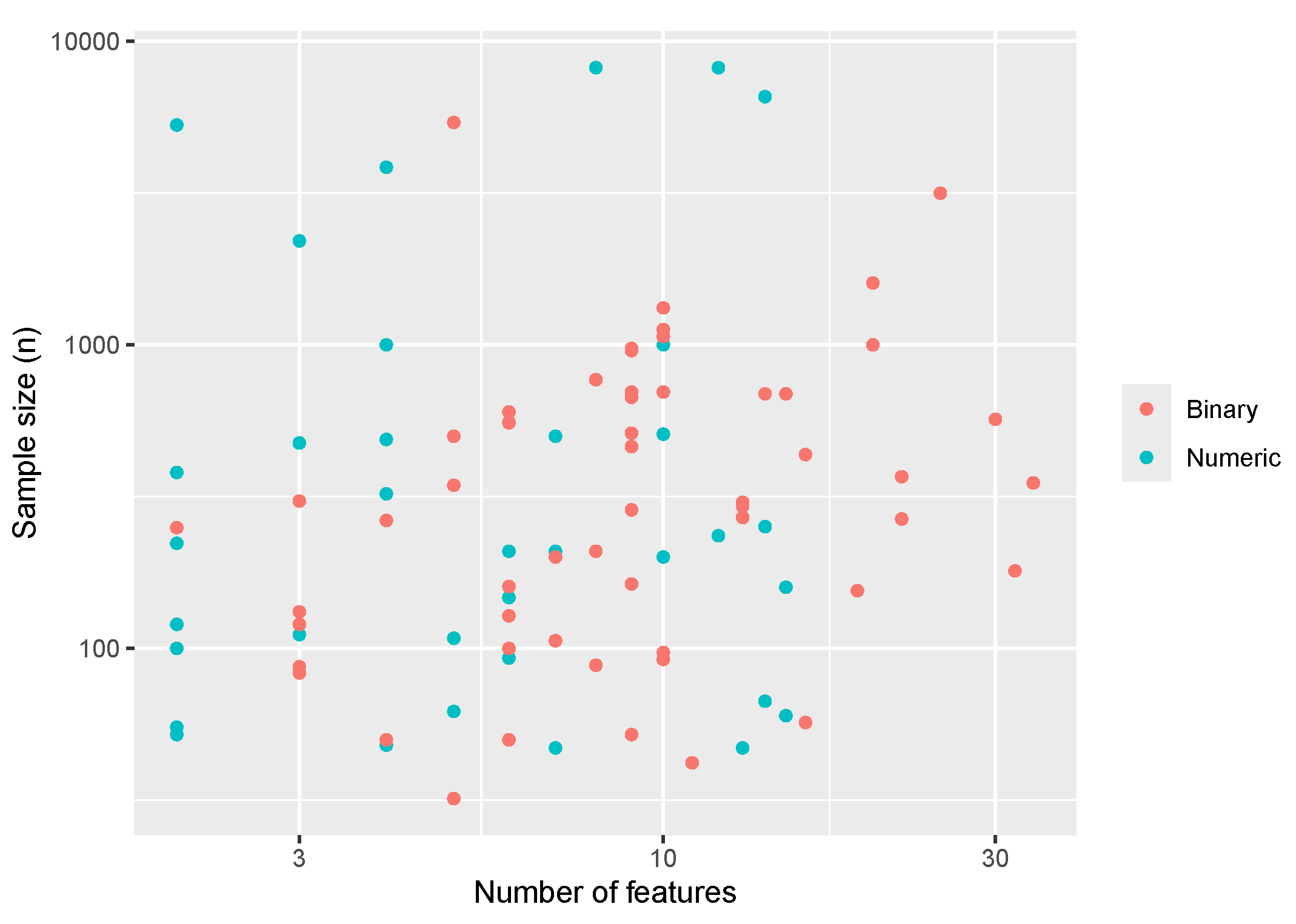
3.3.1. Exemplars
For the
503_wind data set, SRL outperformed all other methods in terms of test R-squared and test RMSE with a notably faster run time than the random forest, SVM, and to a lesser extent neural network methods. Results for the
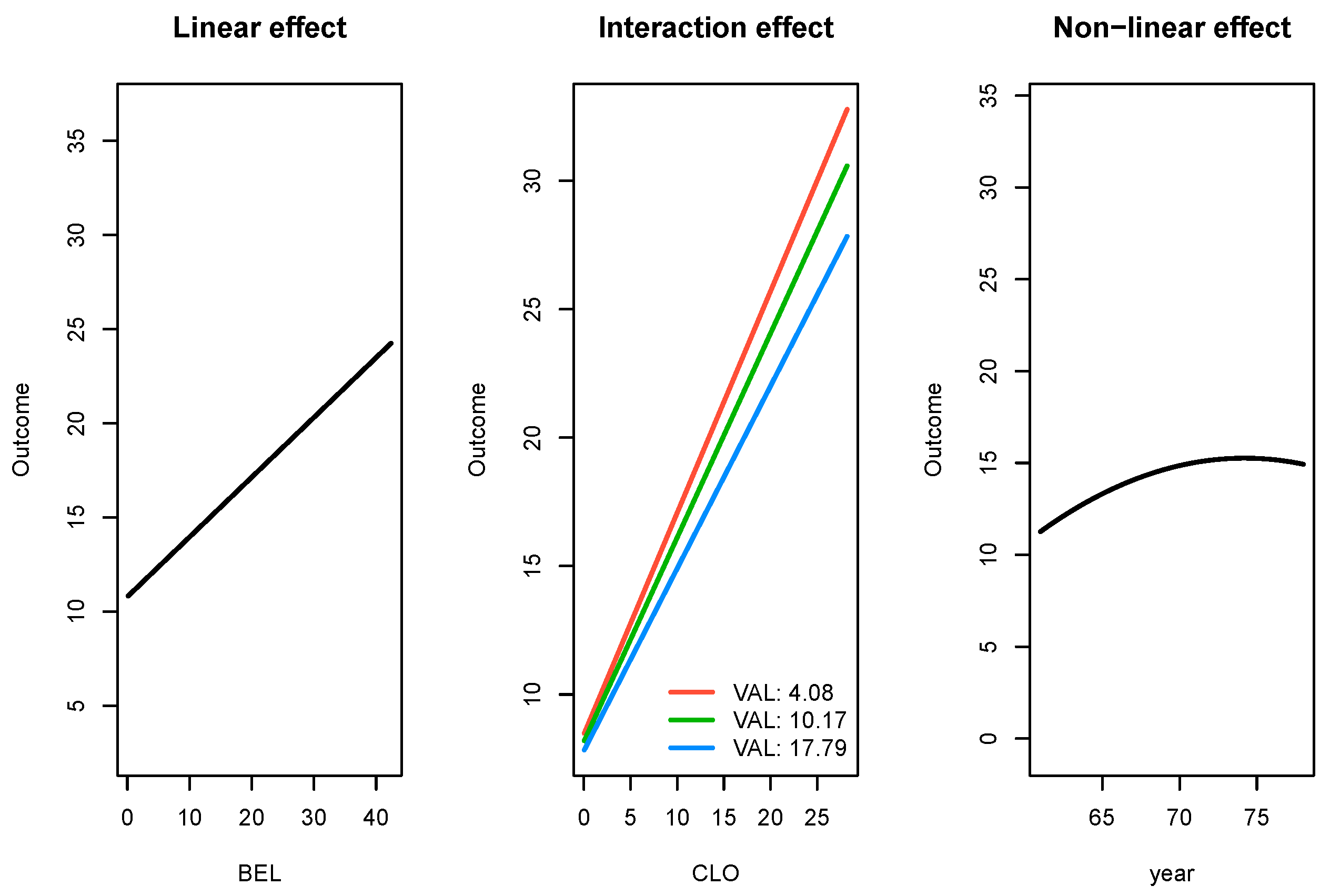
503_wind data set are provided in Table 4. In addition to SRL being the best performer, it also produces parameter estimates which are interpretable. In
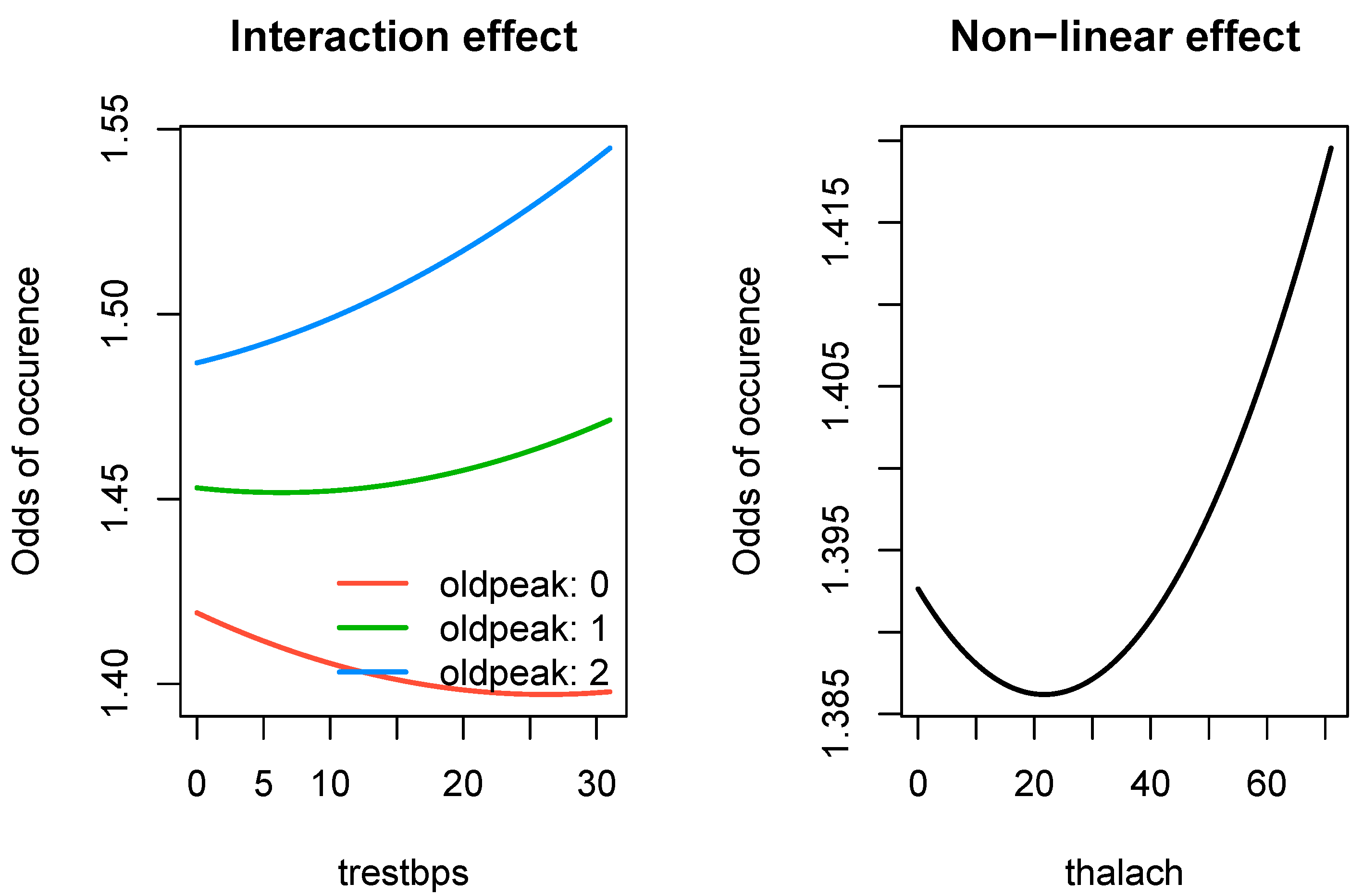
Figure 4, we present the effects for three types of significant relationships found by SRL in the
503_wind data: linear, linear with an interaction effect, and a non-linear effect.
Table 4.
Comparison of performance for the 503_wind set. SRL: sparsity-ranked lasso, NN: neural networks, RF: random forests, SVM: support vector machines, XGB: extreme gradient boosting.
Table 4.
Comparison of performance for the 503_wind set. SRL: sparsity-ranked lasso, NN: neural networks, RF: random forests, SVM: support vector machines, XGB: extreme gradient boosting.
| Model |
Test R-squared |
Test RMSE |
Runtime (s) |
| SRL |
0.773 |
3.12 |
12.8 |
| Lasso |
0.741 |
3.34 |
8.4 |
| RF |
0.766 |
3.17 |
48.7 |
| SVM |
0.744 |
3.32 |
34.8 |
| NN |
0.667 |
3.78 |
17.3 |
| XGB |
0.770 |
3.14 |
4.1 |
For the
hungarian data set, SRL was the fourth best performing model in terms of AUC; however, the performance of the top four models was extremely close with each having an AUC within 0.032 of one another. Results for the
hungarian data set are provided in Table 5. While SRL did not outperform random forest for this data set, it does provide interpretable parameter estimates relative to random forest for only a marginal reduction in performance. In
Figure 5, we present effect of two types of significant nonlinear relationships found by SRL in the
hungarian data: an interaction effect and a quadratic effect.
Table 5.
Comparison of performance for the hungarian data set. SRL: sparsity-ranked lasso, NN: neural networks, RF: random forests, SVM: support vector machines, XGB: extreme gradient boosting.
Table 5.
Comparison of performance for the hungarian data set. SRL: sparsity-ranked lasso, NN: neural networks, RF: random forests, SVM: support vector machines, XGB: extreme gradient boosting.
| Model |
AUC |
Runtime (s) |
| SRL |
0.885 |
5.8 |
| Lasso |
0.894 |
2.5 |
| RF |
0.899 |
7.9 |
| SVM |
0.821 |
9.6 |
| NN |
0.917 |
10.5 |
| XGB |
0.811 |
18.1 |
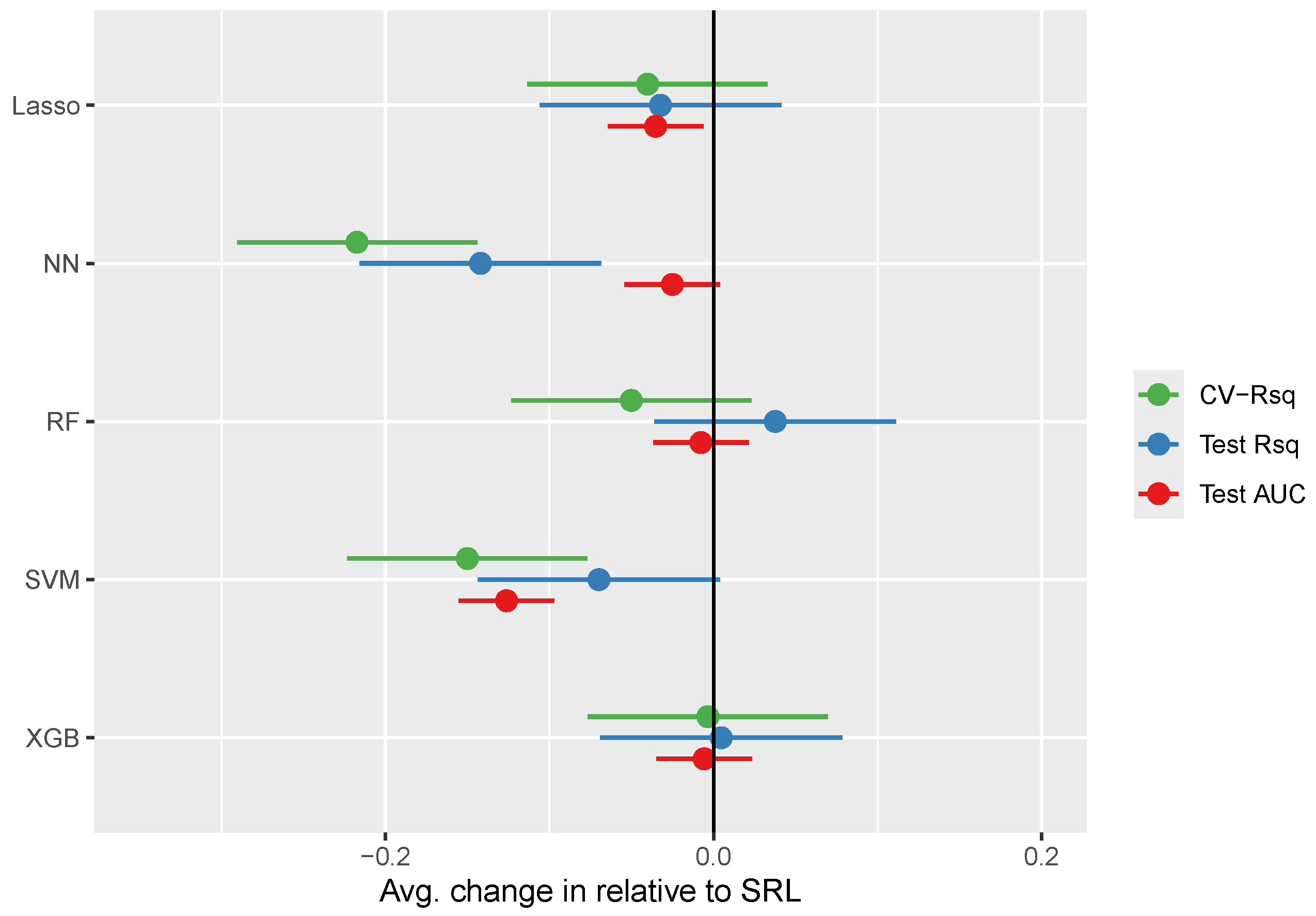
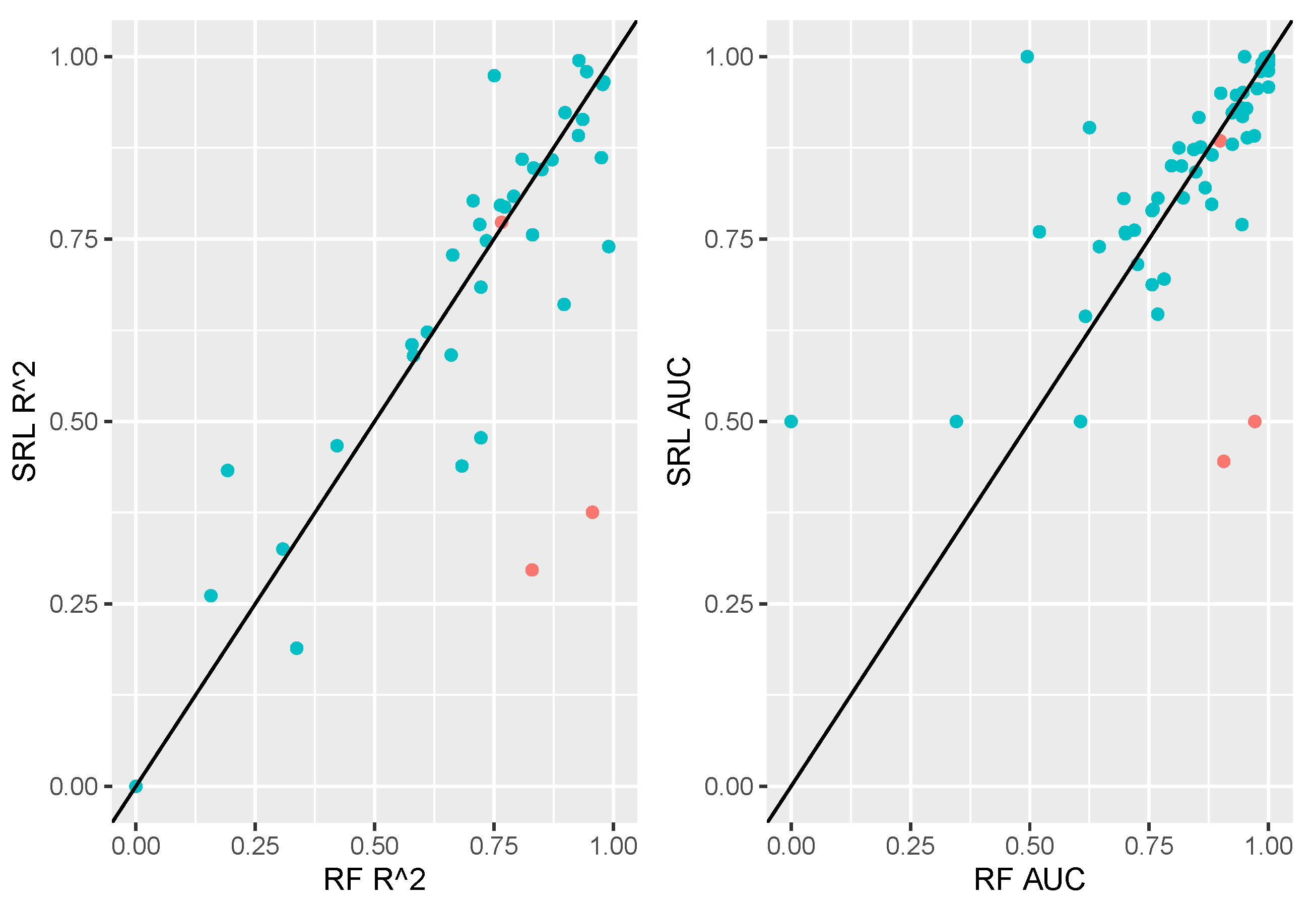
3.3.2. SRL Underperforming RF
In this section we delve more deeply into examples where SRL appears to be performing worse than alternative methods (case studies highlighted in
Figure 3 right of the 45-degree line).
For the sleep apnea data sets
analcatdata_apnea1 and
analcatdata_apnea2, SRL, lasso, and SVM performed considerably worse in terms of test and cross-validated
compared to random forests and XGboost (Table 6). Descriptive statistics for all of the variables included in these data sets are shown in Table S1, and are originally described in Steltner et al. [
28].
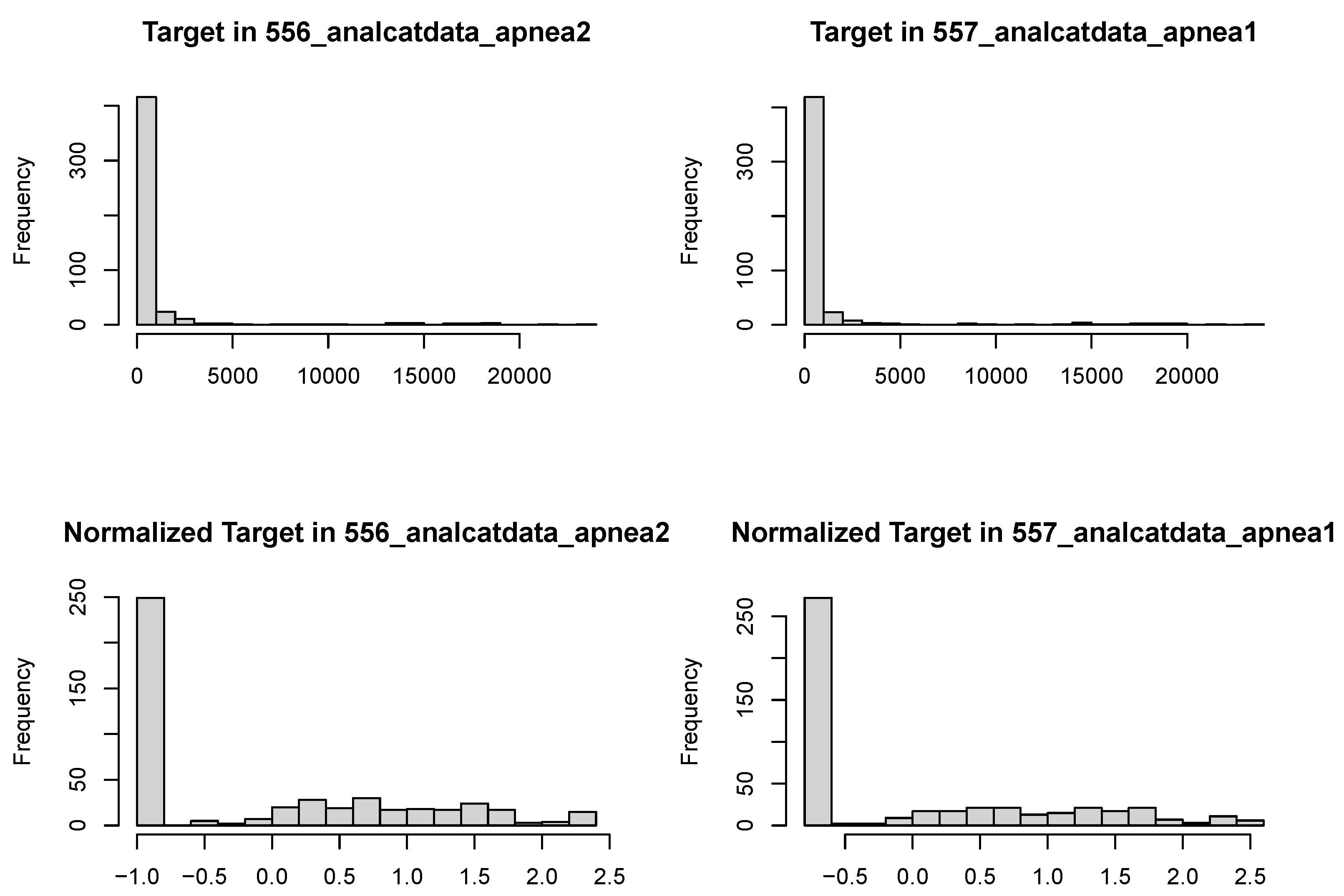
Examining the target outcomes for these data sets (
Figure 6), we see that both outcomes are highly skewed with a point mass at zero, rebutting even normalization methods [
29,
30]. Given these distributions, it makes sense for the models to be fit better by more robust methods. While SRL (and lasso) algorithms could be introduced that adequately capture zero inflation and right skew, this is beyond the scope of this paper.
Upon further inspection, we noticed that the
sparseR package by default removes interactions or other terms with near-zero variance via the
recipes package [
21,
31], which in this case removed all of the candidate interaction features from the model prior to the supervised part of the algorithm. By adding the argument
filter = "zv", only zero-variance variables are removed, and therefore any interactions with variance are retained. The code for applying this solution and its results are shown in the Appendix. Once this is implemented for the
analcatdata_apnea2 data set, the SRL achieves a CV-based R-square of 0.91, and a compact model (within 1 standard error of the RMSE of the best model) achieves a CV-based R-square of 0.88. Coefficients from the latter model and their marginal false discovery rates [
32] can be viewed in the Appendix as well. Briefly, we can interpret the model as follows: observations with
Automatic , or those where
Scorer_1 saw higher values of the target variable. If
Automatic=0 and
Scorer_1=0, there is a multiplicative modest increase in the target, but if both variables are equal to 3, the target jumps up to the extremely high tail of the distribution, increasing by over 13,000 on average. These results are practically identical for the
analcatdata_apnea1 data set.
Table 6.
Comparison of performance for the sleep apnea data sets. SRL: sparsity-ranked lasso, NN: neural networks, RF: random forests, SVM: support vector machines, XGB: extreme gradient boosting, s: seconds
Table 6.
Comparison of performance for the sleep apnea data sets. SRL: sparsity-ranked lasso, NN: neural networks, RF: random forests, SVM: support vector machines, XGB: extreme gradient boosting, s: seconds
| Model |
R-squared (CV) |
R-squared (test) |
Runtime (s) |
| 556_analcatdata_apnea2 |
| SRL |
0.247 |
0.376 |
1.8 |
| Lasso |
0.243 |
0.385 |
1.3 |
| RF |
0.760 |
0.956 |
20.4 |
| SVM |
0.111 |
0.021 |
11.6 |
| NN |
0.292 |
0.719 |
6.2 |
| XGB |
0.684 |
0.930 |
17.9 |
| 557_analcatdata_apnea1 |
| SRL |
0.276 |
0.296 |
1.7 |
| Lasso |
0.297 |
0.299 |
1.1 |
| RF |
0.810 |
0.830 |
17.6 |
| SVM |
0.082 |
0.039 |
8.9 |
| NN |
0.635 |
0.823 |
6.3 |
| XGB |
0.859 |
0.820 |
19.1 |
We also noted two data sets where the SRL underperformed alternative methods in predicting a binary outcome:
analcatdata_boxing1 and
parity5+5. These results are summarized in
Table 7.
Table 7.
Comparison of performance for binary outcome data sets where SRL underperformed. SRL: sparsity-ranked lasso, NN: neural networks, RF: random forests, SVM: support vector machines, XGB: extreme gradient boosting, AUC: Area under the receiver-operator curve, s: seconds
Table 7.
Comparison of performance for binary outcome data sets where SRL underperformed. SRL: sparsity-ranked lasso, NN: neural networks, RF: random forests, SVM: support vector machines, XGB: extreme gradient boosting, AUC: Area under the receiver-operator curve, s: seconds
| Model |
AUC |
Runtime (s) |
| analcatdata_boxing1 |
| SRL |
0.445 |
2.6 |
| Lasso |
0.758 |
1.5 |
| RF |
0.906 |
1.6 |
| SVM |
0.594 |
3.5 |
| NN |
0.727 |
2.3 |
| XGB |
0.898 |
14.4 |
| parity5+5 |
| SRL |
0.500 |
18.0 |
| Lasso |
0.500 |
6.8 |
| RF |
0.971 |
1.2 |
| SVM |
0.500 |
4.1 |
| NN |
0.990 |
31.1 |
| XGB |
0.443 |
12.5 |
The
analcatdata_boxing1 data set contains 120 observations and only three variables:
Official (binary),
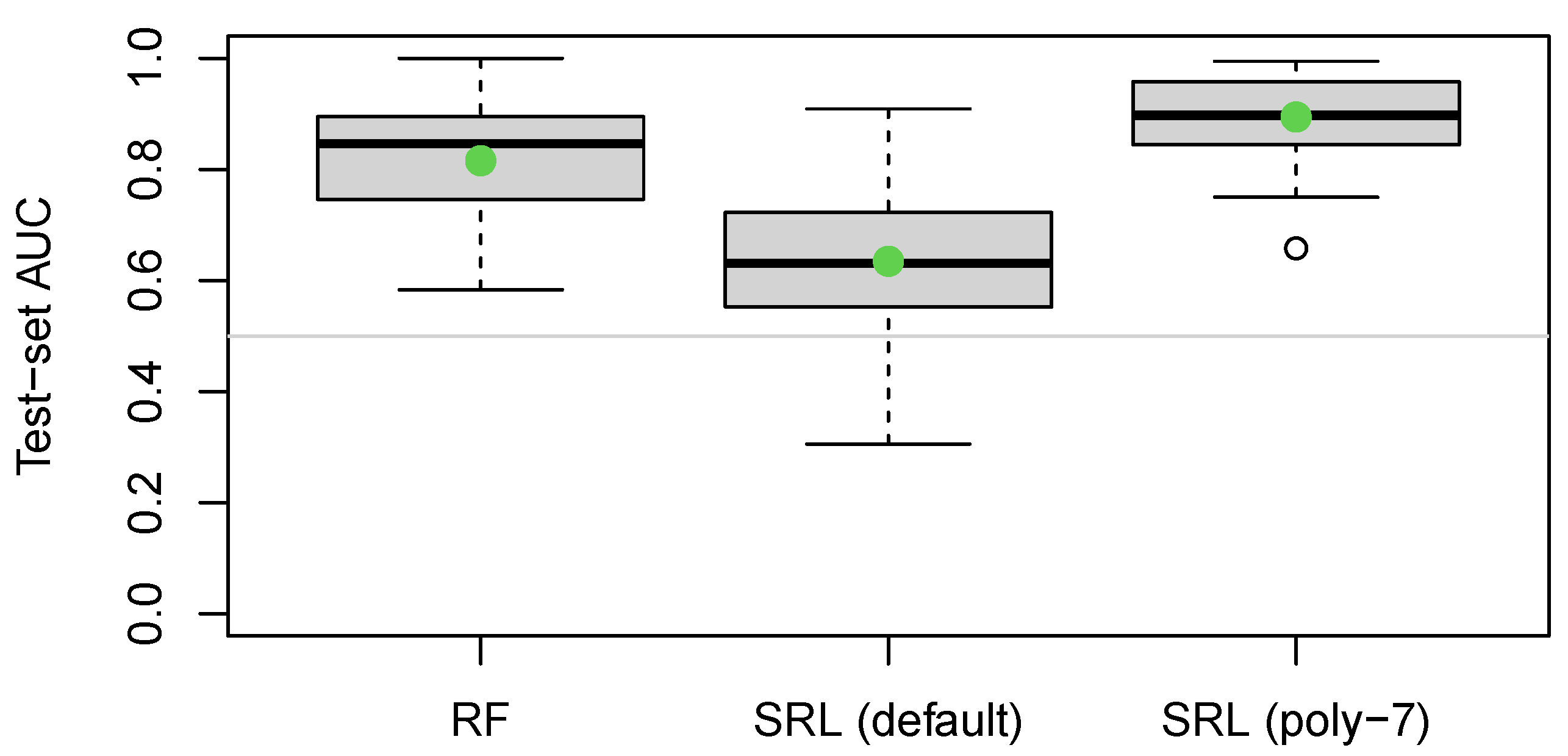
Round (integer from 1-12), and the target. Due to the small sample size, we repeated the train-test split many times and noticed that while there was substantial variability in the test AUC, the SRL still performed worse than the random forest method. We suspected that the difference is due to a nonlinear relationship between
Round and the target. By default,
sparseR only looks for interactions and main effects, but it is readily extendible to search for polynomials as well (increasing skepticism for higher-order polynomials to prefer models with lower order terms; see Peterson and Cavanaugh [
2] and Peterson [
7]). Here we can set
poly = 7 to look for up to 7 orthogonal polynomials in the numeric
Round variable. The results for all three models are shown in
Figure 7.
The parity5+5 data set consists of 1124 observations, 10 binary predictors and a single binary target variable. It seems to us to be designed to showcase a scenario where transparent modeling methods are set up for failure. The target variable for this data set uses the nonlinear parity function based on a random subset of size 5 of the features. In this case, we used the built-in variable importance metrics for the random forest to discover the subset of “important” features were the second, third, fourth, sixth, and eighth features. We could then confirm the importance of these variables by summing these binary features and recognizing that the outcome was always 1 when this subset sum was even, and always 0 otherwise. Finally, we note that adding this summation as a candidate feature to SRL and adding polynomial terms to sparseR does improve the model fit considerably, but as this requires a hybrid approach (i.e. it blends information from random forests and SRL), it does not provide a fair comparison of our method to black-box methods and we do not describe these results.