Submitted:
08 June 2024
Posted:
11 June 2024
You are already at the latest version
Abstract
Keywords:
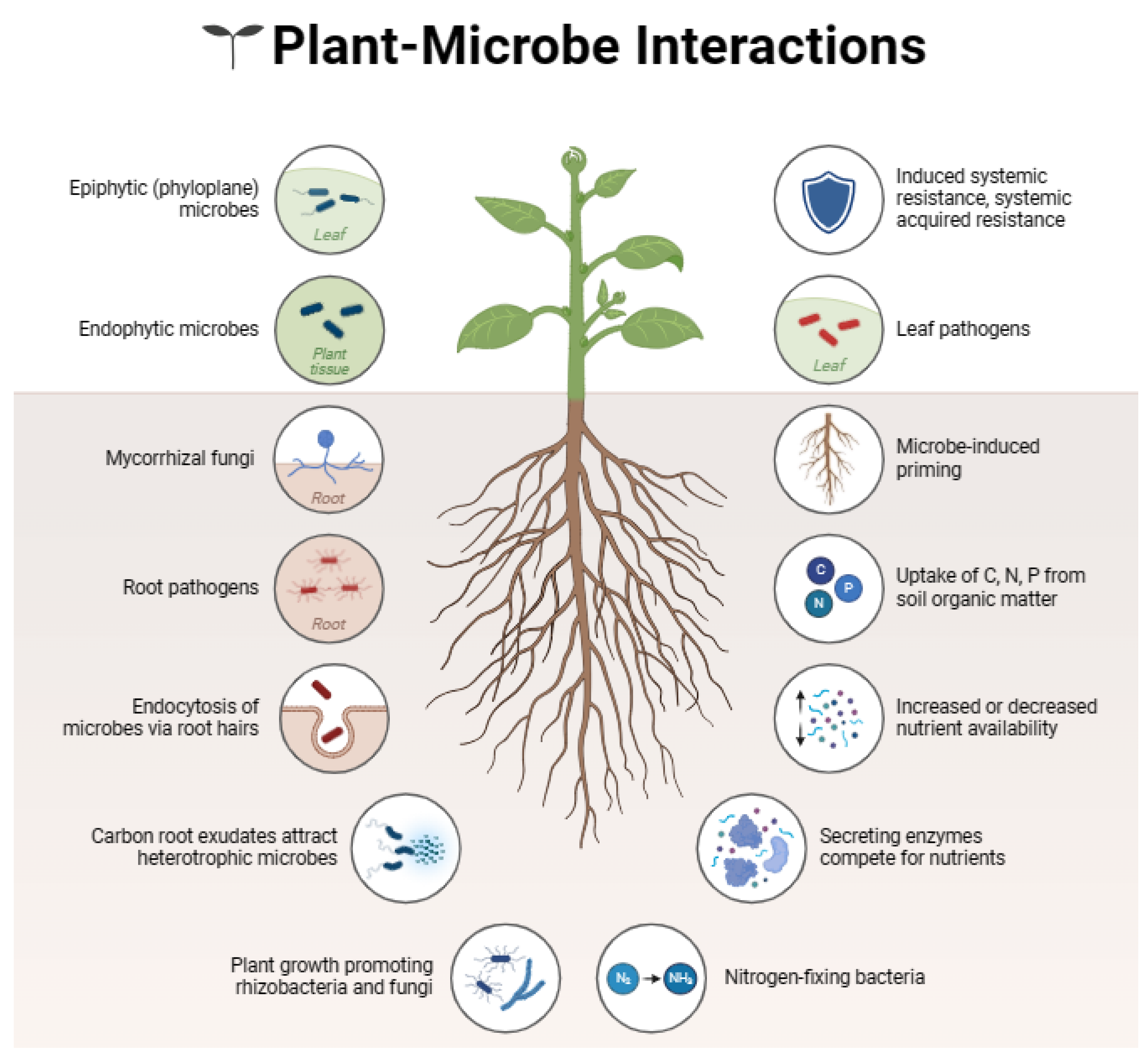
1. Introduction
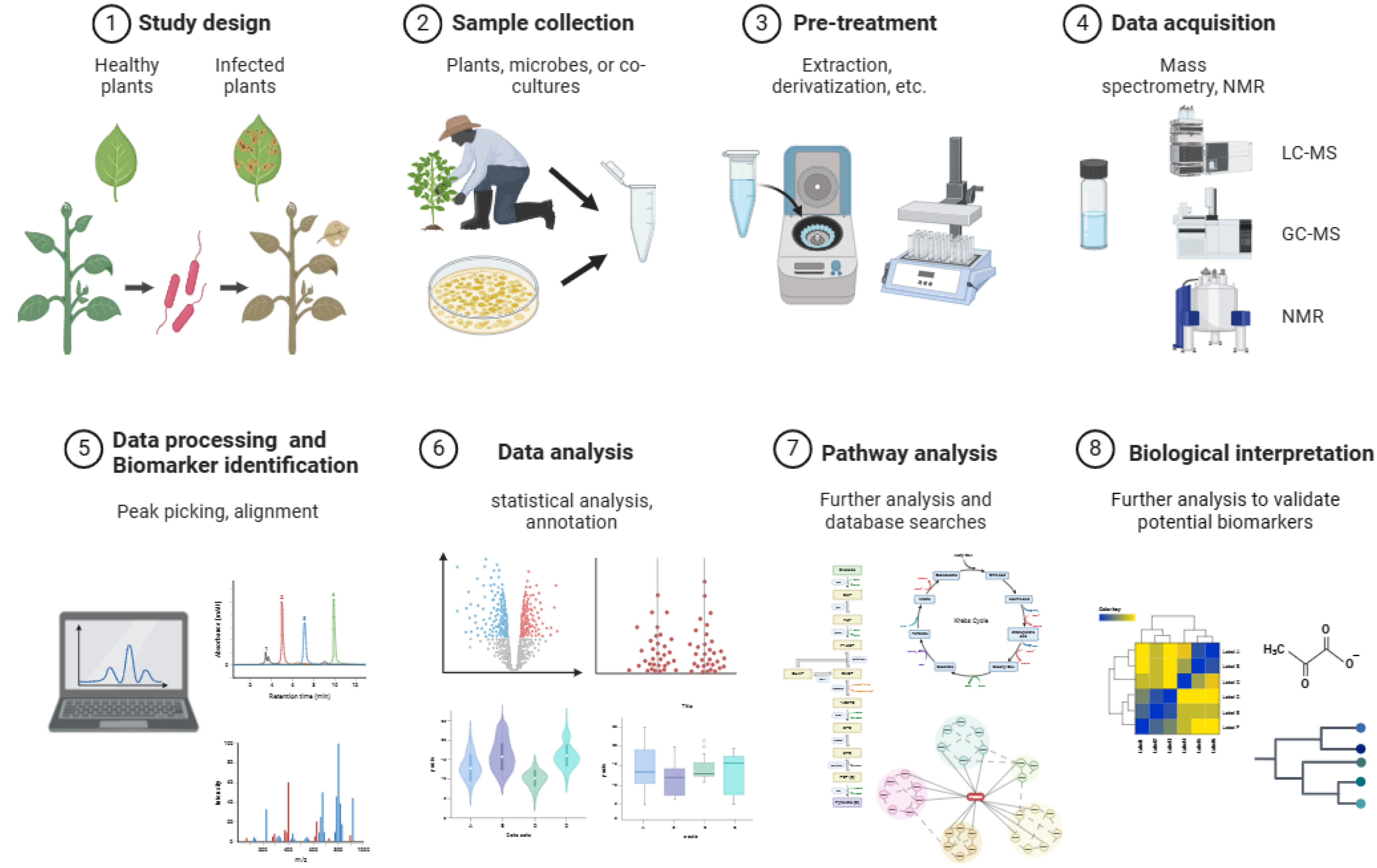
2. Metabolomics Techniques in Plant-Microbe Interaction Studies
2.1. Mass Spectrometry-Based Approaches
- a.
- Gas chromatography-mass spectrometry (GC-MS)
- b.
- Liquid chromatography-mass spectrometry (LC-MS)
- c.
- Capillary electrophoresis-mass spectrometry (CE-MS)
2.2. Nuclear Magnetic Resonance (NMR) Spectroscopy
2.3. Imaging Techniques (e.g., MALDI-MS Imaging, NMR Imaging)
2.4. Bioinformatics Tools and Databases for Metabolomics Data Analysis
2.5. AI and Machine Learning in Metabolomics for Plant-Microbe Interactions
- a.
- Applications of AI and machine learning in metabolomics
- b.
- Machine learning algorithms for metabolomics data analysis
- c.
- Deep learning for metabolomics
- d.
- Challenges and future directions
3. Integration of Metabolomics with Other Omics Approaches
3.1. Transcriptomics and Metabolomics
3.2. Proteomics and Metabolomics
3.3. Metagenomics and Metabolomics
3.4. Fluxomics: A Complementary Approach to Study Plant-Microbe Interactions
3.5. Examples of Multi-Omics Studies in Plant-Microbe Interactions
4. Challenges and Future Perspectives
4.1. Standardization of Metabolomics Protocols and Data Reporting
4.2. Advancements in Computational Tools and Databases
4.3. Omics
5. Conclusion
5.1. Summary of Key Points
5.2. Significance of Metabolomics in Understanding Plant-Microbe Interactions
5.3. Outlook for Future Research Directions
References
- Igarashi, D.; Bethke, G.; Xu, Y.; Tsuda, K.; Glazebrook, J.; Katagiri, F. Pattern-Triggered Immunity Suppresses Programmed Cell Death Triggered by Fumonisin B1. PLoS One 2013, 8, 1–2. [Google Scholar] [CrossRef]
- Bonfante, P.; Anca, I.-A. Plants, mycorrhizal fungi, and bacteria: a network of interactions. Annu. Rev. Microbiol. 2009, 63, 363–383. [Google Scholar] [CrossRef]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; Van Der Putten, W.H. Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, R.P.; Kopriva, S. Metabolic niches in the rhizosphere microbiome: new tools and approaches to analyse metabolic mechanisms of plant–microbe nutrient exchange. J. Exp. Bot. 2019, 70, 1087–1094. [Google Scholar] [CrossRef]
- Mhlongo, M.I.; Piater, L.A.; Madala, N.E.; Labuschagne, N.; Dubery, I.A. The chemistry of plant–microbe interactions in the rhizosphere and the potential for metabolomics to reveal signaling related to defense priming and induced systemic resistance. Front. Plant Sci. 2018, 9, 314703. [Google Scholar] [CrossRef]
- Kessler, A.; Kalske, A. Plant secondary metabolite diversity and species interactions. Annu. Rev. Ecol. Evol. Syst. 2018, 49, 115–138. [Google Scholar] [CrossRef]
- van Dam, N.M.; Bouwmeester, H.J. Metabolomics in the rhizosphere: tapping into belowground chemical communication. Trends Plant Sci. 2016, 21, 256–265. [Google Scholar] [CrossRef]
- Boiteau, R.M.; Hoyt, D.W.; Nicora, C.D.; Kinmonth-Schultz, H.A.; Ward, J.K.; Bingol, K. Structure elucidation of unknown metabolites in metabolomics by combined NMR and MS/MS prediction. Metabolites 2018, 8, 8. [Google Scholar] [CrossRef] [PubMed]
- Mahieu, N.G.; Patti, G.J. Systems-level annotation of a metabolomics data set reduces 25 000 features to fewer than 1000 unique metabolites. Anal. Chem. 2017, 89, 10397–10406. [Google Scholar] [CrossRef]
- Allwood, J.W.; Ellis, D.I.; Goodacre, R. Metabolomic technologies and their application to the study of plants and plant-host interactions. Physiol. Plant. 2008, 132, 117–135. [Google Scholar] [CrossRef]
- Sumner, L.W.; Yang, D.S.; Bench, B.J.; Watson, B.S.; Li, C.; Jones, A.D. Spatially resolved plant metabolomics. Annu. Plant Rev. Vol. 43 Biol. Plant Metabolomics 2011, 43, 343–366. [Google Scholar]
- De Vos, R.C.; Moco, S.; Lommen, A.; Keurentjes, J.J.; Bino, R.J.; Hall, R.D. Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2007, 2, 778–791. [Google Scholar] [CrossRef]
- Tenenboim, H.; Brotman, Y. Omic relief for the biotically stressed: metabolomics of plant biotic interactions. Trends Plant Sci. 2016, 21, 781–791. [Google Scholar] [CrossRef]
- Chen, L.; Schwier, M.; Krumbach, J.; Kopriva, S.; Jacoby, R.P. Metabolomics in plant-microbe interactions in the roots. Adv. Bot. Res. 2021, 98, 133–161. [Google Scholar]
- Lugtenberg, B.; Kamilova, F. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 2009, 63, 541–556. [Google Scholar] [CrossRef] [PubMed]
- Trethewey, R.N. Metabolite profiling as an aid to metabolic engineering in plants. Curr. Opin. Plant Biol. 2004, 7, 196–201. [Google Scholar] [CrossRef]
- Schenk, P.M.; Carvalhais, L.C.; Kazan, K. Unraveling plant–microbe interactions: can multi-species transcriptomics help? Trends Biotechnol. 2012, 30, 177–184. [Google Scholar] [CrossRef]
- Castro-Moretti, F.R.; Gentzel, I.N.; Mackey, D.; Alonso, A.P. Metabolomics as an emerging tool for the study of plant–pathogen interactions. Metabolites 2020, 10, 52. [Google Scholar] [CrossRef]
- Sade, D.; Shriki, O.; Cuadros-Inostroza, A.; Tohge, T.; Semel, Y.; Haviv, Y.; Willmitzer, L.; Fernie, A.R.; Czosnek, H.; Brotman, Y. Comparative metabolomics and transcriptomics of plant response to Tomato yellow leaf curl virus infection in resistant and susceptible tomato cultivars. Metabolomics 2015, 11, 81–97. [Google Scholar] [CrossRef]
- Vacheron, J.; Desbrosses, G.; Bouffaud, M.-L.; Touraine, B.; Moënne-Loccoz, Y.; Muller, D.; Legendre, L.; Wisniewski-Dyé, F.; Prigent-Combaret, C. Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 2013, 4, 57135. [Google Scholar] [CrossRef]
- Pozo, M.J.; Van Der Ent, S.; Van Loon, L.C.; Pieterse, C.M.J. Transcription factor MYC2 is involved in priming for enhanced defense during rhizobacteria-induced systemic resistance in Arabidopsis thaliana. New Phytol. 2008, 180, 511–523. [Google Scholar] [CrossRef] [PubMed]
- Wolfender, J.-L.; Rudaz, S.; Hae Choi, Y.; Kyong Kim, H. Plant metabolomics: from holistic data to relevant biomarkers. Curr. Med. Chem. 2013, 20, 1056–1090. [Google Scholar] [PubMed]
- Worley, B.; Powers, R. Multivariate analysis in metabolomics. Curr. metabolomics 2013, 1, 92–107. [Google Scholar] [PubMed]
- Villas-Bôas, S.G.; Mas, S.; Åkesson, M.; Smedsgaard, J.; Nielsen, J. Mass spectrometry in metabolome analysis. Mass Spectrom. Rev. 2005, 24, 613–646. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Fu, X. Reprogramming of Plant Central Metabolism in Response to Abiotic Stresses: A Metabolomics View. Int. J. Mol. Sci. 2022, 23, 5716. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics by gas chromatography–mass spectrometry: Combined targeted and untargeted profiling. Curr. Protoc. Mol. Biol. 2016, 114, 30–34. [Google Scholar] [CrossRef] [PubMed]
- Lisec, J.; Schauer, N.; Kopka, J.; Willmitzer, L.; Fernie, A.R. Gas chromatography mass spectrometry–based metabolite profiling in plants. Nat. Protoc. 2006, 1, 387–396. [Google Scholar] [CrossRef]
- Fu, X.; Xu, Y. Dynamic metabolic changes in arabidopsis seedlings under hypoxia stress and subsequent reoxygenation recovery. Stresses 2023, 3, 86–101. [Google Scholar] [CrossRef]
- Jud, W.; Winkler, J.B.; Niederbacher, B.; Niederbacher, S.; Schnitzler, J.-P. Volatilomics: a non-invasive technique for screening plant phenotypic traits. Plant Methods 2018, 14, 1–18. [Google Scholar] [CrossRef]
- Sharifi, R.; Lee, S.; Ryu, C. Microbe-induced plant volatiles. New Phytol. 2018, 220, 684–691. [Google Scholar] [CrossRef]
- Allwood, J.W.; Goodacre, R. An introduction to liquid chromatography–mass spectrometry instrumentation applied in plant metabolomic analyses. Phytochem. Anal. An Int. J. Plant Chem. Biochem. Tech. 2010, 21, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Freund, D.M.; Hegeman, A.D.; Cohen, J.D. Metabolic signatures of Arabidopsis thaliana abiotic stress responses elucidate patterns in stress priming, acclimation, and recovery. Stress Biol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Mhlongo, M.I.; Piater, L.A.; Steenkamp, P.A.; Labuschagne, N.; Dubery, I.A. Metabolic profiling of PGPR-treated tomato plants reveal priming-related adaptations of secondary metabolites and aromatic amino acids. Metabolites 2020, 10, 210. [Google Scholar] [CrossRef] [PubMed]
- Rivero, J.; Gamir, J.; Aroca, R.; Pozo, M.J.; Flors, V. Metabolic transition in mycorrhizal tomato roots. Front. Microbiol. 2015, 6, 598. [Google Scholar] [CrossRef] [PubMed]
- Stringlis, I.A.; Yu, K.; Feussner, K.; de Jonge, R.; Van Bentum, S.; Van Verk, M.C.; Berendsen, R.L.; Bakker, P.A.H.M.; Feussner, I.; Pieterse, C.M.J. MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc. Natl. Acad. Sci. 2018, 115, E5213–E5222. [Google Scholar] [CrossRef] [PubMed]
- Ramautar, R.; Somsen, G.W.; de Jong, G.J. CE-MS in metabolomics. Electrophoresis 2009, 30, 276–291. [Google Scholar] [CrossRef]
- Monton, M.R.N.; Soga, T. Metabolome analysis by capillary electrophoresis–mass spectrometry. J. Chromatogr. A 2007, 1168, 237–246. [Google Scholar] [CrossRef]
- Guijas, C.; Montenegro-Burke, J.R.; Domingo-Almenara, X.; Palermo, A.; Warth, B.; Hermann, G.; Koellensperger, G.; Huan, T.; Uritboonthai, W.; Aisporna, A.E. METLIN: a technology platform for identifying knowns and unknowns. Anal. Chem. 2018, 90, 3156–3164. [Google Scholar] [CrossRef] [PubMed]
- Gamir, J.; Pastor, V.; Kaever, A.; Cerezo, M.; Flors, V. Targeting novel chemical and constitutive primed metabolites against P lectosphaerella cucumerina. Plant J. 2014, 78, 227–240. [Google Scholar] [CrossRef]
- Bringel, F.; Couée, I. Pivotal roles of phyllosphere microorganisms at the interface between plant functioning and atmospheric trace gas dynamics. Front. Microbiol. 2015, 6, 144430. [Google Scholar] [CrossRef]
- Kim, H.K.; Choi, Y.H.; Verpoorte, R. NMR-based plant metabolomics: where do we stand, where do we go? Trends Biotechnol. 2011, 29, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, P.; Kruger, N.J.; Ratcliffe, R.G. Metabolite fingerprinting and profiling in plants using NMR. J. Exp. Bot. 2005, 56, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Boughton, B.A.; Thinagaran, D. Mass spectrometry imaging (MSI) for plant metabolomics. Plant Metabolomics Methods Protoc. 2018, 241–252. [Google Scholar]
- Sturtevant, D.; Lee, Y.-J.; Chapman, K.D. Matrix assisted laser desorption/ionization-mass spectrometry imaging (MALDI-MSI) for direct visualization of plant metabolites in situ. Curr. Opin. Biotechnol. 2016, 37, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Veličković, D.; Agtuca, B.J.; Stopka, S.A.; Vertes, A.; Koppenaal, D.W.; Paša-Tolić, L.; Stacey, G.; Anderton, C.R. Observed metabolic asymmetry within soybean root nodules reflects unexpected complexity in rhizobacteria-legume metabolite exchange. ISME J. 2018, 12, 2335–2338. [Google Scholar] [CrossRef] [PubMed]
- Pétriacq, P.; Williams, A.; Cotton, A.; McFarlane, A.E.; Rolfe, S.A.; Ton, J. Metabolite profiling of non-sterile rhizosphere soil. Plant J. 2017, 92, 147–162. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, A.; Kusano, M. Recent progress in the development of metabolome databases for plant systems biology. Front. Plant Sci. 2013, 4, 42957. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y. Metabolomics study on Arabidopsis thaliana abiotic stress responses for priming, recovery, and stress combinations. <bold>2018</bold>.
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 2010, 11, 1–11. [Google Scholar] [CrossRef]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef]
- Smith, C.A.; O’Maille, G.; Want, E.J.; Qin, C.; Trauger, S.A.; Brandon, T.R.; Custodio, D.E.; Abagyan, R.; Siuzdak, G. METLIN: A metabolite mass spectral database. Therapeutic Drug Monitoring 2005, 27, 747–751. [Google Scholar] [CrossRef]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K.; et al. MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef] [PubMed]
- Hall, R.D. Plant metabolomics in a nutshell: potential and future challenges. Annu. Plant Rev. Vol. 43 Biol. Plant Metabolomics 2011, 43, 1–24. [Google Scholar]
- Tugizimana, F.; Piater, L.; Dubery, I. Plant metabolomics: A new frontier in phytochemical analysis. S. Afr. J. Sci. 2013, 109, 1–11. [Google Scholar] [CrossRef]
- Murmu, S.; Sinha, D.; Chaurasia, H.; Sharma, S.; Das, R.; Jha, G.K.; Archak, S. A review of artificial intelligence-assisted omics techniques in plant defense: current trends and future directions. Front. Plant Sci. 2024, 15, 1292054. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Berdine, G. Artificial intelligence in biomedical research. Southwest Respir. Crit. Care Chronicles 2023, 11, 62–65. [Google Scholar] [CrossRef]
- Misra, B.B.; Mohapatra, S. Tools and resources for metabolomics research community: A 2017–2018 update. Electrophoresis 2019, 40, 227–246. [Google Scholar] [CrossRef]
- Salinas, J.L.; Kissinger, J.C.; Jones, D.P.; Galinski, M.R. Metabolomics in the fight against malaria. Mem. Inst. Oswaldo Cruz 2014, 109, 589–597. [Google Scholar] [CrossRef]
- Gromski, P.S.; Muhamadali, H.; Ellis, D.I.; Xu, Y.; Correa, E.; Turner, M.L.; Goodacre, R. A tutorial review: Metabolomics and partial least squares-discriminant analysis–a marriage of convenience or a shotgun wedding. Anal. Chim. Acta 2015, 879, 10–23. [Google Scholar] [CrossRef]
- Mishra, P.; Singh, U.; Pandey, C.M.; Mishra, P.; Pandey, G. Application of student’s t-test, analysis of variance, and covariance. Ann. Card. Anaesth. 2019, 22, 407–411. [Google Scholar] [CrossRef]
- Abdel-Farid, I.B.; Jahangir, M.; Van Den Hondel, C.; Kim, H.K.; Choi, Y.H.; Verpoorte, R. Fungal infection-induced metabolites in Brassica rapa. Plant Sci. 2009, 176, 608–615. [Google Scholar] [CrossRef]
- Jolliffe, I.T.; Cadima, J. Principal component analysis: a review and recent developments. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2016, 374, 20150202. [Google Scholar] [CrossRef]
- Want, E.J.; Wilson, I.D.; Gika, H.; Theodoridis, G.; Plumb, R.S.; Shockcor, J.; Holmes, E.; Nicholson, J.K. Global metabolic profiling procedures for urine using UPLC–MS. Nat. Protoc. 2010, 5, 1005–1018. [Google Scholar] [CrossRef]
- Lecun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Ching, T.; Himmelstein, D.S.; Beaulieu-Jones, B.K.; Kalinin, A.A.; Do, B.T.; Way, G.P.; Ferrero, E.; Agapow, P.-M.; Zietz, M.; Hoffman, M.M. Opportunities and obstacles for deep learning in biology and medicine. J. R. Soc. interface 2018, 15, 20170387. [Google Scholar] [CrossRef]
- Liebal, U.W.; Phan, A.N.T.; Sudhakar, M.; Raman, K.; Blank, L.M. Machine learning applications for mass spectrometry-based metabolomics. Metabolites 2020, 10, 243. [Google Scholar] [CrossRef]
- Uppal, K.; Walker, D.I.; Jones, D.P. xMSannotator: an R package for network-based annotation of high-resolution metabolomics data. Anal. Chem. 2017, 89, 1063–1067. [Google Scholar] [CrossRef] [PubMed]
- Pomyen, Y.; Wanichthanarak, K.; Poungsombat, P.; Fahrmann, J.; Grapov, D.; Khoomrung, S. Deep metabolome: Applications of deep learning in metabolomics. Comput. Struct. Biotechnol. J. 2020, 18, 2818–2825. [Google Scholar] [CrossRef]
- Zhang, A.; Sun, H.; Yan, G.; Wang, P.; Wang, X. Metabolomics for biomarker discovery: moving to the clinic. Biomed Res. Int. 2015, 2015. [Google Scholar] [CrossRef]
- Spicer, R.A.; Salek, R.; Steinbeck, C. Compliance with minimum information guidelines in public metabolomics repositories. Sci. data 2017, 4, 1–8. [Google Scholar] [CrossRef]
- Samek, W.; Montavon, G.; Vedaldi, A.; Hansen, L.K.; Müller, K.-R. <italic>Explainable AI: interpreting, explaining and visualizing deep learning</italic>; Springer Nature, 2019; Vol. 11700; ISBN 3030289540.
- Yang, F.; Du, M.; Hu, X. Evaluating explanation without ground truth in interpretable machine learning. arXiv Prepr. arXiv1907.06831 2019. [Google Scholar]
- Jastrebski, S.F.; Lamont, S.J.; Schmidt, C.J. Chicken hepatic response to chronic heat stress using integrated transcriptome and metabolome analysis. PLoS One 2017, 12, e0181900. [Google Scholar] [CrossRef] [PubMed]
- Weckwerth, W. Metabolomics in systems biology. Annu. Rev. Plant Biol. 2003, 54, 669–689. [Google Scholar] [CrossRef] [PubMed]
- Tang, J. Microbial metabolomics. Curr. Genomics 2011, 12, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Guijas, C.; Montenegro-Burke, J.R.; Warth, B.; Spilker, M.E.; Siuzdak, G. Metabolomics activity screening for identifying metabolites that modulate phenotype. Nat. Biotechnol. 2018, 36, 316–320. [Google Scholar] [CrossRef] [PubMed]
- Camañes, G.; Scalschi, L.; Vicedo, B.; González-Bosch, C.; García-Agustín, P. An untargeted global metabolomic analysis reveals the biochemical changes underlying basal resistance and priming in Solanum lycopersicum, and identifies 1-methyltryptophan as a metabolite involved in plant responses to Botrytis cinerea and Pseudomonas sy. Plant J. 2015, 84, 125–139. [Google Scholar] [CrossRef]
- Fan, K.-T.; Xu, Y.; Hegeman, A.D. Elevated Temperature Effects on Protein Turnover Dynamics in Arabidopsis thaliana Seedlings Revealed by 15N-Stable Isotope Labeling and ProteinTurnover Algorithm. Int. J. Mol. Sci. 2024, 25, 5882. [Google Scholar] [CrossRef]
- Mathesius, U.; Mulders, S.; Gao, M.; Teplitski, M.; Caetano-Anollés, G.; Rolfe, B.G.; Bauer, W.D. Extensive and specific responses of a eukaryote to bacterial quorum-sensing signals. Proc. Natl. Acad. Sci. 2003, 100, 1444–1449. [Google Scholar] [CrossRef] [PubMed]
- Feussner, I.; Polle, A. What the transcriptome does not tell—proteomics and metabolomics are closer to the plants’ patho-phenotype. Curr. Opin. Plant Biol. 2015, 26, 26–31. [Google Scholar] [CrossRef]
- Balmer, D.; de Papajewski, D.V.; Planchamp, C.; Glauser, G.; Mauch-Mani, B. Induced resistance in maize is based on organ-specific defence responses. Plant J. 2013, 74, 213–225. [Google Scholar] [CrossRef]
- Berg, G.; Grube, M.; Schloter, M.; Smalla, K. Unraveling the plant microbiome: looking back and future perspectives. Front. Microbiol. 2014, 5, 71409. [Google Scholar] [CrossRef] [PubMed]
- Sasse, J.; Martinoia, E.; Northen, T. Feed your friends: do plant exudates shape the root microbiome? Trends Plant Sci. 2018, 23, 25–41. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Sun, H.-X.; Xiao, H.; Cui, G.; Hillwig, M.L.; Jackson, A.; Wang, X.; Shen, Y.; Zhao, N.; Zhang, L. Combining metabolomics and transcriptomics to characterize tanshinone biosynthesis in Salvia miltiorrhiza. BMC Genomics 2014, 15, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ratcliffe, R.G.; Shachar-Hill, Y. Measuring multiple fluxes through plant metabolic networks. Plant J. 2006. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Fu, X.; Sharkey, T.D.; Shachar-Hill, Y.; Walker, B.J. The metabolic origins of non-photorespiratory CO2 release during photosynthesis: A metabolic flux analysis. Plant Physiol. 2021, 186, 297–314. [Google Scholar] [CrossRef] [PubMed]
- Schwender, J.; Goffman, F.; Ohlrogge, J.B.; Shachar-Hill, Y. Rubisco without the Calvin cycle improves the carbon efficiency of developing green seeds. Nature 2004, 432, 779–782. [Google Scholar] [CrossRef] [PubMed]
- Salon, C.; Raymond, P.; Pradet, A. Quantification of carbon fluxes through the tricarboxylic acid cycle in early germinating lettuce embryos. J. Biol. Chem. 1988, 263, 12278–12287. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wieloch, T.; Kaste, J.A.M.; Shachar-Hill, Y.; Sharkey, T.D. Reimport of carbon from cytosolic and vacuolar sugar pools into the Calvin–Benson cycle explains photosynthesis labeling anomalies. Proc. Natl. Acad. Sci. 2022, 119, e2121531119. [Google Scholar] [CrossRef] [PubMed]
- Dieuaide-Noubhani, M.; Raffard, G.; Canioni, P.; Pradet, A.; Raymond, P. Quantification of Compartmented Metabolic Fluxes in Maize Root Tips Using Isotope Distribution from 13C-or 14C-Labeled Glucose∗. J. Biol. Chem. 1995, 270, 13147–13159. [Google Scholar] [CrossRef]
- Xu, Y.; Koroma, A.A.; Weise, S.E.; Fu, X.; Sharkey, T.D.; Shachar-Hill, Y. Daylength variation affects growth, photosynthesis, leaf metabolism, partitioning, and metabolic fluxes. Plant Physiol. 2024, 194, 475–490. [Google Scholar] [CrossRef]
- Kruger, N.J.; Masakapalli, S.K.; Ratcliffe, R.G. Strategies for investigating the plant metabolic network with steady-state metabolic flux analysis: lessons from an Arabidopsis cell culture and other systems. J. Exp. Bot. 2012, 63, 2309–2323. [Google Scholar] [CrossRef] [PubMed]
- Schwender, J.; Ohlrogge, J.B.; Shachar-Hill, Y. A flux model of glycolysis and the oxidative pentosephosphate pathway in developing Brassica napus embryos. J. Biol. Chem. 2003. [Google Scholar] [CrossRef] [PubMed]
- Bago, B.; Pfeffer, P.E.; Abubaker, J.; Jun, J.; Allen, J.W.; Brouillette, J.; Douds, D.D.; Lammers, P.J.; Shachar-Hill, Y. Carbon export from arbuscular mycorrhizal roots involves the translocation of carbohydrate as well as lipid. Plant Physiol. 2003, 131, 1496–1507. [Google Scholar] [CrossRef] [PubMed]
- Rich, M.K.; Schorderet, M.; Bapaume, L.; Falquet, L.; Morel, P.; Vandenbussche, M.; Reinhardt, D. The petunia GRAS transcription factor ATA/RAM1 regulates symbiotic gene expression and fungal morphogenesis in arbuscular mycorrhiza. Plant Physiol. 2015, 168, 788–797. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Cheung, C.Y.M.; Hilbers, P.A.J.; Van Riel, N.A.W. Flux balance analysis of plant metabolism: the effect of biomass composition and model structure on model predictions. Front. Plant Sci. 2016, 7, 537. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Schmiege, S.C.; Sharkey, T.D. The oxidative pentose phosphate pathway in photosynthesis: a tale of two shunts. New Phytol. 2024. [Google Scholar] [CrossRef] [PubMed]
- Wilson, N.J.; Smith-Moore, C.M.; Xu, Y.; Edwards, B.; Hovary, C. La; Barampuram, S.; Li, K.; Aslett, D.; Ji, M.; Lin, X. Introduction of a condensed, reverse tricarboxylic acid cycle for additional CO2 fixation in plants. bioRxiv 2022, 2003–2022. [Google Scholar]
- Libourel, I.G.L.; Shachar-Hill, Y. Metabolic Flux Analysis in Plants: From Intelligent Design to Rational Engineering. Annu. Rev. Plant Biol. 2008. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Pang, N.; He, L.; Xu, Y.; Fu, X.; Tang, Y.; Shachar-Hill, Y.; Chen, S. Re-Programing Glucose Catabolism in the Microalga Chlorella sorokiniana under Light Condition. Biomolecules 2022, 12, 939. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, N.; Qiu, M.; Feng, H.; Vivanco, J.M.; Shen, Q.; Zhang, R. Enhanced rhizosphere colonization of beneficial Bacillus amyloliquefaciens SQR9 by pathogen infection. FEMS Microbiol. Lett. 2014, 353, 49–56. [Google Scholar] [CrossRef]
- Schläpfer, P.; Zhang, P.; Wang, C.; Kim, T.; Banf, M.; Chae, L.; Dreher, K.; Chavali, A.K.; Nilo-Poyanco, R.; Bernard, T. Genome-wide prediction of metabolic enzymes, pathways, and gene clusters in plants. Plant Physiol. 2017, 173, 2041–2059. [Google Scholar] [CrossRef] [PubMed]
- Wolfender, J.-L.; Marti, G.; Thomas, A.; Bertrand, S. Current approaches and challenges for the metabolite profiling of complex natural extracts. J. Chromatogr. A 2015, 1382, 136–164. [Google Scholar] [CrossRef] [PubMed]
- Misra, B.B.; van der Hooft, J.J.J. Updates in metabolomics tools and resources: 2014–2015. Electrophoresis 2016, 37, 86–110. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O.; Robertson, D.; Griffin, J.; van der Werf, M.; Nikolau, B.; Morrison, N.; Sumner, L.W.; Goodacre, R.; Hardy, N.W.; Taylor, C. The metabolomics standards initiative (MSI). Metabolomics 2007, 3, 175–178. [Google Scholar] [CrossRef]
- Goodacre, R. Water, water, every where, but rarely any drop to drink. Metabolomics 2014, 10, 5–7. [Google Scholar] [CrossRef]
- Bowen, B.P.; Northen, T.R. Dealing with the unknown: metabolomics and metabolite atlases. J. Am. Soc. Mass Spectrom. 2010, 21, 1471–1476. [Google Scholar] [CrossRef] [PubMed]
- Kind, T.; Wohlgemuth, G.; Lee, D.Y.; Lu, Y.; Palazoglu, M.; Shahbaz, S.; Fiehn, O. FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal. Chem. 2009, 81, 10038–10048. [Google Scholar] [CrossRef] [PubMed]
- Okada, T.; Mochamad Afendi, F.; Altaf-Ul-Amin, M.; Takahashi, H.; Nakamura, K.; Kanaya, S. Metabolomics of medicinal plants: the importance of multivariate analysis of analytical chemistry data. Curr. Comput. Aided. Drug Des. 2010, 6, 179–196. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Gatto, L.; Wang, R.; Sachsenberg, T.; Uszkoreit, J.; Leprevost, F. da V.; Fufezan, C.; Ternent, T.; Eglen, S.J.; Katz, D.S. Ten simple rules for taking advantage of Git and GitHub. PLoS Comput. Biol. 2016, 12, e1004947. [Google Scholar] [CrossRef]
- Mendez, K.M.; Broadhurst, D.I.; Reinke, S.N. The application of artificial neural networks in metabolomics: a historical perspective. Metabolomics 2019, 15, 142. [Google Scholar] [CrossRef]
- Rinaudo, P.; Boudah, S.; Junot, C.; Thévenot, E.A. biosigner: A New Method for the Discovery of Significant Molecular Signatures from Omics Data. Front. Mol. Biosci. 2016, 3, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Toubiana, D.; Puzis, R.; Wen, L.; Sikron, N.; Kurmanbayeva, A.; Soltabayeva, A.; del Mar Rubio Wilhelmi, M.; Sade, N.; Fait, A.; Sagi, M. Combined network analysis and machine learning allows the prediction of metabolic pathways from tomato metabolomics data. Commun. Biol. 2019, 2, 214. [Google Scholar] [CrossRef] [PubMed]
- Sauer, U.; Heinemann, M.; Zamboni, N. Getting closer to the whole picture. Science (80-. ). 2007, 316, 550–551. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, A.; Kusano, M.; Redestig, H.; Arita, M.; Saito, K. Integrated omics approaches in plant systems biology. Curr. Opin. Chem. Biol. 2009, 13, 532–538. [Google Scholar] [CrossRef] [PubMed]
- Coppola, M.; Diretto, G.; Digilio, M.C.; Woo, S.L.; Giuliano, G.; Molisso, D.; Pennacchio, F.; Lorito, M.; Rao, R. Transcriptome and metabolome reprogramming in tomato plants by Trichoderma harzianum strain T22 primes and enhances defense responses against aphids. Front. Physiol. 2019, 10, 745. [Google Scholar] [CrossRef] [PubMed]
- Savoi, S.; Wong, D.C.J.; Degu, A.; Herrera, J.C.; Bucchetti, B.; Peterlunger, E.; Fait, A.; Mattivi, F.; Castellarin, S.D. Multi-omics and integrated network analyses reveal new insights into the systems relationships between metabolites, structural genes, and transcriptional regulators in developing grape berries (Vitis vinifera L.) exposed to water deficit. Front. Plant Sci. 2017, 8, 1124. [Google Scholar] [CrossRef]
- Wanichthanarak, K.; Fahrmann, J.F.; Grapov, D. Genomic, proteomic, and metabolomic data integration strategies. Biomark. Insights 2015, 10, BMI-S29511. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



