Introduction
Lung cancer remains a formidable global health threat due to its high mortality rate and diverse subtypes. Early detection and accurate classification of lung tumors are essential for developing effective treatment plans and improving patient outcomes. Researchers have continuously explored various approaches, from bioinformatics models to advanced machine learning algorithms, to enhance the accuracy and efficiency of lung cancer diagnosis [
1,
2,
3].
Machine learning techniques, such as Support Vector Machines (SVMs) and Naive Bayes classifiers, alongside other predictive models, have become invaluable tools in this fight. These methods utilize protein sequence-derived structural and physicochemical descriptors to excel at classifying proteins and predicting key characteristics associated with different lung cancer types. Past studies have consistently demonstrated the effectiveness of these approaches in various biological domains, including protein classification, protein-protein interactions, subcellular localization, and microarray data analysis.In this vein, our study seeks to build upon previous research endeavors by harnessing machine learning models to develop precise prediction tools for classifying lung cancer types based on essential protein attributes [
4,
5]. By amalgamating feature selection, tree induction, and clustering techniques, we aim to bolster the predictive prowess of these models and contribute to the advancement of lung cancer diagnosis and treatment. Employing established datasets and innovative methodologies, our research endeavors to address the pressing need for early classification and prediction of lung tumor types, thereby facilitating more tailored and efficacious therapeutic interventions.
Furthermore, our study delves into the challenges surrounding clinical and in-situ patient monitoring, which are often fraught with difficulties, time constraints, and exorbitant costs. We aspire to furnish patients with a seamless and dependable treatment environment while unraveling the underlying causes of lung cancer. Machine learning assumes a pivotal role in surmounting these challenges by furnishing cost-effective and time-efficient solutions. Leveraging meticulously curated datasets from reputable sources such as the World Health Organization (WHO) and the Kaggle platform, we strive to ensure reliability, repeatability, precision, and accuracy in identifying the root causes of lung cancer.
In our research, we employ a diverse array of ML models, including conventional and modern algorithms such as DNN, Voting, Bagging, SVC_rbf, SVC_linear, SVC_polynomial, and SVC_sigmoid, to provide comprehensive insights for patients, researchers, and domain experts alike. Moreover, we meticulously address commonly overlooked hyperparameters in ML models, such as min child weights and learning rates, and demonstrate their profound impact on accuracy errors through detailed plots and insightful data analysis. By pushing the boundaries of ML techniques in lung cancer diagnosis and management, our study aims to enhance patient outcomes and streamline decision-making processes in clinical settings.
Methodology
In this research, we employ machine learning (ML) techniques to predict lung cancer types. Acknowledging the difficulties in data collection, especially in clinical settings where it is laborious and expensive, and recognizing the significance of time and cost efficiency across industries, we diligently compiled publicly available data from trusted sources like the World Health Organization (WHO) and Kaggle. This meticulous approach facilitated the creation of a coherent and meaningful dataset, enhancing the accuracy and reliability of our predictions.
Featurization
In our featurization section, we analyze various factors potentially linked to lung cancer, including gender (GENDER), age (AGE), smoking habits (SMOKING), presence of yellow fingers (YELLOW FINGERS), anxiety levels (ANXIETY), peer pressure (PEER PRESSURE), chronic diseases (CHRONIC DISEASE), fatigue (FATIGUE), allergies (ALLERGY), wheezing (WHEEZING), alcohol consumption (ALCOHOL CONSUMING), coughing (COUGHING), shortness of breath (SHORTNESS OF BREATH), swallowing difficulty (SWALLOWING DIFFICULTY), and chest pain (CHEST PAIN). Our target variable, LUNG CANCER, is binary, with 0 representing individuals with lung cancer and 1 indicating those without.
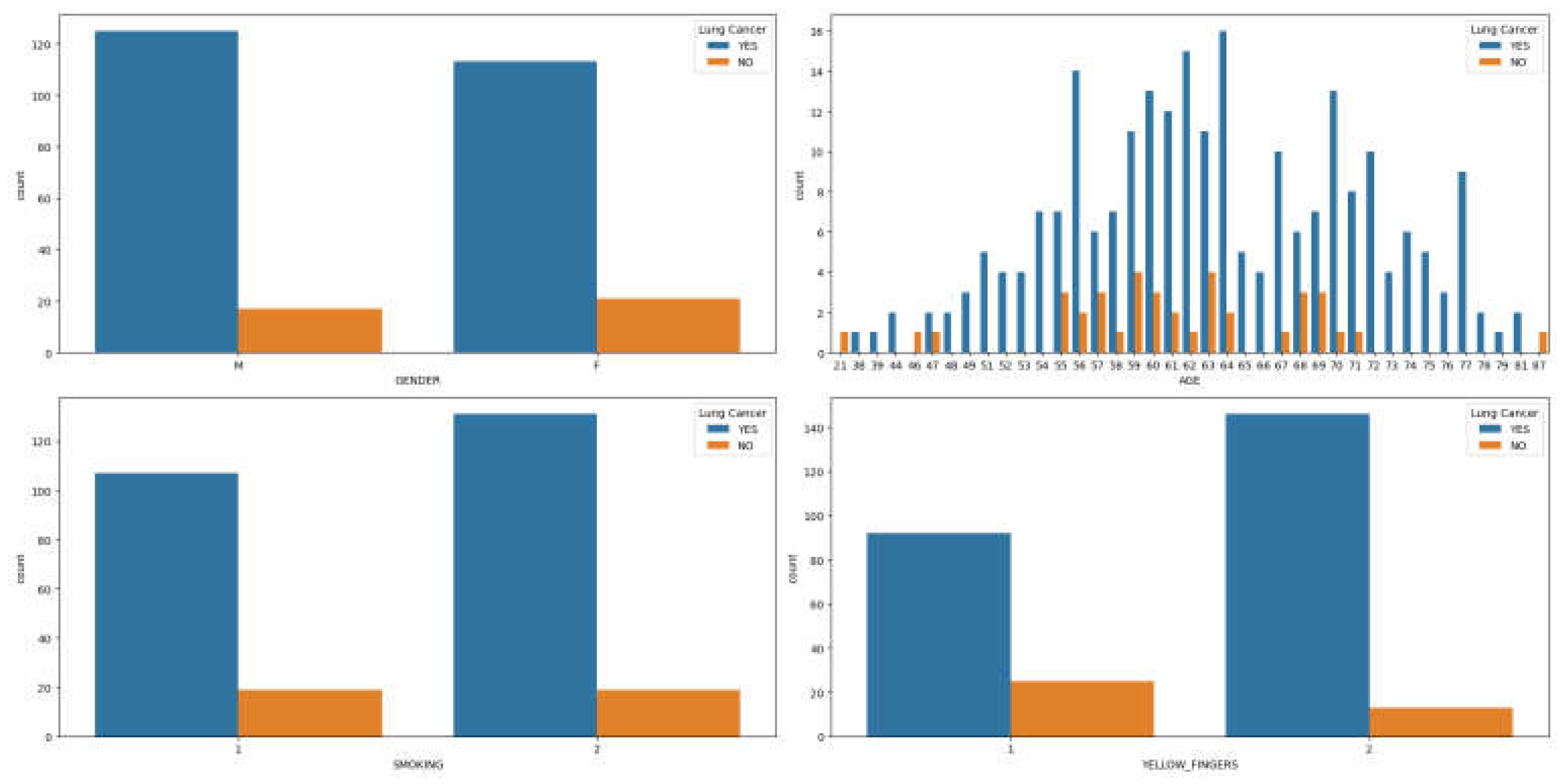
Observations from
Figure 1(a)-(d) suggest a higher prevalence of lung cancer among men, particularly in middle to old age, who are smokers with yellow fingers compared to women. Further analysis, as depicted in
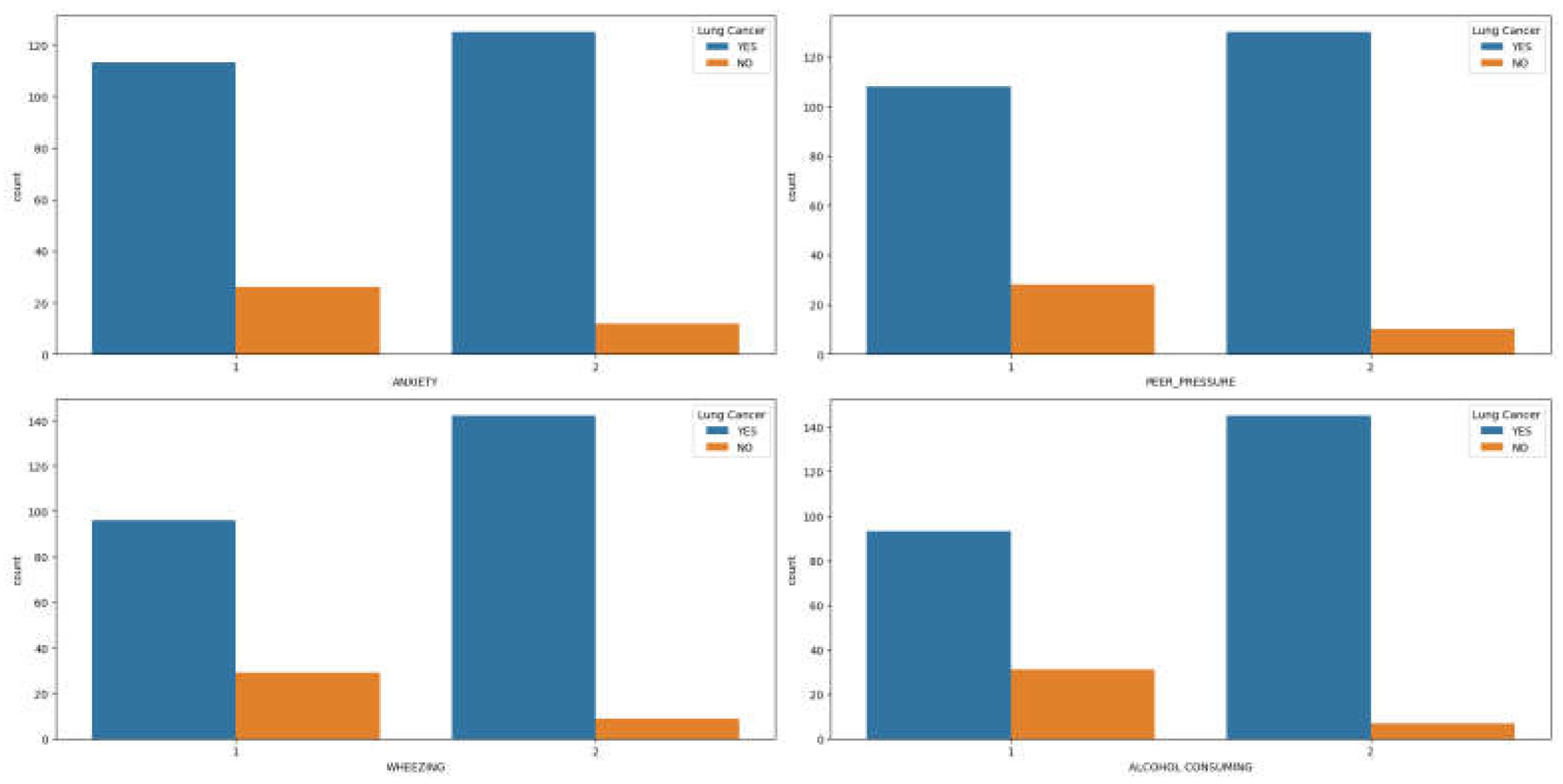
Figure 2(a)-(d), reveals that men with lung cancer tend to experience higher levels of anxiety, peer pressure, wheezing, and alcohol consumption compared to women exhibiting similar symptoms.
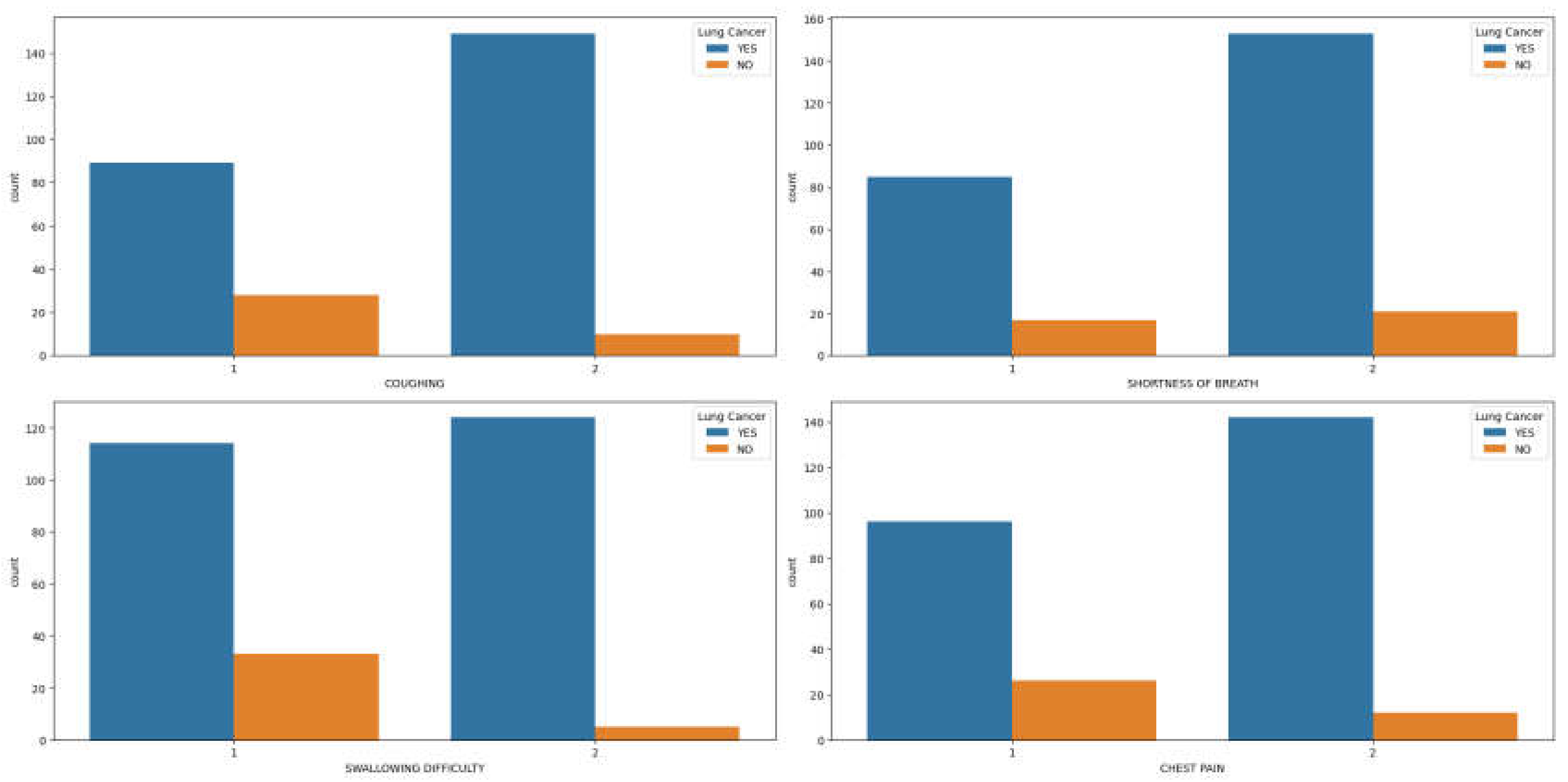
Similarly, symptoms such as coughing, shortness of breath, swallowing difficulty, and chest pain (
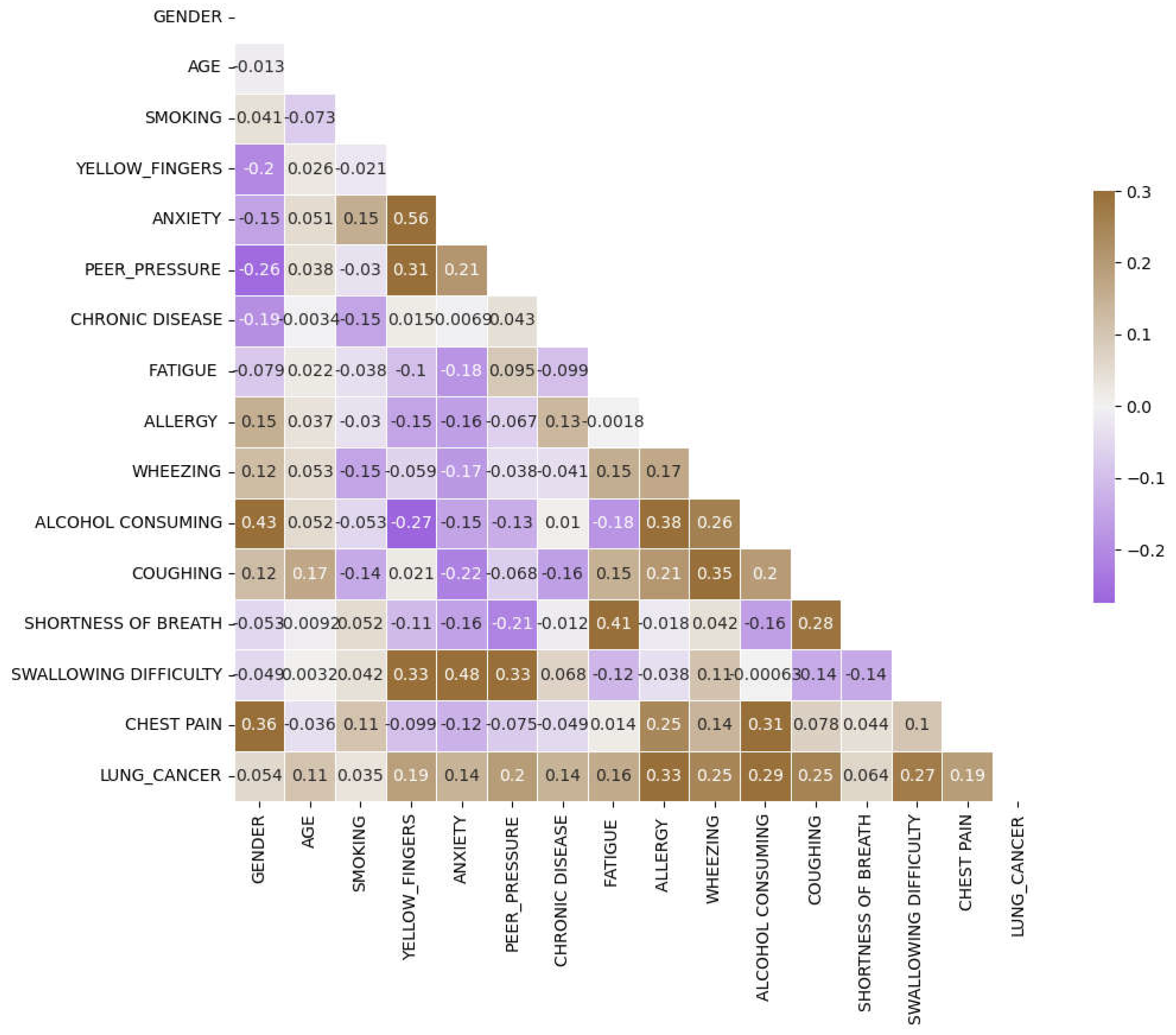
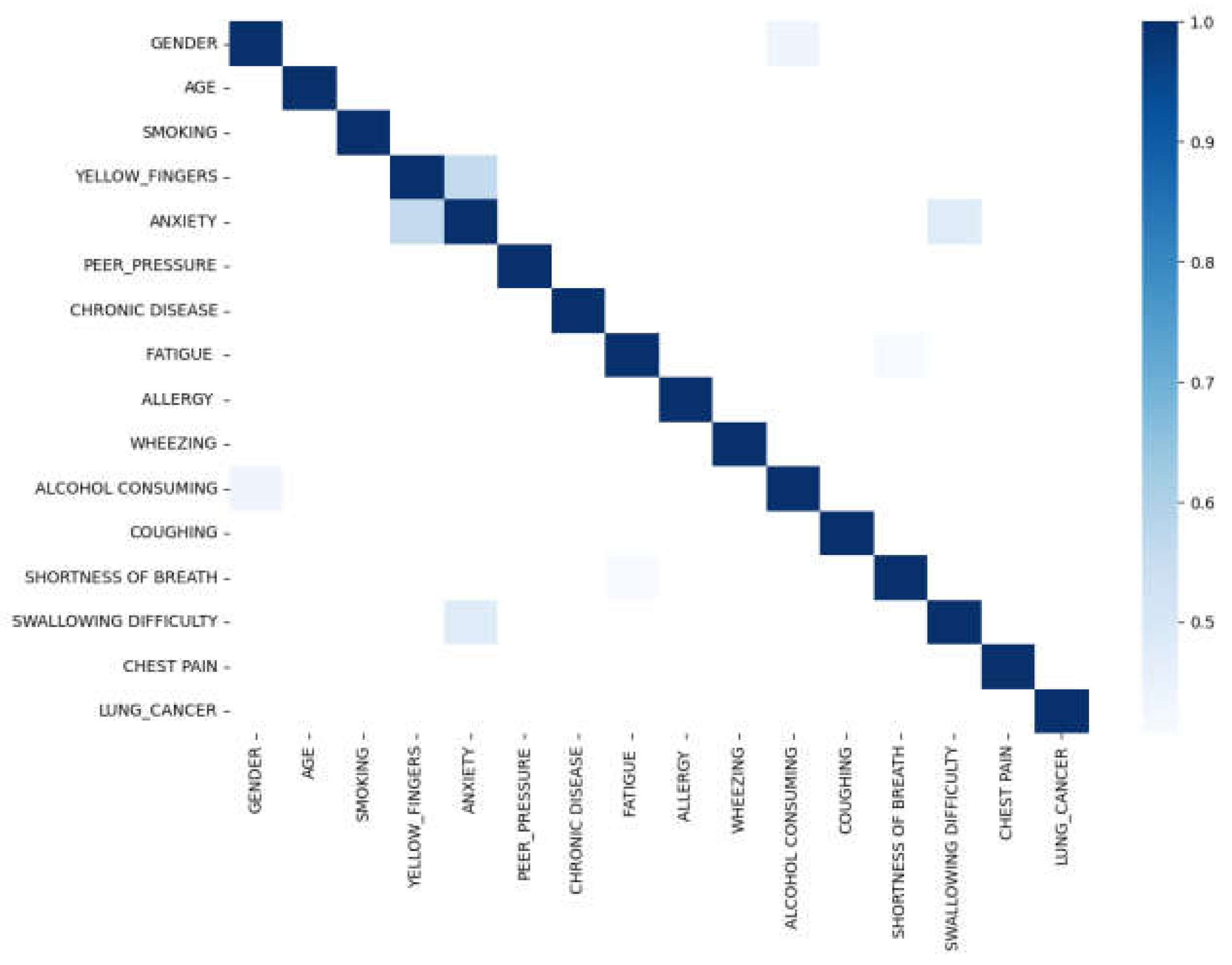
Figure 3(a)-(d)) are more prevalent in men with lung cancer compared to women. Upon examining the correlation plot of features in lung cancer datasets (
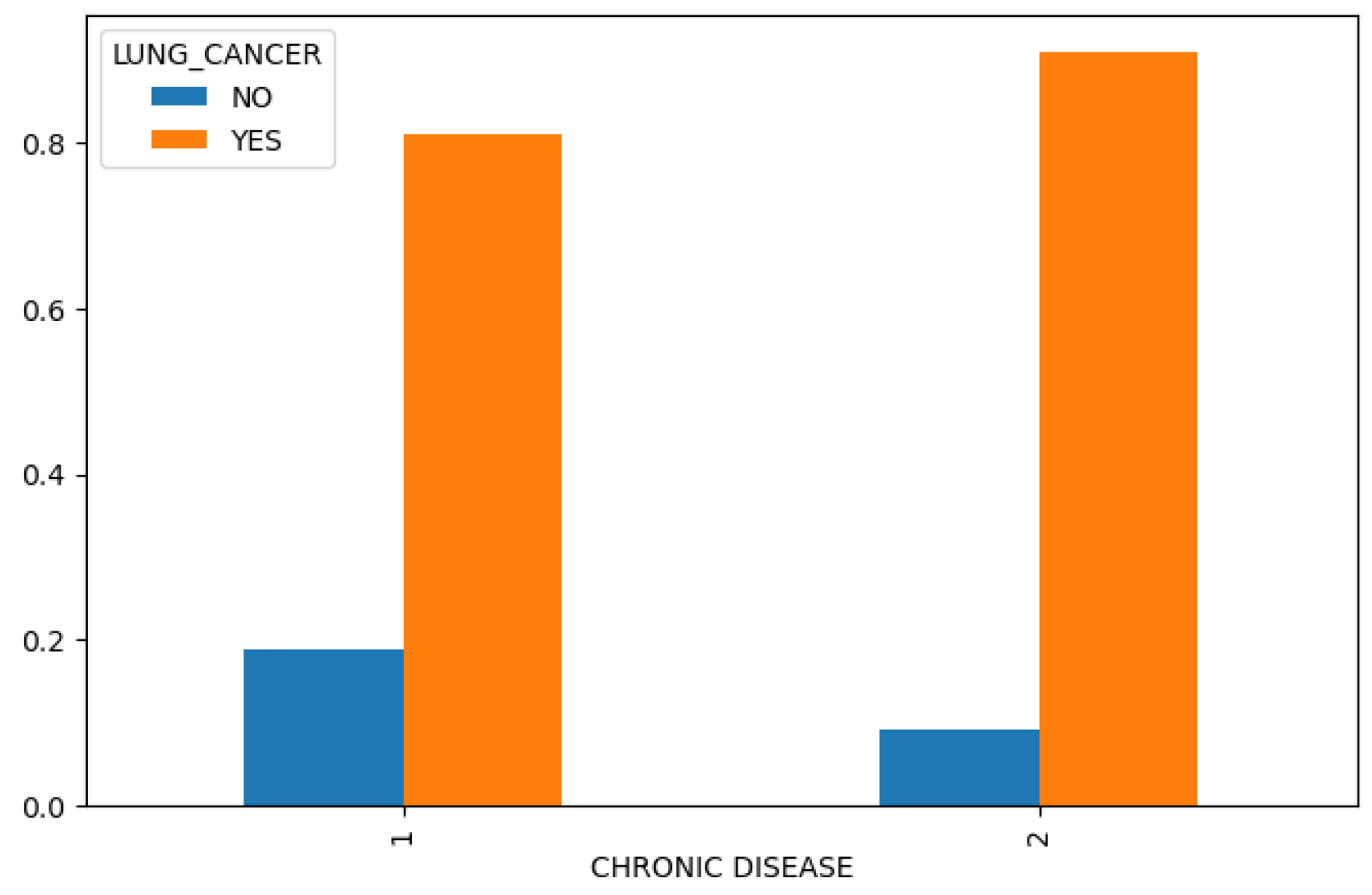
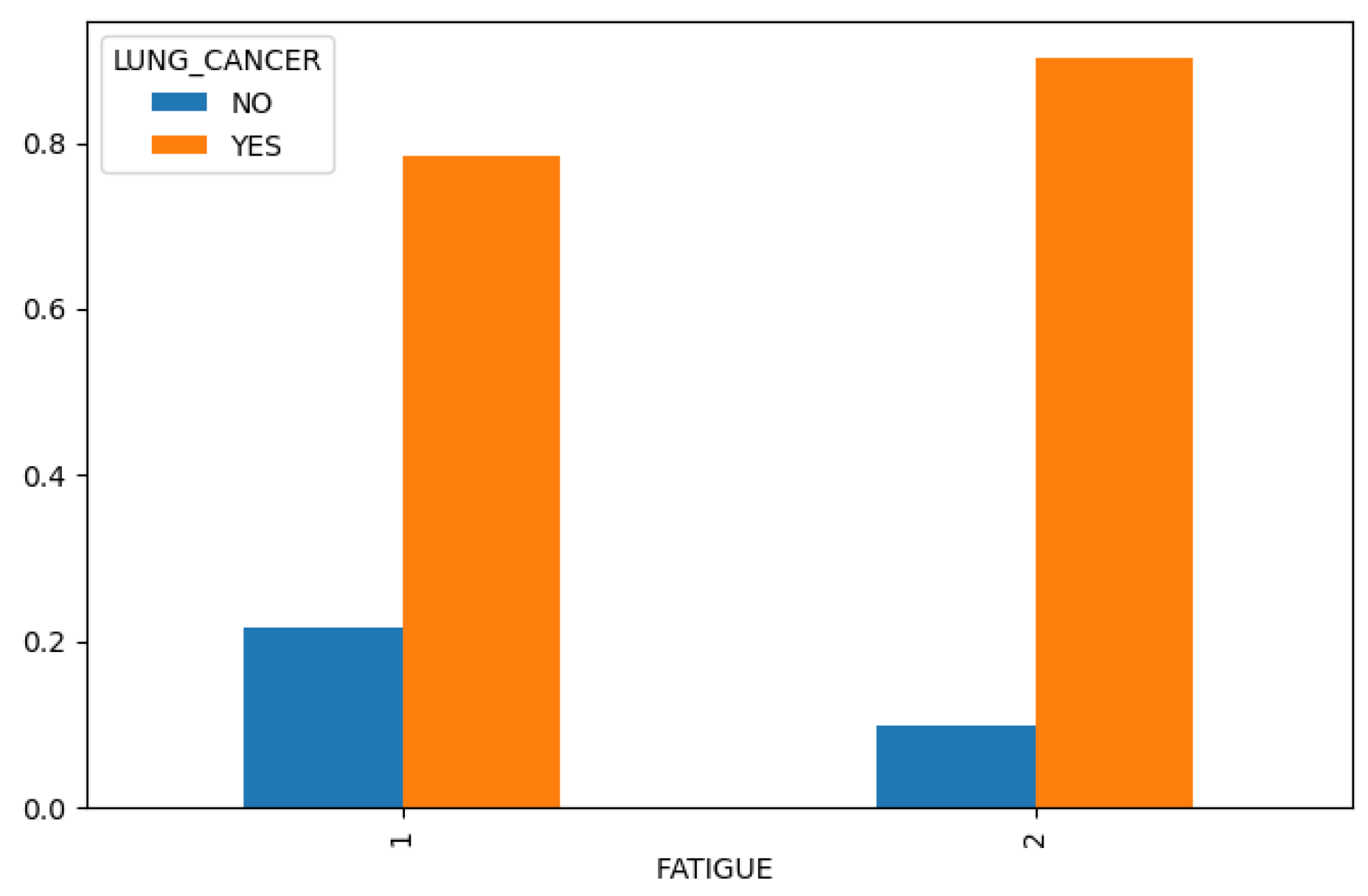
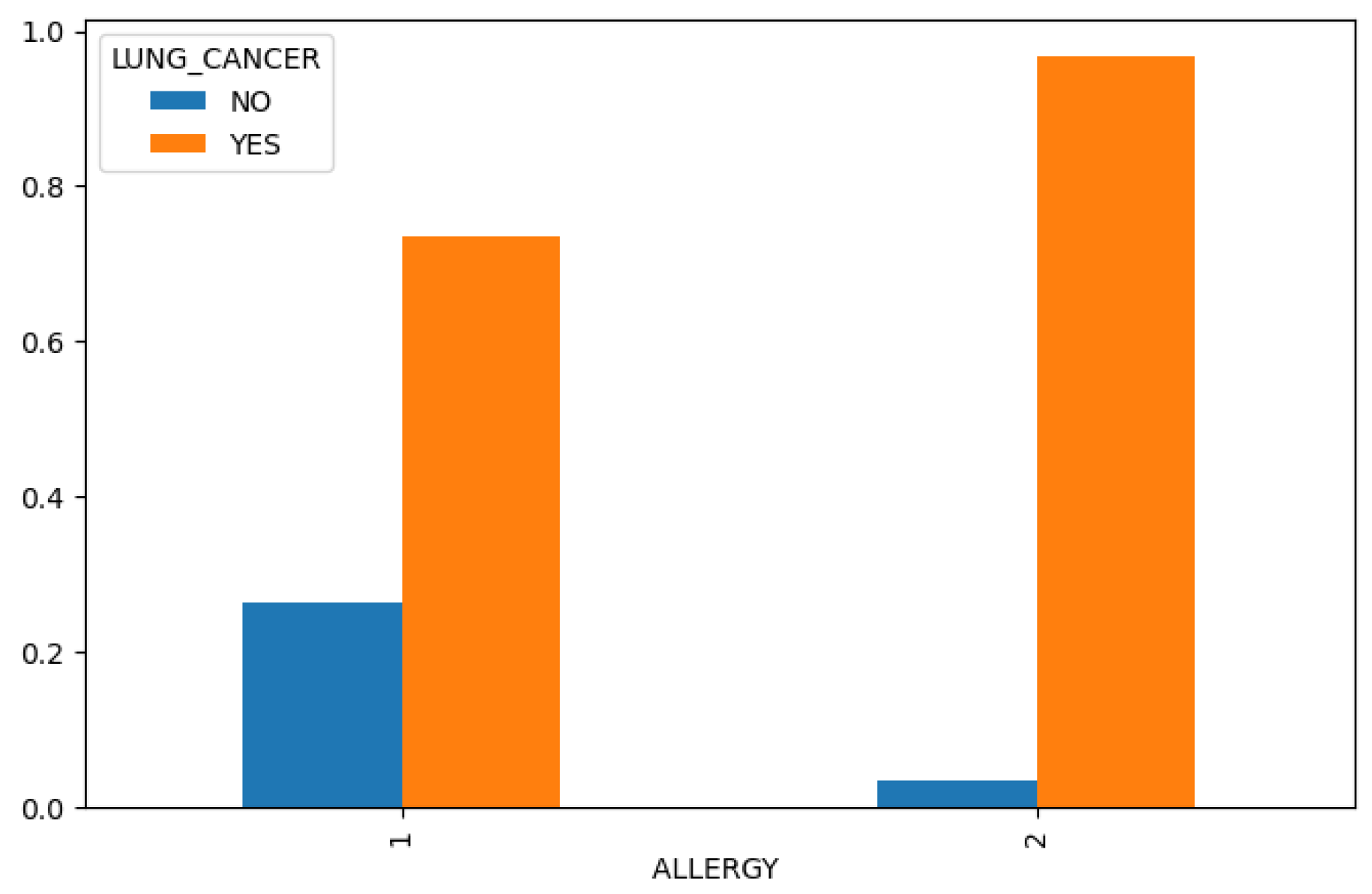
Figure 4), we find that chronic disease, fatigue, and allergies exhibit a higher correlation with lung cancer in women compared to men (
Figure 5,
Figure 6 and
Figure 7).
To delve deeper into the correlation analysis, we identify features with correlations exceeding 0.4 and consider them for combination to enhance predictive performance. As shown in
Figure 8, features such as anxiety-swallowing difficulty, anxiety-yellow fingers, and gender-alcohol consumption exhibit correlations above 0.4. We incorporate these combinations as additional columns, which significantly impact the predictive power of our model.
Data Splitting
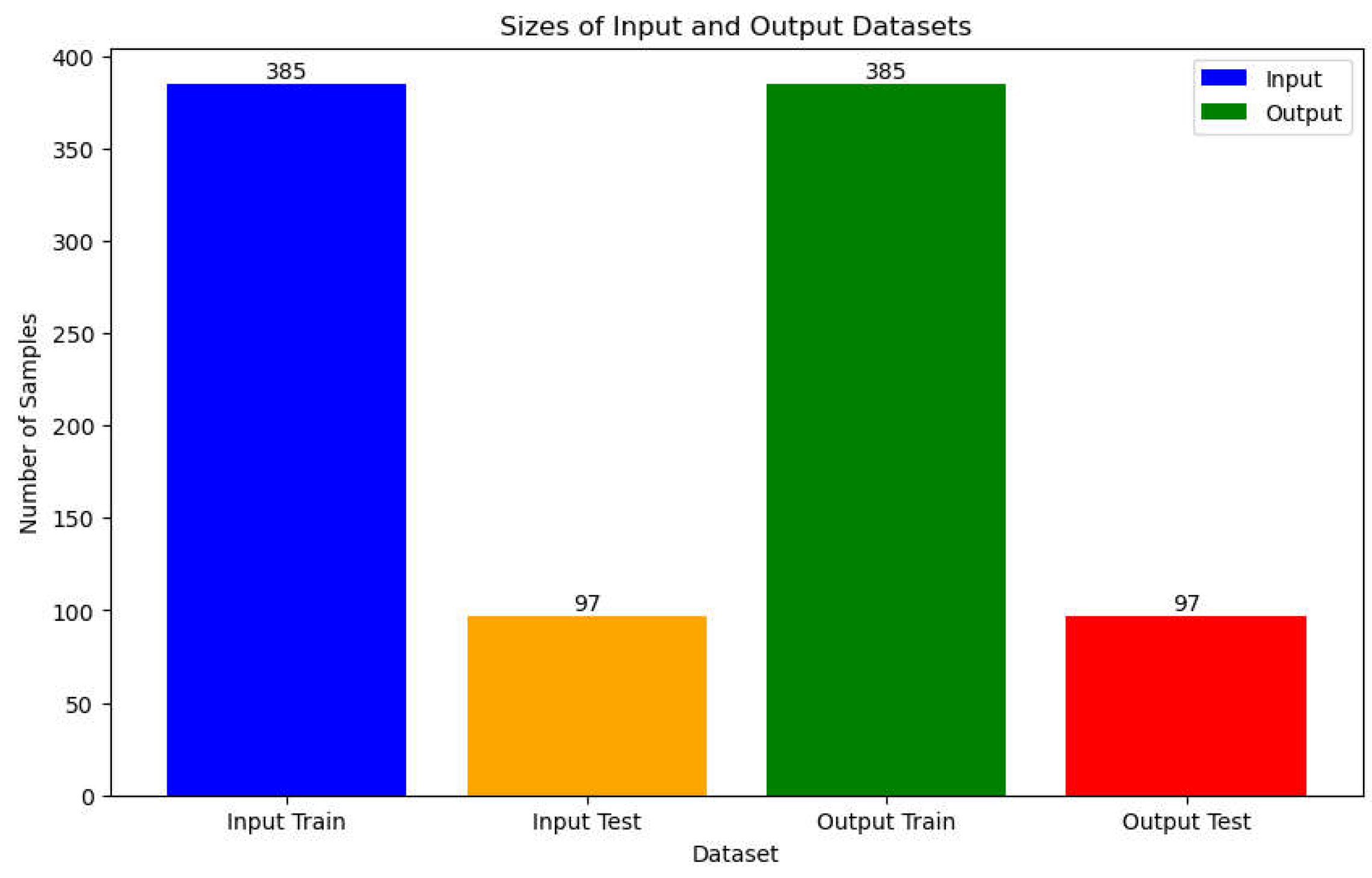
In the data splitting section, our dataset is divided into two subsets: 385 samples for training and 97 samples for testing (
Figure 9). To address potential class imbalance issues, we employ the Adaptive Synthetic Sampling (ADASYN) technique. ADASYN is an oversampling method commonly used in machine learning to balance imbalanced datasets. Unlike traditional oversampling techniques that replicate minority class samples, ADASYN generates synthetic samples for the minority class by focusing on the regions where the class density is low, thereby mitigating the risk of overfitting. Specifically, ADASYN identifies the minority class samples that are difficult to classify correctly and generates synthetic samples along the line segments between them and their nearest neighbors. By doing so, ADASYN effectively augments the minority class without significantly increasing the risk of overfitting. After applying ADASYN, the training dataset is rebalanced to ensure that both classes are adequately represented, thereby enhancing the model's ability to learn from the data.
To further enhance the model's generalizability and reduce the risk of overfitting, we employ k-fold cross-validation with k set to 5. This technique involves dividing the dataset into k equal folds. The model is then trained and evaluated k times, where each iteration utilizes a different fold for validation and the remaining folds for training. This approach provides a more robust assessment of the model's performance on unseen data by evaluating it on various subsets of the dataset. Consequently, k-fold cross-validation helps mitigate overfitting and provides a more reliable estimate of the model's generalizability to new data.
ML Models
In our study, we utilized a diverse array of machine learning (ML) models to address the classification task of predicting lung cancer types. Each of these models offers unique strengths and capabilities, contributing to the overall efficacy of our predictive framework.
Extreme Gradient Boosting (XGBoost)
XGBoost (Extreme Gradient Boosting) stands out as a powerful ensemble learning technique valued for its scalability and efficiency. It achieves this by sequentially training a series of weak learners, progressively enhancing the model's ability to predict. This iterative approach allows XGBoost to effectively capture complex patterns within the data. Furthermore, its ability to handle both numerical and categorical features, coupled with its resistance to overfitting, makes it particularly well-suited for classification tasks.
Light Gradient Boosting Machine (LGBM)
Light Gradient Boosting Machine (LightGBM) stands as another powerful gradient boosting framework, particularly adept at handling large and high-dimensional datasets. Unlike traditional gradient boosting, LightGBM leverages a novel tree-based learning algorithm that focuses on "leaf-wise" growth instead of level-wise growth. This approach significantly improves both computational efficiency and model accuracy. Furthermore, LightGBM's speed, scalability, and capability of handling both categorical features and imbalanced datasets make it a compelling choice for various classification tasks.
Adaptive Boosting (AdaBoost)
AdaBoost (Adaptive Boosting) carves its niche in ensemble learning by iteratively constructing a robust classifier from weaker ones. It achieves this by strategically assigning higher weights to misclassified samples in each pass. This focused approach allows AdaBoost to prioritize improvement in challenging areas of the feature space, ultimately leading to better classification accuracy. Combined with its simplicity, versatility, and ability to adapt to diverse datasets, AdaBoost proves to be a valuable tool for various classification tasks.
Logistic Regression
Logistic regression stands as a cornerstone linear model, prevalent in binary classification tasks. This enduring popularity stems from its simplicity. Even in scenarios where the relationship between features and the target variable isn't perfectly linear, logistic regression can often achieve good performance through linear approximation. Moreover, its interpretability, efficiency, and ease of use make it a popular choice as a baseline model for classification problems.
Decision Trees
Decision trees, a type of non-parametric supervised learning model, excel at interpretability and intuitive decision-making. They achieve this by partitioning the feature space into a hierarchical structure, where each step represents a decision based on a specific feature. This approach makes them well-suited for classification tasks involving non-linear decision boundaries, allowing them to capture complex relationships between features and the target variable. However, decision trees are susceptible to overfitting, particularly with deep structures and noisy datasets.
Random Forest
Random Forest emerges as a robust ensemble learning method built upon decision trees. It tackles overfitting by training a multitude of decision trees independently and then combining their predictions for a final outcome. This aggregation approach enhances the model's ability to generalize to unseen data. Random forests further demonstrate their value in classification tasks through their resistance to noise, scalability, and capability of handling high-dimensional data.
Categorical Boost (CatBoost)
CatBoost carves a distinct path in the realm of gradient boosting frameworks by excelling at handling categorical features seamlessly. Unlike traditional methods that require pre-processing like one-hot encoding, CatBoost utilizes a novel algorithm specifically designed for these types of variables. This innovative approach eliminates the need for such pre-processing steps. Furthermore, CatBoost boasts resistance to overfitting, the ability to tackle large datasets, and delivers high predictive accuracy, making it a compelling choice for classification tasks where categorical features play a significant role.
K-Nearest Neighbors (KNN)
K-Nearest Neighbors (KNN) stands as a straightforward yet powerful classification technique based on instance-based learning. It makes predictions by identifying the k nearest data points (neighbors) in the feature space for a new data point and assigning the most frequent class label amongst them. This simplicity, coupled with its flexibility and ability to capture intricate decision boundaries, makes KNN suitable for various classification tasks, particularly when dealing with non-parametric data distributions.
Deep Neural Networks (DNN)
Deep Neural Networks (DNNs) represent a powerful class of machine learning models known for their ability to learn complex patterns directly from raw data. This capability stems from their multi-layered architecture, where each layer builds upon the previous one to extract increasingly intricate features [
22]. Unlike simpler models, DNNs can automatically learn these hierarchical features, allowing them to capture subtle relationships within the data. This makes them particularly well-suited for classification tasks involving high-dimensional inputs, non-linear data structures, and diverse datasets. Image, text, and speech recognition applications are prime examples where DNNs excel.
Results and discussion
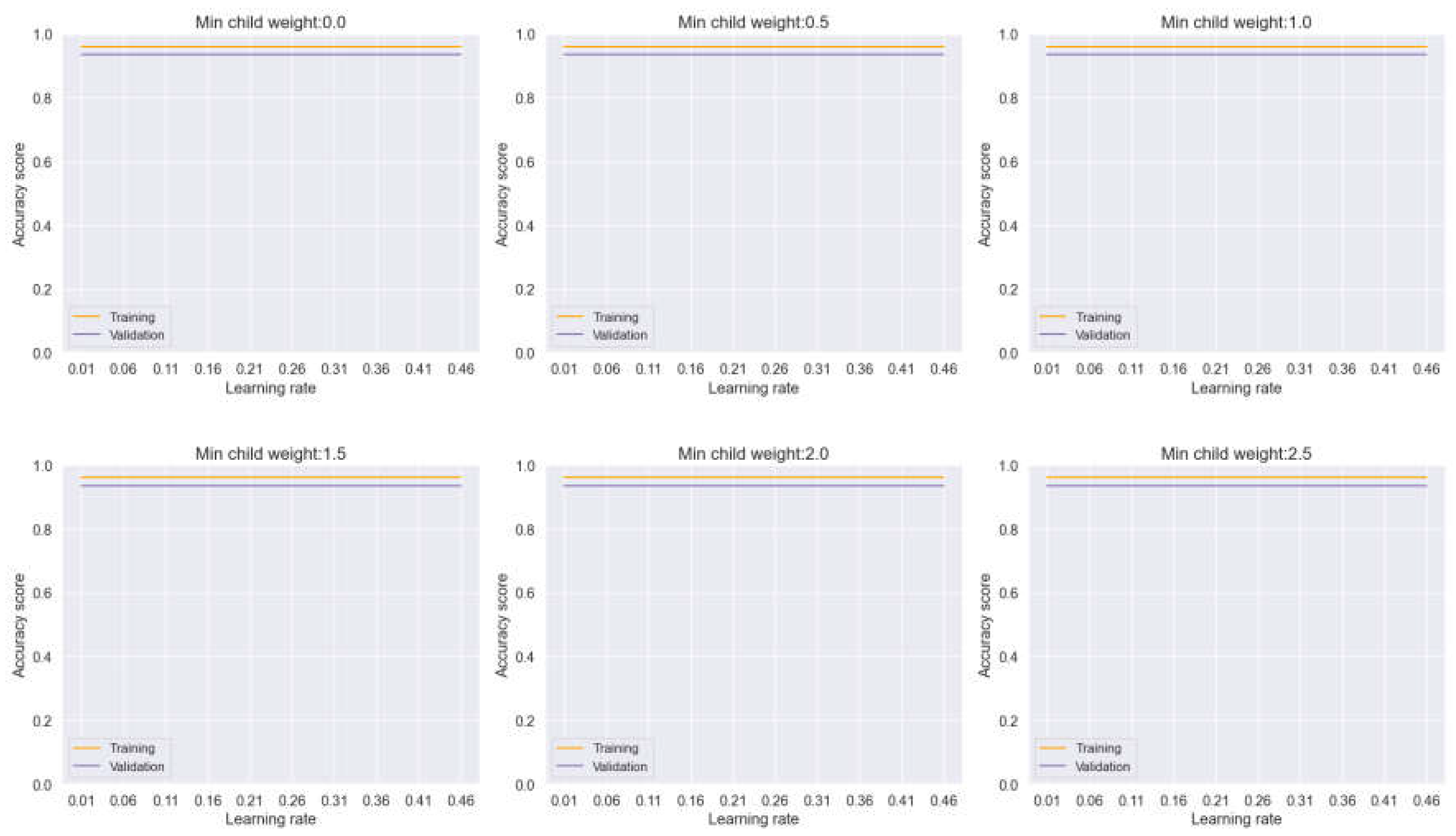
In this section, we undertake a comprehensive analysis of various machine learning (ML) models to evaluate their performance across different hyperparameters, with a particular focus on min child weight and learning rate. These hyperparameters are of paramount importance in addressing the challenges of overfitting and underfitting, thus influencing the overall classification accuracy of the models under consideration. Firstly, we examine the performance of an ensemble model known as Voting, which combines Decision Trees, Logistic Regression, and k-NN algorithms. As illustrated in
Figure 10, our experimentation involves adjusting the min child weight and learning rate to observe their impact on model convergence and overfitting mitigation. Notably, our analysis reveals a gradual convergence in both the training and validation plots as we modify these hyperparameters, indicating a reduction in overfitting tendencies. However, it is noteworthy that the reduction in overfitting appears to be marginal, as evidenced by the consistent movement of both plots. This suggests that while overfitting is present, it may not be as pronounced, posing a less formidable challenge for the model.
Furthermore, our ensemble model demonstrates commendable performance on unseen datasets, as depicted in the confusion matrix. Specifically, the model achieves an accuracy, precision, recall, and F-1 score of 91.75%, underscoring its effectiveness in accurately classifying instances across different classes. These results highlight the robustness and reliability of the Voting ensemble model in handling diverse datasets and complex classification tasks.
Moving forward, it will be imperative to conduct further experimentation and optimization to fine-tune the hyperparameters and enhance the performance of the ML models under consideration. Additionally, exploring alternative ensemble techniques and incorporating feature engineering methods may offer valuable insights into improving model accuracy and generalization capabilities. Overall, our findings contribute to a deeper understanding of the interplay between hyperparameters and model performance, paving the way for more effective utilization of ML algorithms in real-world applications.
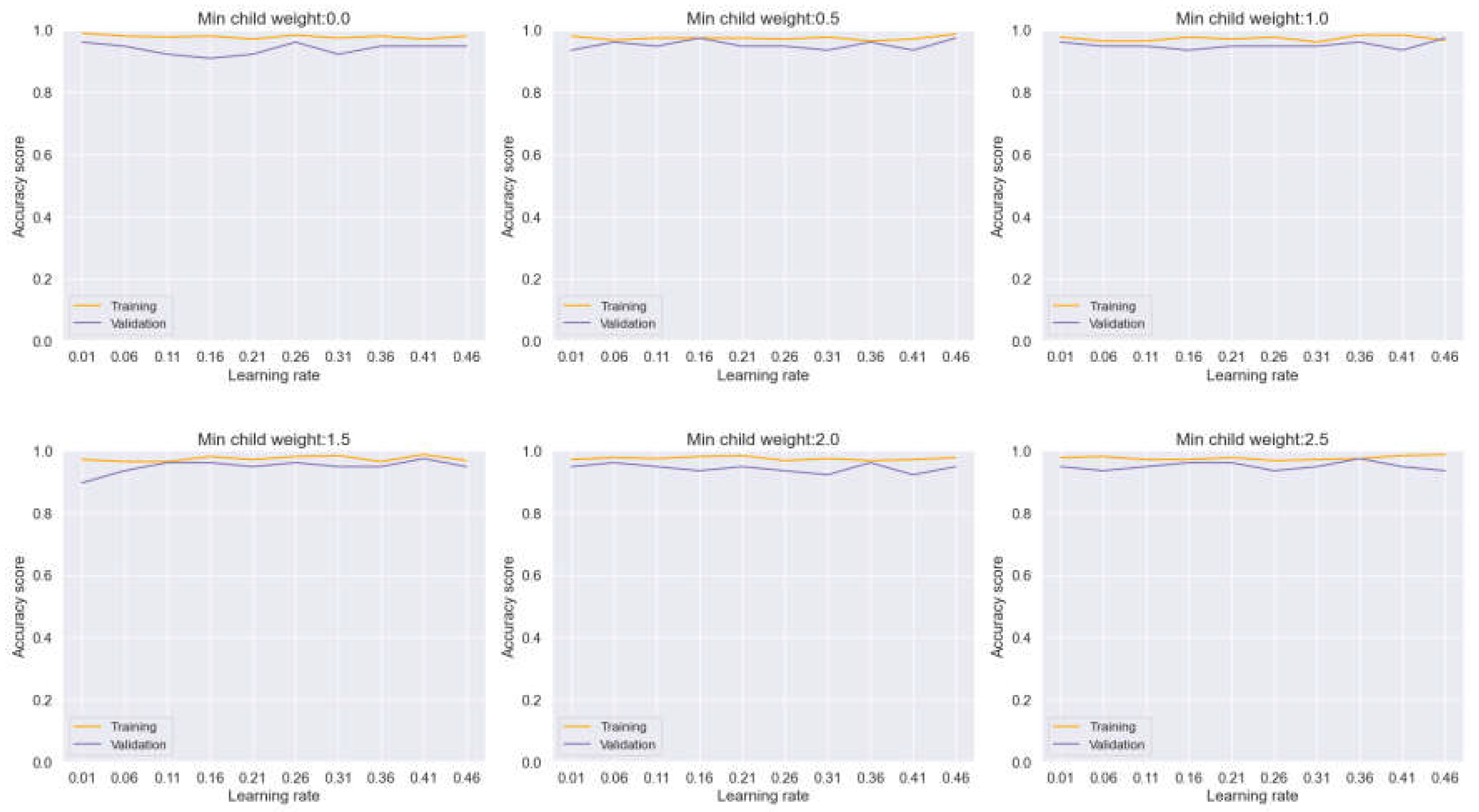
Continuing our analysis, we now turn our attention to Bagging, another ensemble learning technique, as depicted in
Figure 11,. In this evaluation, we closely examine the behavior of the model across varying hyperparameters, with a specific focus on min child weight and learning rate. Notably, our observations reveal alternating changes in the two plots as the hyperparameters fluctuate. This phenomenon results in a widening distance between the training and validation curves, exacerbating overfitting tendencies within the model.
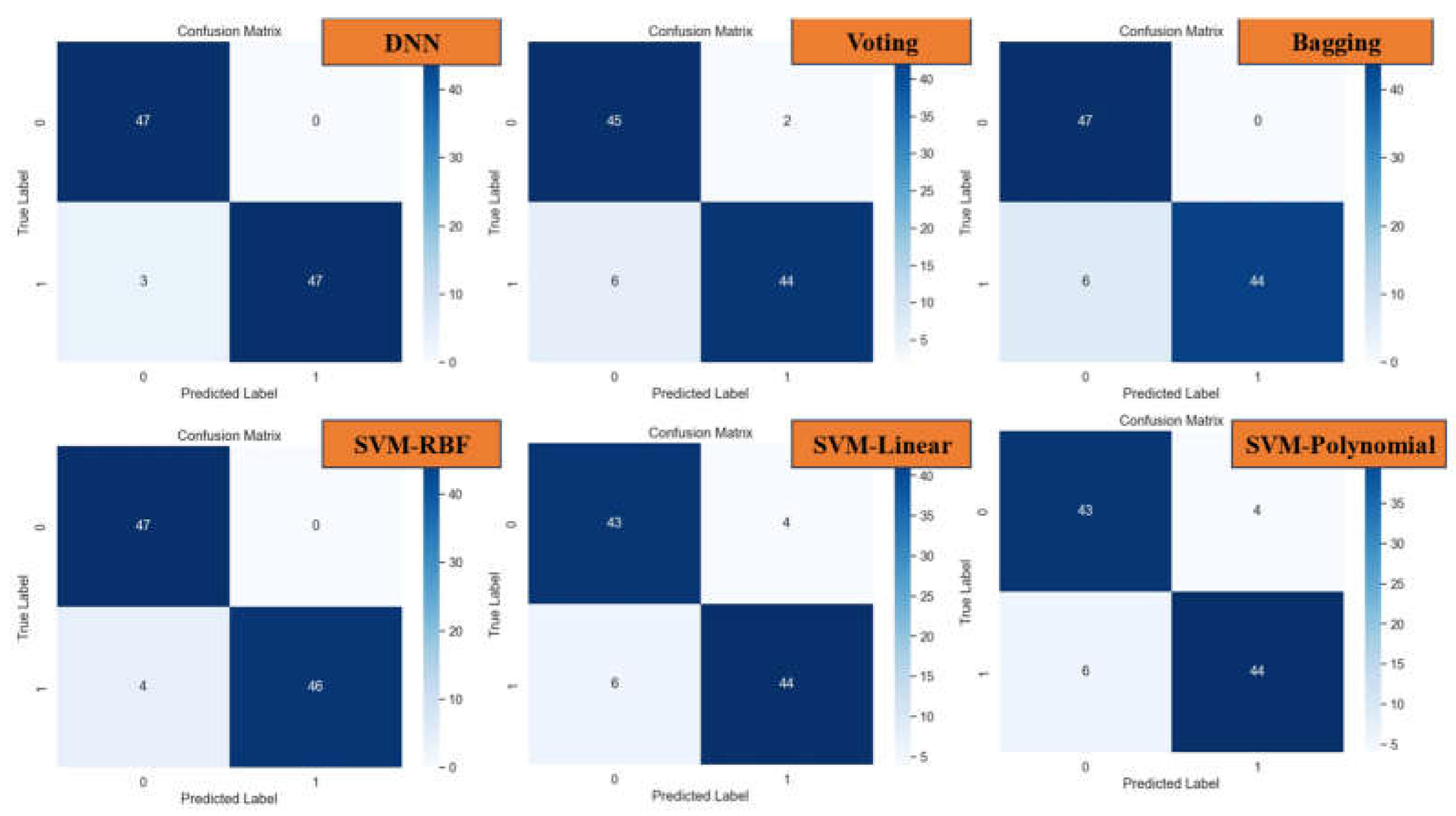
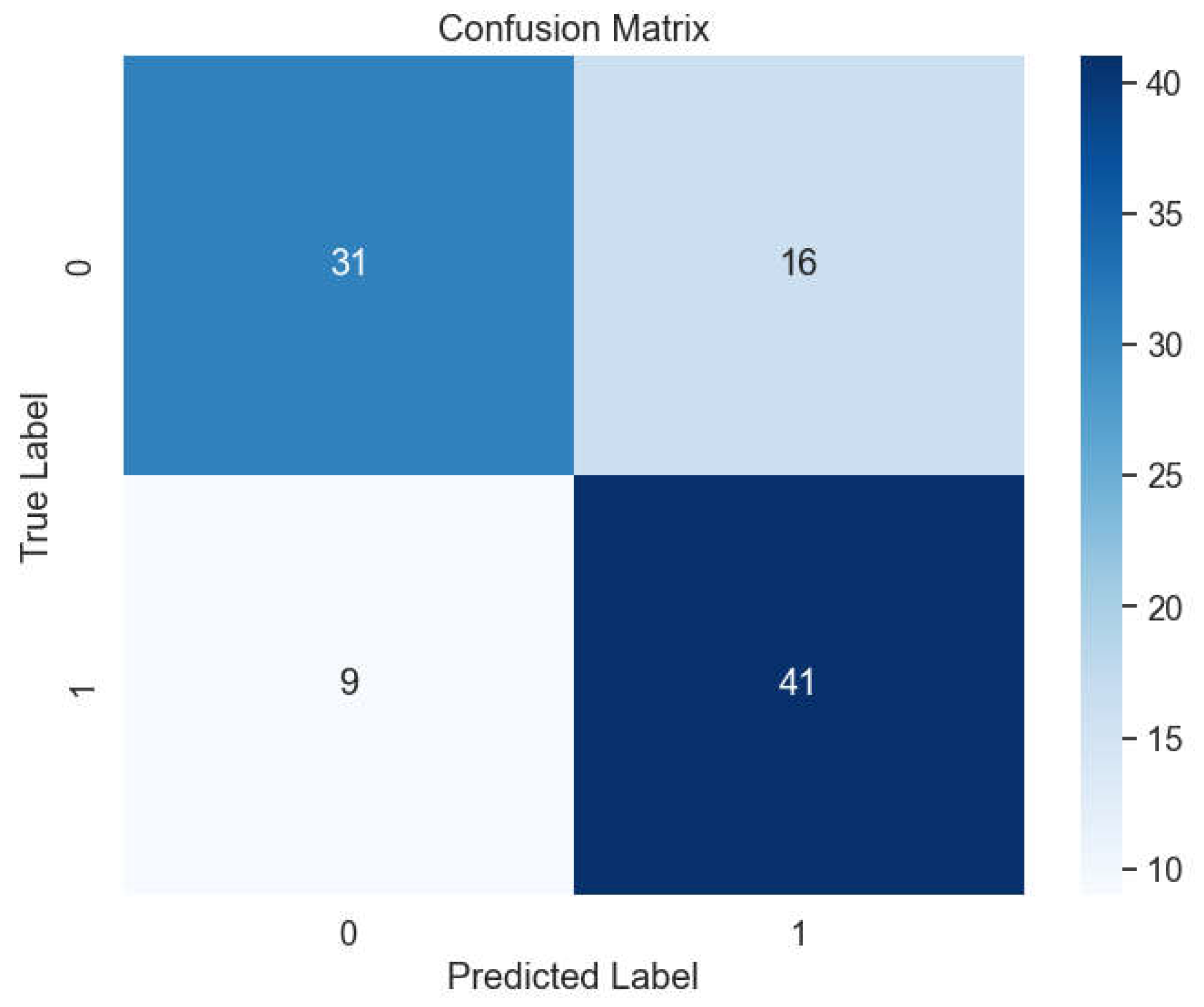
The insights garnered from this analysis play a crucial role in identifying optimal hyperparameters that effectively mitigate overfitting issues while maintaining robust model performance. Despite the challenges posed by overfitting, our experimentation yields promising results, with Bagging achieving an impressive accuracy rate of 93.81%. Furthermore, the model demonstrates precise classification performance across different classes, as evidenced by the detailed insights provided in the confusion matrix (
Figure 12).
Among the array of machine learning models examined, the Deep Neural Network (DNN) model emerges as a standout performer, showcasing remarkable accuracy, precision, recall, and F-1 score metrics, achieving an impressive 96.9%. Following closely behind is the Support Vector Machine (SVM) model with the RBF kernel, attaining a commendable accuracy rate of 95.87%. These findings underscore the robust classification capabilities of both DNN and SVM models, particularly in effectively distinguishing between lung cancer cases and non-cancer cases, as corroborated by the detailed insights provided in the confusion matrix (
Figure 12).
However, it is noteworthy that the SVM models employing Linear, Polynomial, and Sigmoid kernels (
Figure 13) exhibit comparatively lower accuracy rates, ranging from 74.22% to 89.69%. Despite these variations in performance, these SVM models contribute valuable insights to the classification task, albeit with differing degrees of effectiveness. Their inclusion in the analysis enriches our understanding of the diverse landscape of machine learning models and their applicability in addressing complex classification challenges.
These observations highlight the importance of carefully selecting and fine-tuning machine learning models to suit the specific requirements of the classification task at hand. While certain models may excel in certain scenarios, others may offer complementary strengths or insights that contribute to a comprehensive understanding of the problem domain. Moving forward, further exploration and experimentation with different model architectures, hyperparameters, and feature engineering techniques will be essential to optimize classification performance and enhance the reliability and robustness of machine learning-based lung cancer classification systems. By leveraging a diverse array of machine learning models and methodologies, researchers can continue to advance the field of medical diagnostics and contribute to improved patient outcomes in the realm of lung cancer detection and treatment.
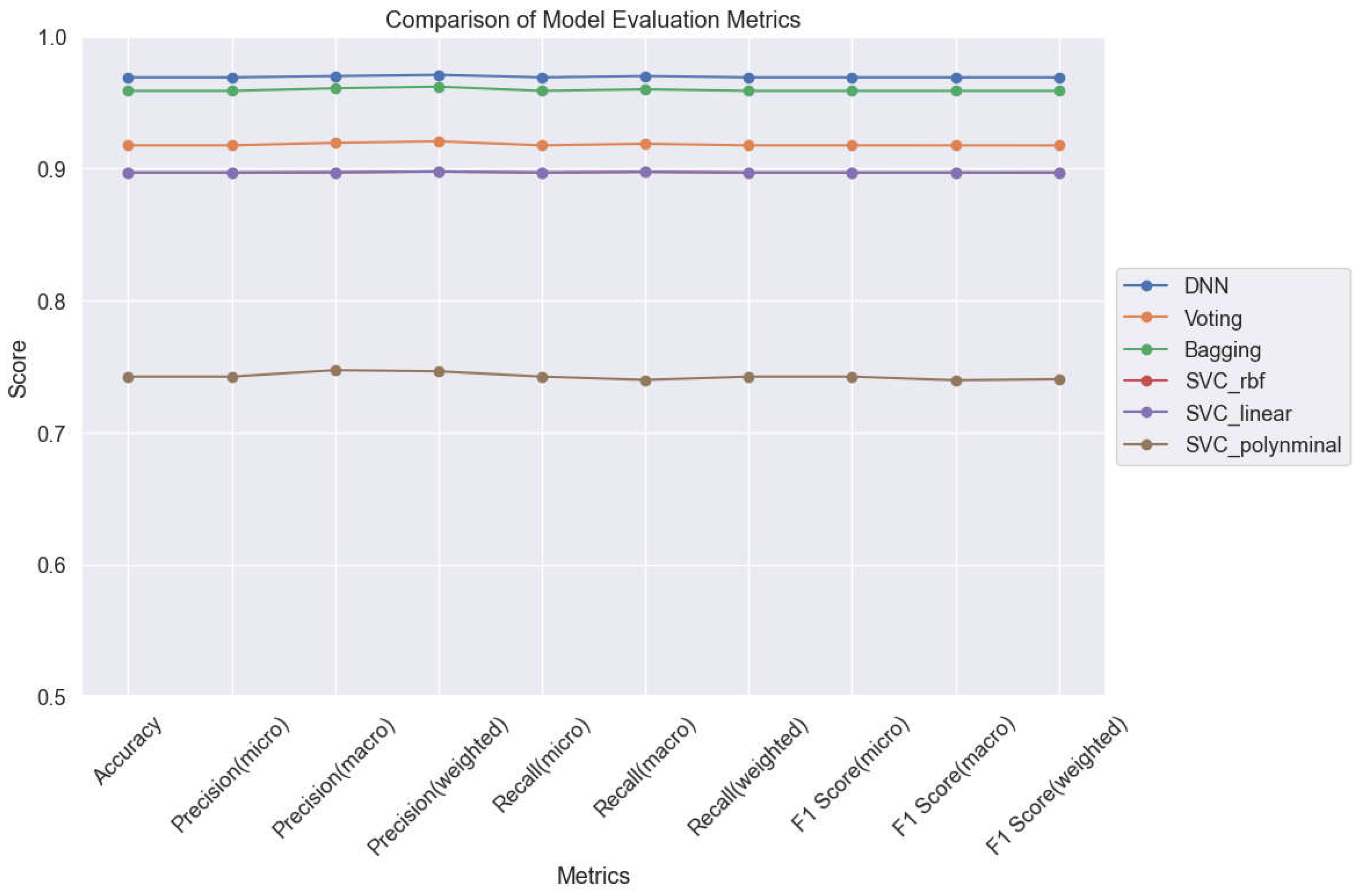
Among the diverse range of machine learning models evaluated in our study, the Deep Neural Network (DNN) and Bagging techniques emerge as standout performers in the realm of lung cancer classification. Notably, these models showcase exceptional performance metrics, demonstrating their efficacy in accurately distinguishing between different levels of lung cancer. A key highlight of both the DNN and Bagging models is their robust adaptability to hyperparameters, enabling them to effectively mitigate overfitting issues while maintaining high levels of classification accuracy. This adaptability is exemplified in
Figure 14, where both models exhibit remarkable stability and consistency in their classification performance across varying hyperparameter configurations.
The heightened stability and consistency observed in the classification accuracy of the DNN and Bagging models underscore their potential as promising candidates for further exploration and real-world application in clinical settings. By leveraging these models, healthcare practitioners can benefit from more accurate and reliable diagnostic tools, ultimately leading to improved patient outcomes and enhanced clinical decision-making.
Conclusion
This study explored the efficacy of machine learning algorithms in classifying lung cancer types based on protein characteristics. Deep Neural Networks (DNNs) emerged as the frontrunner, achieving an impressive 96.91% accuracy. Ensemble methods like Voting and Bagging also yielded promising results, exceeding 91% accuracy.
Our investigation further highlighted the importance of parameter optimization for Support Vector Machines (SVMs) with different kernel functions. By meticulously analyzing confusion matrices, we provided insights into the strengths and limitations of each model, aiding informed decision-making in clinical settings. These findings underscore the immense potential of machine learning to revolutionize lung cancer diagnosis and treatment. By leveraging these computational techniques, we can pave the way for personalized interventions, ultimately improving patient outcomes and advancing oncology research.
Our study also emphasizes the value of machine learning in deciphering complex patterns within protein data for lung cancer classification. This computational analysis can reveal subtle nuances that might be missed by traditional methods, leading to more accurate and targeted interventions. The exploration of various models showcased the versatility of machine learning in handling diverse datasets and classification tasks. The DNN's robust performance, in particular, highlights the potential of deep learning in extracting valuable insights from complex biological data.
Looking ahead, integrating machine learning into clinical practice holds immense promise for lung cancer diagnosis and treatment. By incorporating these techniques into healthcare workflows, we can empower clinicians with tools for more personalized and precise patient care. In essence, this study serves as a testament to the transformative impact of machine learning in oncology. Continued research and innovation can unlock the full potential of these technologies to combat lung cancer and improve patient outcomes globally.
Reference
- E. Dritsas, M. Trigka, Lung Cancer Risk Prediction with Machine Learning Models, Big Data and Cognitive Computing 2022, Vol. 6, Page 139 6 (2022) 139. [CrossRef]
- N. Banerjee, S. Das, Prediction Lung Cancer- in Machine Learning Perspective, 2020 International Conference on Computer Science, Engineering and Applications, ICCSEA 2020 (2020). [CrossRef]
- R. Patra, Prediction of lung cancer using machine learning classifier, Communications in Computer and Information Science 1235 CCIS (2020) 132–142. [CrossRef]
- S.S. Raoof, M.A. Jabbar, S.A. Fathima, Lung Cancer Prediction using Machine Learning: A Comprehensive Approach, 2nd International Conference on Innovative Mechanisms for Industry Applications, ICIMIA 2020 - Conference Proceedings (2020) 108–115. [CrossRef]
- I. El Naqa, Machine learning methods for predicting tumor response in lung cancer, Wiley Interdiscip Rev Data Min Knowl Discov 2 (2012) 173–181. [CrossRef]
- A. Sadhwani, H.W. Chang, A. Behrooz, T. Brown, I. Auvigne-Flament, H. Patel, R. Findlater, V. Velez, F. Tan, K. Tekiela, E. Wulczyn, E.S. Yi, C.H. Mermel, D. Hanks, P.H.C. Chen, K. Kulig, C. Batenchuk, D.F. Steiner, P. Cimermancic, Comparative analysis of machine learning approaches to classify tumor mutation burden in lung adenocarcinoma using histopathology images, Scientific Reports 2021 11:1 11 (2021) 1–11. [CrossRef]
- Z.S. Zubi, R.A. Saad, Z.S. Zubi, R.A. Saad, Improves Treatment Programs of Lung Cancer Using Data Mining Techniques, Journal of Software Engineering and Applications 7 (2014) 69–77. [CrossRef]
- S. Hussein, P. Kandel, C.W. Bolan, M.B. Wallace, U. Bagci, Lung and Pancreatic Tumor Characterization in the Deep Learning Era: Novel Supervised and Unsupervised Learning Approaches, IEEE Trans Med Imaging 38 (2019) 1777–1787. [CrossRef]
- S. Zaza, M. Al-Emran, Mining and exploration of credit cards data in UAE, Proceedings - 2015 5th International Conference on e-Learning, ECONF 2015 (2016) 275–279. [CrossRef]
- S. Huang, I. Arpaci, M. Al-Emran, S. Kılıçarslan, M.A. Al-Sharafi, A comparative analysis of classical machine learning and deep learning techniques for predicting lung cancer survivability, Multimed Tools Appl 82 (2023) 34183–34198. [CrossRef]
- M. Goldszmidt, Bayesian Network Classifiers, Wiley Encyclopedia of Operations Research and Management Science (2011). [CrossRef]
- I. Arpaci, S. Huang, M. Al-Emran, M.N. Al-Kabi, M. Peng, Predicting the COVID-19 infection with fourteen clinical features using machine learning classification algorithms, Multimed Tools Appl 80 (2021) 11943–11957. [CrossRef]
- S.A. Ajagbe, O.A. Oki, M.A. Oladipupo, A. Nwanakwaugwum, Investigating the Efficiency of Deep Learning Models in Bioinspired Object Detection, International Conference on Electrical, Computer, and Energy Technologies, ICECET 2022 (2022). [CrossRef]
- Q. Wu, W. Zhao, Small-Cell Lung Cancer Detection Using a Supervised Machine Learning Algorithm, Proceedings - 2017 International Symposium on Computer Science and Intelligent Controls, ISCSIC 2017 2018-February (2017) 88–91. [CrossRef]
- J.B. Awotunde, S.A. Ajagbe, M.A. Oladipupo, J.A. Awokola, O.S. Afolabi, T.O. Mathew, Y.J. Oguns, An Improved Machine Learnings Diagnosis Technique for COVID-19 Pandemic Using Chest X-ray Images, Communications in Computer and Information Science 1455 CCIS (2021) 319–330. [CrossRef]
- J. Xu, P. Wu, Y. Chen, Q. Meng, H. Dawood, M.M. Khan, A Novel Deep Flexible Neural Forest Model for Classification of Cancer Subtypes Based on Gene Expression Data, IEEE Access 7 (2019) 22086–22095. [CrossRef]
- A.K. Dwivedi, Artificial neural network model for effective cancer classification using microarray gene expression data, Neural Comput Appl 29 (2018) 1545–1554. [CrossRef]
- S. Tarek, R. Abd Elwahab, M. Shoman, Gene expression based cancer classification, Egyptian Informatics Journal 18 (2017) 151–159. [CrossRef]
- Z. Yang, B. Liu, T. Lin, Y. Zhang, L. Zhang, M. Wang, Multiomics analysis on DNA methylation and the expression of both messenger RNA and microRNA in lung adenocarcinoma, J Cell Physiol 234 (2019) 7579–7586. [CrossRef]
- C. Park, J. Ha, S. Park, Prediction of Alzheimer’s disease based on deep neural network by integrating gene expression and DNA methylation dataset, Expert Syst Appl 140 (2020) 112873. [CrossRef]
- A. Kutlay, Y. Aydin Son, Integrative Predictive Modeling of Metastasis in Melanoma Cancer Based on MicroRNA, mRNA, and DNA Methylation Data, Front Mol Biosci 8 (2021) 637355. [CrossRef]
- R. Hosseini Rad, S. Baniasadi, P. Yousefi, H. Morabbi Heravi, M. Shaban Al-Ani, M. Asghari Ilani, Presented a Framework of Computational Modeling to Identify the Patient Admission Scheduling Problem in the Healthcare System, J Healthc Eng 2022 (2022). [CrossRef]
Figure 1.
Features (Gender, Age, Smoking and Yellow Finger) Distribution in hue of Lung Cancer.
Figure 1.
Features (Gender, Age, Smoking and Yellow Finger) Distribution in hue of Lung Cancer.
Figure 2.
Features (Anxiety, Per-pressure, Wheezing and alcohol Consuming) Distribution in hue of Lung Cancer.
Figure 2.
Features (Anxiety, Per-pressure, Wheezing and alcohol Consuming) Distribution in hue of Lung Cancer.
Figure 3.
Features (Coughing, Shortness of Breath, Swallowing Difficulty and Chest Pain) Distribution in hue of Lung Cancer.
Figure 3.
Features (Coughing, Shortness of Breath, Swallowing Difficulty and Chest Pain) Distribution in hue of Lung Cancer.
Figure 4.
Lung Cancer Dataset Correlation.
Figure 4.
Lung Cancer Dataset Correlation.
Figure 5.
Chronic Disease in Lung Cancer Dataset.
Figure 5.
Chronic Disease in Lung Cancer Dataset.
Figure 6.
Fatigue in Lung Cancer Dataset.
Figure 6.
Fatigue in Lung Cancer Dataset.
Figure 7.
Allergy in Lung Cancer Dataset.
Figure 7.
Allergy in Lung Cancer Dataset.
Figure 8.
Lung Cancer Correlation Bigger than 0.4.
Figure 8.
Lung Cancer Correlation Bigger than 0.4.
Figure 9.
Splitting Datasets into Training and Test as Input and Output.
Figure 9.
Splitting Datasets into Training and Test as Input and Output.
Figure 10.
Training and Validation plots under consideration of Min child Weight and Learning Rate in Ensemble Methods, Voting.
Figure 10.
Training and Validation plots under consideration of Min child Weight and Learning Rate in Ensemble Methods, Voting.
Figure 11.
Training and Validation plots under consideration of Min child Weight and Learning Rate in Ensemble Methods, Bagging.
Figure 11.
Training and Validation plots under consideration of Min child Weight and Learning Rate in Ensemble Methods, Bagging.
Figure 12.
Confusion Matrix of DNN, Voting, Bagging, SVM with kernel of RBF, Linear and Polynomial.
Figure 12.
Confusion Matrix of DNN, Voting, Bagging, SVM with kernel of RBF, Linear and Polynomial.
Figure 13.
Confusion Matrix of SVM with kernel of Sigmoid.
Figure 13.
Confusion Matrix of SVM with kernel of Sigmoid.
Figure 14.
Comparison of 6 ML Models for Lung Cancer Prediction.
Figure 14.
Comparison of 6 ML Models for Lung Cancer Prediction.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).