Submitted:
05 June 2024
Posted:
06 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Methodological Approach
2.1. Structural Phylogenomic Reconstructions with Alignment-free Methods
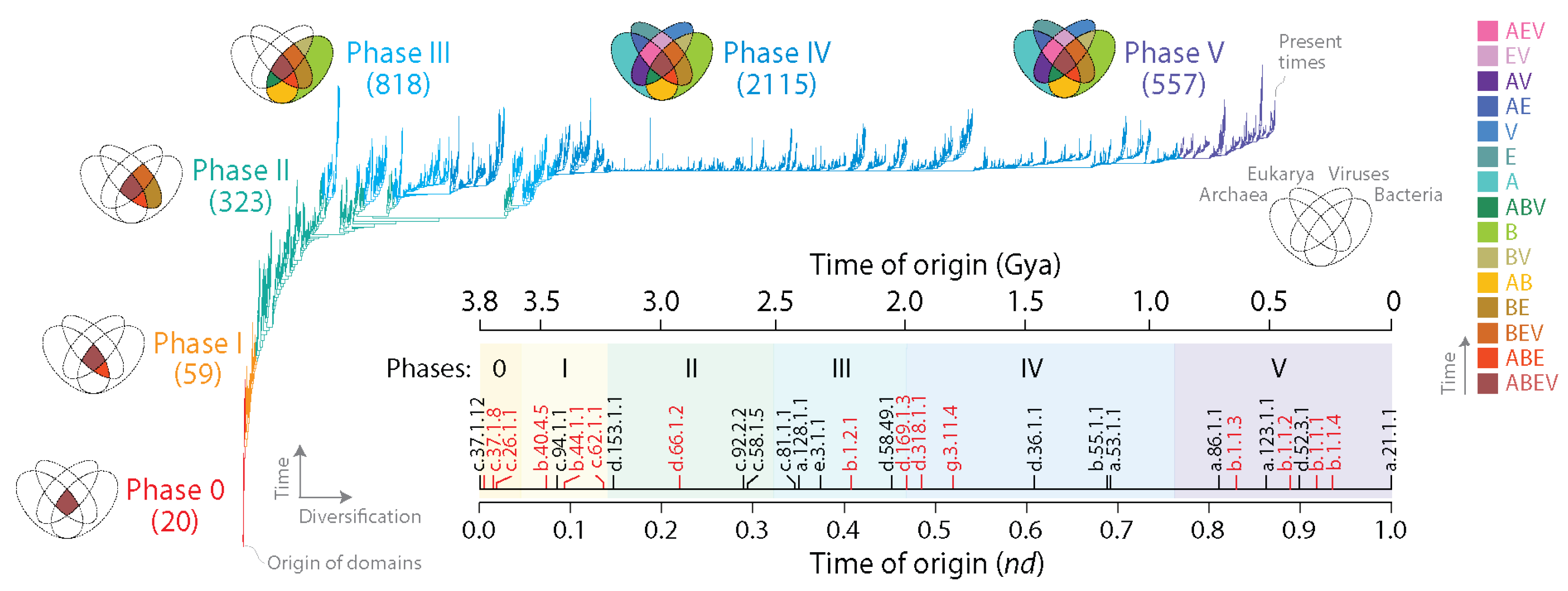
2.2. Evolutionary Chronologies
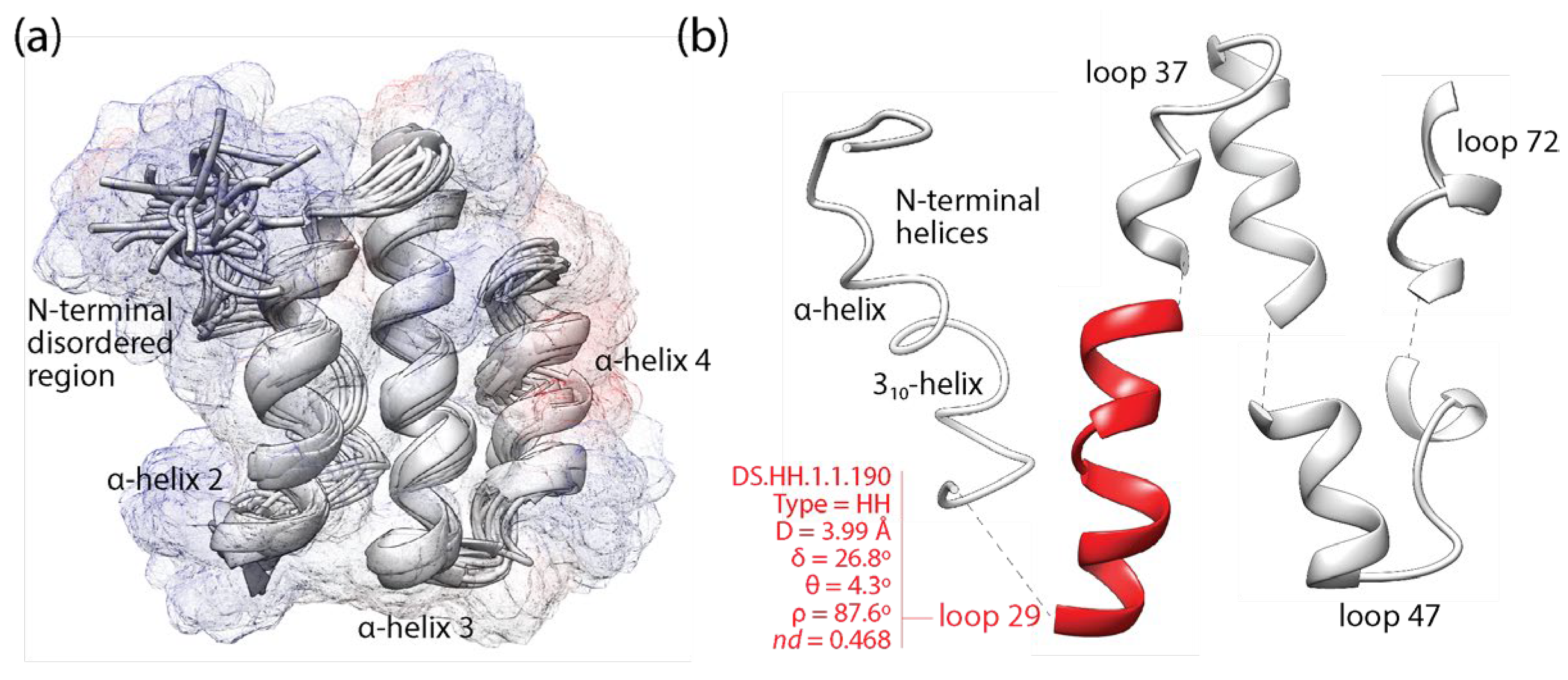
2.3. Loops: Architects of Protein Domains
3. Phylogenomic Analysis of Protein Structural Domains and Loops
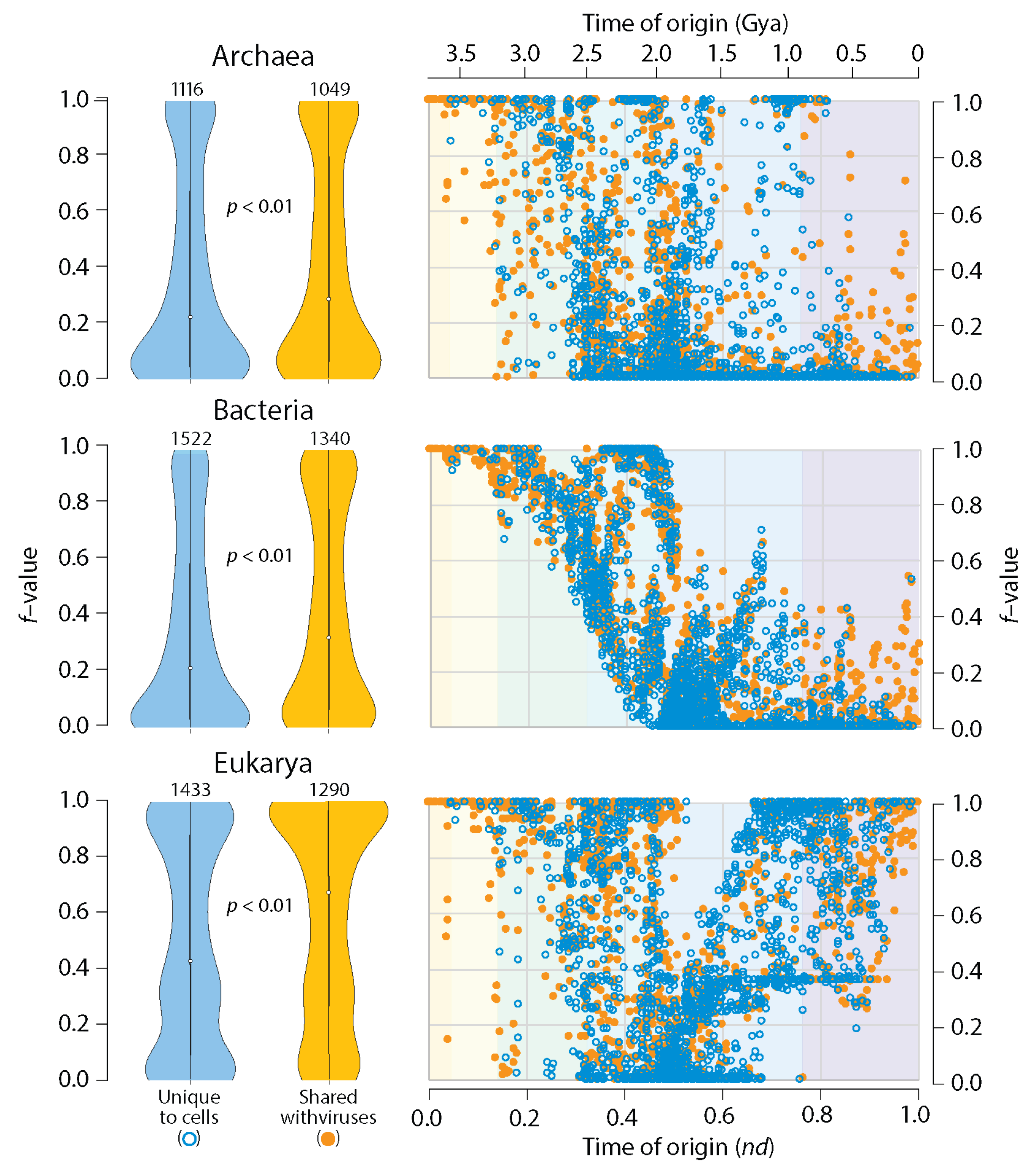
3.1. Viruses Help Spread Domain Wealth in the Proteomes of Superkingdoms
3.2. Viruses and the Evolutionary Primacy of Horizontal Transfer of Prior Molecular States
3.3. Microbial Supergroup-Specific Prior Molecular States Fail to Remain Persistent
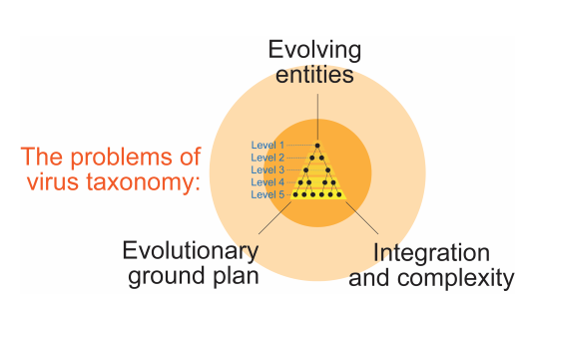
4. Are Viruses Taxonomic Units?
4.1. Taxonomic Classification is Hampered by Conceptual Difficulties
4.1.1. Most Species Co-Evolve with Others
4.1.2. Evolution is Reticulated
4.1.3. Independent Origins Break Up Monophyletic Relationships
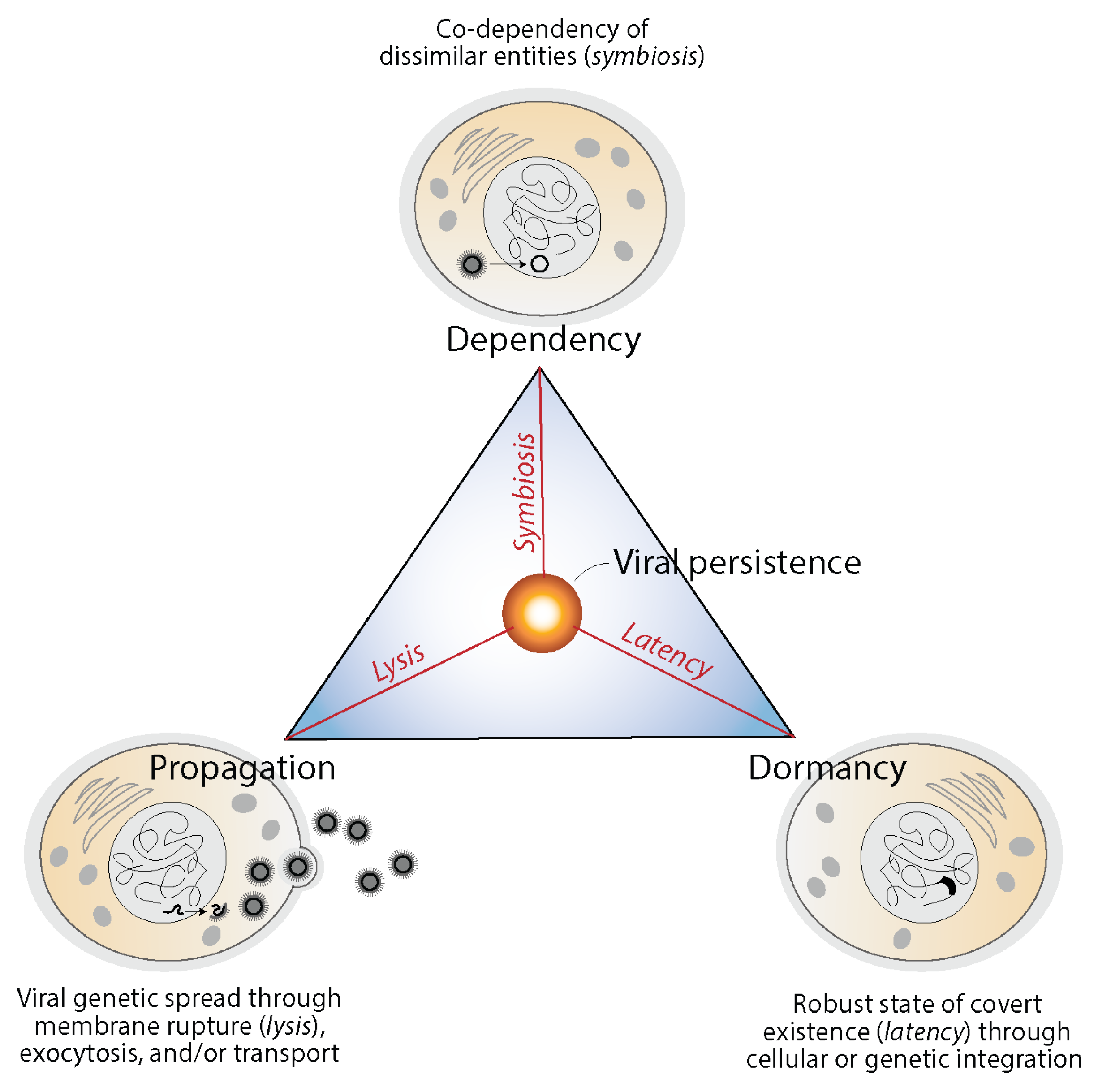
4.2. Are Viruses Self-Standing or Fully-Integrated Biological Systems?
4.3. Are Viruses Mobile Genetic Elements, Microbes or Cellularly-Integrated Processes?
4.4. A Phylogenomic-Centric Assessment
5. Conclusions and Recommendations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
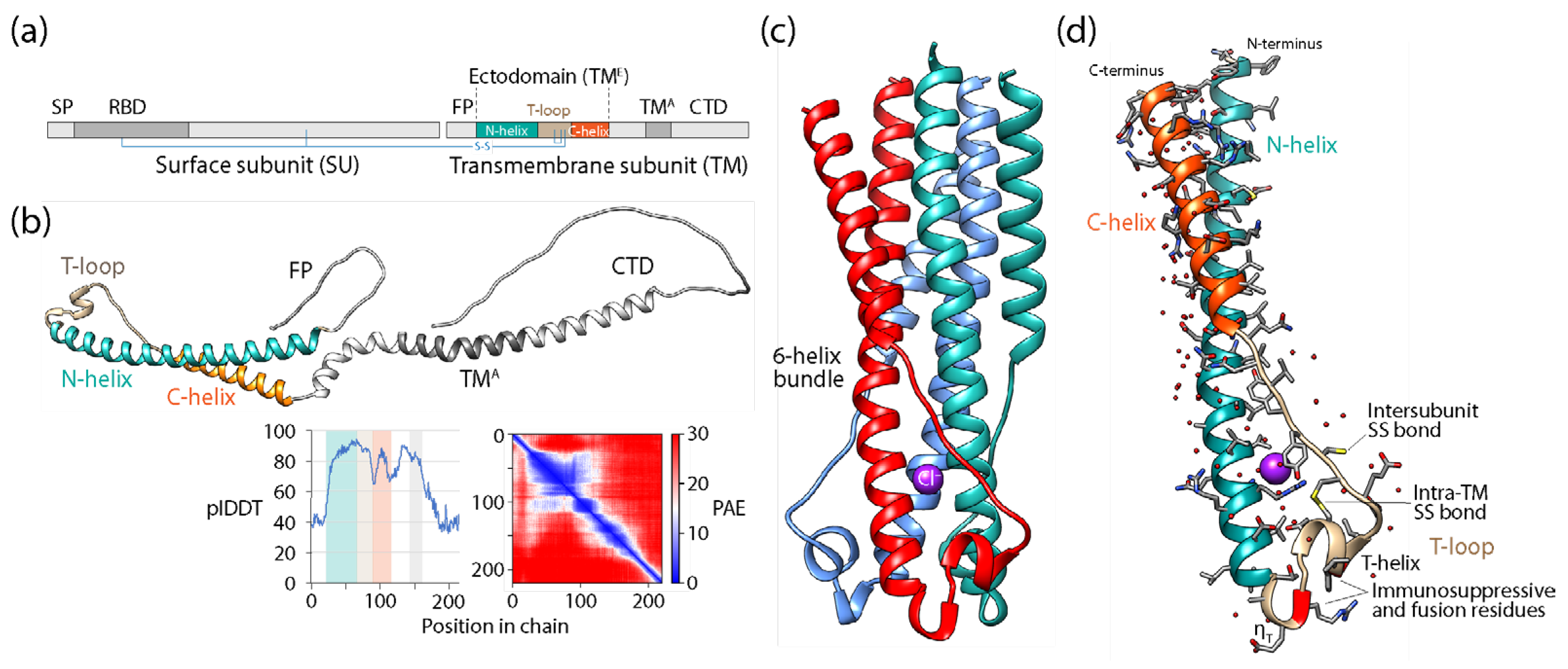
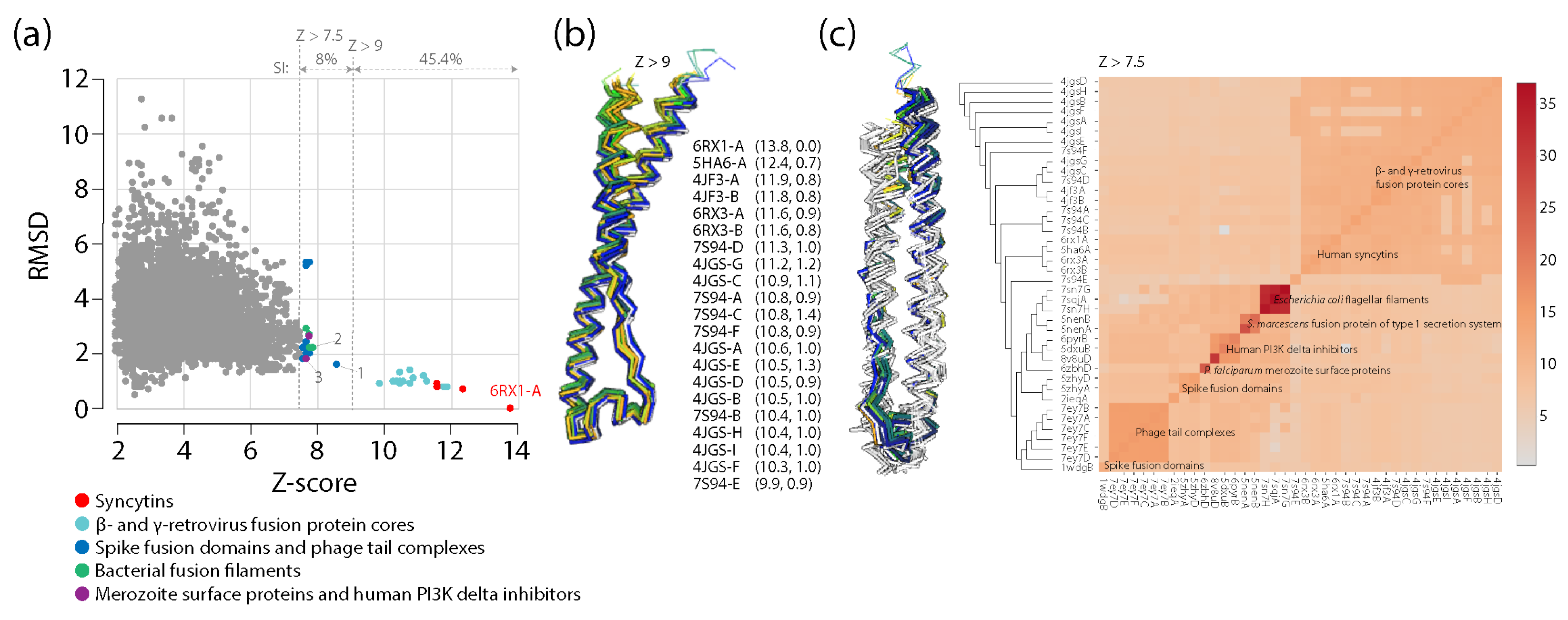
- The structural neighborhood of human syncytin-1
References
- La Scola, B.; Audic, S.; Robert, C.; Jungang, L.; de Lamballerie, X.; Drancourt, M.; Birtles, R.; Claverie, J.-M.; Raoult, D. A giant virus of amoeba. Science 2003, 299, 2033. [Google Scholar] [CrossRef] [PubMed]
- Nasir, A.; Romero-Severson, E.; Claverie, J.-M. Investigating the concept and origin of viruses. Trends Microbiol. 2020, 28, 959–967. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Dolja, V.V.; Krupovic, M.; et al. Global organization and proposed megataxonomy of the virus world. Microbiol. Mol. Biol. Rev. 2020, 84, e00061–e00019. [Google Scholar] [CrossRef] [PubMed]
- Colson, P.; La Scola, B.; Levasseur, A.; Caetano-Anollés, G.; Raoult, D. Mimivirus: leading the way in the discovery of giant viruses of amoebae. Nature Rev. Microbiol. 2017, 15, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Schulz, F.; Abergel, C.; Woyke, T. Giant virus biology and diversity in the era of genome-resolved metagenomics. Nature Rev. Microbiol. 2022, 20, 721–736. [Google Scholar] [CrossRef] [PubMed]
- Philippe, N.; Legendre, M.; Doutre, G.; Couté, Y.; Poirot, O.; Lescot, M.; Arslan, D.; Seltzer, V.; Bertaux, L.; Bruley, C.; Garin, J.; Claverie, J.-M.; Abergel, C. Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science 2013, 341, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Abrahão, J.; Silva, L.; Santos Silva, L.; Yaacoub Bou Khalil, J.; Rodrigues, R.; Arantes, T.; Assis, F.; Boratto, P.; Andrade, M.; Kron, E.G.; Ribeiro, B. , Bergier, I.; Seligmann, H.; Ghigo, E.; Colson, P.; Levasseur, A.; Kroemer, G.; Raoult, D.; La Scola, B. Tailed giant Tupanvirus possesses the most complete translational apparatus of the known virosphere. Nat. Commun. 2018, 9, 749. [Google Scholar] [CrossRef]
- Fischer, M.G.; Allen, M. J.; Wilson, W.H.; Suttle, C.A. Giant virus with a remarkable complement of genes infects marine zooplankton. Proc. Natl Acad. Sci. USA 2010, 107, 19508–19513. [Google Scholar] [CrossRef]
- Liu, Y.; Bisio, H.; Toner, C.M.; Jeudy, S.; Philippe, N.; Zhou, K.; Bowerman, S.; White, A.; Edwards, G.; Abergel, C.; Luger, K. Virus-encoded histone doublets are essential and form nucleosome-like structures. Cell 2021, 184, 4237–4250.e19. [Google Scholar] [CrossRef]
- La Scola, B.; Desnues, C.; Pagneier, I.; Robert, C.; Barrasi, L.; Fournous, G.; Merchat, M.; Suzan-Monti, P.; Forterre, P.; Koonin, E.; Raoult, D. The virophage as a unique parasite of the giant mimivirus. Nature 2008, 455, 100–104. [Google Scholar] [CrossRef]
- Desnues, C.; La Scola, B.; Yutin, N.; Fournous, G.; Robert, C.; Azza, S.; Jardot, P.; Monteil, S.; Campocasso, A.; Koonin, E.V. , Raoult, D. Provirophages and transpovirons as the diverse mobilome of giant viruses. Proc. Natl Acad. Sci. USA 2012, 109, 18078–18083. [Google Scholar] [CrossRef] [PubMed]
- Jeudy, S.; Bertaux, L.; Alempic, J.-M.; Lartigue, A.; Legendre, M.; Belmudes, L.; Santini, S.; Philippe, N.; Beucher, L.; Biondi, E.G.; Juul, S.; Turner, D.J.; Couté, Y.; Claverie, J.-M.; Abergel, C. Exploration of the propagation of transpovirons within Mimiviridae reveals a unique example of commensalism in the viral world. ISME J. 2020, 14, 727–739. [Google Scholar] [CrossRef] [PubMed]
- Caetano-Anollés, K.; Aziz, M.F.; Mughal, F.; Caetano-Anollés, G. On protein loops, prior molecular states and common ancestors of life. J. Mol. Evol. [CrossRef] [PubMed]
- Caetano-Anollés, G. Agency in evolution of biomolecular communication. Ann. N.Y. Acad. Sci. 2023, 1525, 88–103. [Google Scholar] [CrossRef] [PubMed]
- Caetano-Anollés, G.; Claverie, J.-M; Nasir, A. A critical analysis of the current state of virus taxonomy. Front. Microbiol. 2023, 14, 1240993. [Google Scholar] [CrossRef] [PubMed]
- Caetano-Anollés, G.; Nasir, A.; Kim, K.M.; Caetano-Anollés, D. Rooting phylogenies and the Tree of Life while minimizing ad hoc and auxiliary assumptions. Evol. Bioinformatics 2018, 14, 1176934318805101. [Google Scholar] [CrossRef] [PubMed]
- Caetano-Anollés, G.; Nasir, A. Benefits of using molecular structure and abundance in phylogenomic analysis. Front. Genet. 2012, 3, 172. [Google Scholar] [CrossRef]
- Murzin, A.G.; Brenner, S.E.; Hubbard, T.; Chothia, C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995, 247, 536–540. [Google Scholar] [CrossRef]
- Orengo, C.; Michie, A.; Jones, S.; Jones, D.; Swindells, M.; Thornton, J. CATH - a hierarchic classification of protein domain structures. Structure 1997, 5, 1093–1109. [Google Scholar] [CrossRef]
- Caetano-Anollés, G.; Aziz, M.F.; Mughal, F.; Caetano-Anollés, D. Tracing protein and proteome history with chronologies and networks: folding recapitulates evolution. Exp. Rev. Proteomics 2021, 18, 863–880. [Google Scholar] [CrossRef]
- Fox, N.K.; Brenner, S.E.; Chandonia, J.M. SCOPe: Structural Classification of Proteins—extended, integrating SCOP and ASTRAL data and classification of new structures. Nucleic Acids Res. 2014, 42, D304–D309. [Google Scholar] [CrossRef] [PubMed]
- Chandonia, J.-M.; Guan, L.; Lin, S.; Yu, C.; Fox, N.K.; Brenner, S.E. SCOPe: improvements to the structural classification of proteins – extended database to facilitate variant interpretation and machine learning. Nucleic Acids Res. 2022, 50, D553–D559. [Google Scholar] [CrossRef] [PubMed]
- Nasir, A.; Caetano-Anollés, G. A phylogenomic data-driven exploration of viral origins and evolution. Sci. Adv. 2015, 1, e1500527. [Google Scholar] [CrossRef] [PubMed]
- Gough, J. Convergent evolution of domain architectures (is rare). Bioinformatics 2005, 21, 1464–1471. [Google Scholar] [CrossRef] [PubMed]
- Mughal, F.; Nasir, A.; Caetano-Anollés, G. The origin and evolution of viruses inferred from fold family structure. Arch. Virol. 2020, 165, 2177–2191. [Google Scholar] [CrossRef] [PubMed]
- Swofford, D.L. PAUP*: Phylogenomic Analysis Using Parsimony (*and other methods). Ver 4.0b10. Sinauer, Sunderland, MA, 2004.
- Kolaczkowski, B.; Thornton, J.W. Performance of maximum parsimony and likelihood phylogenetics when evolution is heterogeneous. Nature 2004, 431, 980–984. [Google Scholar] [CrossRef] [PubMed]
- Goloboff, P.A.; Torres, A.; Arias, J.S. Weighted parsimony outperforms other methods of phylogenetic inference under models appropriate for morphology. Cladistics 2018, 34, 407–437. [Google Scholar] [CrossRef] [PubMed]
- Brower, A.V.Z. Statistical consistency and phylogenetic inference: a brief review. Cladistics 2018, 34, 562–567. [Google Scholar] [CrossRef] [PubMed]
- FigTree. Available online: https://github.com/rambaut/figtree/) (accessed on 1 March 2024).
- Lundberg, J. Wagner networks and ancestors. Syst. Zool. 1972, 18, 1–32. [Google Scholar] [CrossRef]
- Weston, P.H. Indirect and direct methods in systematics. In Ontogeny and Systematics; Humphries, C.J., Ed. Columbia University Press: New York, NY, 1988; pp. 27–56. [Google Scholar]
- Weston, P.H. Methods for rooting cladistic trees. In Models in Phylogeny Reconstruction (Systematics association special volume No. 52); Siebert, D.J., Scotland, R.W., Williams, D.M., Eds.; Clarendon Press: Oxford, UK, 1994; pp. 125–155. [Google Scholar]
- Caetano-Anollés, D.; Nasir, A.; Kim, K.M.; Caetano-Anollés, G. Testing empirical support for evolutionary models that root the tree of life. J. Mol. Evol. 2019, 87, 131–142. [Google Scholar] [CrossRef]
- Caetano-Anollés, G. Universal sharing patterns in proteomes and evolution of protein fold architecture and life. J. Mol. Evol. 2005, 60, 484–498. [Google Scholar] [CrossRef]
- Wang, M.; Jiang, Y.-Y.; Kim, K.M.; Wu, G.; Ji, H.-F.; Mittenthal, J.E.; Zhang, H.-Y.; Caetano-Anollés, G. A universal molecular clock of protein folds and its power in tracing the early history of aerobic metabolism and planet oxygenation. Mol. Biol. Evol. 2011, 28, 567–582. [Google Scholar] [CrossRef]
- Nasir, A.; Kim, K.M.; Caetano-Anollés, G. Giant viruses coexisted with the cellular ancestors and represent a distinct supergroup along with superkingdoms Archaea, Bacteria and Eukarya. BMC Evol Biol. 2012, 12, 156. [Google Scholar] [CrossRef] [PubMed]
- Nasir, A.; Sun, F.J.; Kim, K.M.; Caetano-Anollés, G. Untangling the origin of viruses and their impact on cellular evolution. Ann. N.Y. Acad. Sci. 2015, 1341, 61–74. [Google Scholar] [CrossRef]
- Oliva, B.; Bates, P.A.; Querol, E.; Aviles, F.X.; Sternberg, M.J. An automated classification of the structure of protein loops. J. Mol. Biol. 1997, 266, 814–830. [Google Scholar] [CrossRef]
- Fernandez-Fuentes, N.; Oliva, B.; Fiser, A. A supersecondary structure library and search algorithm for modeling loops in protein structures. Nucleic Acids Res. 2006, 34, 2085–2097. [Google Scholar] [CrossRef] [PubMed]
- Alva, V.; Söding, J.; Lupas, A.N. A vocabulary of ancient peptides at the origin of folded proteins. eLife 2015, 4, e09410. [Google Scholar] [CrossRef]
- Goncearenco, A.; Berezovsky, I.N. Prototypes of elementary functional loops unravel evolutionary connections between protein functions. Bioinformatics 2010, 26, i497–i503. [Google Scholar] [CrossRef] [PubMed]
- Goncearenco, A.; Berezovsky, I.N. Protein function from its emergence to diversity in contemporary proteins. Phys. Biol. 2015, 12, 045002. [Google Scholar] [CrossRef]
- Goncearenco, A.; Berezovsky, I.N. Exploring the evolution of protein function in Archaea. BMC Evol. Biol. 2012, 12, 75. [Google Scholar] [CrossRef]
- Aziz, M.F.; Caetano-Anollés, K.; Caetano-Anollés, G. The early history and emergence of molecular functions and modular scale-free network behavior. Sci. Rep. 2016, 6, 25058. [Google Scholar] [CrossRef]
- Nepomnyachiy, S.; Ben-Tal, N.; Kolodny, R. Complex evolutionary footprints revealed in an analysis of reused protein segments of diverse lengths. Proc. Natl. Acad. Sci. USA 2017, 114, 11703–11708. [Google Scholar] [CrossRef] [PubMed]
- Aziz, M.F.; Mughal, F.; Caetano-Anollés, G. Tracing the birth of structural domains from loops during protein evolution. Sci. Rep. 2023, 13, 14688. [Google Scholar] [CrossRef]
- Mughal, F.; Caetano-Anollés, G. (2023) Evolution of intrinsic disorder in protein loops. Life 2023, 13, 2055. [Google Scholar] [CrossRef]
- Bonet, J.; Planas-Iglesias, J.; Garcia-Garcia, J.; Marín-López, M.A.; Fernandez-Fuentes, N.; Oliva, B. ArchDB 2014: Structural classification of loops in proteins. Nucleic Acids Res. 2014, 42, D315–D319. [Google Scholar] [CrossRef]
- Bonet, J.; Fiser, A.; Oliva, B.; Fernandez-Fuentes, N. (2014b) S motifs as structural local descriptors of supersecondary elements: classification, completeness and applications. Bio-Algorithms Med. Syst, 2014; 10, 195–212. [Google Scholar] [CrossRef]
- Skolnick, J.; Zhou, H.; Brylinski, M. Further evidence for the likely completeness of the library of solved single domain protein structures. J. Phys. Chem. B 2012, 116, 6654–6664. [Google Scholar] [CrossRef] [PubMed]
- Peti, W.; Johnson, M.A.; Herrmann, T.; Neuman, B.W.; Buchmeier, M.J.; Nelson, M.; Joseph, J.; Page, R.; Stevenes, R.C.; Kuhn, P.; Wüthrich, K. Structural genomics of the severe acute respiratory syndrome coronavirus: Nuclear magnetic resonance structure of the protein nsP7. J. Virol. 2005, 79, 12905–12913. [Google Scholar] [CrossRef]
- Zhang, C.; Li, L.; He, J.; Chen, C.; Su, D. Nonstructural protein 7 and 8 complexes of SARS-CoV-2. Protein Sci. 2021, 30, 873–881. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yafremava, L.S.; Caetano-Anollés, D.; Mittenthal, J.E.; Caetano- Anollés, G. Reductive evolution of architectural repertoires in proteomes and the birth of the tripartite world. Genome Res. 2007, 17, 1572–1585. [Google Scholar] [CrossRef]
- Wang, M.; Caetano-Anollés, G. The evolutionary mechanics of domain organization in proteomes and the rise of modularity in the protein world. Structure 2009, 17, 66–78. [Google Scholar] [CrossRef]
- Kim, K.M.; Caetano-Anollés, G. The evolutionary history of protein fold families and proteomes confirms that the archaeal ancestor is more ancient than the ancestors of other superkingdoms. BMC Evol. Biol. 2012, 12, 13. [Google Scholar] [CrossRef] [PubMed]
- Bukhari, S.A.; Caetano-Anollés, G. Origin and evolution of protein fold designs inferred from phylogenomic analysis of CATH domain structures in proteomes. PLoS Comput. Biol. 2013, 9, e1003009. [Google Scholar] [CrossRef] [PubMed]
- Aziz, M.F.; Caetano-Anollés, G. Evolution of networks of protein domain organization. Sci. Rep. 2021, 11, 12075. [Google Scholar] [CrossRef] [PubMed]
- Nasir, A.; Kim, K.M.; Caetano-Anollés, G. Global patterns of protein domain gain and loss in superkingdoms. PLoS Comput. Biol. 2014, 10, e1003452. [Google Scholar] [CrossRef] [PubMed]
- Nasir, A.; Kim, K.M.; Caetano-Anollés, G. Phylogenetic tracings of proteome size support the gradual accretion of protein structural domains and the early origin of viruses from primordial cells. Front. Microbiol. 2017, 8, 1178. [Google Scholar] [CrossRef] [PubMed]
- Salthe, S. N. Hierarchical structures. Axiomathes 2012, 22, 355–383. [Google Scholar] [CrossRef]
- Godfray, H.C.J. Challenges for taxonomy. Nature 2002, 417, 17–19. [Google Scholar] [CrossRef] [PubMed]
- De Queiroz, K. Species concepts and species delimitation. Syst. Biol. 2007, 56, 879–886. [Google Scholar] [CrossRef] [PubMed]
- Hey, J.; Fitch, W.M.; Ayala, F. J. Systematics and the origin of species: on Ernst Mayr's 100th anniversary; The National Academies Press: Washington DC, 2005. [Google Scholar] [CrossRef]
- Shapiro, B.J.; Leducq, J.-B.; Mallet, J. What is speciation? PloS Genet. 2016, 12, e1005860. [Google Scholar] [CrossRef]
- Bobay, L.-M.; Ochman, H. Biological species in the viral world. Proc. Natl. Acad. Sci. USA 2018, 115, 6040–6045. [Google Scholar] [CrossRef]
- Rosenberg, E.; Zilber-Rosenberg, I. The hologenome concept: Human, animal and plant microbiota; Springer International Publishing: Cham, Switzerland, 2013. [Google Scholar] [CrossRef]
- Meyer-Abich, A. Beiträge zur Theorie der Evolution der Organismen. I. Das typologische Grundgesetz und seine Folgerungen für Phylogenie und Entwicklungsphysiologie. Acta Biotheor. 1943, 7, 1–80. [Google Scholar] [CrossRef]
- Bordenstein, S.R.; Theis, K.R. Host biology in light of the microbiome: ten principles of holobionts and hologenomes. PLoS Biol. 2015, 13, e1002226. [Google Scholar] [CrossRef] [PubMed]
- Theis, K.R.; Dheilly, N.M.; Klassen, J.L.; Brucker, R.M.; Baines, J.F.; Bosch, T.C.G.; Cryan, J.F.; Gilbert, S.F.; Goodnight, C.J.; Lloyd, E.A.; Sapp, J.; Vandenkoornhuyse, P.; Zilberg-Rosenberg, I.; Rosenberg, E.; Bordenstein, S.R. Getting the hologenome concept right: an eco-evolutionary framework for hosts and their microbiomes. mSystems 2016, 1, e00028–e00016. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, S.F.; Sapp, J.; Tauber, A.I. A symbiotic view of life: we have never been individuals. Q. Rev. Biol. 2012, 87, 325–341. [Google Scholar] [CrossRef] [PubMed]
- Queller, D. C.; Strassmann, J.E. Beyond society: the evolution of organismality. Phil. Trans. R. Soc. B. 2009, 364, 3143–3155. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, E. Microbiomes: Current Knowledge and Unanswered Questions; Springer Nature Switzerland AG: Cham Switzerland, 2021. [Google Scholar] [CrossRef]
- Sender, R.; Fuchs, S.; Milo, R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.; Arif, B.; Caetano-Anollés, G.; Kim, K.M.; Nasir, A. Horizontal gene transfer in human-associated microorganisms inferred by phylogenetic reconstruction and reconciliation. Sci. Rep. 2019, 9, 5953. [Google Scholar] [CrossRef] [PubMed]
- Ku, C.; Martin, W.F. A natural barrier to lateral gene transfer from prokaryotes to eukaryotes revealed from genomes: the 70% rule. BMC Bio.ogy 2016, 14, 89. [Google Scholar] [CrossRef] [PubMed]
- Crisp, A.; Boschetti, C.; Perry, M.; Tunnacliffe, A.; Micklem, G. Expression of multiple horizontally acquired genes is a hallmark of both vertebrate and invertebrate genomes. Genome Biol. 2015, 16, 50. [Google Scholar] [CrossRef]
- Huang, W.; Tsai, L.; Li, Y.; Hua, N.; Sun, C.; Wei, C. Widespread of horizontal gene transfer in the human genome. BMC Genomics 2017, 18, 274. [Google Scholar] [CrossRef]
- Sieber, K.B.; Bromley, R.E.; Dunning Hotopp, J.C. Lateral gene transfer between prokaryotes and eukaryotes. Exp. Cell Res. 2017, 358, 421–426. [Google Scholar] [CrossRef] [PubMed]
- Boothby, T.C.; Tenlen, J.R.; Smith, F.W.; Wang, J.R.; Patanella, K.A.; Osborne Nishimura, E.; Tintori, S.C.; Li, Q.; Jones, C.D.; Yandell, M.; Mesina, D.N.; Glasscock, J.; Goldstein, B. Evidence for extensive horizontal gene transfer from the draft genome of a tardigrade. Proc. Natl. Acad. Sci USA 2015, 112, 15976–15981. [Google Scholar] [CrossRef]
- Koutsovoulos, G.; Kumar, S.; Laetsch, D.R.; Stevens, L.; Daub, J.; Conlon, C.; Maroon, H.; Thomas, F.; Aboobaker, A.; Blaxter, M. No evidence for extensive horizontal gene transfer in the genome of the tardigrade Hypsibius dujardini. Proc. Natl. Acad. Sci USA 2016, 113, 5053–5058. [Google Scholar] [CrossRef] [PubMed]
- Lerner, A.; Matthias, T.; Aminov, R. Potential effects of horizontal gene exchange in the human gut. Front. Immunol. 2017, 8, 1630. [Google Scholar] [CrossRef] [PubMed]
- Suttle, C. A. Marine viruses—Major players in the global ecosystem. Nature Rev. Microbiol. 2007, 5, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Kavagutti, V.S.; Andrei, A.-S.; Mehrshad, M.; Salcher, M.M.; Ghai, R. Phage-centric ecological interactions in aquatic ecosystems revealed through ultra-deep metagenomics. Microbiome 2019, 7, 135. [Google Scholar] [CrossRef] [PubMed]
- Grasis, J.A. The intra-dependence of viruses and the holobiont. Front. Immunol. 2017, 8, 1501. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.M.; Green, J.A.; Schulz, L.C. The evolution of the placenta. Reproduction 2016, 152, R179–R189. [Google Scholar] [CrossRef]
- Lavialle, C.; Cornelis, G.; Dupressoir, A.; Esnault, C.; Heidmann, O.; Vernochet, C.; Heidmann, T. Paleovirology of ‘syncytins’, retroviral env genes exapted for a role in placentation. Phil. Trans. R. Soc. B 2013, 368, 20120507. [Google Scholar] [CrossRef]
- Cornelis, G.; Funk, M.; Vernochet, C.; Leal, F.; Tarazona, A.; Meurice, G.; Heidman, O.; Dupressoire, A.; Miralles, A.; Ramirez-Pinilla, M.P.; Heidmann, T. An endogenous retroviral envelope syncytin and its cognate receptor identified in the viviparous placental Mabuya lizard. Proc. Natl. Acad. Sci. USA 2017, 114, E10991–E11000. [Google Scholar] [CrossRef]
- Bell, P.J.L. Evidence supporting a viral origin of the eukaryotic nucleus. Virus Res. 2020, 289, 198168. [Google Scholar] [CrossRef] [PubMed]
- Chaikeeratisak, V.; Nguyen, K.; Khanna, K.; Brilot, A.F.; Erb, M. L.; Coker, J.K.C.; Vavilina, A.; Newton, G.L.; Buschauer, R.; Pogliano, K.; Villa, E.; Agard, D.A.; Pogliano, J. Assembly of a nucleus-like structure during viral replication in bacteria. Science 2017, 355, 194–197. [Google Scholar] [CrossRef] [PubMed]
- Mallet, J.; Besansky, N.; Hahn, M.W. How reticulated are species? Bioessays 2016, 38, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Merhej, V.; Raoult, D. Rhizome of life, catastrophes, sequence exchanges, gene creations, and giant viruses: how microbial genetics challenges Darwin. Front. Cell. Infect. Microbiol. 2012, 2, 113. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, A.; Colson, P.; Merhej, V.; Zgheib, R.; Maatouk, M.; Naud, S.; Bittar, F.; Raoult, D. Rhizomal reclassification of living organisms. Int. J. Mol. Sci. 2021, 22, 5643. [Google Scholar] [CrossRef] [PubMed]
- Caetano-Anollés, G.; Aziz, M.F.; Mughal, F.; Gräter, F.; Koç, I.; Caetano-Anollés, K.; Caetano-Anollés, D. Emergence of hierarchical modularity in evolving networks uncovered by phylogenetic analysis. Evol. Bioinform. 2019, 15, 1176934319872980. [Google Scholar] [CrossRef]
- Vernikos, G.; Medini, D.; Riley, D.R.; Tettelin, H. Ten years of pan-genome analyses. Curr. Opin. Microbiol. 2015, 23, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Gabaldón, T. Patterns and impacts of nonvertical evolution in eukaryotes: a paradigm shift. Ann. N.Y. Acad. Sci. 2020, 1476, 78–92. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.-W.; et al. A draft human pangenome reference. Nature 2023, 617, 312–324. [Google Scholar] [CrossRef]
- Gong, Y.; Li, Y.; Liu, X.; Ma, Y.; J., L. A review of the pangenome: how it affects our understanding of genomic variation, selection and breeding in domestic animals? J. Animal Sci. Biotechnol. 2023, 14, 73. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C. A.; Blake, J. A.; Botstein, D.; Butler, H.; Cherry, J. M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; Harris, M.A.; Hill, D.P.; Issel-Tarver, L.; Karaskis, A.; Lewis, S.; Matese, J.C.; Richardson, J.E.; Ringwald, M.; Rubin, G.M.; Sherlock, G. (2000). Gene ontology: tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Rupp, R.; Scornavacca, C. Phylogenetic networks: concepts, algorithms and applications; Cambridge University Press: Cambridge, 2010. [Google Scholar] [CrossRef]
- Morrison, D.A. An introduction to phylogenetic networks; RJR Productions: Uppsala, Sweden, 2011. [Google Scholar]
- Bryant, D.; Moulton, V. Neighbor-net: an agglomerative method for the construction of phylogenetic networks. Mol. Biol. Evol. 2004, 21, 255–265. [Google Scholar] [CrossRef]
- Yang, J.; Grünewald, S.; Wan, X. F. Quartet-net: a quartet-based method to reconstruct phylogenetic networks. Mol. Biol. Evol. 2013, 30, 1206–1217. [Google Scholar] [CrossRef]
- Wheeler, W.C. Phylogenetic network analysis as a parsimony optimization problem. BMC Bioinformatics 2015, 16, 296. [Google Scholar] [CrossRef] [PubMed]
- Podani, J. Monophyly and paraphyly: a discourse without end? Taxon 2010, 59, 1011–1015. [Google Scholar] [CrossRef]
- Hinchliff, C.E.; Smith, S.A.; Allman, J.F.; Burleigh, J.G.; Chaudary, R.; Coghill, L.M.; Crandall, K.A.; Deng, J.; Drew, B.T.; Grazis, R.; Gude, K.; Hibbett, D.S.; Katz, L.A.; Laughinghouse IV, H.D.; McTavish, E.J.; Midford, P.E.; Owen, C.L.; Ree, R.H.; Rees, J.A.; Soltis, D.E.; Williams, T.; Cranston, K.A. Synthesis of phylogeny and taxonomy into a comprehensive tree of life. Proc. Natl. Acad. Sci. USA 2015, 112, 12764–12769. [Google Scholar] [CrossRef] [PubMed]
- Nasir, A.; Kim, K.M.; Caetano-Anollés, G. Long-term evolution of viruses: A Janus-faced balance. Bioessays 2017, 39, 1700026. [Google Scholar] [CrossRef] [PubMed]
- Roossinck, M.J.; Bazán, E.R. Symbiosis: viruses as intimate partners. Annu. Rev. Virol. 2017, 4, 123–139. [Google Scholar] [CrossRef] [PubMed]
- Luganini, A.; Gribaudo, G. Retroviruses of the human virobiota: The recycling of viral genes and the resulting advantages for human hosts during evolution. Front. Microbiol. 2020, 11, 1140. [Google Scholar] [CrossRef]
- Grinde, B. Herpesviruses: latency and reactivation –viral strategies and host response. J. Oral Microbiol. 2013, 5, 22766. [Google Scholar] [CrossRef]
- Barton, E.; White, D.; Cathelyn, J.; Brett-McClelland, K.A.; Engle, M.; Diamond, M.S.; Miller, V.L.; Virgin IV, H.W. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature 2007, 447, 326–329. [Google Scholar] [CrossRef]
- Liu, L.; Gong, T.; Tao, W.; Lin, B.; Li, C.; Zzheng, X.; Zhu, S.; Jiang, W.; Zhou, R. Commensal viruses maintain intestinal intraephitelial lymphocites via noncanonical RIG-I signaling. Nature Immunol. 2019, 20, 1681–1691. [Google Scholar] [CrossRef]
- Roetman, J.J.; Apostolova, M.K.I.; Philip, M. Viral and cellular oncogenes promote immune evasion. Oncogene 2022, 41, 921–929. [Google Scholar] [CrossRef] [PubMed]
- Tsai, K.; Cullen, B. R. Epigenetic and epitranscriptomic regulation of viral replication. Nature Revs. Microbiol. 2020, 18, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Willbanks, A.; Leary, M.; Greenshields, M.; Tyminski, C.; Heerboth, S.; Lapinska, K.; Haskins, K.; Sarkar, S. The evolution of epigenetics: From prokaryotes to humans and its biological consequences. Gen. Epigen. 2016, 8, 25–36. [Google Scholar] [CrossRef]
- Buschle, A.; Hammerschmidt, W. Epigenetic lifestyle of Epstein-Barr virus. Sem. Immunopathol. 2020, 42, 131–142. [Google Scholar] [CrossRef]
- Hoelzer, K.; Shackelton, L.A.; Parrish, C. R. Presence and role of cytosine methylation in DNA viruses of animals. Nucleic Acids Res. 2008, 36, 2825–2837. [Google Scholar] [CrossRef]
- Jeudy, S.; Rigou, S.; Alempic, J.-M.; Claverie, J.-M.; Abergel, C.; Legendre, M. The DNA methylation landscape of giant viruses. Nature Commun. 2020, 11, 2657. [Google Scholar] [CrossRef] [PubMed]
- International Committee on Taxonomy of Viruses (ICTV). The International Code of Virus Classification and Nomenclature (ICVCN), March 2021 edition. Available online: https://ictv.global/about/code (accessed on 14 May 2024).
- Frost, L.S.; Leplae, R.; Summers, A.O.; Toussaint, A. Mobile genetic elements: the agents of open source evolution. Nature Rev. Microbiol. 2005, 3, 722–732. [Google Scholar] [CrossRef]
- Van Regenmortel, M.H.V. Solving the species problem in viral taxonomy: recommendations on non-Latinized binomial species names and on abandoning attempt to assign metagenomic viral sequences to species taxa. Arch. Virol. 2019, 164, 2223–2229. [Google Scholar] [CrossRef]
- Burnet, F. M. The structure of the influenza virus. Sci. Amer. 1957, 196, 37. [Google Scholar] [CrossRef]
- Lwoff, A. The concept of virus – The Third Marjory Stephenson Memorial Lecture. J. Gen. Microbiol. 1957, 17, 239–253. [Google Scholar] [PubMed]
- Claverie, J.M.; Abergel, C. Mimivirus: the emerging paradox of quasi- autonomous viruses. Trends Genet. 2010, 26, 431–437. [Google Scholar] [CrossRef]
- Dupré, J.; Guttinger, S. Viruses as living processes. Stud. Hist. Phil. Biol. Biomed. Sci. 2016, 59, 109–116. [Google Scholar] [CrossRef]
- Nasir, A.; Romero-Severson, E.; Claverie, J.-M. Investigating the concept and origin of viruses. Trends Microbiol. 2020, 28, 959–967. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023, 49, D545–D551. [Google Scholar] [CrossRef]
- Pringle, C.R. The 20th meeting of the executive committee of the ICTV. Virus species, higher taxa, a universal database and other matters. Arch Virol. 1991, 119, 303–304. [Google Scholar] [CrossRef]
- Ruigrok, K.; Vaney, M.-C.; Buchrieser, J.; Baquero, E.; Hellert, J.; Baron, B.; England, P.; Schwartz, O.; Rey, F.A.; Backovic, M. X-ray structures of the post-fusion 6-helix bundle of the human syncytin and their functional implications. J. Mol. Biol. 2019, 431, 4922–4940. [Google Scholar] [CrossRef] [PubMed]
- Holm, L. Dali server: structural unification of protein families. Nucleic Acids Res. 2020, 50, W210–W215. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nature Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Zheng, Q.; Deng, Y.; Liu, J.; Hoek, L.V.; Berkout, B.; Liu, M. Core structure of S2 from the human coronavirus NL63 spike glycoprotein. Biochemisty 2006, 45, 15205–15215. [Google Scholar] [CrossRef] [PubMed]
- Kreutzberger, M.A.B.; Sobe, R.C.; Sauder, A.B.; Chatterjee, S.; Pena, A.; Wang, F.; Giron, J.A.; Kiessling., V.; Costa, T.R.D.; Conticello, V.P.; Frankel, G.; Kendall, M.M.; Scharf, B.E.; Egelman, E.H. Flagellin outer domain dimerization modulates motility in pathogenic and soil bacteria from viscous environments. Nature Commun. 2022, 13, 1422–1422. [Google Scholar] [CrossRef] [PubMed]
- Dijkman, P.M.; Marzluf, T.; Zhang, Y.; Chang, S.S.; Helm, D.; Lanzer, M.; Bujard, H.; Kudryashev, M. Structure of the merozoite surface protein 1 from Plasmodium falciparum. Sci. Adv. 2021, 7, eabg0465. [Google Scholar] [CrossRef] [PubMed]
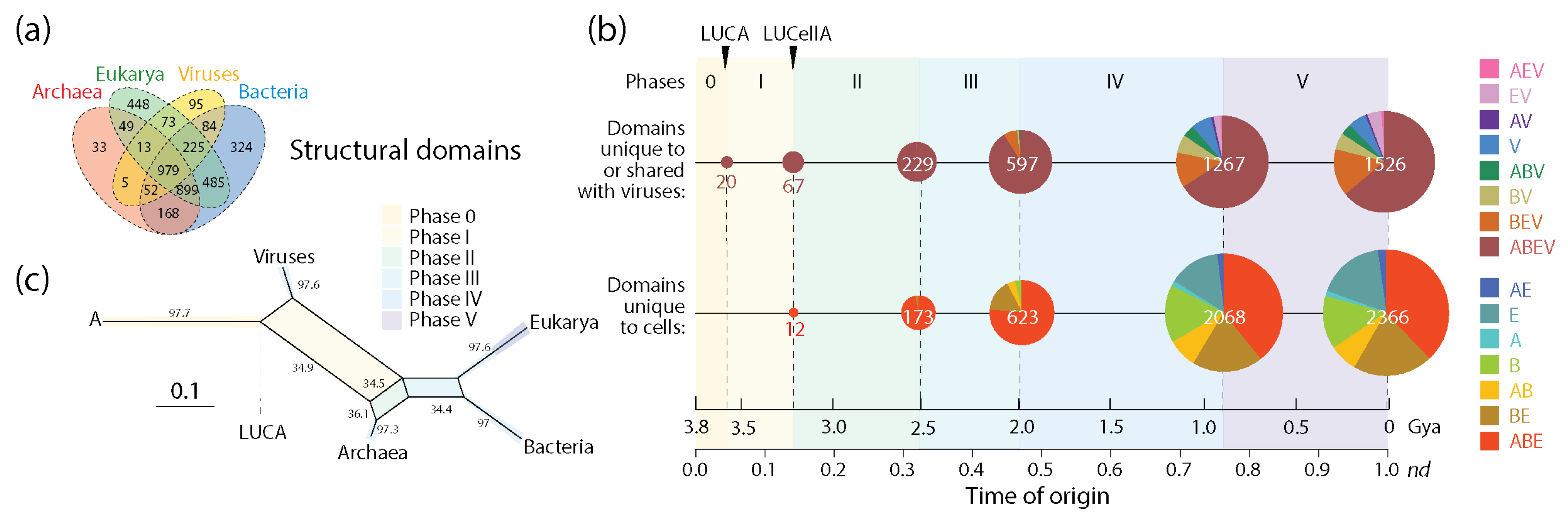
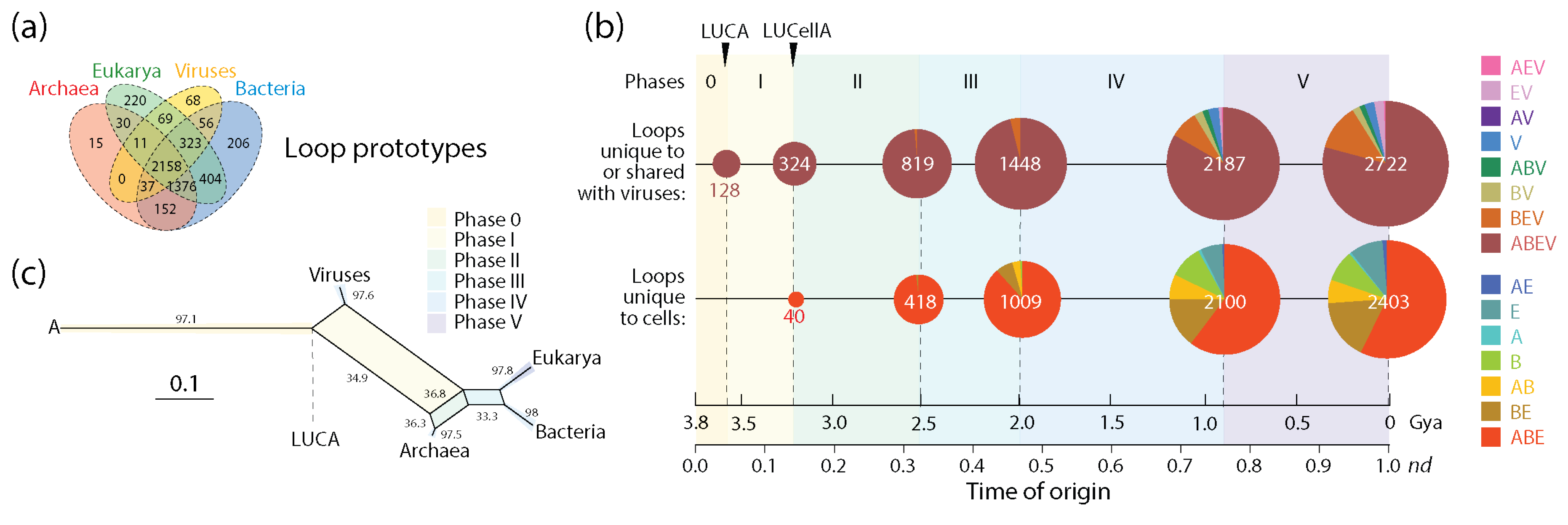
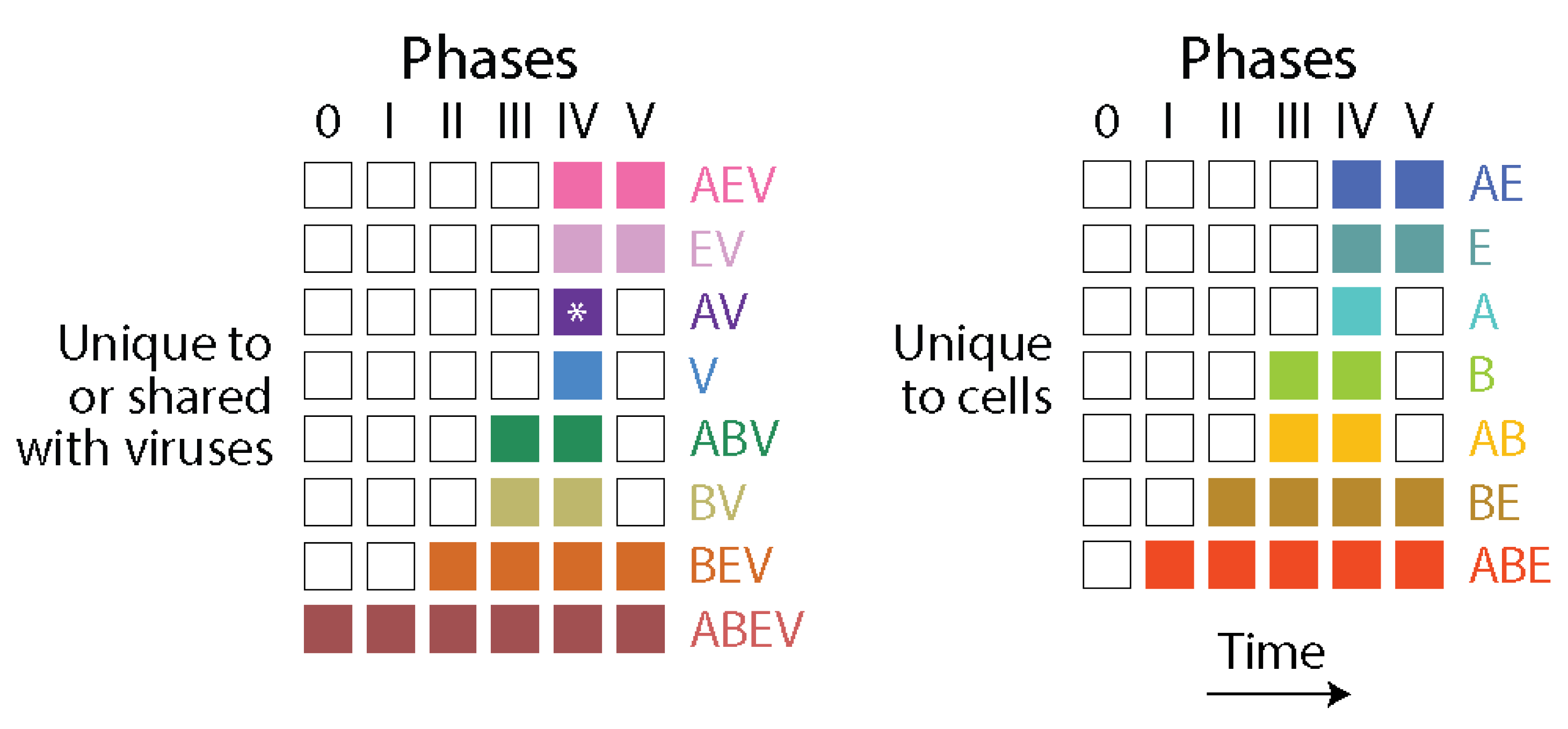
| Evolutionary phases | |||||||
| 0 | I | II | III | IV | V | ||
| Unique to or shared with viruses | Prototypes | 128 | 196 | 495 | 629 | 879 | 535 |
| Domains | 20 | 47 | 162 | 368 | 670 | 259 | |
| Ratio | 6.40 | 4.17 | 3.06 | 1.71 | 1.31 | 2.07 | |
| Unique to cells | Prototypes | 0 | 40 | 378 | 656 | 1146 | 303 |
| Domains | 0 | 12 | 161 | 450 | 1445 | 298 | |
| Ratio | – | 3.33 | 2.35 | 1.46 | 0.79 | 1.02 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





