A.; Alshammari, T.K.; Nenov, M.N.; Scala, F.; Laezza, F. Fibroblast Growth Factor 14 Modulates the Neurogenesis of Granule Neurons in the Adult Dentate Gyrus. Mol. Neurobiol. 2015, 53, 7254–7270. [Google Scholar] [CrossRef] [PubMed]
- Alshammari, T.K.; A Alshammari, M.; Nenov, M.N.; Hoxha, E.; Cambiaghi, M.; Marcinno, A.; James, T.F.; Singh, P.; Labate, D.; Li, J.; et al. Genetic deletion of fibroblast growth factor 14 recapitulates phenotypic alterations underlying cognitive impairment associated with schizophrenia. Transl. Psychiatry 2016, 6, e806. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Xia, C.; Jiang, Y.; Chen, Y.; Zhou, J.; Dai, R.; Han, C.; Mao, Z.; Liu, C.; PsychENCODE, Consortium; et al. Transcriptomic sex differences in postmortem brain samples from patients with psychiatric disorders. Sci. Transl. Med 2024, eadh9974. [Google Scholar] [CrossRef] [PubMed]
- Paucar, M.; Lundin, J.; Alshammari, T.; Bergendal, Å.; Lindefeldt, M.; Alshammari, M.; Solders, G.; Di Re, J.; Savitcheva, I.; Granberg, T.; et al. Broader phenotypic traits and widespread brain hypometabolism in spinocerebellar ataxia 27. J. Intern. Med. 2020, 288, 103–115. [Google Scholar] [CrossRef] [PubMed]
| Condition | Peak density | Activation | Steady-state Inactivation |
Tau (τ) |
|---|---|---|---|---|
| pA/pF | mV | mV | ms | |
| GFP Vehicle | -129.6 ± 5.5 (11) | -26.15 ± 1.03 (11) | -57.85 ± 0.85 (9) | 0.94 ± 0.07 (11) |
| GFP Wee1 | -119.8 ± 10.5 (10)#ns | -25.42 ± 1.73 (9)#ns | -55.6 ± 1.3 (10)#ns | 1.05 ± 0.06 (10)#ns |
| FGF14 Vehicle | -83.85 ± 6.0 (10)#a | -21.0 ± 0.9 (10)#h | -68.5 ± 3.1 (8)#n | 1.21 ± 0.07 (10)#t |
| FGF14 Wee1 | -31.34 ± 7.0 (9)$b,#c | -17.3 ± 1.0 (10)$#i | -55.14 ± 1.1 (9)$o | 2.1 ± 0.32 (10)$u |
| FGF14 Tri | -78.8 ± 18.16 (7)#d | -26.95 ± 1.4 (7)$j | -49.2 ± 0.77 (7)$p | 1.25 ± 0.06 (7)$ns |
| FGF14 Tri + Wee1 | -26.3 ± 5.5 (8)$%e | -22.3 ± 0.8 (8)%k | -63.1 ± 2.7 (11)%q | 2.12 ± 0.27 (8)$v |
| FGF14 GSK3 inh. XIII | -127.6 ± 7.2 (6)$f | -27.75 ± 0.9 (6)$l | -48.5 ± 0.4 (6)$r | 1.26 ± 0.07 (6)$ns |
| FGF14 GSK3 inh. XIII + Wee1 | -40.7 ± 4.2 (6)$@g | -24.5 ± 0.7 (6)#m | -50.8 ± 1.4 (6)$s | 1.33 ± 0.07 (6)@ns |
| Condition | LTI (% Maximal Na+ current) | ||
|---|---|---|---|
| 2nd Pulse | 3rd Pulse | 4th Pulse | |
| GFP Vehicle | 96.13 ± 1.0 (9) | 93.75 ± 1.0 (9) | 92.44 ± 1.0 (9) |
| GFP Wee1 | 97.73 ± 1.56 (10)#ns | 96.6 ± 1.4 (10)#ns | 93.76 ± 1.9 (10)#ns |
| FGF14 Vehicle | 99.54 ± 1.6 (10)#ns | 100.1 ± 0.8 (10)#ns | 99.24 ± 1.9 (14)#ns |
| FGF14 Wee1 | 114.4 ± 4.7 (10)$a,#b | 117.2 ± 4.4 (10)$f,#g | 118.34 ± 4.6 (10)$j,#k |
| FGF14 Tri | 98.5 ± 5.5 (6)$,#ns | 101.86 ± 6.5 (6)$,#ns | 101.5 ± 6.1 (6)$,#ns |
| FGF14 Tri + Wee1 | 110.7 ± 3.4 (8)$c,%d,#e | 112.65 ± 1.8 (8)$h,#i | 115.8 ± 2.8 (8)$l,%m |
| FGF14 GSK3 XIII | 100.2 ± 1.0 (6)$,#ns | 103.7 ± 1.5 (6)$,#ns | 101.1 ± 3.6 (6)$,#ns |
| FGF14 GSK3 XIII + Wee1 | 95.94 ± 1.5 (6)$,#ns | 93.24 ± 1.6 (6)$,#,@ns | 92.82 ± 2.0 (6)$,#,@ns |
| Cumulative use-dependency | |||
| Condition | 10th Pulse | 15th Pulse | 20th Pulse |
| GFP Vehicle | 1.01 ± 0.04 (7) | 0.99 ± 0.03 (7) | 1.0 ± 0.04 (7) |
| GFP Wee1 | 1.09 ± 0.05 (7)#ns | 1.14 ± 0.09 (7)#ns | 1.15 ± 0.08 (7)#ns |
| FGF14 Vehicle | 1.03 ± 0.04 (9)#ns | 1.05 ± 0.04 (9)#ns | 1.07 ± 0.05 (9)#ns |
| FGF14 Wee1 | 1.26 ± 0.06 (10)$n,#o | 1.31 ± 0.07 (10)$t,#u | 1.4 ± 0.04 (10)$z,#aa |
| FGF14 Tri | 0.92 ± 0.01 (7)$,#ns | 0.89 ± 0.02 (7)$,#ns | 0.89 ± 0.02 (7)$ab |
| FGF14 Tri + Wee1 | 1.2 ± 0.05 (8)$p,#q,%r | 1.22 ± 0.04 (8)$v,#w,%x | 1.24 ± 0.01 (8)$ac,%ad |
| FGF14 GSK3 XIII | 1.02 ± 0.01 (6)$,#ns | 1.0 ± 0.01 (6)$,#ns | 1.0 ± 0.04 (6)$,#ns |
| FGF14 GSK3 XIII + Wee1 | 0.89 ± 0.03 (6)$s,#,@ns | 0.87 ± 0.04 (6)$y,#,@ns | 0.87 ± 0.04 (6)$ae,#,@ns |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





