1. Introduction
The human brain stands as one of the body’s vital and intricate organs, containing billions of neurons with tons of complex connections, commonly named as synapses [
1]. Serving as the central command and control center of the nervous system, it regulates the functions of various bodily organs. Consequently, any abnormalities in the brain can have severe implications for human health [
2]. Cancer ranks among the deadliest diseases, representing the second leading cause of death globally, with approximately 10 million deaths recorded in 2020, as reported by the World Health Organization (WHO) [
3]. Consequently, if cancer infiltrates the brain, it can inflict damage on critical areas responsible for essential bodily functions, resulting in significant disabilities and, in extreme cases, death [
4]. Thus, brain cancer poses a substantial threat to overall well-being, underscoring the importance of early detection and treatment. Early diagnosis significantly increases the likelihood of a patient’s survival [
5].
In contrast to cancer, a brain tumor denotes an irregular and unregulated proliferation of cells within the human brain. Determining whether a brain tumor is malignant or benign hinges on several factors, such as its characteristics, developmental stage, rate of advancement, and its specific location [
6,
7]. Benign brain tumors are less likely to invade surrounding healthy cells, exhibiting a slower progression rate and well-defined borders, as seen in cases like meningioma and pituitary tumors. Conversely, malignant tumors can invade and damage nearby healthy cells, such as those in the spinal cord or brain, featuring a rapid growth rate with extensive borders, as observed in glioma. The classification of a tumor is also influenced by its origin, wherein a tumor originating in the brain tissue is referred to as a primary tumor, and a tumor developing elsewhere in the body and spreading to the brain through blood vessels is identified as a secondary tumor [
8].
Various diagnostic techniques, both invasive and non-invasive, are utilized in the detection of human brain cancer [
9,
10,
11,
12]. An invasive method, such as biopsy, involves extracting a tissue sample through incision for microscopic examination by physicians to determine malignancy. Commonly, in contrast to tumors found in other bodily regions, a biopsy is generally not performed before definitive brain surgery for brain tumors. Hence, computed tomography (CT), positron emission tomography (PET), and magnetic resonance imaging (MRI), recognized as non-invasive imaging techniques, are embraced as quicker and safer alternatives for promptly diagnosing patients with brain cancer. Patients and caretakers largely prefer these techniques due to their comparative safety and efficiency.
Among these, MRI is particularly favored for its capability to offer comprehensive information about the size, shape, progression, and location of brain tumors in both 2D and 3D formats [
13,
14]. The manual analysis of MRI images can be a laborious and error-prone endeavor for healthcare professionals, particularly given the substantial patient load. Thanks to the swift progress of advanced learning algorithms, computer-aided diagnosis (CAD) systems have made notable advancements in supporting physicians in the diagnosis of brain tumors [
15,
16,
17]. Numerous methods, ranging from conventional to modern AI-based deep learning approaches, have been proposed for the timely diagnosis of conditions, including potential brain tumors [
18].
Conventional machine learning methods predominantly rely on extracting pertinent features to ascertain classification accuracy. These features are broadly classified into global or low-level features and local or high-level features. Low-level features encompass elements like texture, first-order, and second-order statistical features, commonly employed in training traditional classifiers like Naïve Bayes, support vector machine (SVM), and trees. For instance, in a study by [
19], the SVM model was trained using the gray-level co-occurrence matrix for binary classification (normal or abnormal) of brain MRI images, achieving a reasonably high accuracy level but necessitating substantial training time.
In a subsequent study [
20], the authors employed principal component analysis to reduce dimensionality on the training features, effectively decreasing the training time. However, in the domain of multiclass classification, models trained on global features often demonstrate lower accuracy due to the similarities among various types of brain tumors in aspects such as texture, intensity, and size. To address this issue, some researchers have redirected their attention to features extracted at the local level in images, such as fisher vector [
21], SIFT (scale-invariant feature transformation) [
22], and an algorithm based on the bag-of-words approach [
23]. However, these techniques are susceptible to errors as their accuracy relies heavily on prior information regarding the tumor’s location in brain MRI images.
Recent advancements in machine learning algorithms have empowered the development of deep learning-based methods that can automatically learn optimal data features. Deep neural networks, including convolutional neural networks (CNNs) and fully convolutional networks (FCNs), are now widely employed in the classification of MRI images to assist in the diagnosis of brain tumors [
24]. CNNs can be utilized for brain tumor classification by either employing a pre-trained network or designing a network specifically tailored for the particular problem. Taking such considerations into account, Pereira et al. [
25] conducted a study where they developed a CNN for classifying whole-brain and brain mask images into binary classes, achieving an accuracy of 89.5% and 92.9%, respectively.
In an alternative study [
26], a straightforward CNN architecture was formulated by the authors for a three-class classification of brain tumors (pituitary, meningioma, and glioma), achieving an accuracy of 84.19%. Furthermore, a 3D deep convolutional neural network with multiscale capabilities was created to classify images into low-grade or high-grade subcategories of glioma, achieving an accuracy of 96.49%. Another research work involved the use of a 22-layer network for a three-class classification of brain tumors. The researchers validated their model using an online MRI brain-image dataset [
27] and implemented data augmentation to triple the dataset size (from 3064 to 9192) for enhanced model training. Their approach also incorporated 10-fold cross-validation during training, resulting in an accuracy of 96.56%.
Making further progress, in another study [
28], researchers introduced two separate convolutional neural network models—one with 13 layers and another with 25 layers—to perform two-class and five-class classifications of brain tumors, respectively. However, as the number of classes increased, the proposed model exhibited diminished performance, resulting in a reduced accuracy of 92.66%. A notable drawback of this approach was the utilization of two distinct models for brain tumor classification and detection. Similar to the to the authors in [
28], Deepak et al. [
29] employed a pre-trained network (GoogleNet) to classify three classes of brain tumors, achieving an impressive accuracy of 98% on an available online dataset. Furthermore, in [
30], the authors assessed the performance of various pre-trained models using a transfer learning approach on a brain MRI dataset. Their findings revealed that ResNet-50 achieved a notable accuracy of 97.2% for binary classification, despite the dataset’s limited set of brain images.
Despite producing better results, the pre-trained network required a substantial amount of time. To address this issue, some authors utilized pre-trained neural networks for feature extraction from brain tumors and subsequently trained a traditional classifier. By combining features extracted from ShuffleNet V2, DenseNet-169, and MnasNet with SVM, the authors reported to achieve 93.72% accuracy in a four-class classification (pituitary, meningioma, glioma, and no tumor) during testing. They also observed that validating the model after applying data augmentation further improved accuracy. Although prior research has presented that augmentation approaches can improve classification accuracy, their effectiveness in real-time applications remains unverified. Consequently, additional investigation is necessary to detect and classify brain tumors.
In this study, a lightweight dual-stream network is introduced for the enhanced detection of brain tumors in MRI images, surpassing the accuracy achieved by current state-of-the-art methods. The contributions of the proposed work are outlined as follows:
Propose a lightweight dual-stream network.
Employ dual inputs that comprise pre-processing with CLAHE and the White Patch Retinex Algorithm for rich feature learning.
Employ a two-fold margin loss for better and effective feature learning.
The successful execution of studies focusing on the detection of brain tumors on MRI images hinges on the seamless integration of three vital components: data pre-processing, deep learning model training, and result visualization. Each of these elements plays a pivotal role in contributing to the overall efficacy and reliability of the investigative process.
In the initial phase, meticulous data pre-processing is undertaken to refine and optimize the raw input data. This involves tasks such as noise reduction, image normalization, and resolution standardization, ensuring that the data fed into the subsequent stages of the study are of high quality and devoid of any potential confounding factors. The effectiveness of the subsequent analysis heavily relies on the quality of the pre-processed data, making this step a critical precursor to the overall success of the study [
26].
Subsequently, the deep learning model undergoes a comprehensive training process, where it learns intricate features essential for accurate brain tumor detection. The model is exposed to a diverse set of MRI images, enabling it to discern patterns, textures, and subtle nuances indicative of tumor presence. The optimization of model parameters and the fine-tuning of neural network architectures contribute to the model’s ability to generalize well across varied datasets, a crucial attribute for real-world applicability.
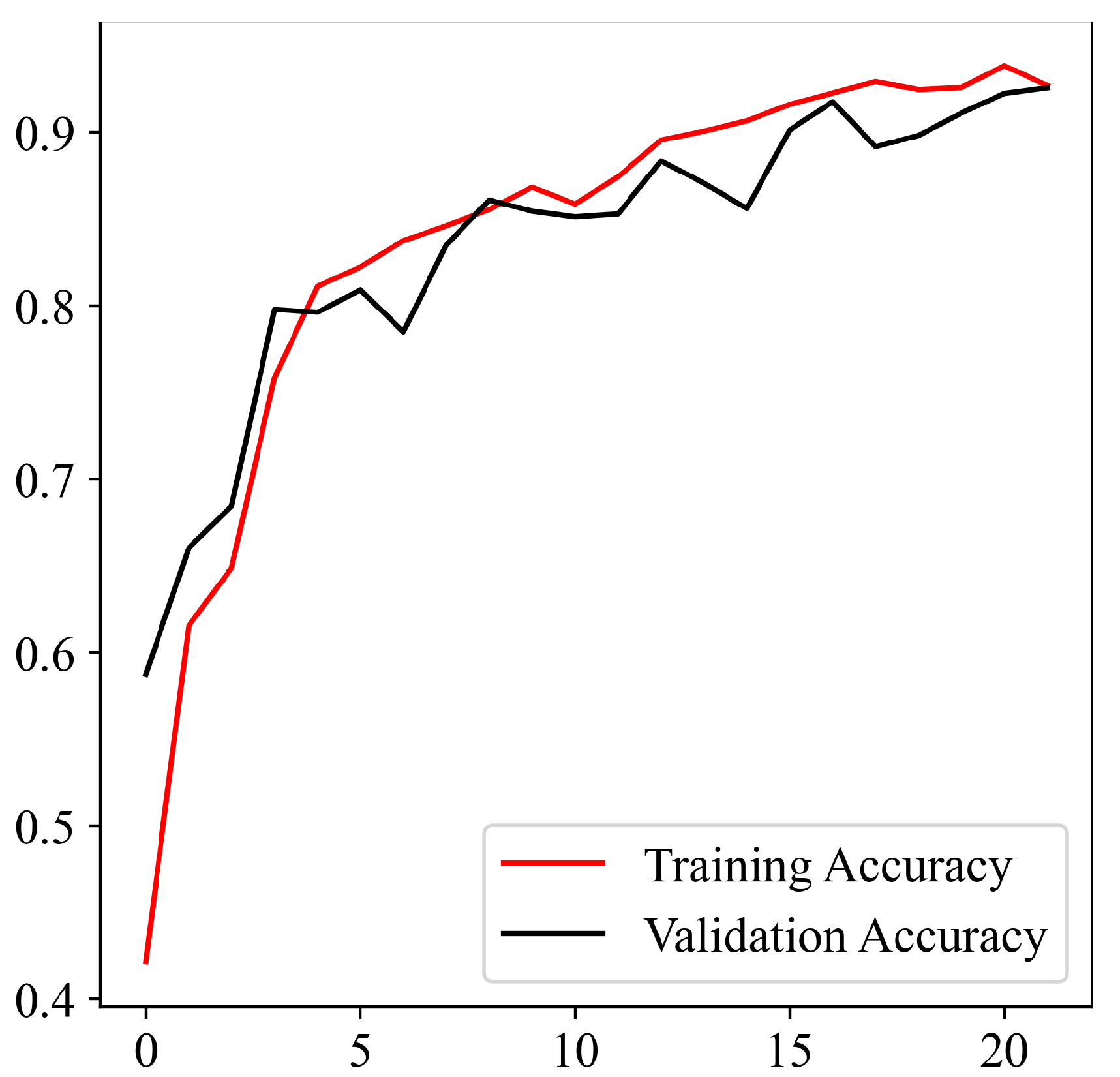
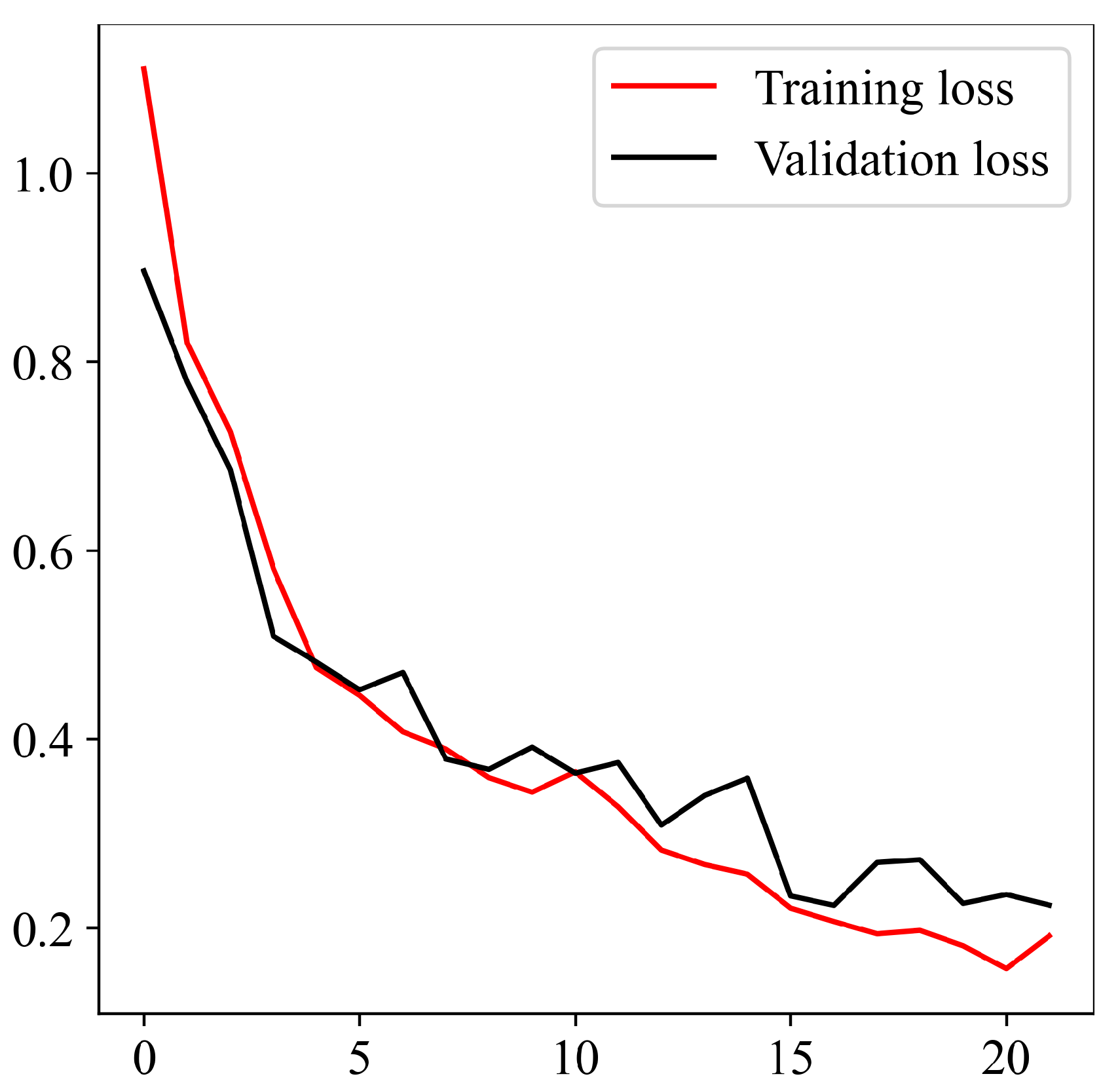
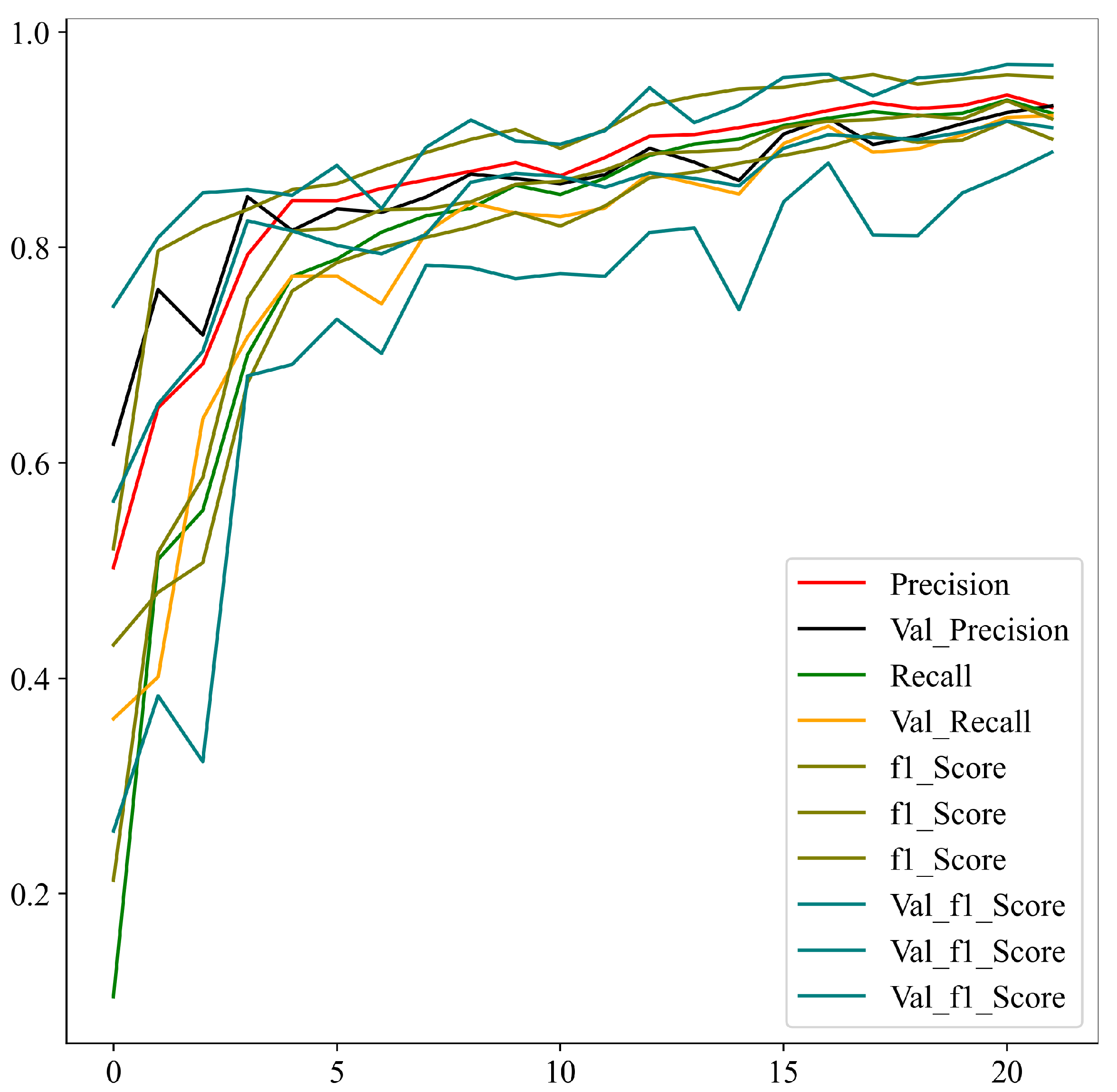
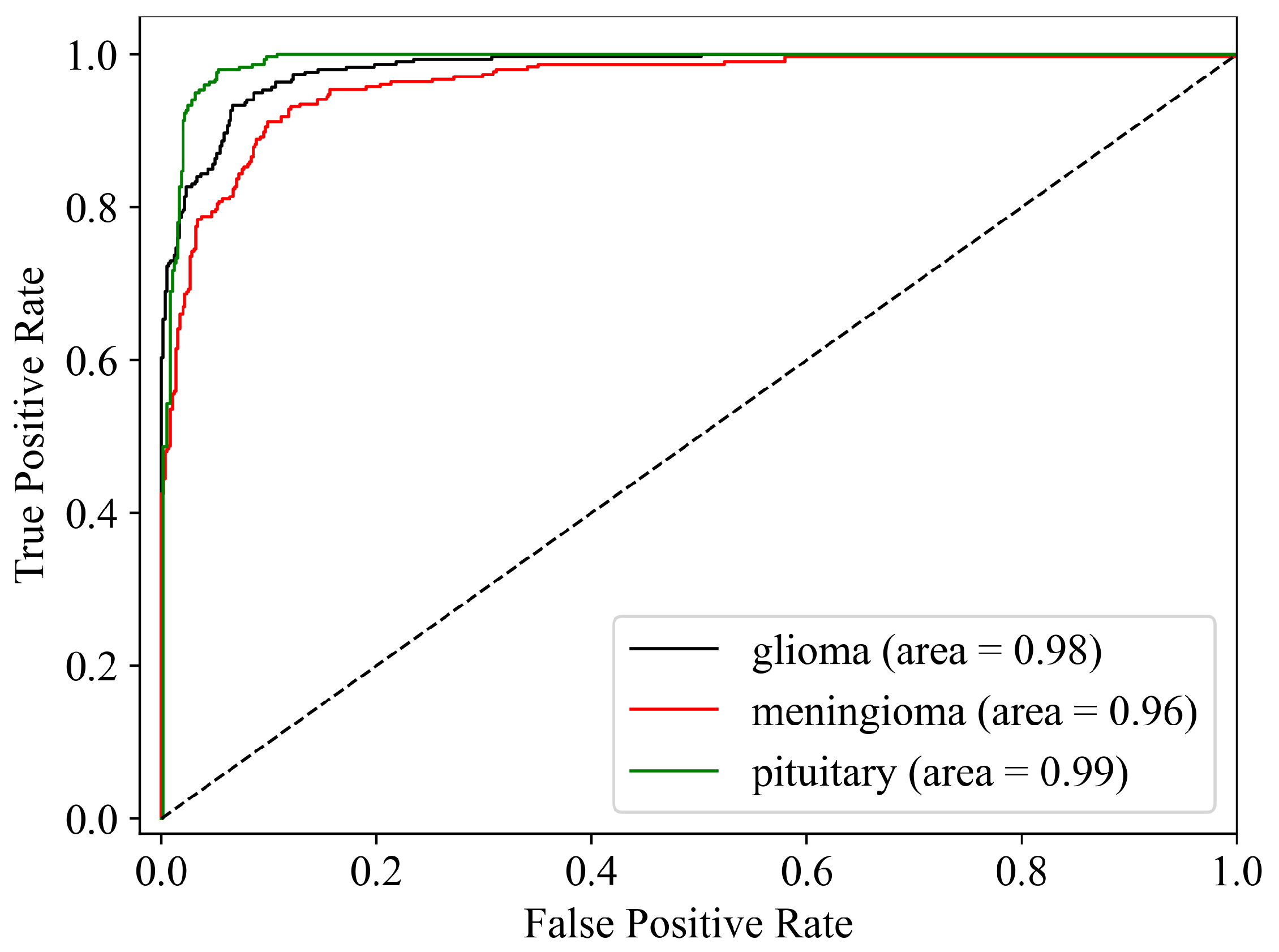
Upon successful model training, the focus shifts to the third component: result visualization. The presentation and interpretation of the model’s outcomes are crucial for conveying this study’s findings effectively. Visualizations may include accuracy metrics and comparative analyses of the model’s performance across different classes of brain tumors. These visual aids not only facilitate a deeper understanding of the model’s capabilities but also enhance the transparency and interpretability of the study’s outcomes.
The proposed work (see
Table 1) achieves significant results in classifying three types of brain tumors: glioma, meningioma, and pituitary tumors. This success not only underscores the robustness of our approach but also paves the way for further experimentation and comparison with state-of-the-art methods. By building on this foundation, future research can highlight the significance of our findings and explore potential improvements, thereby contributing to the advancement of brain tumor classification techniques.
The overarching goal of the proposed work is to harness the power of deep learning to learn features effectively, enabling accurate detection and subsequent classification of brain tumors in MRI images. By delving into the intricate details of each component data pre-processing, model training, and result visualization—this study aspires to contribute valuable insights to the burgeoning field of medical image analysis, paving the way for advancements in early diagnosis and treatment planning for individuals affected by brain tumors [
19].
1.1. Background
In recent years, deep convolutional neural networks (DCNNs) have gained widespread recognition for their significant advancements across various applications, especially in tasks related to image classification. The success of DCNNs can be primarily attributed to their complex architectural design and end-to-end learning approach, allowing them to derive meaningful feature representations from input data. Furthermore, ongoing research in the realm of DCNNs is focused on refining networks and training algorithms to extract even more discriminative features [
25].
There has been a significant interest in enhancing the capabilities of DCNNs, frequently achieved by either building deeper or wider networks. In this investigation, we embrace a wider network approach by conceptualizing a DCNN as the amalgamation of two subnetworks’ feature extractors positioned alongside each other, constituting the feature extractor of a dual-stream network [
13]. Consequently, when presented with a specific input image, two streams of features are extracted and then merged to create a unified representation for the final classifier in the overall classification process.
Dual-stream convolutional neural networks (CNNs) refer to a specific architecture that involves two parallel streams of information processing within the network. This concept is commonly employed in computer vision tasks, particularly in scenarios where information from different sources or modalities needs to be integrated for improved performance. Dual-stream CNNs typically consist of two parallel branches or streams that process different types of input data independently. These streams can handle different modalities, such as visual and spatial information, or they may process different aspects of the same modality. The main purpose of using dual streams is to capture and integrate complementary information from different sources. Fusion mechanisms are employed to combine features or representations extracted by each stream, enhancing the overall understanding of the input data [
30].
In some cases, dual-stream architectures are designed to process spatial and temporal information separately. For instance, in video analysis, one stream may focus on spatial features within individual frames, while the other stream considers the temporal evolution of these features across frames. Dual-stream networks are commonly used when dealing with multi-modal data, such as combining RGB and depth information in computer vision tasks. Each stream is specialized in processing a specific modality, and the information is fused to provide a more comprehensive representation. In object recognition tasks, one stream may be dedicated to recognizing the category of an object, while the other stream focuses on localizing the object within the image. This helps in achieving both accurate classification and precise localization [
19].
Pre-processing techniques, aside from deep learning, significantly improve feature learning by enhancing the quality of the input data. The White Patch Retinex Algorithm and CLAHE are employed in this work to achieve improved feature learning capabilities. The White Patch Retinex Algorithm enhances image quality by adjusting illumination, ensuring consistent brightness across the image and thereby improving color constancy. CLAHE, or Contrast-Limited Adaptive Histogram Equalization, further refines image quality by enhancing local contrast and making features in different regions more distinguishable. By applying these techniques, the data become uniform and representative, facilitating more accurate and efficient feature extraction in subsequent processing stages.
In digital image processing, the White Patch Retinex Algorithm is a color constancy algorithm used to adjust colors under different lighting circumstances. The method works on the premise that other colors can be adjusted in accordance with the brightest color in an image, which is presumed to be white. To equalize the effect of lighting disparities, the algorithm determines the highest intensity value in each color channel and then scales all pixel values in the image. This technique aids in producing a constant appearance of colors, which makes it helpful in fields like computer vision and photography where accurate color reproduction is essential.
Similarly, Contrast-Limited Adaptive Histogram Equalization, or CLAHE, is an advanced image processing method used to improve contrast in pictures. In contrast to conventional histogram equalization, CLAHE works on specific areas or tiles within the picture as opposed to the full image. This method reduces noise amplification and avoids oversaturation while enhancing local contrasts to help detect details in lighter or darker areas. CLAHE efficiently enhances the visibility of features in a variety of imaging applications, including medical imaging, photography, and video processing, by reducing contrast amplification.
By integrating the White Patch Retinex Algorithm and CLAHE, this work leverages the strengths of both techniques to improve the quality and consistency of the input data. This combined approach ensures that the processed images have uniform brightness and enhanced contrast, facilitating better feature learning and extraction in subsequent analysis stages.
1.2. Proposed Method
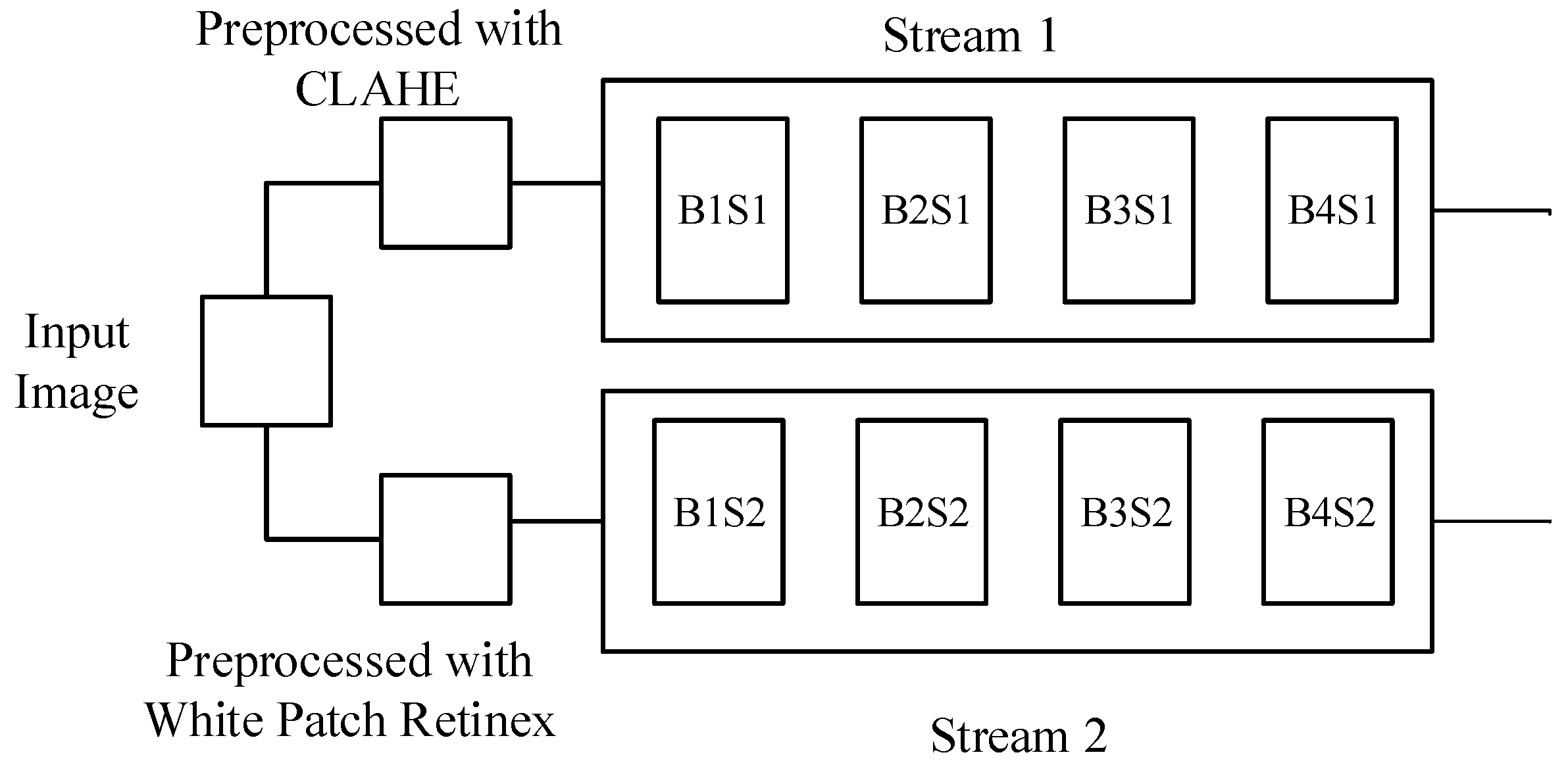
The proposed technique is based on a dual-stream network, illustrated in
Figure 1 and
Figure 2, respectively. Since both streams consist of a limited number of layers, their primary focus is on learning a variety of features, which, in our context, represent structural features. The dual-stream technique facilitates the model in acquiring a multitude of these features, subsequently amalgamating them to construct a comprehensive feature bank.
The proposed model incorporates two streams
and
which are based on CNNs as illustrated conceptually on
Figure 1 and
Figure 2. The streams
and
take two different inputs
and
. The stream, S1 consists of four blocks:
,
,
and
. Similarly, the stream
also comprises four blocks:
,
,
and
. Each block is based on two convolutional layers, a batch normalization layer, and a dropout layer. Stream
employs
filters, while
utilizes
filters; mathematically, we can write
in
and
in
. The features learned by each stream, formally denoted as
, and
, respectively, are generated using filter sizes of
and
, respectively. In each block, both streams
and
incorporate batch normalization and dropout layers, along with a max-pooling layer using a
filter.
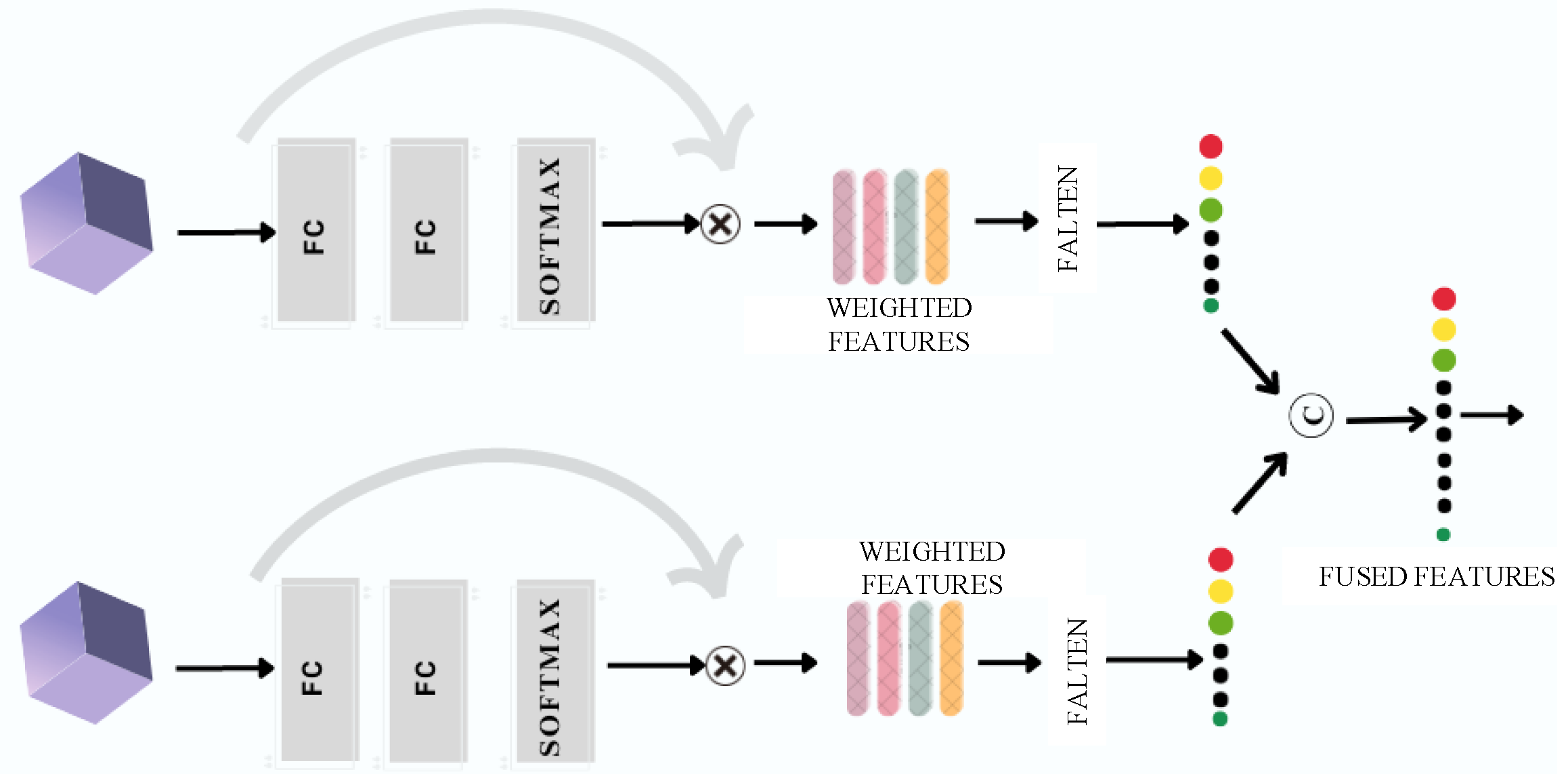
The resulting features,
and
, are then concatenated into
, as expressed in Equation (
3).
The streams,
and
, receive distinct inputs labeled as
and
. Input
undergoes pre-processing with Contrast-Limited Adaptive Histogram Equalization (CLAHE) [
20], while input
undergoes pre-processing with the White Patch Retinex Algorithm [
31]. Mathematically, we can write
The proposed model is depicted in two parts in
Figure 1 and
Figure 2. Part one (
Figure 1) shows the earlier part of the model, where, the network is primarily focused to learn features in a better way. Similarly, part two (
Figure 2) acquires the extracted features from an earlier part of the network and performs a fusion operation to combine the features acquired from both the streams. Hence, a comprehensive features bank is built to facilitate the identification process for the tumors.
The dual-stream CNN architecture is designed to leverage distinct convolutional pathways to process different inputs, extract meaningful features, and concatenate them for a comprehensive representation. The provided mathematical expressions offer a detailed insight into the operations occurring in each block and the overall flow of information through the network.
1.3. Image Pre-Processing
In order to identify better features within MRI images, the model is designed to comprehend a variety of features encompassing both anatomical and generic aspects. To enhance this objective, improved pre-processing approaches have been implemented. Upon training the model with pre-processed images, it demonstrates an enhanced ability to learn features, as reflected in the empirical results showcased in the Results section.
1.3.1. Contrast-Limited Adaptive Histogram Equalization (CLAHE)
The input
is the input to stream
; it is based on Contrast-Limited Adaptive Histogram Equalization (CLAHE) [
30]. CLAHE is an image processing technique employed to boost the contrast of an image without excessively amplifying noise. It serves as an extension of traditional histogram equalization, which seeks to evenly distribute intensity levels across an image to encompass the entire available range. However, conventional histogram equalization may inadvertently intensify noise, particularly in areas characterized by low contrast.
CLAHE addresses this issue by applying histogram equalization locally, in small regions of the image, rather than globally. Additionally, it introduces a contrast-limiting mechanism to prevent the over-amplification of intensity differences. Here is a brief explanation of CLAHE mathematically:
- (a)
Image Partitioning: The input image is divided into non-overlapping tiles or blocks. Each tile is considered as a local region.
- (b)
Histogram Equalization for Each Tile: Histogram equalization is applied independently to the intensity values within each tile. This is typically achieved using the cumulative distribution function
of the pixel intensities. For a given tile, let
be the histogram of intensities,
be the cumulative distribution function, and
be the intensity at pixel
. The transformed intensity
for each pixel is given by
- (c)
-
Clip Excessive Contrast: After histogram equalization, some intensity values may still be amplified significantly. To limit the contrast, a clipping mechanism is applied to the transformed intensities. If
exceeds a certain threshold, it is scaled back to the threshold value. The contrast-limited intensity
is given by
The threshold is a user-defined parameter that determines the maximum allowed contrast enhancement.
- (d)
Recombining Tiles: Finally, the processed tiles are recombined to form the output image. By applying histogram equalization locally and limiting the contrast, CLAHE enhances the image contrast effectively while avoiding the drawbacks associated with global histogram equalization. The algorithm is widely used in medical image processing and other applications where local contrast enhancement is crucial.
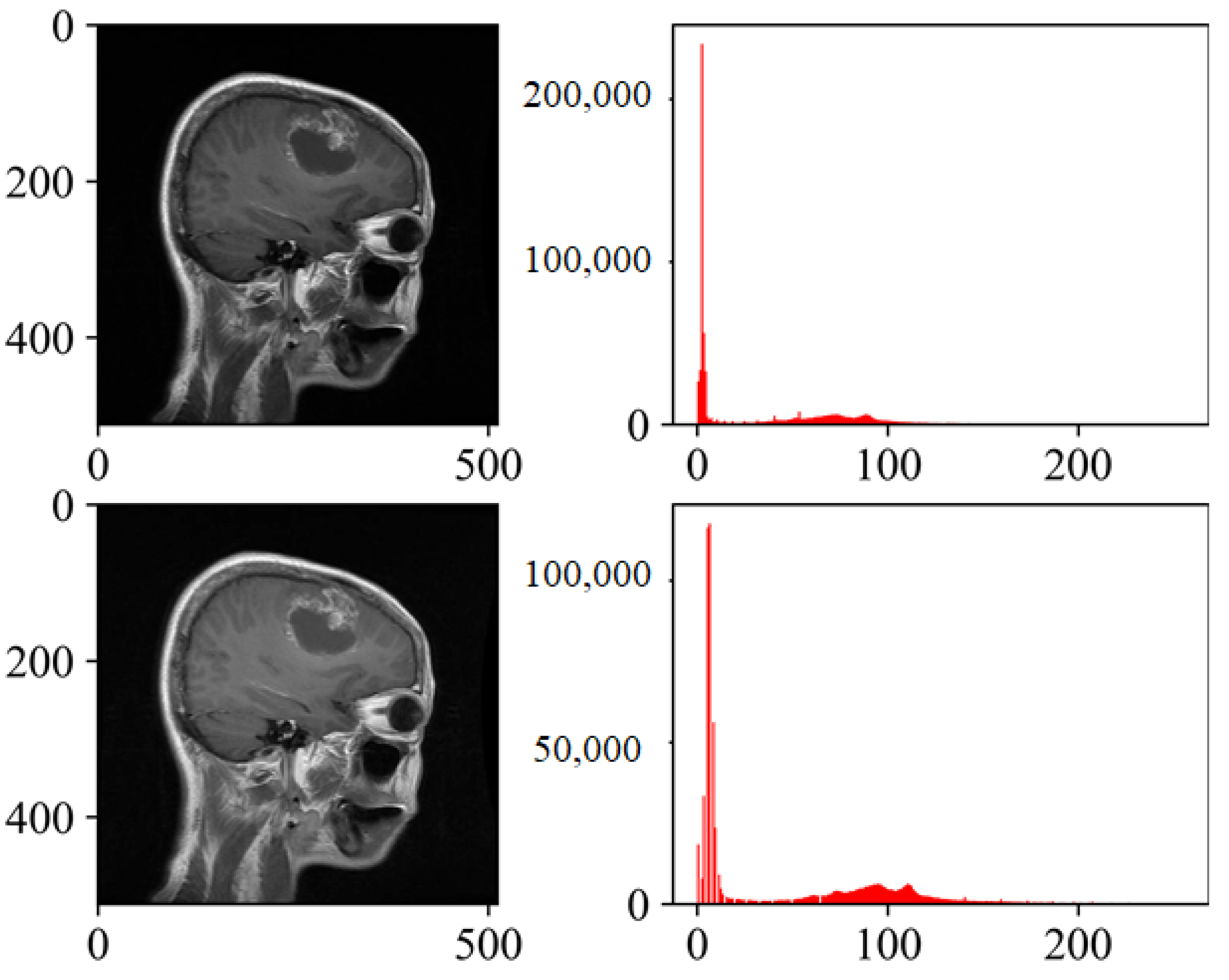
Figure 3 presents the histogram visualizations both pre- and post-processing using CLAHE.
1.3.2. White Patch Retinex Algorithm
The input
is the input to stream
; it is based on the White Patch Retinex Algorithm [
32]. It is a color correction technique used to enhance the color balance of an image. The basic idea is to assume that the color of the brightest object in the scene (often a white patch or object) should be achromatic (neutral) and to use this assumption to correct the color of the entire image [
32]. The Retinex algorithm, on which White Patch Retinex is based, is designed to correct for variations in lighting conditions.
The algorithm is based on few computational steps, which include computing the illuminance map, chromaticity map, maximum chromaticity, and scaling factor, adjusting the color channels, and clipping the adjusted color values. These computations are conducted as follows:
- (a)
-
Compute Illuminance Map: For each pixel in the image, calculate the log-average of the pixel’s color values. This is carried out separately for each color channel
.
Here, represents the illuminance map at pixel .
- (b)
-
Compute Chromaticity Map: The calculation of the chromaticity of each pixel is performed by subtracting the log-average value from each color channel.
The term represents the chromaticity value of color channel c at pixel .
- (c)
Find Maximum Chromaticity: For each pixel, finding the maximum chromaticity value across the color channels is accomplished by computing
.
- (d)
Compute Scaling Factor: Computation of a scaling factor for each color channel is based on the maximum chromaticity value.
- (e)
-
Adjust Color Channels: Each color channel is scaled by its corresponding scaling factor.
The symbols represent the adjusted color values.
- (f)
Clip Values: Clip the adjusted color values to ensure they are within the valid color range (0 to 255 for 8-bit images). The output of the algorithm is the color-corrected image with improved color balance. The White Patch Retinex Algorithm assumes that the whitest object in the scene is achromatic, and it corrects the image by scaling the color channels based on the chromaticity information. It is a computationally efficient method for color correction and has been widely used in image processing applications.
Figure 4 depicts the histogram visualizations before and after pre-processing with the White Patch Retinex Algorithm.
1.4. Two-Fold Margin Loss
In this work, a two-fold margin loss is used to boost the feature learning process for brain tumor detection.
Equation (
8) represents the mathematical notation of the two-fold margin loss, and the complete details are shown in Equations (2)–(8). Here,
represents the corresponding class, and
I is the indicator function. If a sample corresponds to
, then
; otherwise,
. The symbols
and
represent the upper bounds. Similarly, the symbols
and
represent the lower bounds. In this work, the
,
and
have the values of
,
,
and
, respectively. The term
indicates the Euclidean distance of the resulting vector. The symbols
and
represent the lower bounds of the non-existent classes. Both of these parameters help to maintain the length of the activity vector during the learning process of the neurons, which are fully connected. In this work, the values of
and
are
and
, respectively.
is the total calculated loss after the amalgamation of
and
.
1.5. Performance Metric
In the context of medical image classification, accuracy is frequently deemed a less reliable performance metric. Imagine a scenario where only 5% of the training dataset represents the positive class, and the objective is to classify every case as the negative class. In such a situation, the model would achieve a 95% accuracy rate. While 95% accuracy on the entire dataset might seem impressive, this approach overlooks the crucial detail that the model misclassified all positive samples. Consequently, accuracy fails to offer meaningful insights into the model’s effectiveness in this particular classification task [
31]. Hence, in addition to accuracy, we incorporate sensitivity, specificity, f1-score, and AUC_ROC curves for evaluating performance. The performance metrics employed in this analysis are outlined below: