1. Introduction
Fluorescence microscope images are obtained by imaging cells or tissues labeled with fluorescent dyes and fluorescent proteins. It contains many biologically relevant phenotypic features, which allows people to characterize experimentally gene expression, protein expression, and molecular interactions in a living cell [
1] . The application of cell analysis based on fluorescence microscopy images is diverse, including identifying disease phenotypes, gene functions, and mechanisms of action, toxicity, or targets of drugs [
2]. Analysis methods based on fluorescence microscope images, such as cell classification, segmentation, colocalization analysis, and morphological analysis, require high-quality microscopic images. However, due to the protein binds to an excessive amount of fluorescent dye, long exposure times, and inhomogeneous illumination [
3], there are usually artifacts such as blur, boundary shadows, and saturation artifacts in microscopic images. These artifacts may severely affect the analysis results. For example, uneven illumination increased the error detection and missed detection of yeast cell images by 35% by CellProfiler [
1], and saturation artifacts will make the measurement of protein position invalid in colocalization location analysis.
At present, the research on processing fluorescence image artifacts mainly focuses on inhomogeneous illumination, super-resolution reconstruction, and denoising. Smith et al. [
4], Goswami et al.[
5] and wang et al.[
6] use prospective methods or retrospective methods to correct illumination between different fluorescence images and reduce abiotic structural differences between different images. These methods reduce abiotic structural differences between different images or remove the artifact noise on a single microscopic image. However, none of them can eliminate saturation artifacts in a single microscopic image. Saturation artifacts can be regarded as extreme illumination imbalances. Excessive exposure makes the artifact area blank, and a large area of biological structure information is missing. Often these microphotographs with a large amount of missing biological structure information will be screened out in quantitative analysis experiments [
7]. There is still no method that can eliminate the influence of saturation artifacts.
Generative adversial networks (GAN) were proposed by Goodfellow et al. [
8] in 2014 as a tool for generating data. GANs and improved GANs algorithms are widely used in image generation, image inpainting, and other fields by data-driven approaches in recent years and have excellent performance. Zhang et al. [
9] used GAN to provide an effective method for medical image data enhancement and forgery detection, effectively improving the accuracy and reliability of computer-aided diagnostic tasks. GANs have also had stunning success in image processing of fluorescence microscopy. Chen et al. [
10] use the GAN method to realize super-resolution reconstruction of fluorescence microscope images, making the biological structure information stand out clearly from the artifact. In this paper, we proposed a method to restore the missing biological structure information caused by saturation artifacts in each image. To our best knowledge, this is the first study to deal with the lost biological information. Belthangady et al. [
11] showed that CNN-based techniques for inpainting missing image regions are well-positioned to address the problem of losing information. Their work inspired us to believe that the deep learning method is a good way to solve the problem of losing biological information by saturation artifacts.
In this work, we further explore GAN-based methods to solve the problem of missing biological information due to saturation artifacts in the fluorescence microscope images. The method is based on EdgeConnect GAN [
12], we call it Two-stage Cell image GAN (TC-GAN). To obtain more stable and credible inpainting results, the model adopts a two-step progressive repair method. In the first stage, the shape features of the cell and the context features between cells are restored by the proposed Edge-GAN. In the second part, the texture features and intensity-based features of the cell are restored by the proposed Content-GAN based on the edge information. We introduce the contextual attention [
13] architecture into the model to learn where to borrow or copy feature information from known background patches to generate missing patches. With this model, images with losing information can restore the phenotypic features according to their existing phenotypic features and make up for the scarce samples in morphological analysis experiments.
The structure of the paper is as follows.
Section 2 describes the structure and loss function of the model.
Section 3 introduces the data and processing methods. The image inpainting experiment and verification experiment are presented in detail in
Section 4. The conclusion is made in
Section 5.
2. Methodology
This chapter describes in detail the structure of the proposed fluorescence microscope cell image inpainting model and the loss function used.
2.1. Generative Adversarial Networks
GAN was proposed by Goodfellow et al. [
8] in 2014 to generate signals with the same feature distribution as the training set. A typical GAN consists of a generator and a discriminator, where the generator tries to generate data that matches the distribution of the training set; the discriminator determines whether the input signal is the original signal or the signal generated by the generator.
However, the content generated based on GAN usually has the problem of blurred edge of restored content or semantic mismatch between restored content and background content. We focus on the performance of the restoration results on four numerical features of a cell fluorescence microscopic image and use a two-stage GAN with a contextual attention layer to restore different feature contents.
2.2. Feature Restoration
The phenotypic features of cells provide the raw data for profiling. It can be extracted to quantitatively describe complex cell-morphology phenotypes. Here, these phenotypic features can be separated into four categories [
1], [
14]: (1) Shape features, which represent boundaries, size, or shape of nuclei, cells, or other organelles. (2) Microenvironment and context features, including the distribution among cells and subcellular structures in the field of view. (3) Texture features, which describe the distribution of pixel-intensity values within the cellular structure. These features can intuitively display the fluorescent protein structure of a single cell. (4) Intensity-based features, which are computed from actual intensity values on a single-cell basis. Intensity-based features are closely related to texture features. The intensity-based features dominate when analyzing between a few pixels, as the number of distinguishable discrete intensities increases within a small area, the texture features will dominate [
15]. In the fluorescence microscope image, saturation artifacts will cause sparse texture features.
We used a two-stage network (from Edge-GAN to Content-GAN) to restore the above four features from saturated artifacts. Edge-GAN is used to generate the phenotypes of shape features and context features, which including the cell shape and centroid direction, established the basic morphology of cell phenotype. After determining this most basic and important information in saturation artifacts, the texture features can be further restored by using the Content-GAN, which are usually shown as the protein structure, organelle structure, and cytoplasm of the cell in our eyes. Contextual attention architecture [
13] is added to the network structure, to make the boundary and texture features of the patched area consistent with the surrounding cells in morphology.
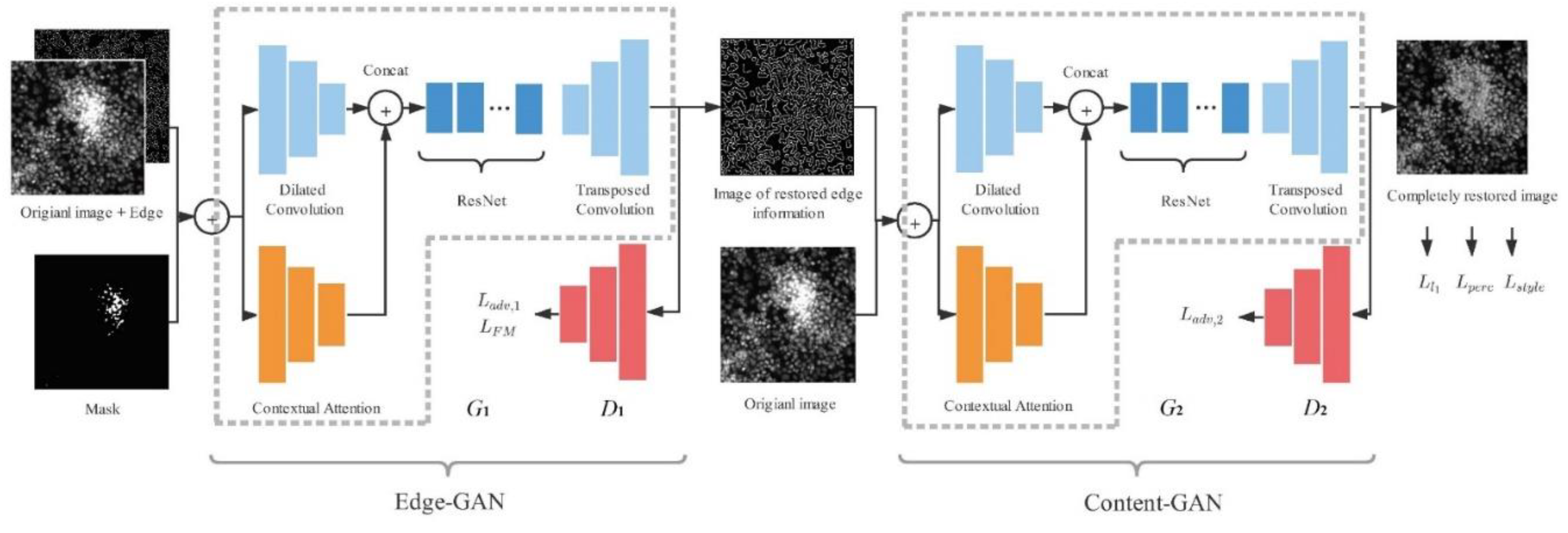
2.3. Model Strutcture
The modules of our network are shown in
Figure 1. The model is divided into two parts, we use Edge-GAN and Content-GAN respectively in these two parts. The Edge-GAN consists of generator
and discriminator
. The original grayscale image, the imaging mask, and masked edge image obtained from the region of saturation artifacts by the Canny operator are the input of the Edge-GAN, generator
and discriminator
. By learning the distribution of the features extracted from the input image, the Edge-GAN outputs the edge image. The Content-GAN consists of
and
. The original grayscale image and edge grayscale image are the input of Content-GAN. By learning the texture features from the original image and the shape features from the edge image, the output is the restored image without saturation artifacts.
The generator of Edge-GAN,
, is composed of an encoder-decoder convolution architecture with a contextual attention architecture [
13]. Specifically, the encoder-decoder architecture consists of the encoder, ResNet module, and decoder. The contextual attention architecture is parallel to the encoder architecture. The discriminator of Edge-GAN,
, follows the same architecture of 70×70 PatchGAN [
16], the detailed structure of
and
are shown in
Table 1.
The architecture of the generator of Content-GAN, , is the same as , except all the spectral normalization is removed from . And the architecture of the discriminator of Content-GAN, , is the same as . is used to judge whether the semantic information of the generated content by is reasonable or not.
2.4. Contextual Attention
Contextual attention architecture is proposed by Yu et al. [
13] to learn where to borrow or copy feature information from known background patches to generate missing patches. Its detailed structure is shown in the contextual attention architecture of
Table 1. We use the contextual attention layer to accelerate the convergence rate of model training and enhance the semantic rationality of the generating region. The similarity of a patch centered in the patch to be restored
and the background patch
is defined as:
According to the calculated similarity score , contextual attention layer can learn which part of the background features should be used from the repaired texture information.
2.5. Edge-GAN Loss Function
The Edge-GAN is trained with adversarial loss and feature-matching loss [
17] as:
where
is adversarial loss,
is feature-matching loss,
and
are regularization parameters. The adversarial loss
is defined as:
where
is the ground truth images, E is the edge map of
.
is predicted edge map for the masked region.
The feature-matching loss
extracts the middle feature layer of the discriminator for comparison. The
is defined as:
where
means the number of feature layers,
is the final layer of
,
is the number of elements in
th layer,
is the ith layer of
.
In our experiments, and .
2.6. Content-GAN Loss Function
The Content-GAN is trained by four losses. The overall loss function is to
, which is defined as:
where
is
loss,
is adversarial loss,
is perceptual loss [
18],
is style loss [
19].
,
,
and
are regularization parameter.
Adversarial loss,
is defined as:
where the composite edge map
, the inpainting color image
.
The perceptual loss is similar to feature-matching loss, which extracts the middle feature layer for comparison in
. Using perceptual loss can avoid generating final content that is the same as the input image, as long as the abstract features are the same. It is defined as:
where
is the activated feature map of the ith layer of
. Here we use the VGG-19 pre-trained parameter on the ImageNet dataset [
20] to be the parameter of
.
The style loss is used to punish the non-intensity affine transformation and reduce the distortion of cell morphological transformation. It is defined as:
where
is a Gram matrix constructed of
from feature maps
,
.
3. Data and Processing
3.1. Data of Fluorescence Microscope Image
There are 1,108 different small interference RNAs (siRNAs) introduced into four types of cells to create distinct genetic conditions. The experiment uses a modified cell painting staining protocol which uses 6 different stains to adhere to different parts of the cell. The stains fluoresce at different wavelengths and are therefore captured by different imaging channels. Thus there are 6 channels per imaging site in a well.
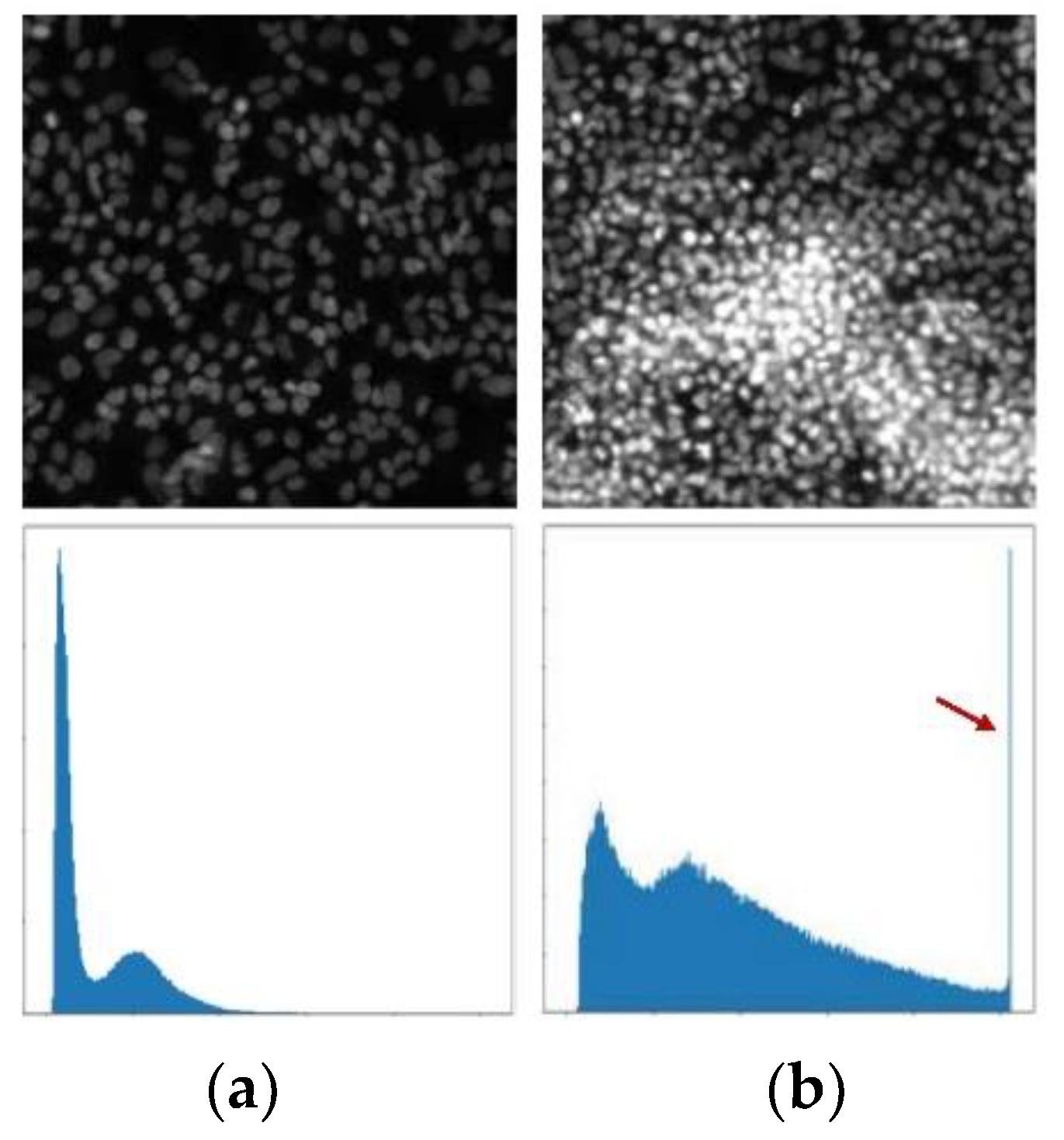
Different types of cell information are reflected in the morphological differences of fluorescence microscope images. The morphological analysis of cells is usually based on these morphological features. The most significant influence on the features of morphological differences is the saturation artifacts, which is shown in
Figure 2.
This section may be divided by subheadings. It should provide a concise and precise description of the experimental results, their interpretation, as well as the experimental conclusions that can be drawn.
3.2. Training Set Preparation
The data used in this study were divided into 2 groups (T1 and SET1). The data set T1 is used to train TC-GAN models, which are used to restore with the saturation artifacts in the image. SET1 is used to evaluate the validity of the restored feature.
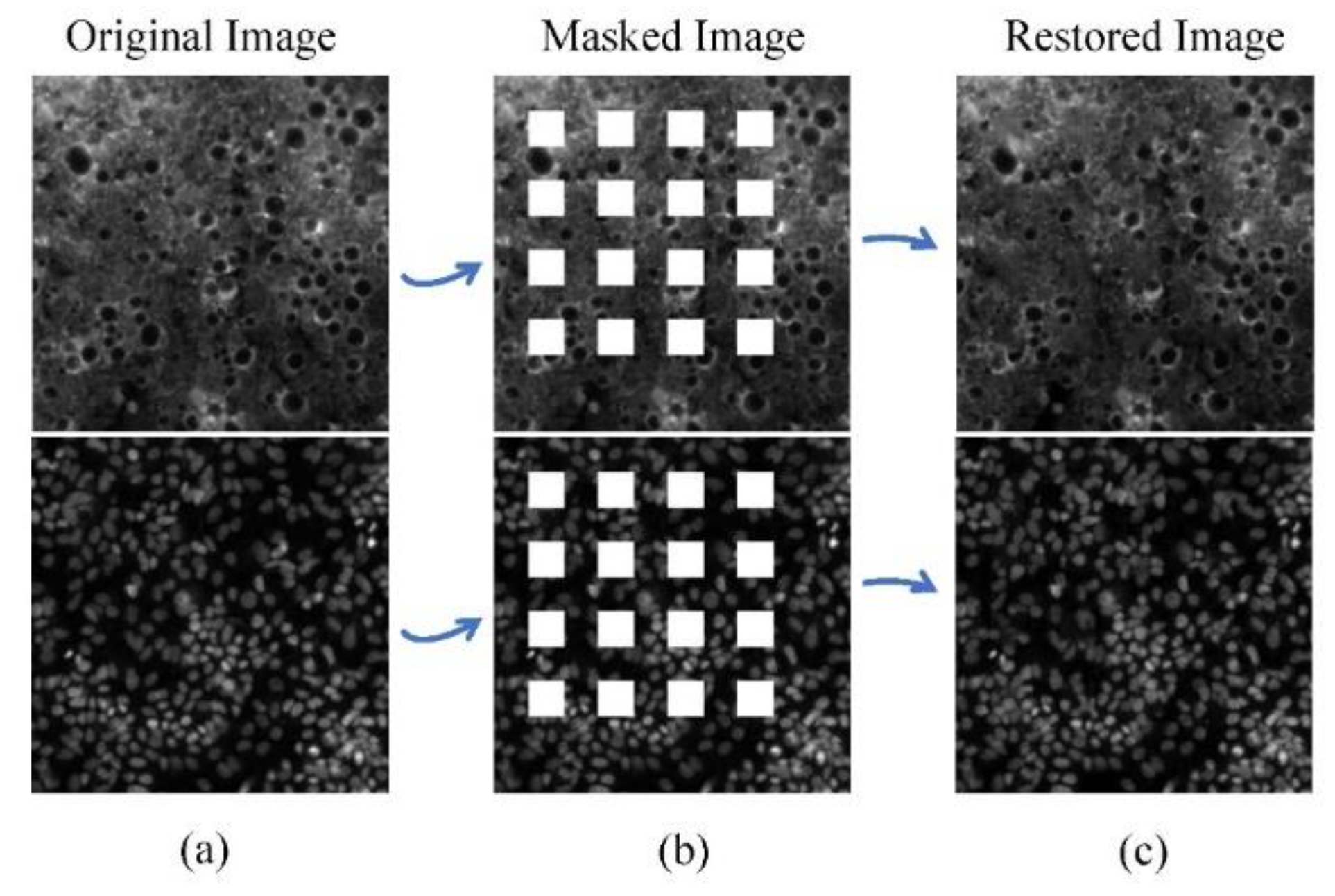
The data in T1 were selected from RxRx1 that did not contain saturation artifacts and were morphologically rich. This ensures that the trained algorithm can fill with rich textures. SET1 includes 5 images without saturation artifact selected in original RxRx1 data, 5 masked images with a regularly distributed rectangular mask added to simulate saturation artifacts in 20% of the area, and 5 restored images restored by TC-GAN.
4. Training Strategy and Analysis
In this section, we first introduce the experimental progressive training strategy and its ablation experiment results. and the training process and experimental results of TC-GAN are introduced. We used Peak Signal-to-Noise Ratio (PSNR), Structural Similarity Index (SSIM), and Frechet Inception Distance (FID) are used to evaluate the validity of restoration results.
4.1. Training Strategy
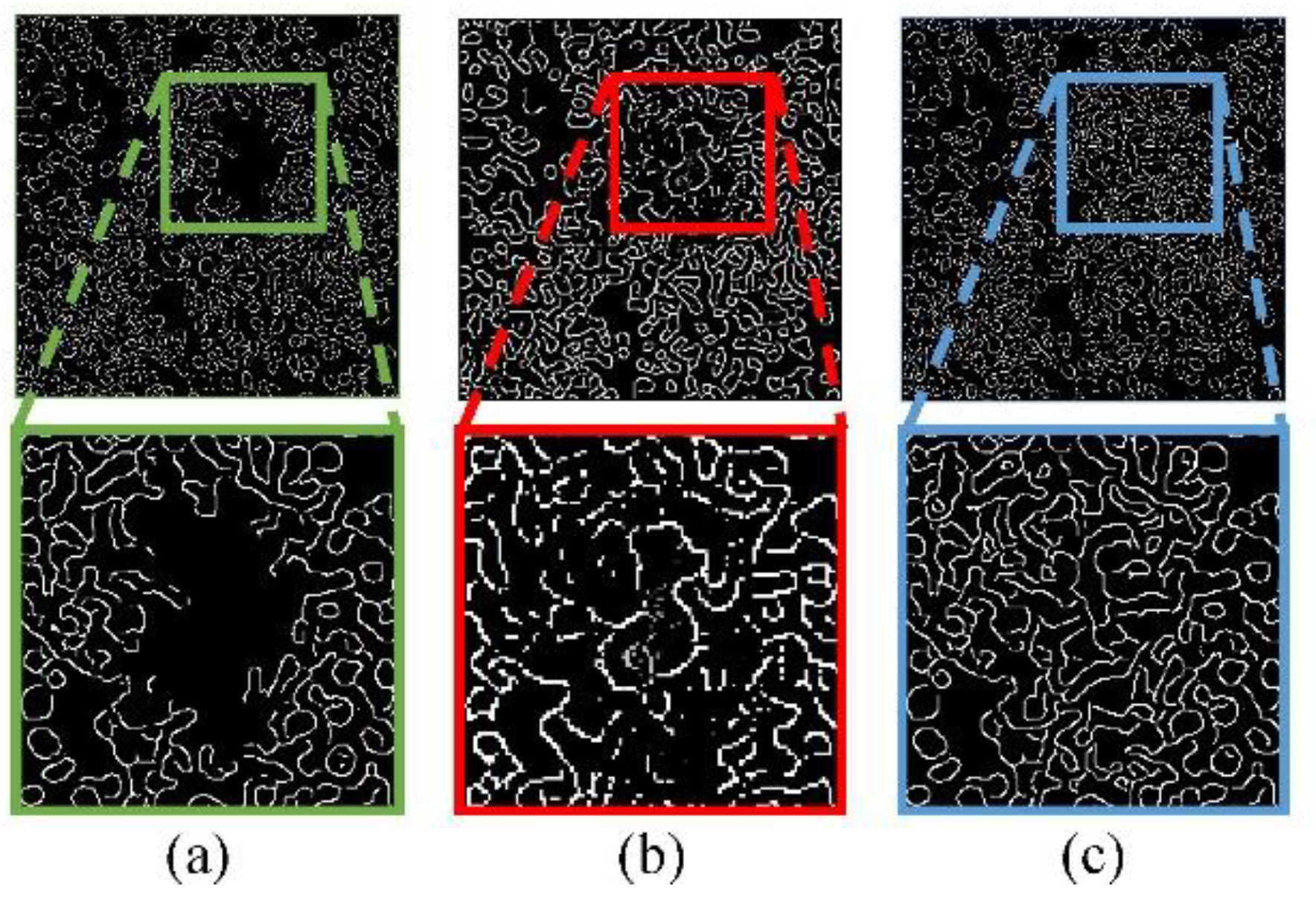
It is challenging to restore the phenotypic features directly, especially shape features and context features of cells in a large area of saturation artifacts. We use the method of progressive generation in the process of Edge-GAN training. And by using the results of Edge-GAN, Content-GAN can restore apparent texture features.
Specifically, the Edge-GAN trains on low-resolution images for pre-training, then we use transfer learning based on pre-training to train Edge-GAN on high-resolution images. The results of shape features and context features restoration in stages are shown in
Figure 3. As it shows, the phenotypic features between cells are gradually restored.
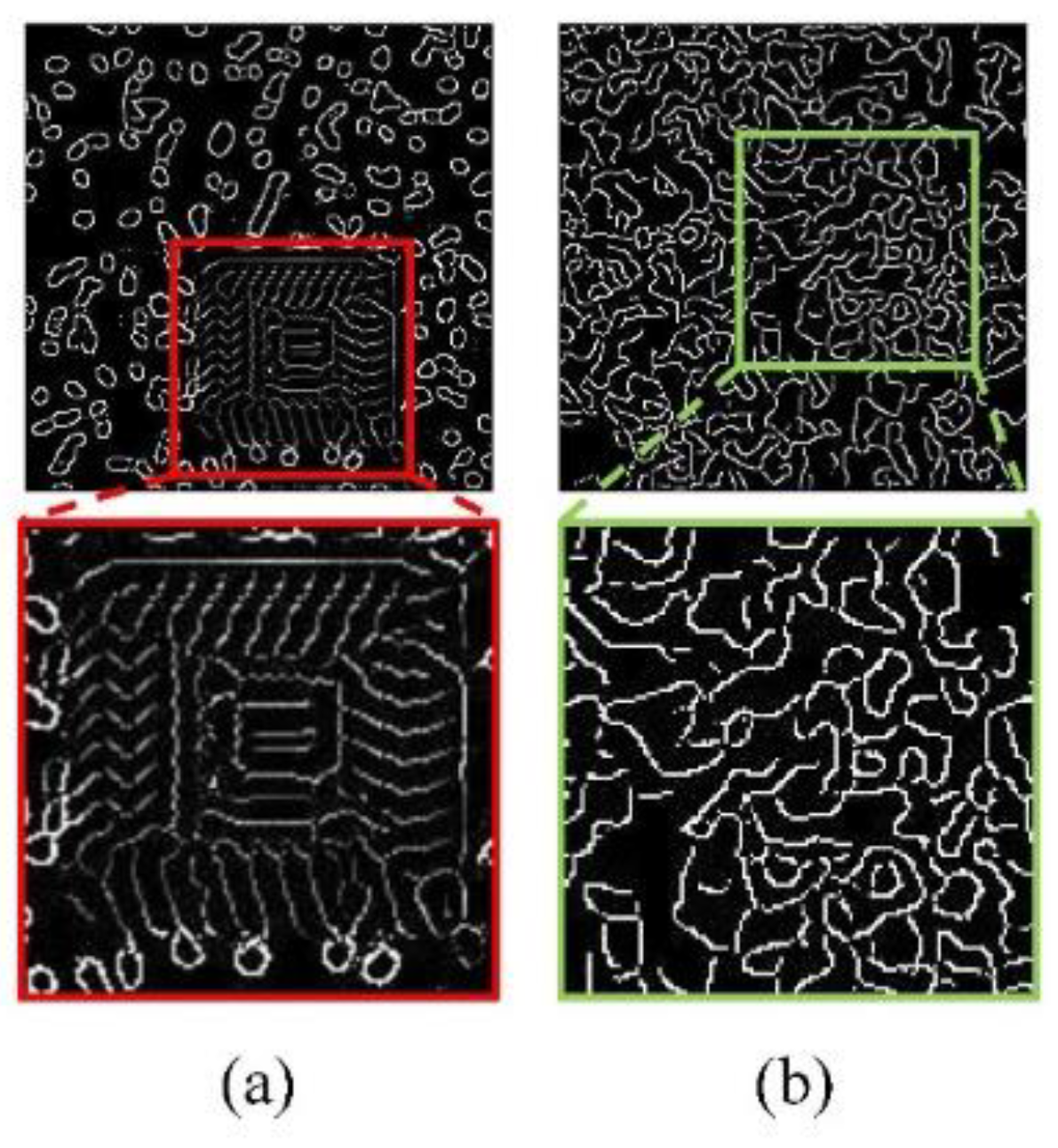
In this study, an ablation experiment was carried out, and the experimental results of progressive restore and no progressive restore were compared, and the restore results were shown in
Figure 4 showing the restore results of missing edge information at the 50,000th step in Edge-GAN.
4.2. Model Training and Sesult
The restoration TC-GAN models are trained by T1, which have rich phenotypic features without saturation artifacts. Images in the training set, imaging mask, and the masked edge consist of the input of TC-GAN. And the final output of TC-GAN is the restored image without saturation artifacts.
For Edge-GAN, the optimizer is Adam [
21] with a learning rate of
0.0001,
= 0,
= 0.9. The total training iteration is 1,000,000. For Content-GAN, the optimizer is the same as Edge-GAN, and the training iteration is 200,000.
The result of the two-stage restoration of the network is shown in
Figure 5. As was shown, the restored image fills the lost phenotypic features in the original saturation artifacts area.
4.3. Evaluation of Validity
4.3.1. Evaluation Indicators
We use several quantitative statistical features to verify the effectiveness of the method. The peak signal-to-noise ratio (PSNR), the structural similarity index (SSIM) [
22], and Fréchet Inception Distance (FID) [
23] are used to evaluate the quality of generation features quantitatively.
PSNR evaluates the quality of the generated features compared with the original features. The higher the PSNR, the smaller the distortion of the generated features.
SSIM is an index to measure the similarity of features in two images. The closer the SSIM is to 1, the closer the patched features are to the original cell.
4.3.2. Evaluation Methods
By comparing the verification metrics of the mask group image with artificial artifacts and the restoration group image, we intend to verify whether the restoration result can effectively restore the image information. We calculate the PSNR, SSIM, and FID between the 5 original images without saturation artifacts and the 5 masked images after they are artificially covered in SET1 to obtain the data of the mask group. Then we calculate the PSNR, SSIM, and FID between the 5 original images and the 5 restored images after being covered in SET1 to obtain the restoration group data. Calculation results of the mask group and restoration group can show the image quality before and after restoration. In addition, they can reflect the similarity between the restoration area and the original cell.
4.3.3. Result
We use PSNR, SSIM, and FID to evaluate the validity of generation features in SET1. The index difference between the restored image and the masked image is shown in
Table 2. The PSNR and SSIM of the restored image are higher than those of the masked image. This means the restored phenotypic features can effectively fill the gap of saturation artifacts and make the restored image closer to the original image than the masked image. The FID of the restored image are lower than that of the masked image, which means the similarity between the restored image and the original image is higher than that between the masked image and the original image. Two examples of the original image, the masked image, and the restored image are shown in
Figure 6.
5. Conclusions
This paper introduces the TC-GAN model, a two-stage phenotypic feature restoration approach addressing saturation artifacts in fluorescence microscopy images. The model separately restores the shape and texture features of cells. Through ablation studies and quantitative and qualitative experiments, the effectiveness of the network under progressive training is validated. The results demonstrate the model's practical significance and its potential to enhance qualitative and quantitative analysis of cell fluorescence microscopy images.
References
- Caicedo, J.; et al. Data-analysis strategies for image-based cell profiling. Nat. Methods 2017, 14, 849–863. [Google Scholar] [CrossRef]
- Bougen-Zhukov, N.; Loh, S.; Hwee-Kuan, L.; Loo, L.-H. Large-scale image-based screening and profiling of cellular phenotypes: Phenotypic Screening and Profiling. Cytom. Part A 2016, 91. [Google Scholar]
- Ettinger, A.; Wittmann, T.J.M. Fluorescence live cell imaging. Methods Cell Biol. 2014, 123, 77–94. [Google Scholar]
- Smith, K.; et al. CIDRE: An illumination-correction method for optical microscopy. Nat. Methods 2015, 12. [Google Scholar] [CrossRef] [PubMed]
- Goswami, S.; Singh, A. A Simple Deep Learning Based Image Illumination Correction Method for Paintings. Pattern Recognit. Lett. 2020, 138. [Google Scholar] [CrossRef]
- Wang, Q.; Li, Z.; Zhang, S.; Chi, N.; Dai, Q. A versatile Wavelet-Enhanced CNN-Transformer for improved fluorescence microscopy image restoration. Neural Netw. 2024, 170, 227–241. [Google Scholar] [CrossRef] [PubMed]
- Aladeokin, A. ; et al. Network-guided analysis of hippocampal proteome identifies novel proteins that colocalize with Aβ in a mice model of early-stage Alzheimer’s disease. Neurobiol. Dis. 2019, 132, 104603. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, I.; et al. Generative Adversarial Networks. Adv. Neural Inf. Process. Syst. 2014, 3. [Google Scholar] [CrossRef]
- Zhang, J.; Huang, X.; Liu, Y.; Han, Y.; Xiang, Z. GAN-based medical image small region forgery detection via a two-stage cascade framework. PLoS ONE 2024, 19, e0290303. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, C.; Zhao, J.; Xiong, Z.; Zha, Z.J.; Wu, F. Weakly Supervised Neuron Reconstruction From Optical Microscopy Images With Morphological Priors. IEEE Trans. Med. Imaging 2021, 40, 3205–3216. [Google Scholar] [CrossRef] [PubMed]
- Belthangady, C.; Royer, L. Applications, Promises, and Pitfalls of Deep Learning for Fluorescence Image Reconstruction. Nat. Methods 2018, 16, 1215–1225. [Google Scholar] [CrossRef] [PubMed]
- Nazeri, K.; Ng, E.; Joseph, T.; Qureshi, F.; Ebrahimi, M. EdgeConnect: Generative Image Inpainting with Adversarial Edge Learning. arXiv preprint arXiv 2019.
- J. Yu, Z. J. Yu, Z. Lin, J. Yang, X. Shen, and X. Lu, Generative Image Inpainting with Contextual Attention,”IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2018, pp. 5505–5514.
- Boutros, M.; Heigwer, F.; Laufer, C. Microscopy-Based High-Content Screening. Cell 2015, 163. [Google Scholar] [CrossRef] [PubMed]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973. SMC-3. [Google Scholar] [CrossRef]
- P. Isola, J. Y. Zhu, T. Zhou, and A. A. Efros, "Image-to-Image Translation with Conditional Adversarial Networks. IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2017, pp. 5967–5976.
- T.-C. Wang, M.-Y. Liu, J.-Y. Zhu, A. Tao, J. Kautz, and B. Catanzaro, "High-Resolution Image Synthesis and Semantic Manipulation with Conditional GANs. IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2017; pp. 8798–8807.
- Johnson, J.; Alahi, A.; Li, F. Perceptual Losses for Real-Time Style Transfer and Super-Resolution. European Conference on Computer Vision, 2016.
- Gatys, L.A.; Ecker, A.S.; Bethge, M. Image Style Transfer Using Convolutional Neural Networks. in 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2016, pp. 2414–2423.
- Russakovsky, O.; et al. ImageNet Large Scale Visual Recognition Challenge. International Journal of Computer Vision 2014, 115. [Google Scholar]
- Kingma, D.; Ba, J. Adam: A Method for Stochastic Optimization. Int. Conf. Learn. Represent. 2014.
- O. Elharrouss, N. Almaadeed, S. Al-Maadeed, and Y. Akbari, "Image Inpainting: A Review. Neural Process. Lett. 2020, 51, 2007–2028. [CrossRef]
- Bynagari, N. GANs Trained by a Two Time-Scale Update Rule Converge to a Local Nash Equilibrium. Asian J. Appl. Sci. Eng. 2019, 8. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).