Submitted:
04 June 2024
Posted:
05 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials
3. Methods
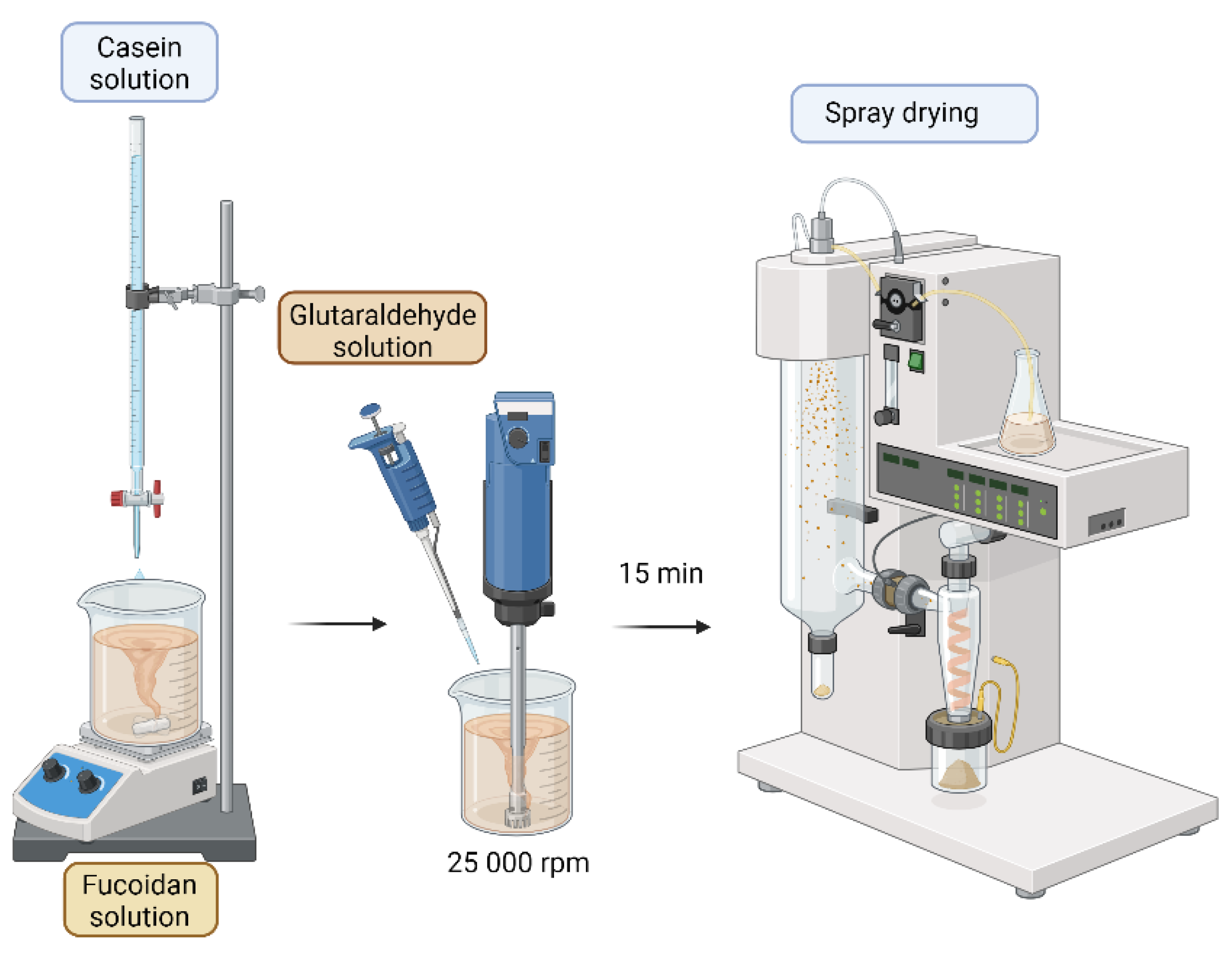
3.1. Preparation of Casein/Fucoidan Composite Nanoparticles
3.2. Characterization of Physicochemical Properties
3.2.1. Production Yield
3.2.2. Particle Size Analysis, Size Distribution, and Zeta Potential
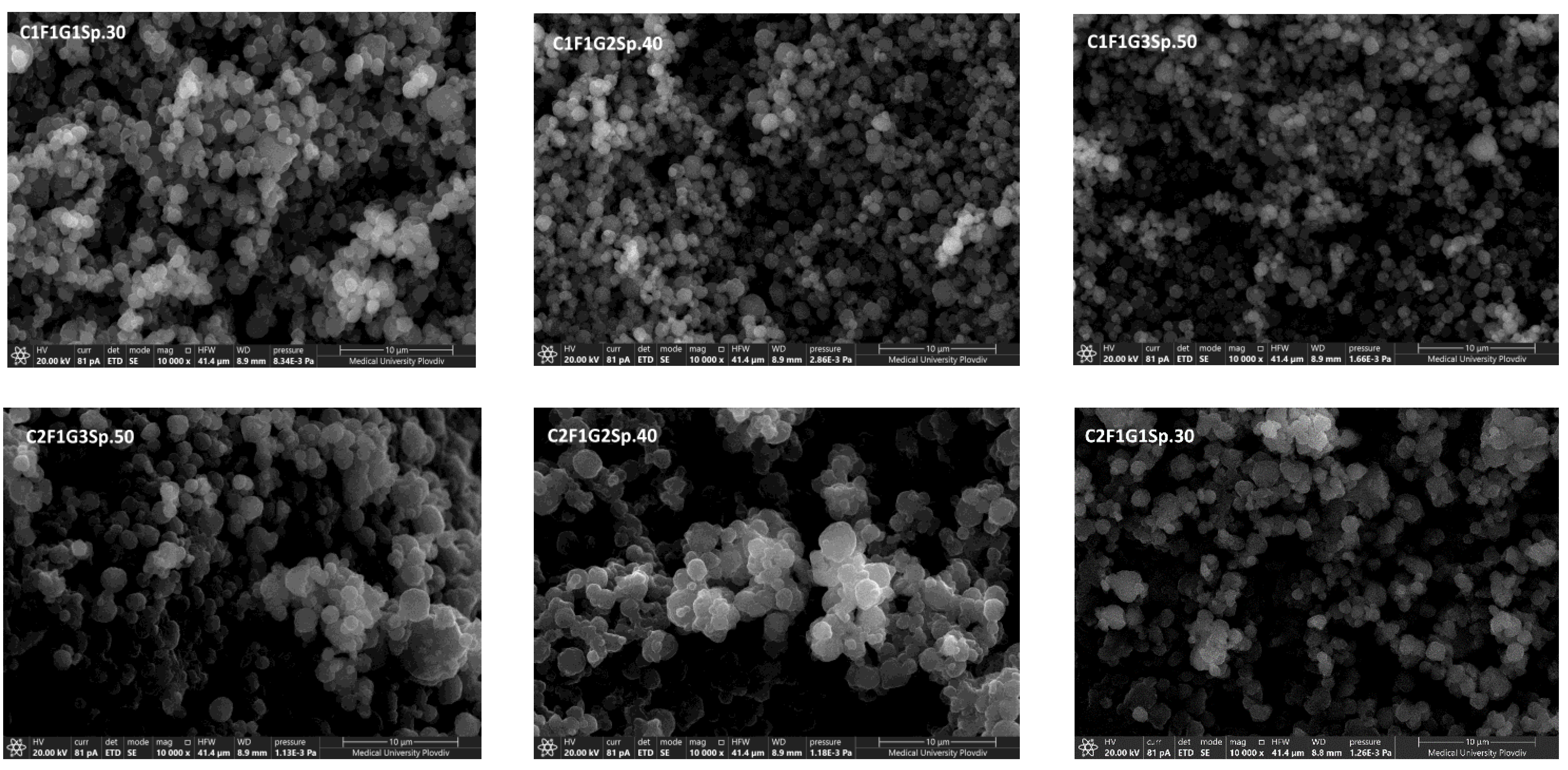
3.2.3. Scanning Electron Microscopy
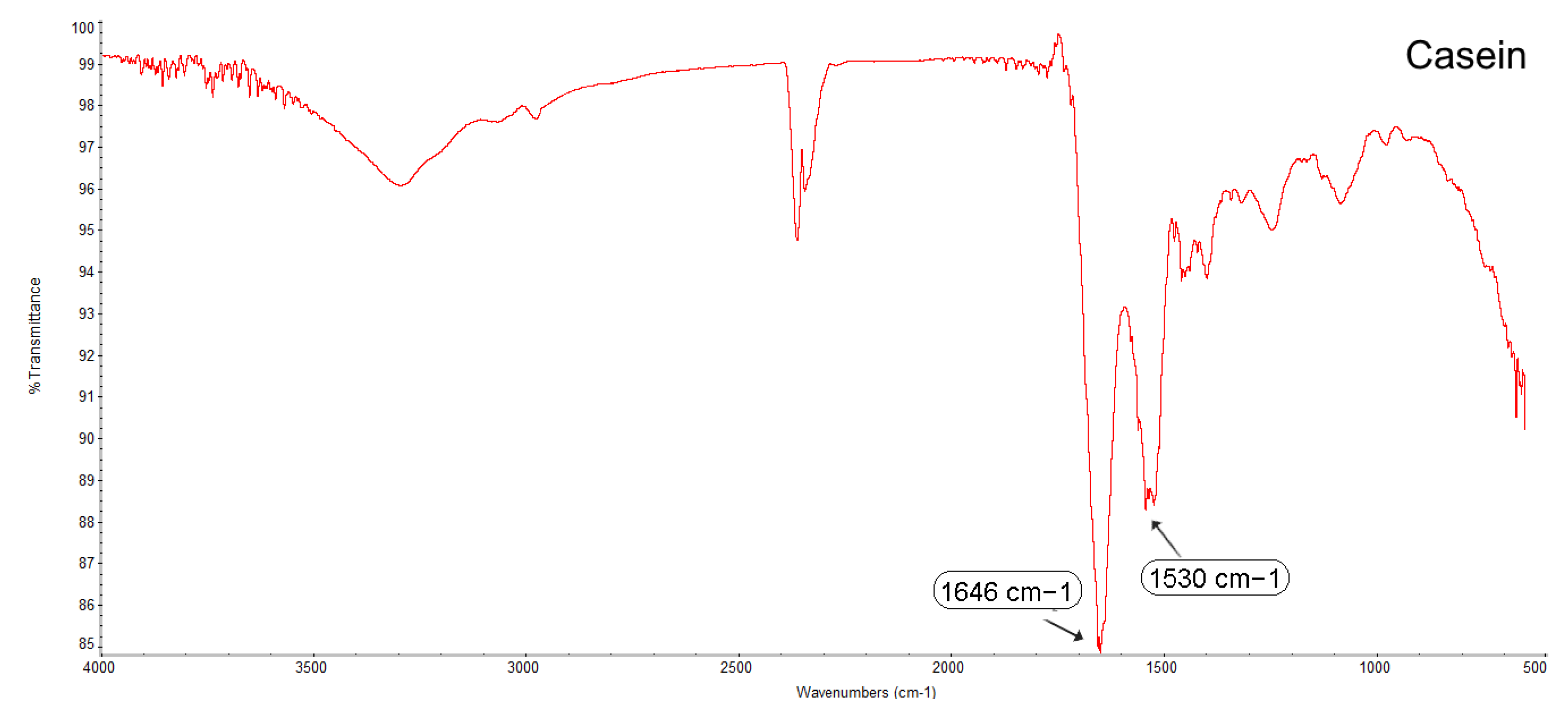
3.2.4. Fourier Transform Infrared Spectroscopy (FTIR)
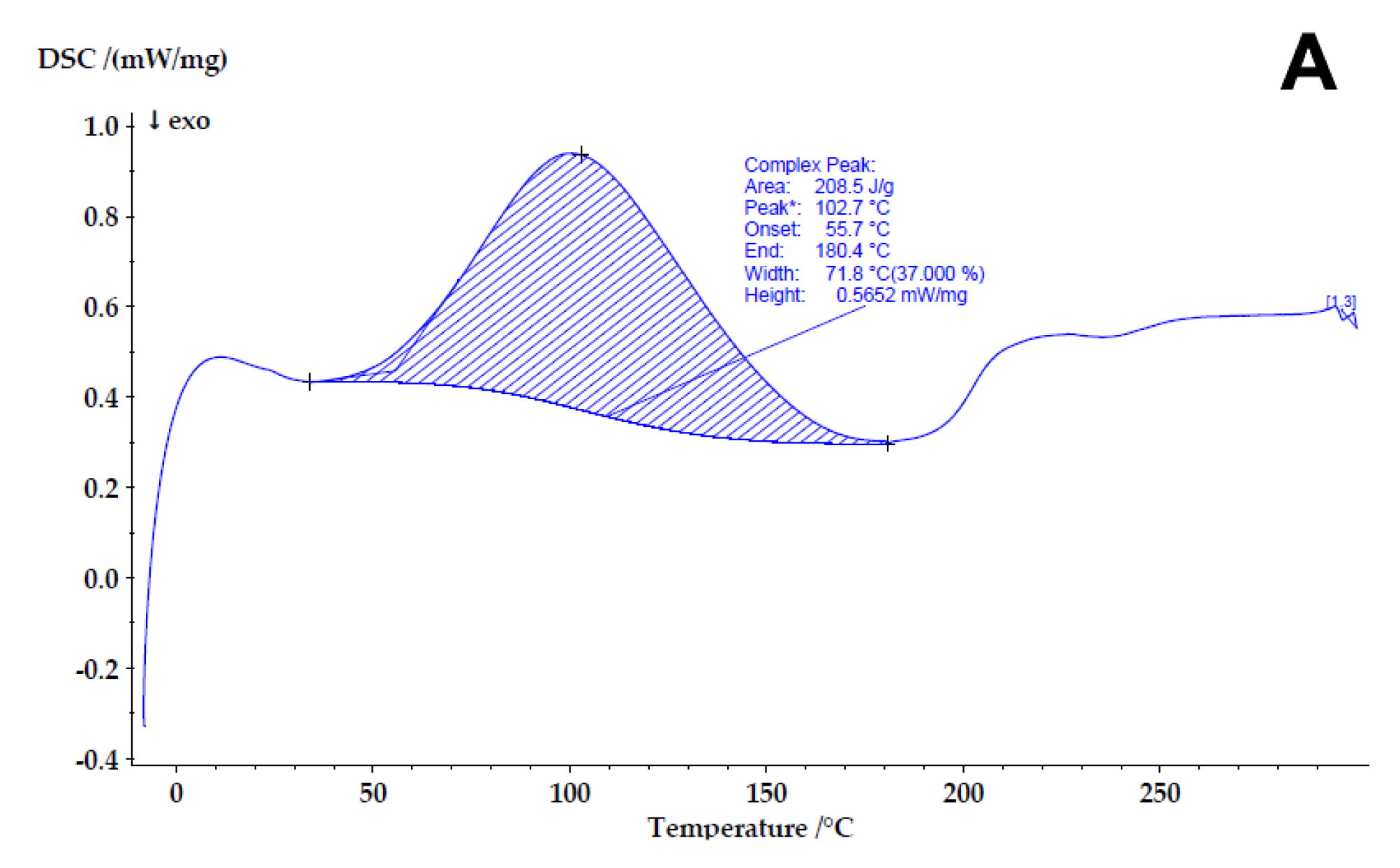
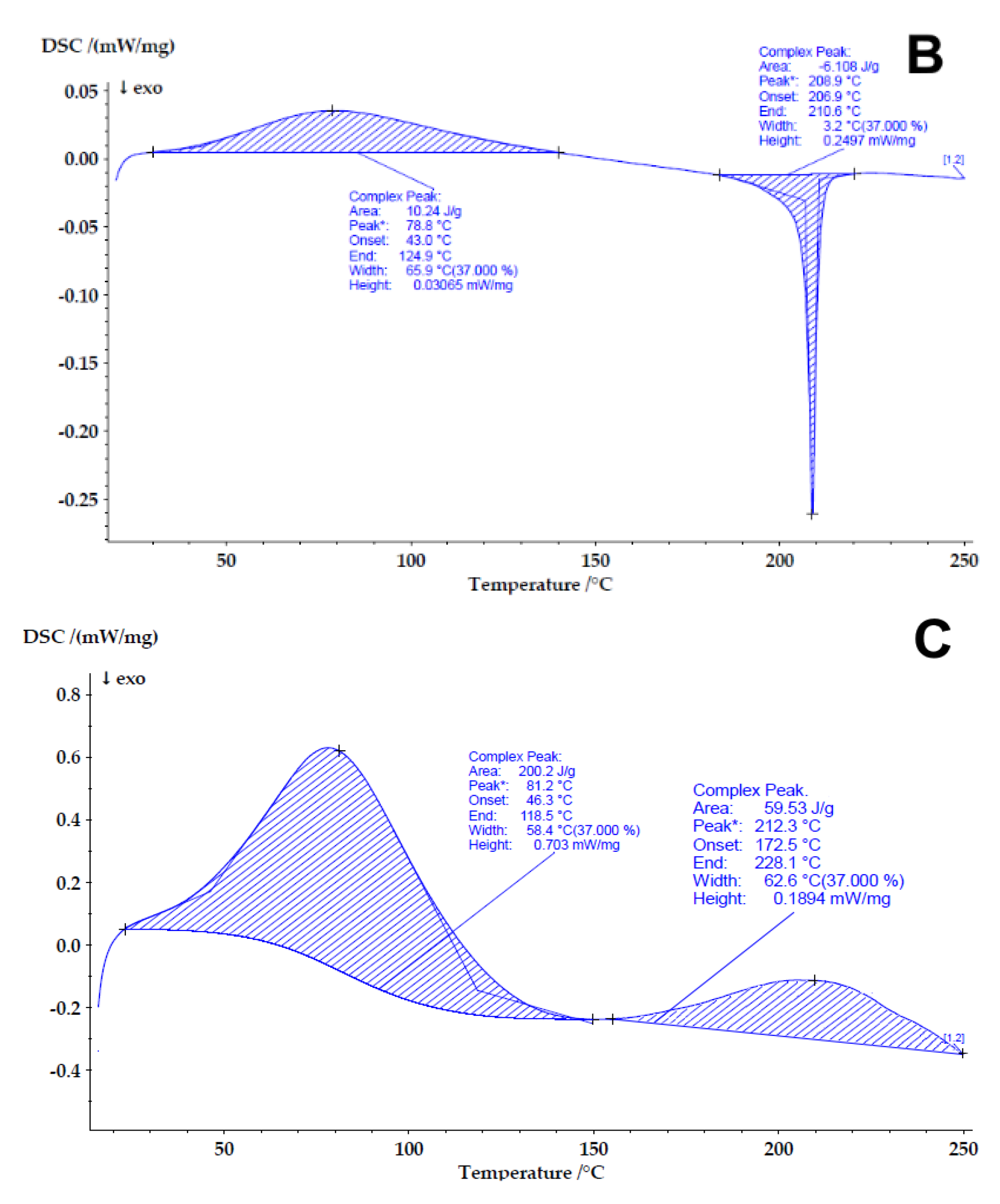
3.2.5. Differential Scanning Calorimetry (DSC)
3.3. Experimental Design and Statistical Analysis
4. Results
4.1. Preliminary Experiments
4.1.1. Optimal Spray Head Mesh Size
4.1.2. Inlet Temperature and Solution Feed Rate
4.1.3. Gas Flow Rate
4.1.4. Spray Intensity
4.2. Experimental Design
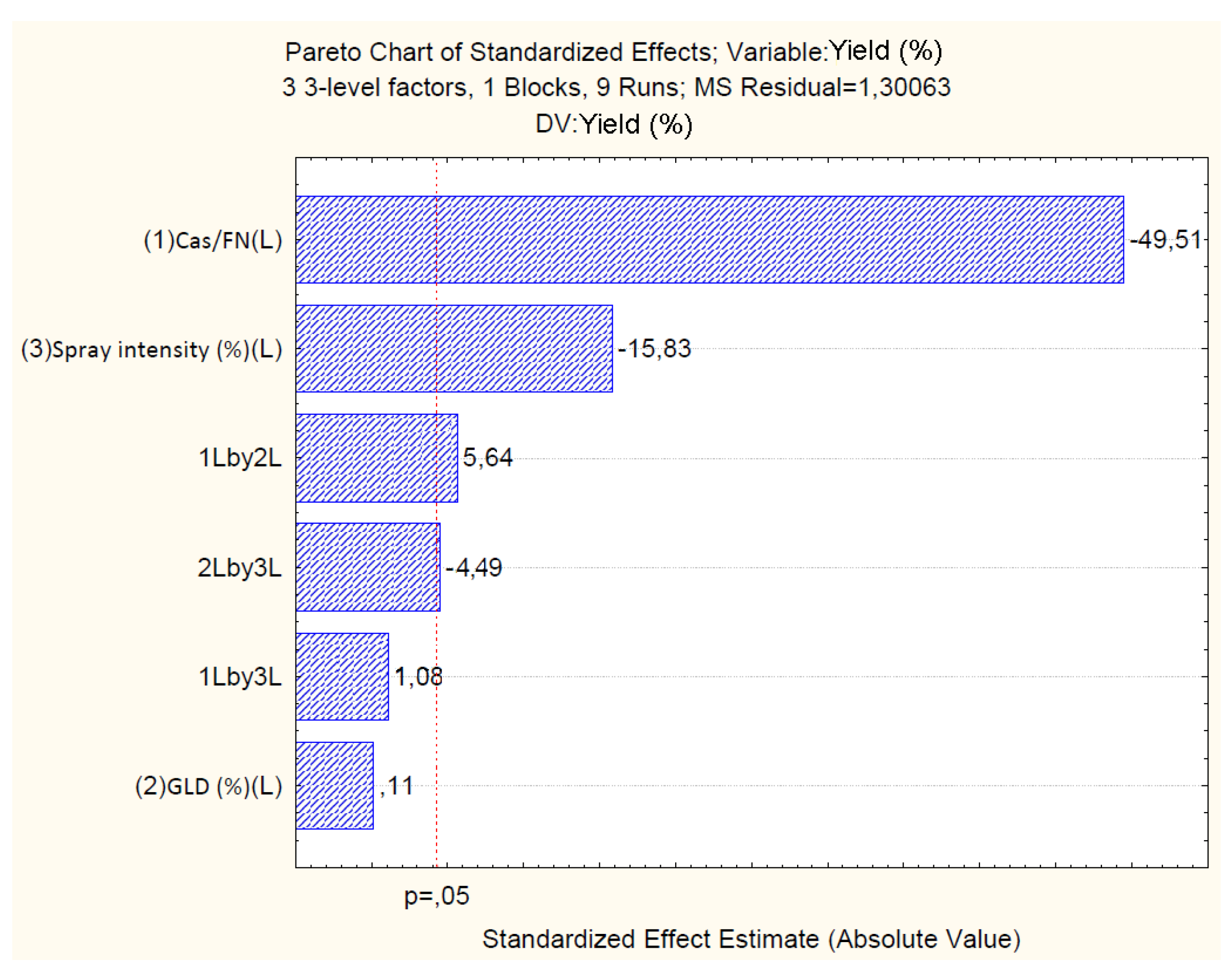
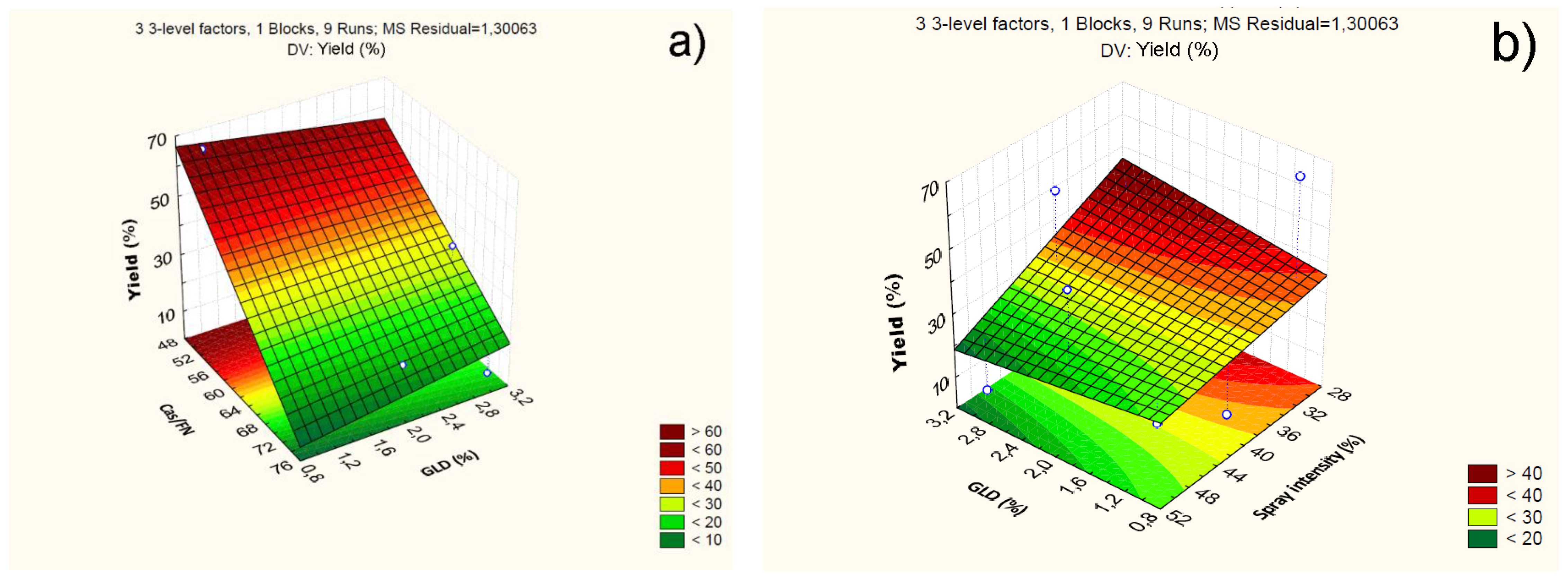
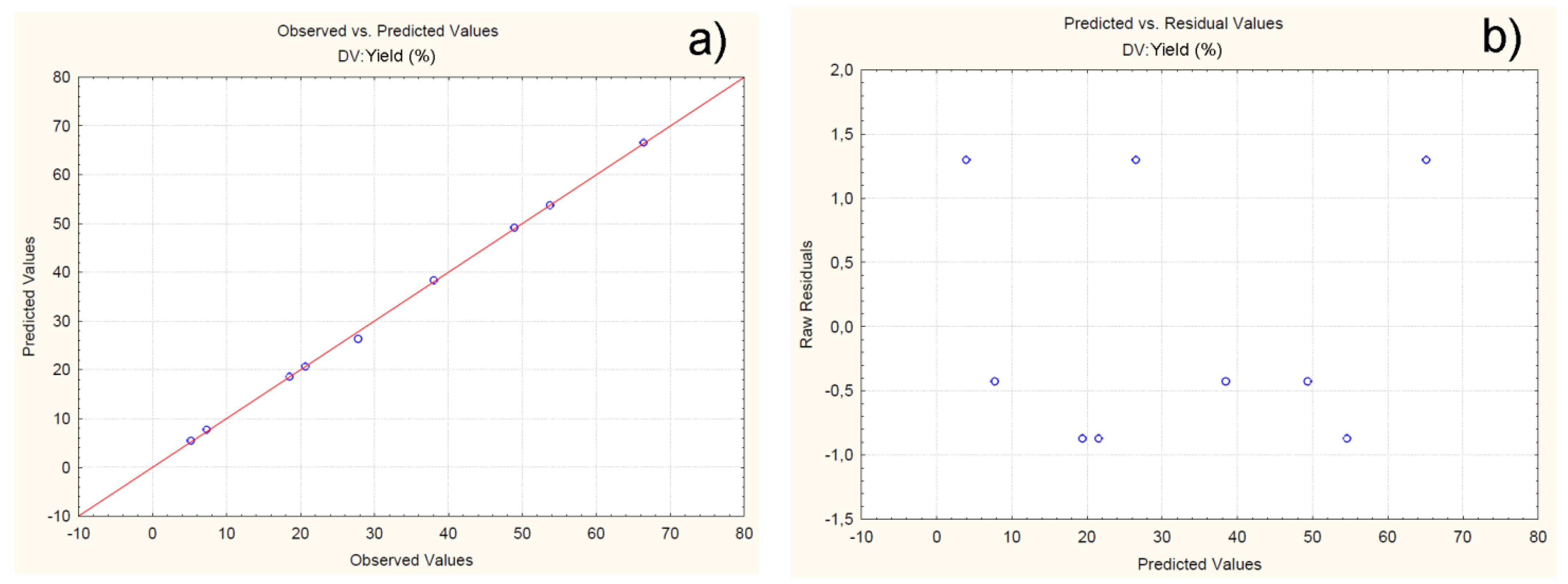
4.2.1. Effect of Independent Variables on Particle Yield
| R2 | R2adj. | SS | df | MS | SS | df | MS | F | p |
|---|---|---|---|---|---|---|---|---|---|
| 0.999 | 0.997 | 3629.92 | 6 | 604.98 | 2.601 | 2 | 1.300 | 465.148 | 0.002 |
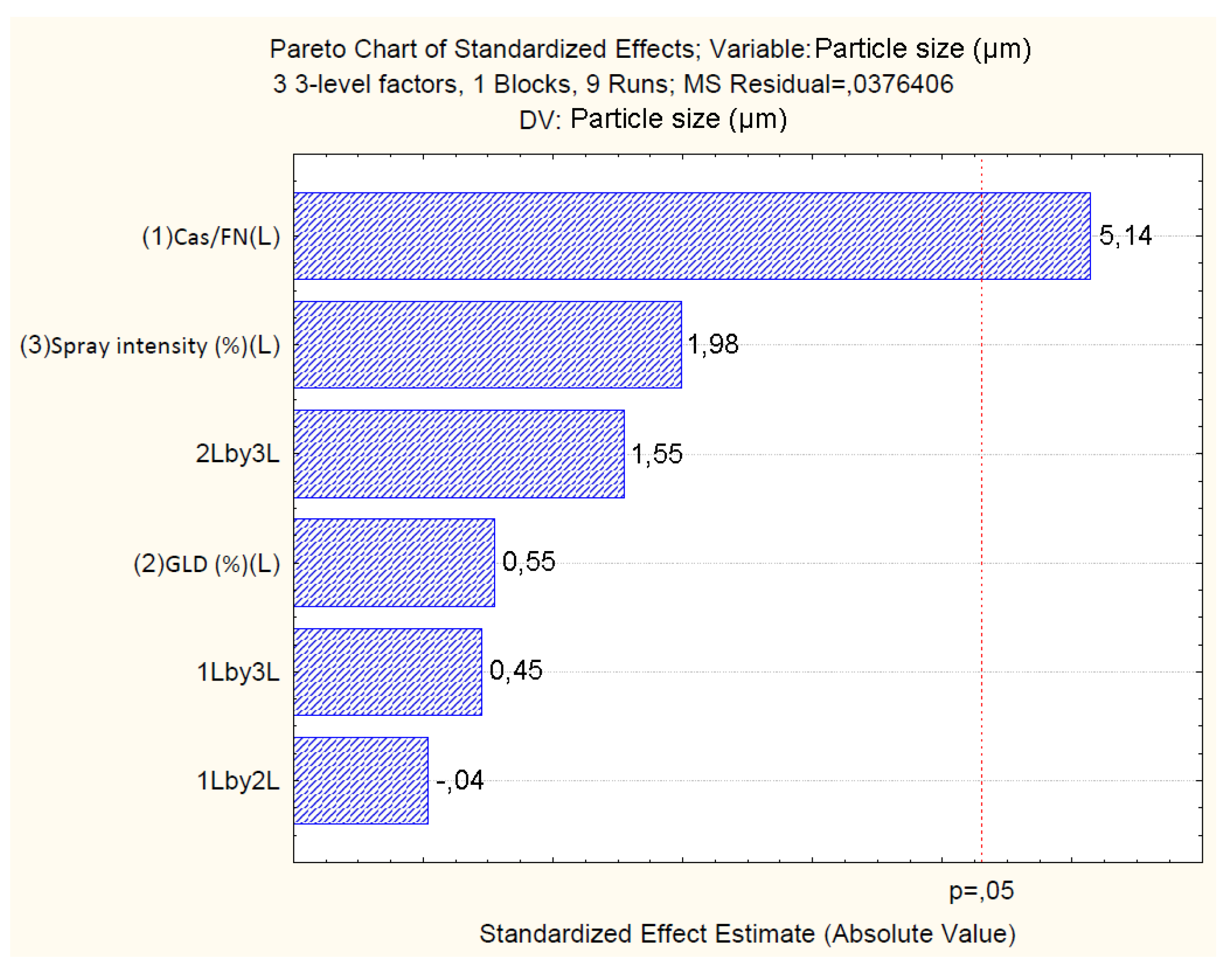
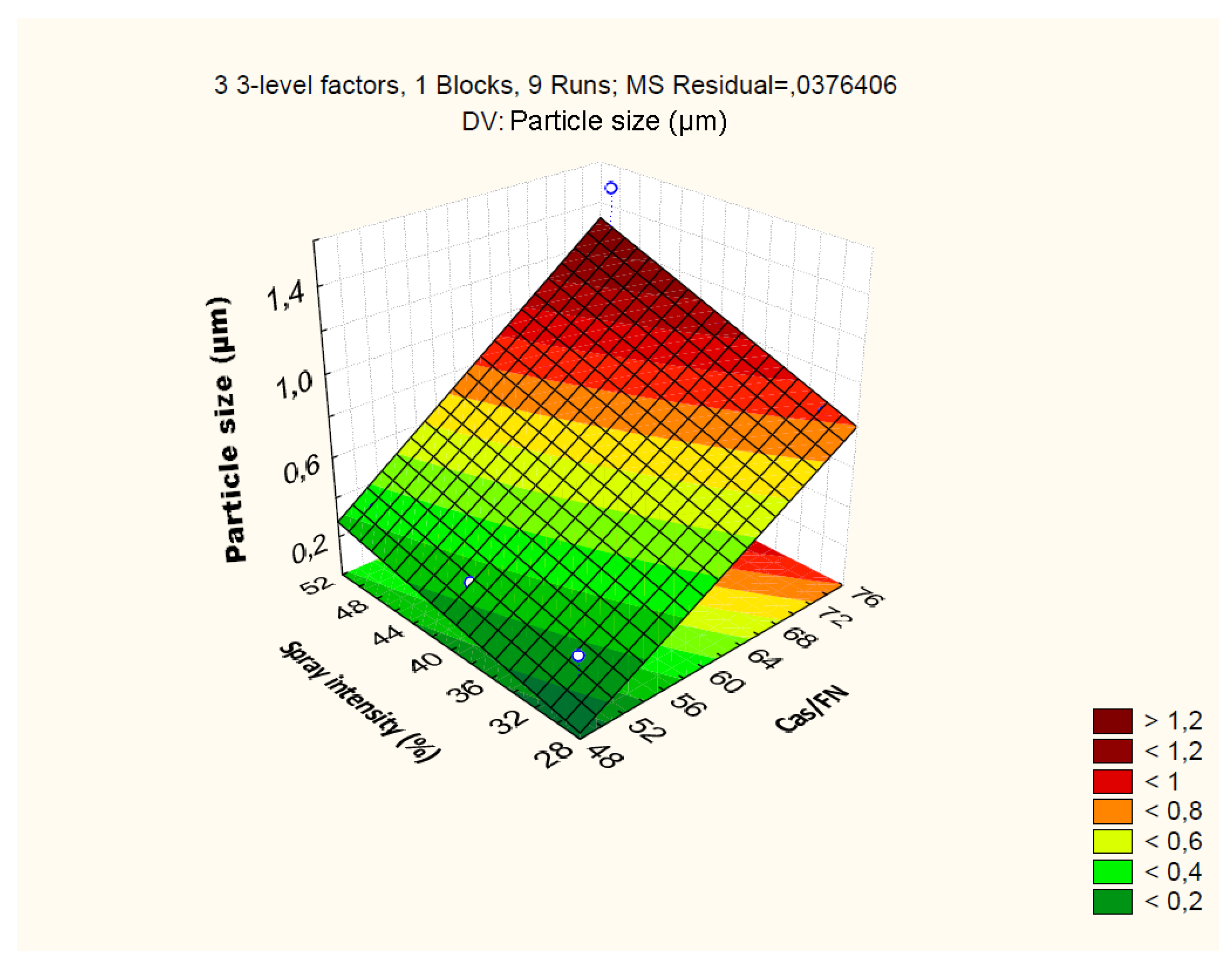
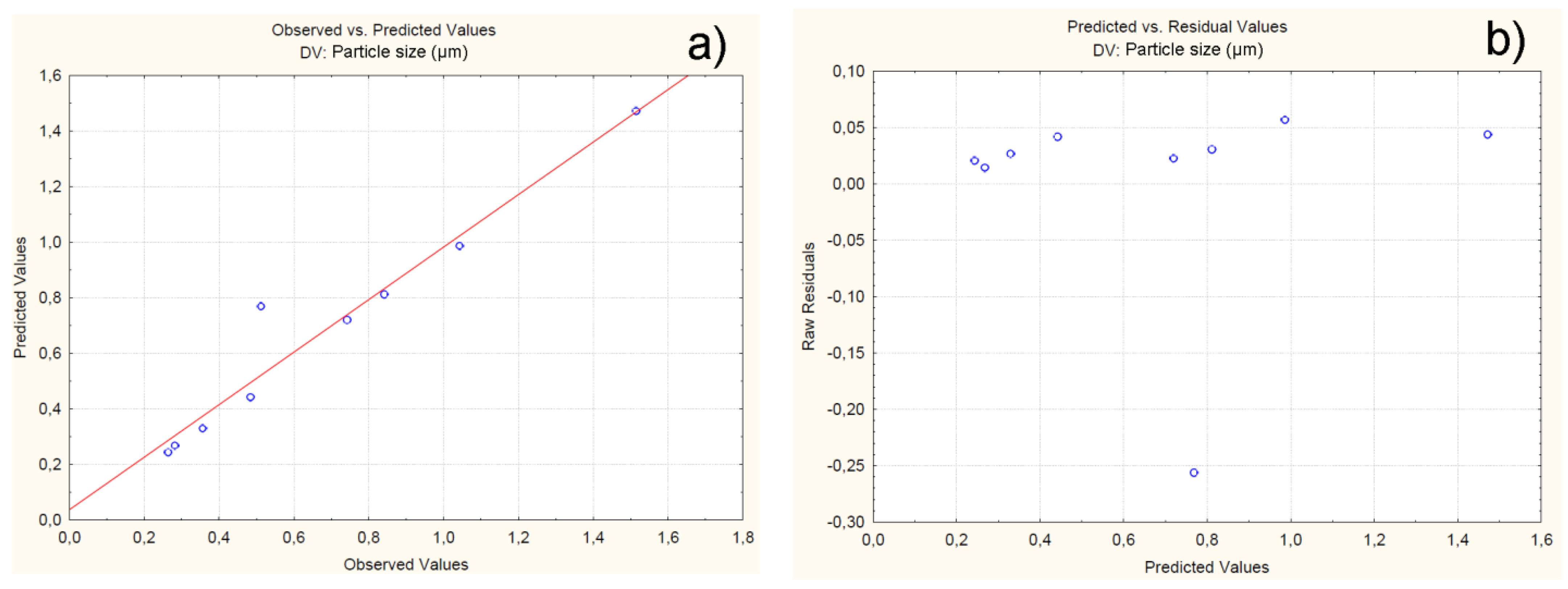
4.2.2. Effect of Independent Variables on Particle Size
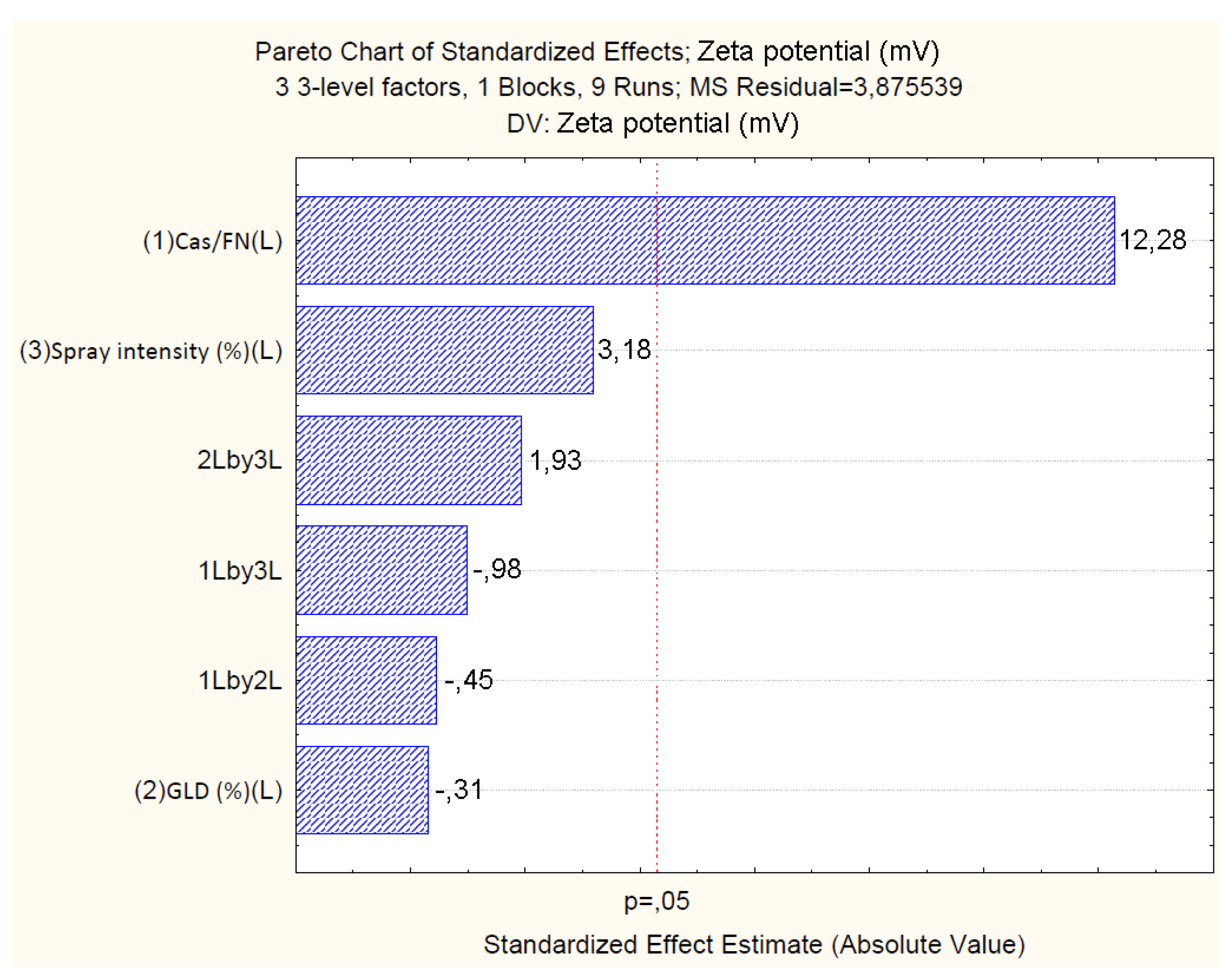
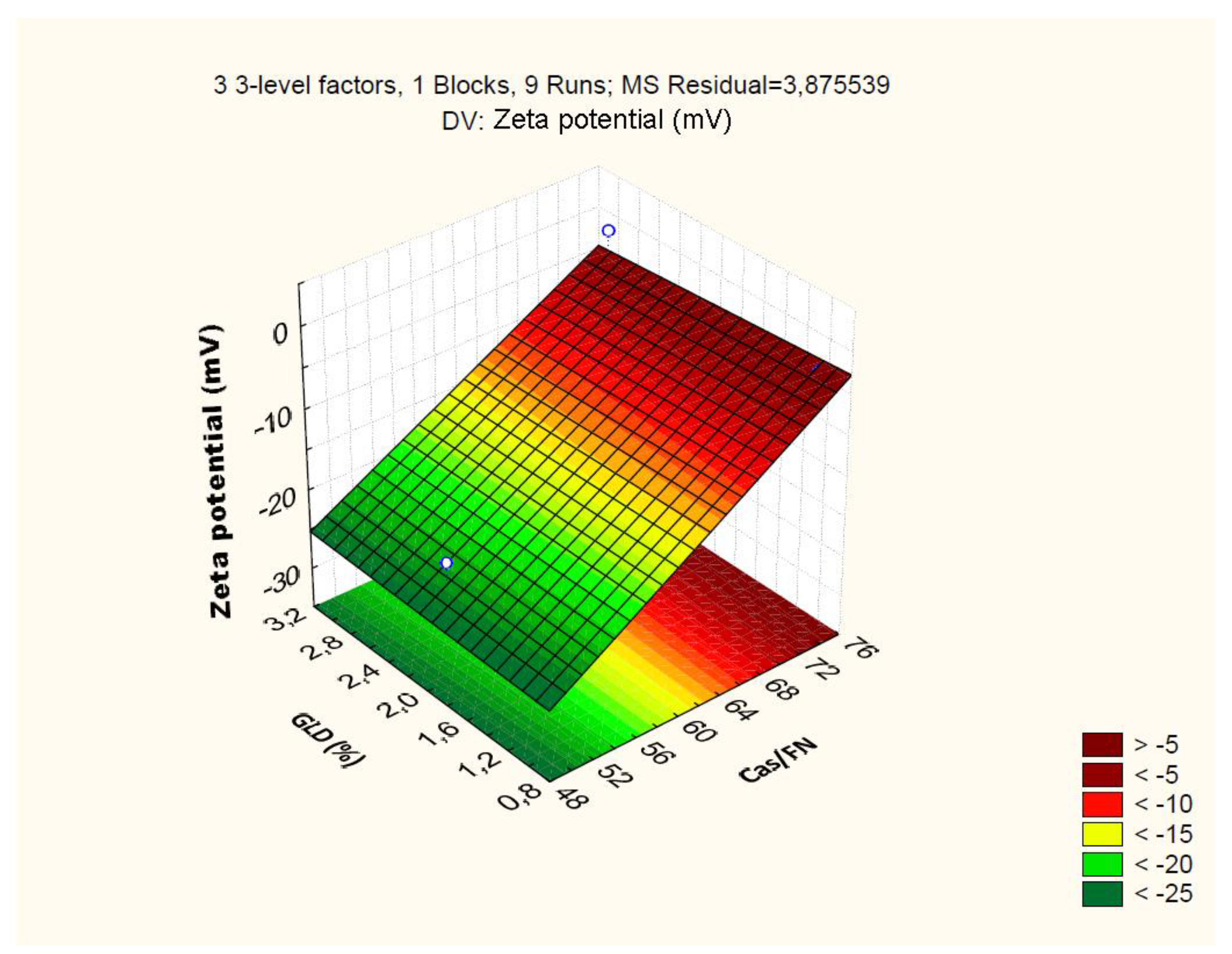
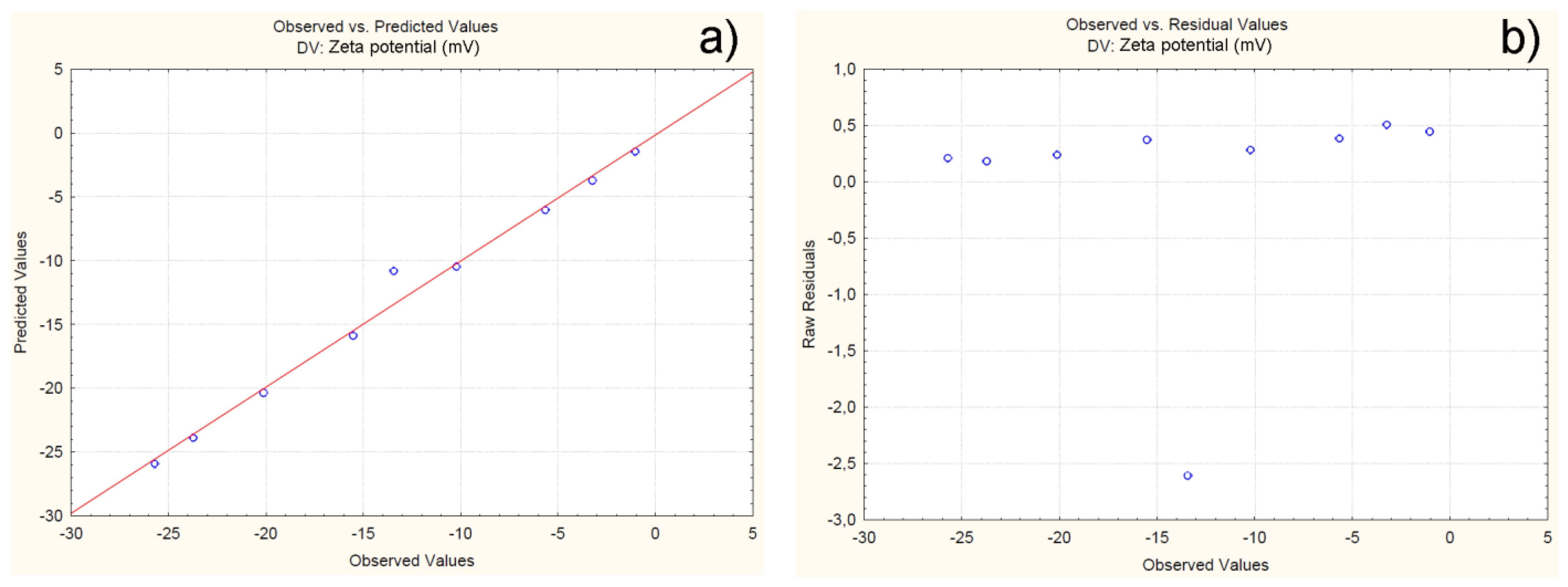
4.2.3. Effect of Independent Variables on Particle Zeta Potential
4.3. Determining the Compatibility and Physical State of Casein/Fucoidan Composite Nanoparticles
5. Conclusion
6. Patents
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Kučuk, N.; Primožič, M.; Knez, Ž.; Leitgeb, M. Sustainable Biodegradable Biopolymer-Based Nanoparticles for Healthcare Applications. Int. J. Mol. Sci. 2023, 24, 3188. [Google Scholar] [CrossRef] [PubMed]
- Vodyashkin, A.A.; Kezimana, P.; Vetcher, A.A.; Stanishevskiy, Y.M. Biopolymeric Nanoparticles–Multifunctional Materials of the Future. Polymers 2022, 14, 2287. [Google Scholar] [CrossRef]
- Han, M.; Liu, K.; Liu, X.; Rashid, M.T.; Zhang, H.; Wang, M. Research Progress of Protein-Based Bioactive Substance Nanoparticles. Foods 2023, 12, 2999. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Zhang, Y.; Hao, X.; Xie, D.; Wang, C.; Zhang, H.; Jin, P.; Du, Q. Improved Stability and In Vitro Anti-Arthritis Bioactivity of Curcumin–Casein Nanoparticles by Ultrasound-Driven Encapsulation. Nutrients 2022, 14, 5192. [Google Scholar] [CrossRef] [PubMed]
- Zahariev, N.; Draganova, M.; Zagorchev, P.; Pilicheva, B. Casein-Based Nanoparticles: A Potential Tool for the Delivery of Daunorubicin in Acute Lymphocytic Leukemia. Pharmaceutics 2023, 15, 471. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Hsu, F.-Y.; Lin, W.-T.; Cha, C.-Y.; Ho, Y.-C.; Mi, F.-L. Biodegradable Nanoparticles Prepared from Chitosan and Casein for Delivery of Bioactive Polysaccharides. Polymers 2022, 14, 2966. [Google Scholar] [CrossRef] [PubMed]
- Sadiq, U.; Gill, H.; Chandrapala, J. Casein Micelles as an Emerging Delivery System for Bioactive Food Components. Foods 2021, 10, 1965. [Google Scholar] [CrossRef] [PubMed]
- van de Langerijt, T.M.; O’Mahony, J.A.; Crowley, S.V. Structural, Binding and Functional Properties of Milk Protein-Polyphenol Systems: A Review. Molecules 2023, 28, 2288. [Google Scholar] [CrossRef] [PubMed]
- Jao, D.; Xue, Y.; Medina, J.; Hu, X. Protein-Based Drug-Delivery Materials. Materials 2017, 10, 517. [Google Scholar] [CrossRef]
- Luo, Y.; Pan, K; Zhong, Q. Casein/pectin nanocomplexes as potential oral delivery vehicles. Int. J. Pharm. 2015, 486, 59–68. [Google Scholar] [CrossRef]
- Thongkaew, C.; Gibis, M.; Hinrichs, J; Weiss, J. Polyphenol interactions with whey protein isolate and whey protein isolate− pectin coacervates. Food Hydrocolloids. 2014, 41, 103–112. [Google Scholar] [CrossRef]
- Wei, Z.; Yang, W.; Fan, R.; Yuan, F.; Gao, Y. Evaluation of structural and functional properties of protein−EGCG complexes and their ability of stabilizing a model β-carotene emulsion, Food Hydrocolloids. 2015, 45, 337−350. [CrossRef]
- Bealer, EJ.; Onissema-Karimu, S.; Rivera-Galletti, A.; Francis, M.; Wilkowski, J.; Salas-de la Cruz, D.; Hu, X. Protein-Polysaccharide Composite Materials: Fabrication and Applications. Polymers. 2020, 12, 464. [Google Scholar] [CrossRef]
- Sun, X.; Wang, H.; Li, S.; Song, C.; Zhang, S.; Ren, J.; Udenigwe, C.C. Maillard-Type Protein–Polysaccharide Conjugates and Electrostatic Protein–Polysaccharide Complexes as Delivery Vehicles for Food Bioactive Ingredients: Formation, Types, and Applications. Gels 2022, 8, 135. [Google Scholar] [CrossRef]
- Dai, K.; Wang, J.; Luo, Y.; Tu, Y.; Ren, F.; Zhang, H. Characteristics and Functional Properties of Maillard Reaction Products from α-Lactalbumin and Polydextrose. Foods 2023, 12, 2866. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Mu, M.; Hu, B.; Yao, P.; Jiang, M. Micellization of casein-graft-dextran copolymer prepared through Maillard reaction. Biopolymers. 2006, 81, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Di Pierro, P.; Chico, B.; Villalonga, R.; Mariniello, L.; Masi, P. Porta, R. Transglutaminase-catalyzed preparation of chitosan− ovalbumin films. Enzyme Microb. Technol. 2007, 40, 437–441. [Google Scholar] [CrossRef]
- Liu, Y.; Ma, L.; Gao, C. Facile fabrication of the glutaraldehyde cross-linked collagen/chitosan porous scaffold for skin tissue engineering. Mater. Sci. Eng.C. 2012, 32, 2361–2366. [Google Scholar] [CrossRef]
- Takigawa, T.; Endo, Y. Effects of glutaraldehyde exposure on human health. J Occup Health. 2006, 48, 75–87. [Google Scholar] [CrossRef]
- Dodero, A.; Scarfi, S.; Mirata, S.; Sionkowska, A.; Vicini, S.; Alloisio, M.; Castellano, M. Effect of Crosslinking Type on the Physical-Chemical Properties and Biocompatibility of Chitosan-Based Electrospun Membranes. Polymers. 2021, 13, 831. [Google Scholar] [CrossRef]
- Kuo, HH.; Chan, C.; Burrows, LL.; Deber, CM. Hydrophobic interactions in complexes of antimicrobial peptides with bacterial polysaccharides. Chem Biol Drug Des. 2007, 69, 405–1. [Google Scholar] [CrossRef]
- Liu, F.; McClements, DJ.; Ma, C.; Liu, X. Novel Colloidal Food Ingredients: Protein Complexes and Conjugates. Annu Rev Food Sci Technol. 2023, 14, 35–61. [Google Scholar] [CrossRef] [PubMed]
- Markman, G.; Livney, YD. Maillard-conjugate based core-shell co-assemblies for nanoencapsulation of hydrophobic nutraceuticals in clear beverages. Food Funct. 2012, 3, 262–70. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Ting, Y.; Yang, X.; Tang, W.; Zeng, X.; Huang, Q. Nanochemoprevention by encapsulation of (-)-epigallocatechin-3-gallate with bioactive peptides/chitosan nanoparticles for enhancement of its bioavailability. Chem Commun. 2012, 48, 2421–3. [Google Scholar] [CrossRef] [PubMed]
- Elbialy, NS.; Mohamed, N. Alginate-coated caseinate nanoparticles for doxorubicin delivery: Preparation, characterisation, and in vivo assessment. Int J Biol Macromol. 2020, 154, 114–122. [Google Scholar] [CrossRef] [PubMed]
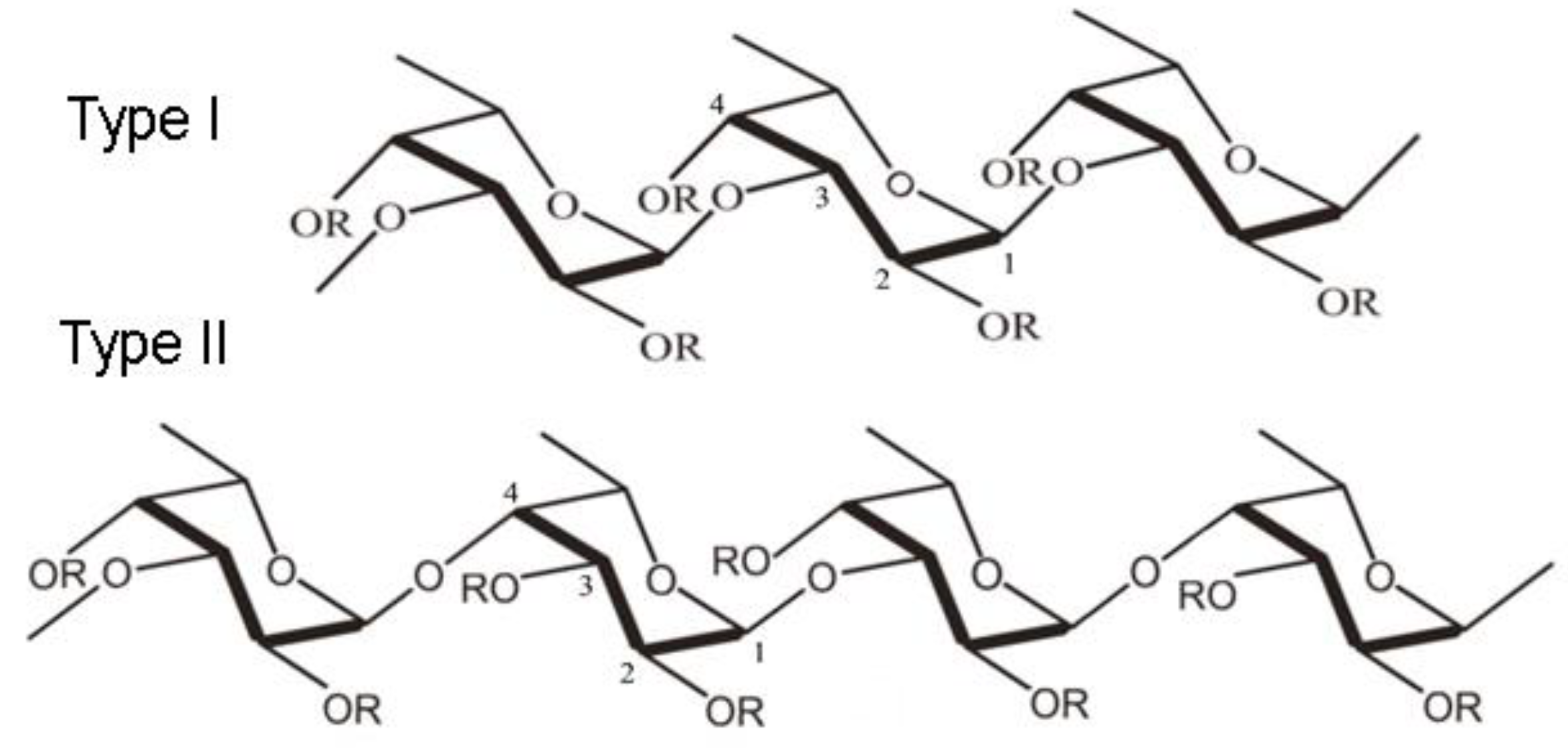
- Haggag, Y.A.; Abd Elrahman, A.A.; Ulber, R.; Zayed, A. Fucoidan in Pharmaceutical Formulations: A Comprehensive Review for Smart Drug Delivery Systems. Mar. Drugs 2023, 21, 112. [Google Scholar] [CrossRef] [PubMed]
- Citkowska, A.; Szekalska, M.; Winnicka, K. Possibilities of Fucoidan Utilization in the Development of Pharmaceutical Dosage Forms. Mar Drugs. 2019, 17, 458. [Google Scholar] [CrossRef] [PubMed]
- Zayed, A,; El-Aasr, M. ; Ibrahim, AS.; Ulber, R. Fucoidan Characterization: Determination of Purity and Physicochemical and Chemical Properties. Mar Drugs. 2020, 18, 571. [Google Scholar] [CrossRef] [PubMed]
- Luthuli, S.; Wu, S.; Cheng, Y.; Zheng, X.; Wu, M.; Tong, H. Therapeutic Effects of Fucoidan: A Review on Recent Studies. Mar Drugs. 2019, 17, 487. [Google Scholar] [CrossRef]
- Kurczewska, J. Recent Reports on Polysaccharide-Based Materials for Drug Delivery. Polymers, 2022; 14, 4189. [Google Scholar] [CrossRef]
- Sezer, AD.; Cevher, E. Fucoidan: A versatile biopolymer for biomedical applications. In Active Implants and Scafolds for Tissue Regeneration. Editor(s): Zilberman M. Springer: Berlin/Heidelberg, Germany. 2011; 8:377–406. ISBN: 978-3-642-18064-4.
- Jeon, M.S.; Han, SI.; Park, Y.H. Rapid green synthesis of silver nanoparticles using sulfated polysaccharides originating from Porphyridium cruentum UTEX 161: evaluation of antibacterial and catalytic activities. J Appl Phycol 2021, 33, 3091–3101. [Google Scholar] [CrossRef]
- Huang, YC.; Yang, YT. Effect of basic fibroblast growth factor released from chitosan-fucoidan nanoparticles on neurite extension. J Tissue Eng Regen Med. 2016, 10, 418–27. [Google Scholar] [CrossRef]
- Lee, HM.; Kim, JK.; Cho, TS. Applications of ophthalmic biomaterials embedded with fucoidan. Ind. Eng. Chem. 2012, 18, 1197–1201. [Google Scholar] [CrossRef]
- Fan, L.; Lu, Y.; Ouyang, XK.; Ling, J. Development and characterization of soybean protein isolate and fucoidan nanoparticles for curcumin encapsulation. Int J Biol Macromol. 2021, 169, 194–205. [Google Scholar] [CrossRef]
- Park, HW.; Kim, DY.; Shin, WS. Fucoidan improves the structural integrity and the molecular stability of β-lactoglobulin. Food Sci Biotechnol. 2018, 27, 1247–1255. [Google Scholar] [CrossRef] [PubMed]
- Chollet, L.; Saboural, P.; Chauvierre, C.; Villemin, JN.; Letourneur, D.; Chaubet, F. Fucoidans in Nanomedicine. Mar Drugs. 2016, 14, 145. [Google Scholar] [CrossRef]
- Aleksiev, A.; Kostova, B. Development and Optimization of the Reservoir-type Oral Multiparticulate Drug Delivery Systems of Galantamine Hydrobromide. Indian Journal of Pharmaceutical Sciences. 2016, 78. [Google Scholar] [CrossRef]
- Penkov, D.; Lukova, P.; Manev, H.; Dimitrova, S.; Kassarova, M. Polymer Tablet Matrix Systems for the Controlled Release of Dry Betula pendula Leaf Extract. Polymers 2023, 15, 3558. [Google Scholar] [CrossRef]
- Dimer, F.; de Souza Carvalho-Wodarz, C.; Haupenthal, J.; Hartmann, R.; Lehr, CM. Inhalable Clarithromycin Microparticles for Treatment of Respiratory Infections. Pharm Res. 2015, 32, 3850–61. [Google Scholar] [CrossRef]
- Fontana, MC.; Durli, TL.; Pohlmann, AR.; Guterres, SS.; Beck, RCR. Polymeric controlled release inhalable powder produced by vibrational spray-drying: one-step preparation and in vitro lung deposition. Powder Technol. 2014, 258, 49–59. [Google Scholar] [CrossRef]
- Elazazy, M.; Issa, A.; Al-Mashreky, M.; Al-Sulaiti, M.; Al-Saad, K. Application of fractional factorial design for green synthesis of cyano-modified silica nanoparticles: Chemometrics and multifarious response optimization. Advanced Powder Technology. 2018, 29. [Google Scholar] [CrossRef]
- Figueroa, H.; Aristizabal, J.; Reinoso-Guerra, E.; Arce, B.; Vargas-Straube, M.J.; Gentil, D.; Ramírez, C.; Cordero, J.; Barrera, N.P.; Parra, C. Fractional Factorial Design to Evaluate the Synthesis and Electrochemical Transfer Parameters of h-BN Coatings. Nanomaterials 2023, 13, 2992. [Google Scholar] [CrossRef]
- Burova, T.; Grinberg, N.; Dubovik, A.; Plashchina, I.; Usov, A.; Grinberg, V. β-Lactoglobulin–fucoidan nanocomplexes: Energetics of formation, stability, and oligomeric structure of the bound protein. Food Hydrocolloids. 2022, 129, 107666. [Google Scholar] [CrossRef]
- Lakshmanan, A.; Balasubramanian, B.; Maluventhen, V.; Malaisamy, A.; Baskaran, R.; Liu, W-C. ; Arumugam, M. Extraction and Characterization of Fucoidan Derived from Sargassum ilicifolium and Its Biomedical Potential with In Silico Molecular Docking. Applied Sciences. 2022, 12, 13010. [Google Scholar] [CrossRef]
- Saravana, PS.; Cho, YN.; Patil, MP.; Cho, YJ.; Kim, GD.; Park, YB.; Woo, HC.; Chun, BS. Hydrothermal degradation of seaweed polysaccharide: Characterization and biological activities. Food Chem. 2018, 268, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, N.; Mori, K.; Yoshida, H.; Kamiuchi, N.; Kitaura, R.; Hirasawa, R.; Yamashita, H. Thermal Stability of High-Entropy Alloy Nanoparticles Evaluated by In Situ TEM Observations. Nano Lett. 2024. [Google Scholar] [CrossRef] [PubMed]
- Szyk-Warszyńska, L.; Raszka, K.; Warszyński, P. Interactions of Casein and Polypeptides in Multilayer Films Studied by FTIR and Molecular Dynamics. Polymers 2019, 11, 920. [Google Scholar] [CrossRef]
- Soto-Vásquez, M.R.; Alvarado-García, P.A.A.; Youssef, F.S.; Ashour, M.L.; Bogari, H.A.; Elhady, S.S. FTIR Characterization of Sulfated Polysaccharides Obtained from Macrocystis integrifolia Algae and Verification of Their Antiangiogenic and Immunomodulatory Potency In Vitro and In Vivo. Mar. Drugs 2023, 21, 36. [Google Scholar] [CrossRef]


| Variables | Levels | ||
|---|---|---|---|
| Independent variables | -1 | 0 | +1 |
| X1: Casein:Fucoidan ratio | 1:1 | 2:1 | 3:1 |
| X2: Glutaraldehyde concentration (%) | 3 | 2 | 1 |
| X3: Spray intensity (%) | 30 | 40 | 50 |
| Dependent variables | |||
| Y1: Yield (%) | |||
| Y2: Particle size (µm) Y3: Zeta potential (mV) |
|||
| Models | Independent variables | Dependent variables | |||||
|---|---|---|---|---|---|---|---|
| Cas/FN | GLD (%) | Sp.int.(%) | Size ± SD (µm) |
ζ ± SD (mV) |
Yield ± SD (%) |
||
| C1F1G1Sp.30 | -1 | -1 | -1 | 264 ± 32 | -25.71 ± 1.0 | 66.41 ± 2.2 | |
| C1F1G2Sp.40 | -1 | 0 | +1 | 356 ± 11 | -20.12 ± 0.9 | 48.91 ± 2.9 | |
| C1F1G3Sp.50 | -1 | +1 | 0 | 282 ± 25 | -23.72 ± 1.8 | 53.72 ± 1.9 | |
| C2F1G1Sp.50 | 0 | -1 | +1 | 742 ± 252 | -10.21 ± 1.5 | 20.66 ± 1.5 | |
| C2F1G2Sp.40 | 0 | 0 | 0 | 512 ± 360 | -13.43 ± 1.6 | 27.81 ± 2.6 | |
| C2F1G3Sp.30 | 0 | +1 | -1 | 484 ± 121 | -15.51 ± 1.4 | 38.03 ± 1.8 | |
| C3F1G1Sp.40 | +1 | -1 | 0 | 1043 ± 986 | -3.23 ± 0.7 | 7.31 ± 3.4 | |
| C3F1G2Sp.30 | +1 | 0 | -1 | 842 ± 263 | -5.65 ± 0.9 | 18.54 ± 2.7 | |
| C3F1G3Sp.50 | +1 | +1 | +1 | 1515 ± 353 | -1.03 ± 1.9 | 5.21 ± 2.2 | |
| Factor | Yield (%) | Particle size (µm) | ζ (mV) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Effect | SE | p | β | Effect | SE | p | β | Effect | SE | p | β | |
| Mean/Int. | 34.59 | 0.38 | 0.00 | 207.54 | 0.62 | 0.06 | 0.01 | 0.34 | -14.35 | 0.66 | 0.00 | -83.330 |
| Cas/Fn (1) | -45.86 | 0.92 | 0.00 | -2.68 | 0.80 | 0.15 | 0.03 | 0.01 | 19.64 | 1.59 | 0.00 | 1.244 |
| GLD (2) | 0.11 | 0.96 | 0.91 | -6.40 | 0.09 | 0.16 | 0.63 | -0.68 | -0.15 | 1.67 | 0.78 | -7.010 |
| Sp.Int. (3) | -15.32 | 0.96 | 0.00 | -0.49 | 0.32 | 0.16 | 0.18 | -0.04 | 5.31 | 1.67 | 0.08 | 0.374 |
| 1 by 2 | 7.86 | 1.36 | 0.02 | 0.30 | 0.00 | 0.23 | 0.97 | 0.00 | -1.08 | 2.35 | 0.69 | -0.040 |
| 1 by 3 | 1.48 | 1.36 | 0.38 | 0.00 | 0.10 | 0.23 | 0.69 | 0.00 | -2.32 | 2.35 | 0.42 | -0.010 |
| 2 by 3 | -6.37 | 1.41 | 0.04 | -0.31 | 0.37 | 0.24 | 0.26 | 0.01 | 4.72 | 2.44 | 0.19 | 0.236 |
| Factor | SS | df | MS | F | p | |||
|---|---|---|---|---|---|---|---|---|
| Yield: R2=0.99; R2Adj.=0.99; MS=1.30 | ||||||||
| Cas/Fn (1) | 3188.93 | 1 | 3188.93 | 2451.84 | 0.00 | |||
| GLD (2) | 0.01 | 1 | 0.01 | 0.01 | 0.91 | |||
| Spray.Int (3) | 326.21 | 1 | 326.21 | 250.81 | 0.00 | |||
| 1 by 2 | 41.43 | 1 | 41.43 | 31.86 | 0.02 | |||
| 1 by 3 | 1.54 | 1 | 1.54 | 1.18 | 0.38 | |||
| 2 by 3 | 26.25 | 1 | 26.25 | 20.18 | 0.04 | |||
| Error | 2.60 | 2 | ||||||
| Total SS | 3632.52 | 8 | ||||||
| Particle size: R2=0.94; R2Adj.=0.77; MS=0.03 | ||||||||
| Cas/Fn (1) | 0.99 | 1 | 0.99 | 26.40 | 0.03 | |||
| GLD (2) | 0.01 | 1 | 0.01 | 0.30 | 0.63 | |||
| Spray.Int (3) | 0.14 | 1 | 0.14 | 3.95 | 0.18 | |||
| 1 by 2 | 0.00 | 1 | 0.00 | 0.00 | 0.97 | |||
| 1 by 3 | 0.00 | 1 | 0.00 | 0.20 | 0.69 | |||
| 2 by 3 | 0.09 | 1 | 0.09 | 2.40 | 0.26 | |||
| Error | 0.07 | 2 | ||||||
| Total SS | 1.36 | 8 | ||||||
| Zeta potential: R2=0.98; R2Adj.=0.95; MS=3.87 | ||||||||
| Cas/Fn (1) | 585.15 | 1 | 585.15 | 150.98 | 0.00 | |||
| GLD (2) | 0.37 | 1 | 0.37 | 0.09 | 0.78 | |||
| Spray.Int (3) | 39.31 | 1 | 39.31 | 10.14 | 0.08 | |||
| 1 by 2 | 0.82 | 1 | 0.81 | 0.21 | 0.69 | |||
| 1 by 3 | 3.79 | 1 | 3.79 | 0.97 | 0.42 | |||
| 2 by 3 | 14.44 | 1 | 14.44 | 3.72 | 0.19 | |||
| Error | 7.75 | 2 | 3.87 | |||||
| Total SS | 633.89 | 8 | ||||||
| Batch | Observed | Predicted | Residuals | Error (%) |
|---|---|---|---|---|
| C1F1G1Sp.30 | 66.41 | 66.52 | -0.11 | -0.16 |
| C1F1G2Sp.40 | 48.91 | 49.12 | -0.21 | -0.42 |
| C1F1G3Sp.50 | 53.72 | 53.74 | -0.02 | -0.03 |
| C2F1G1Sp.50 | 20.66 | 20.69 | -0.03 | -0.14 |
| C2F1G2Sp.40 | 27.81 | 26.34 | 1.46 | 5.28 |
| C2F1G3Sp.30 | 38.03 | 38.36 | -0.33 | -0.86 |
| C3F1G1Sp.40 | 7.31 | 7.74 | -0.45 | -5.88 |
| C3F1G2Sp.30 | 18.54 | 18.58 | -0.04 | -0.21 |
| C3F1G3Sp.50 | 5.21 | 5.49 | -0.24 | -5.37 |
| Batch | Observed | Predicted | Residuals | Error (%) |
|---|---|---|---|---|
| C1F1G1Sp.30 | 0.264 | 0.243 | 0.02 | -0.21 |
| C1F1G2Sp.40 | 0.356 | 0.329 | 0.02 | -0.23 |
| C1F1G3Sp.50 | 0.282 | 0.267 | 0.01 | -0.17 |
| C2F1G1Sp.50 | 0.742 | 0.719 | 0.02 | -0.28 |
| C2F1G2Sp.40 | 0.512 | 0.768 | -0.25 | 2.60 |
| C2F1G3Sp.30 | 0.484 | 0.442 | 0.04 | -0.37 |
| C3F1G1Sp.40 | 1.043 | 0.986 | 0.05 | -0.50 |
| C3F1G2Sp.30 | 0.842 | 0.811 | 0.03 | -0.38 |
| C3F1G3Sp.50 | 1.515 | 1.471 | 0.04 | -0.44 |
| R2 | R2adj | SS | df | MS | SS | df | MS | F | p |
|---|---|---|---|---|---|---|---|---|---|
| 0.810 | 0.696 | 1.102 | 3 | 0.367 | 0.258 | 5 | 0.0517 | 7.105 | 0.0297 |
| Batch | Observed | Predicted | Residuals | Error (%) |
|---|---|---|---|---|
| C1F1G1Sp.30 | -25.71 | -25.91 | 0.20 | -0.81 |
| C1F1G2Sp.40 | -20.12 | -20.35 | 0.23 | -1.18 |
| C1F1G3Sp.50 | -23.72 | -23.89 | 0.17 | -0.75 |
| C2F1G1Sp.50 | -10.21 | -10.49 | 0.28 | -2.75 |
| C2F1G2Sp.40 | -13.43 | -10.82 | 2.60 | -19.4 |
| C2F1G3Sp.30 | -15.51 | -15.88 | 0.37 | -2.39 |
| C3F1G1Sp.40 | -3.23 | -3.73 | 0.50 | -15.63 |
| C3F1G2Sp.30 | -5.65 | -6.03 | 0.38 | -6.75 |
| C3F1G3Sp.50 | -1.03 | -1.47 | 0.44 | -43.05 |
| R2 | R2adj. | SS | df | MS | SS | df | MS | F | p |
|---|---|---|---|---|---|---|---|---|---|
| 0.9642 | 0.9642 | 611.22 | 3 | 203.74 | 22.66 | 5 | 4.533 | 44.94 | 0.00048 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





