Submitted:
08 June 2024
Posted:
11 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
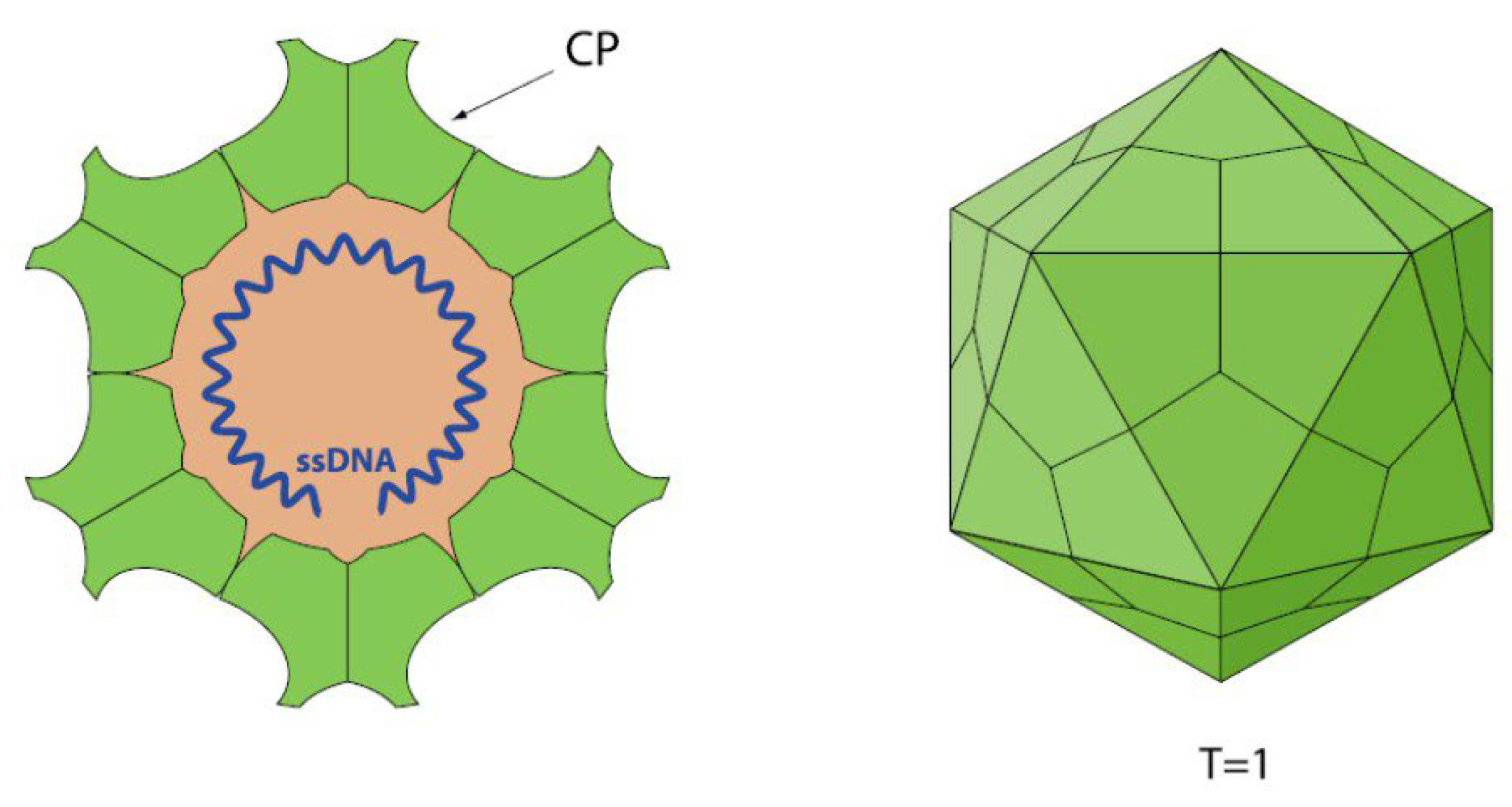
1.1. Parvovirus Classification and Virion Properties
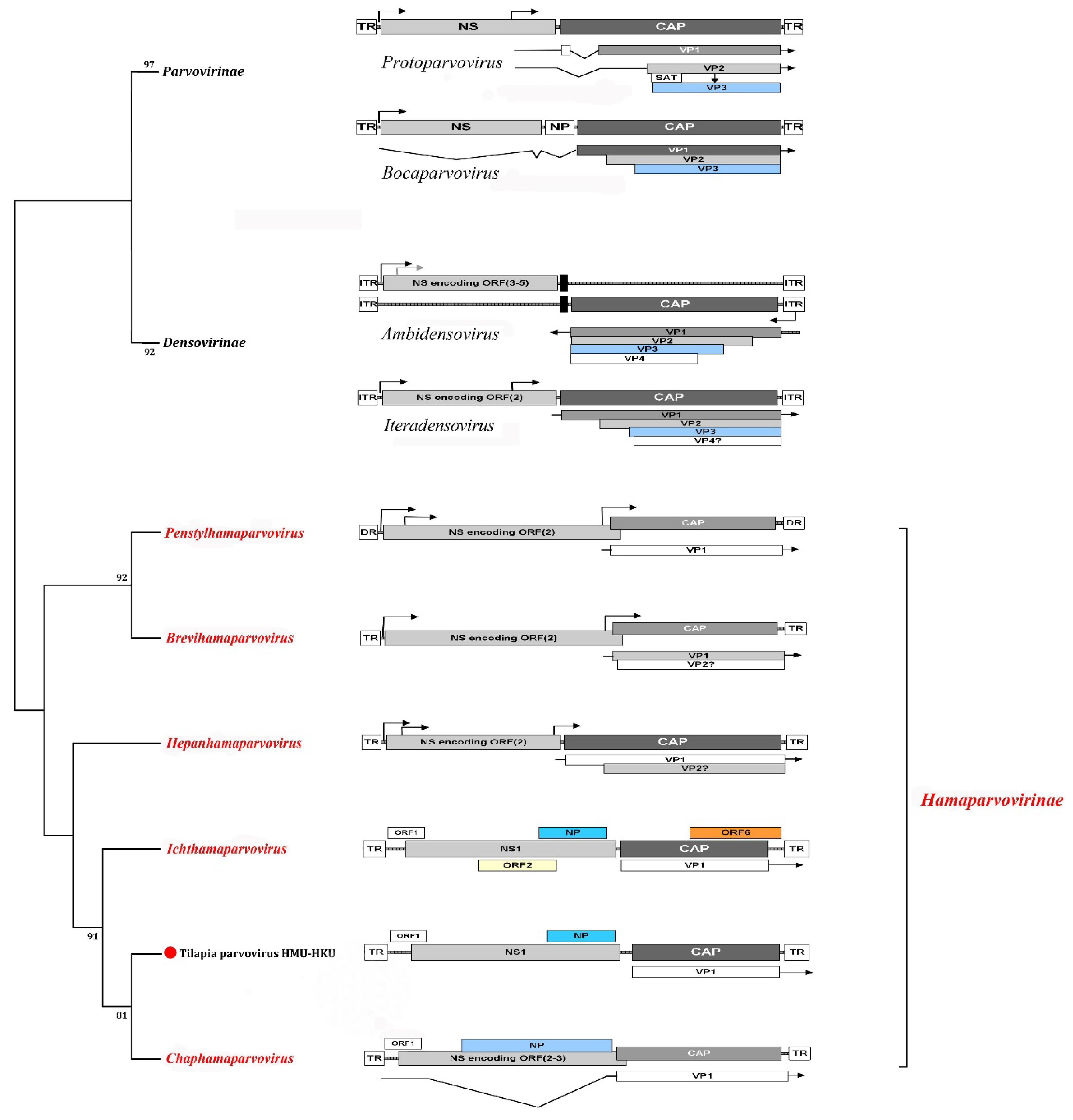
1.1.1. Parvovirus Genome Organization
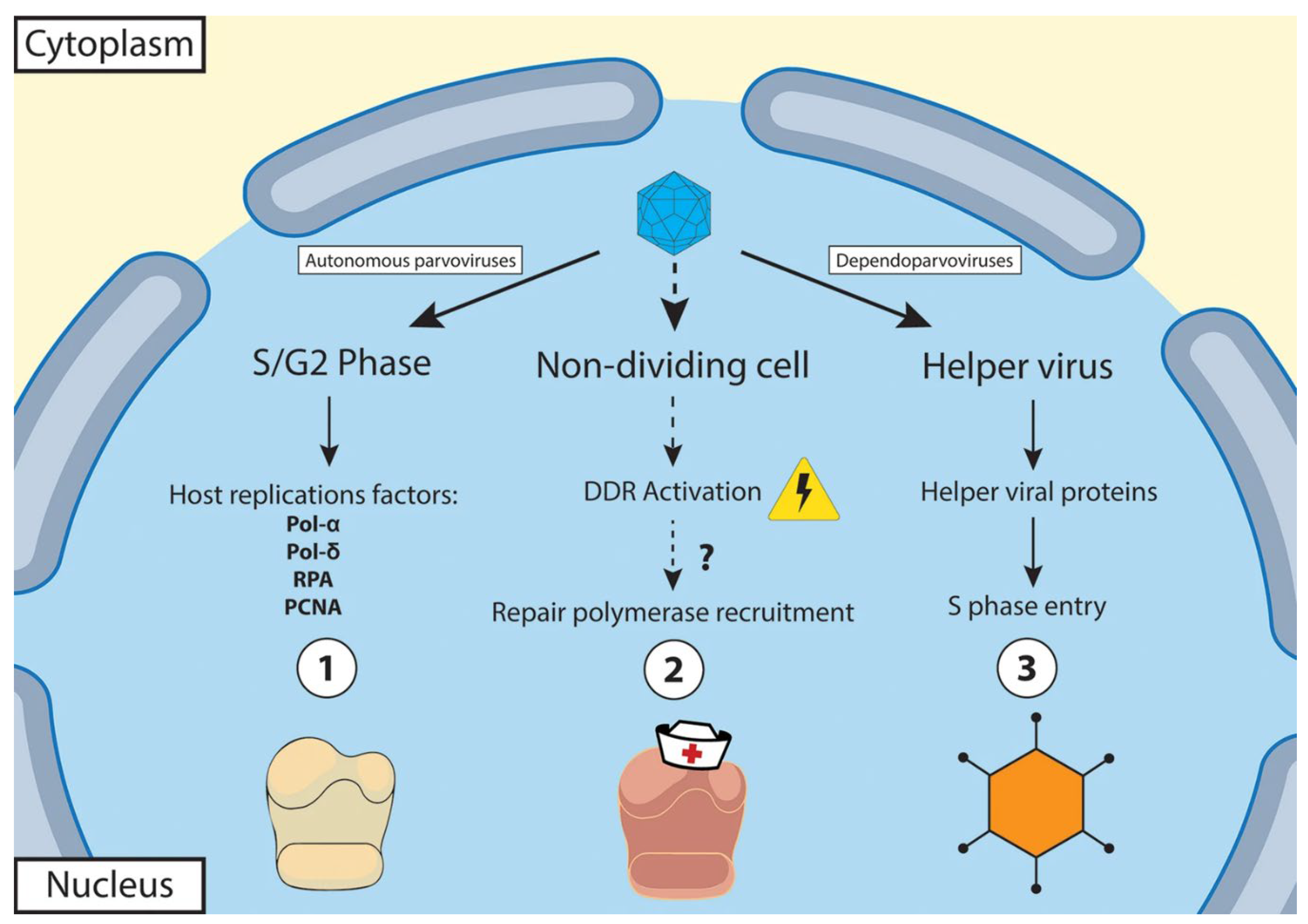
1.1.2. Virus Replication
1.2. General Pathogenesis of Parvoviruses
2. Genus Aquambidensovirus
2.1. Decapod aquambidensovirus 1 (Cherax Quadricarinatus Densovirus (CqDV))
2.2. Asteroid aquambidensovirus 1 (Sea Star-Associated Densovirus (SSaDV))
2.3. Clinch Densovirus 1
3. Genus Hepanhamaparvovirus
3.1. Decapod hepanhamaparvovirus 1 (DHPV-1) (Hepatopancreatic Parvovirus (HPV))
4. Genus Penstylhamaparvovirus
5. Genus Ichthamaparvovirus
5.1. Syngnathid ichthamaparvovirus 1
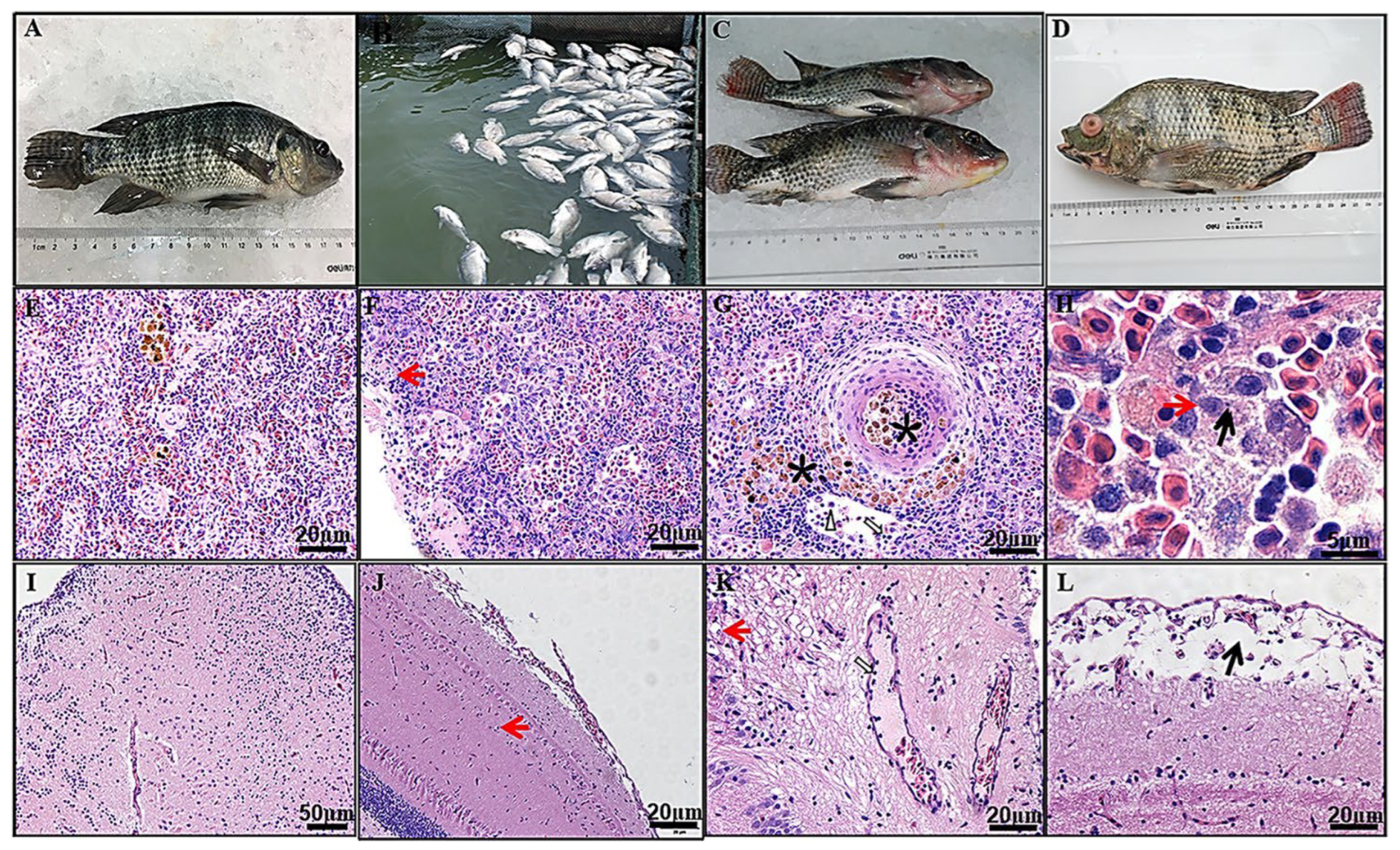
5.2. Tilapia Parvovirus (TiPV)
6. Unassigned
6.1. Novel Salmon Parvovirus from Sockeye Salmon
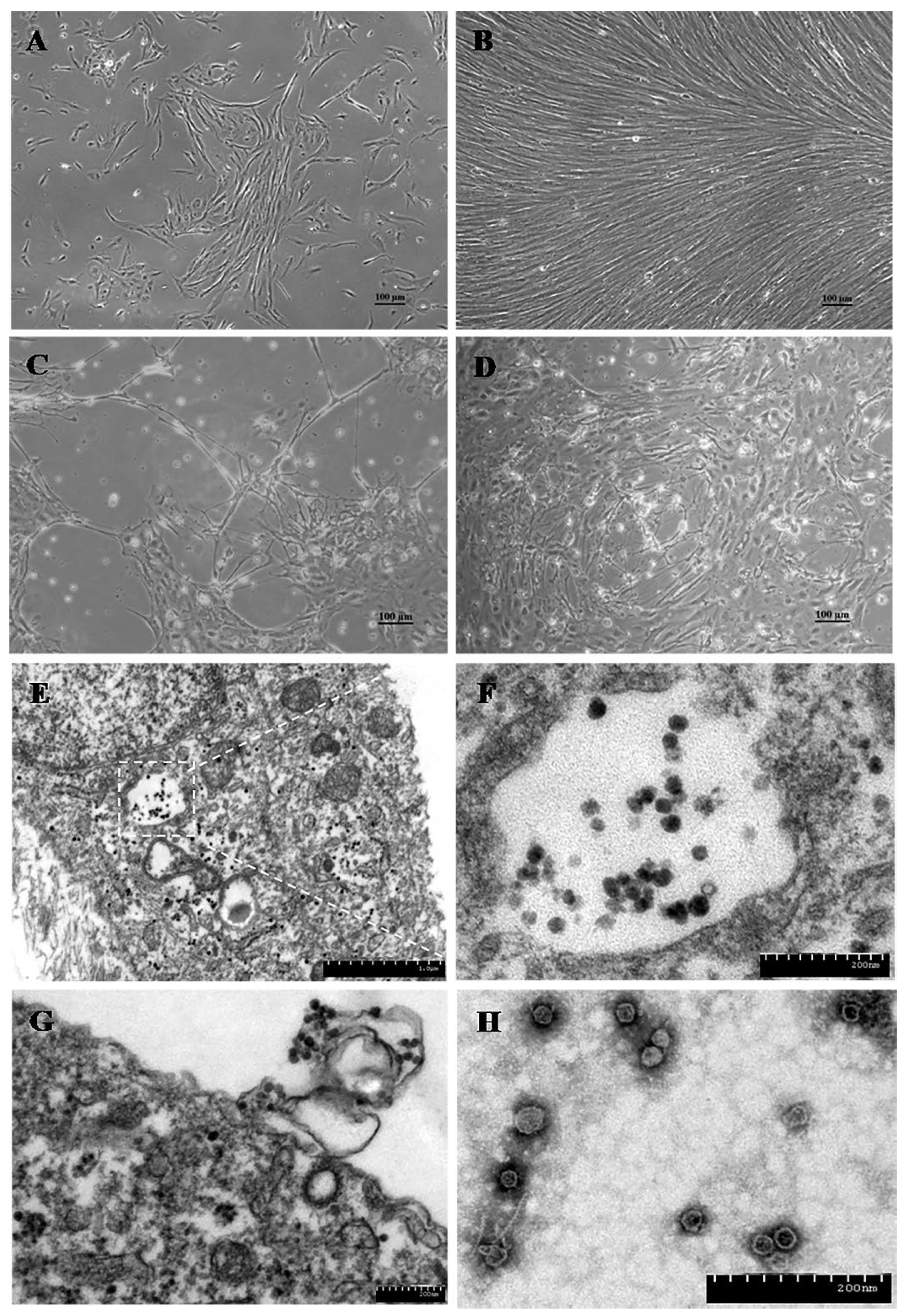
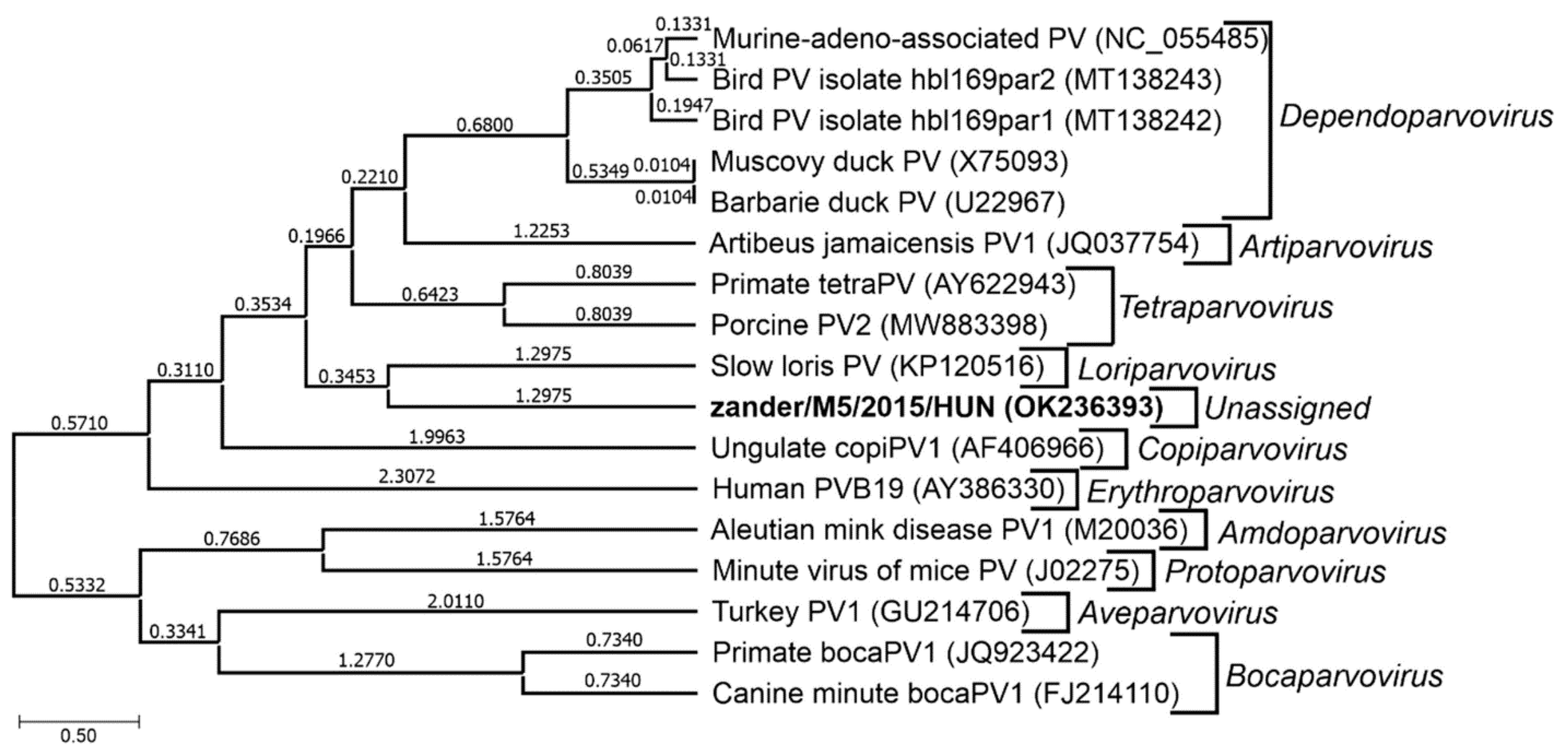
6.2. Novel Zander Parvovirus
6.3. Spawner-Isolated Mortality Virus (SMV)
6.4. Crangon crangon Parvo-Like Virus (CcPaLV)
7. Concluding Remarks
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pénzes, J.J.; Söderlund-Venermo, M.; Canuti, M.; Eis-Hübinger, A.M.; Hughes, J.; Cotmore, S.F.; Harrach, B. 2020. Reorganizing the family Parvoviridae: a revised taxonomy independent of the canonical approach based on host association. Archives of Virology 165, 2133-2146. [CrossRef]
- Cotmore, S.F.; Agbandje-McKenna, M.; Canuti, M.; Chiorini, J.A.; Eis-Hubinger, A.-M.; Hughes, J.; Mietzsch, M.; Modha, S.; Ogliastro, M.; Penzes, J.J.; Pintel, D.J.; Qiu, J.; Soderlund-Venermo, M.; Tattersall, P.; Tijssen, P. ; ICTV Report Consortium, 2019. ICTV Virus Taxonomy Profile: Parvoviridae. Journal of General Virology 100, 367-368. [CrossRef]
- MacLachlan, N.J.; Dubovi, E.J. 2017. Chapter 12, Parvoviridae. In N. J. MacLachlan and E. J. Dubovi (ed.), Fenner’s Veterinary Virology, 5th ed. Academic Press, pp. 245-257. [CrossRef]
- Qiu, J.; Söderlund-Venermo, M.; Young, N.S. 2017. Human parvoviruses. Clinical Microbiology Reviews 30, 43-113. [CrossRef]
- Reuter, G.; Boros, Á.; Mátics, R.; Altan, E.; Delwart, E.; Pankovics, P. 2022. A novel parvovirus (family Parvoviridae) in a freshwater fish, zander (Sander lucioperca). Archives of Virology 167, 1163-1167. [CrossRef]
- Fédière, G. 2000. Epidemiology and pathology of Densovirinae, p. 1-11. In S. Faisst and J. Rommelaere (eds), Parvoviruses. From molecular biology to pathology and therapeutic uses. Karger, Basel, Switzerland. https://horizon.documentation.ird.fr/exl-doc/pleins_textes/pleins_textes_7/b_fdi_59-60/010025973.pdf.
- Edgerton, B.F.; Webb, R.; Anderson, I.G.; Kulpa, E.C. 2000. Description of a presumptive hepatopancreatic reovirus, and a putative gill parvovirus, in the freshwater crayfish Cherax quadricarinatus. Diseases of Aquatic Organisms 41, 83-90. [CrossRef]
- Bowater, R.O.; Wingfield, M.; Fisk, A.; Condon, K.M.L.; Reid, A.; Prior, H.; Kulpa, E.C. 2002. A parvo-like virus in cultured redclaw crayfish Cherax quadricarinatus from Queensland, Australia. Diseases of Aquatic Organisms 50, 79-86. [CrossRef]
- Hewson, I.; Button, J.B.; Gudenkauf, B.M.; Miner, B.; Newton, A.L.; Gaydos, J.K.; Wynne, J.; Groves, C.L.; Hendler, G.; Murray, M.; Fradkin, S.; Breitbart, M.; Fahsbender, E.; Lafferty, K.D.; Kilpatrick, A.M.; Miner, C.M.; Raimondi, P.; Lahner, L.; Friedman, C.S.; Daniels, S.; Haulena, M.; Marliave, J.; Burge, C.A.; Eisenlord, M.E.; Harvell, C.D. 2014. Densovirus associated with sea-star wasting disease and mass mortality. Proceedings of the National Academy of Science, USA, 111, 17278-17283. [CrossRef]
- Richard, J.C.; Leis, E.; Dunn, C.D.; Agbalog, R.; Waller, D.; Knowles, S.; Putnam, J.; Goldberg, T.L. 2020. Mass mortality in freshwater mussels (Actinonaias Pectorosa) in the Clinch River, USA, linked to a novel densovirus. Sci. Rep. 10, 14498. [CrossRef]
- Kalagayan, H. et al. 1991. IHHN virus as an etiological factor in runt-deformity syndrome (RDS) of juvenile Penaeus vannamei cultured in Hawaii. Journal of the World Aquaculture Society 22, 235-243. [CrossRef]
- Lightner, D.V.; Redman, R.M.; Bell, T.A. 1983. Infectious hypodermal and hematopoietic necrosis, a newly recognized virus disease of penaeid shrimp. Journal of Invertebrate Pathology 42, 62-70. [CrossRef]
- Lightner, D.V.; Redman, R.M.; Bell, T.A.; Brock, J.A. 1983b. Detection of IHHN virus in Penaeus stylirostris and P. vannamei imported into Hawaii. Journal of World Mariculture Society 14, 212-225. [CrossRef]
- Tang, K.F.J.; Durand, S.; White, B.L.; Redman, R.M.; Pantoja, C.R.; Lightner, D.V. 2000. Postlarvae and juveniles of a selected line of Penaeus stylirostris are resistant to infectious hypodermal and hematopoietic necrosis virus infection. Aquaculture 190, 203-210. [CrossRef]
- Liu, W.; Zhang, Y.; Ma, J.; Jiang, N.; Fan, Y.; Zhou, Y.; Cain, K.; Yi, M.; Jia, K.; Wen, H.; Liu, W.; Guan, W.; Zeng, L. 2020. Determination of a novel parvovirus pathogen associated with massive mortality in adult tilapia. PLoS Pathogens 16, e1008765. [CrossRef]
- Yamkasem, J.; Tattiyapong, P.; Gorgoglione, B.; Surachetpong, W. 2021. Uncovering the first occurrence of Tilapia parvovirus in Thailand in tilapia during co-infection with Tilapia tilapinevirus. Transboundary and Emerging Diseases 00:1– 9. [CrossRef]
- Piewbang, C.; Tattiyapong, P.; Khemthong, M.; Lachroje, S.; Boonrungsiman, S.; Kasantikul, T.; Surachetpong, W.; Techangamsuwan, S. 2022. Dual infections of tilapia parvovirus (TiPV) and tilapia lake virus (TiLV) in multiple tilapia farms: Their impacts, genetic diversity, viral tropism, and pathological effects. Aquaculture 550, 737887. [CrossRef]
- Du, J.; Wang, W.; Chan, J.F.-W.; Wang, G.; Huang, Y.; Yi, Y.; Zhu, Z.; Peng, R.; Hu, X.; Wu, Y.; Zeng, J.; Zheng, J.; Cui, X.; Niu, L.; Zhao, W.; Lu, G.; Yuen, K.-Y.; Yin, F. 2019. Identification of a novel ichthyic parvovirus in marine species in Hainan Island, China. Frontiers in Microbiology 10:2815. [CrossRef]
- ViralZone <italic>Parvoviridae</italic>. Available at: https://viralzone.expasy.org/103?outline=all_by_species (accessed May 27, 2024).
- Gordon, J.C.; Angrick, E.J. 1986. Canine parvovirus: environmental effects on infectivity. Am J Vet Res. 47(7), 1464-1467.
- Cotmore, S.F.; Tattersall, P. 2014. Parvoviruses: Small does not mean simple. Annu Rev Virol. 1(1), 517-537. [CrossRef]
- Cotmore, S.F.; Tattersall, P. 2005. Genome packaging sense is controlled by the efficiency of the nick site in the right-end replication origin of parvoviruses minute virus of mice and LuIII. Journal of Virology 79, 2287-2300. [CrossRef]
- Jager, M.C.; Tomlinson, J.E.; Lopez-Astacio, R.A.; Parrish, C.P.; Van de Walle, G.R. 2021. Small but mighty: old and new parvoviruses of veterinary significance. Virology Journal 18, 210. [CrossRef]
- Chen, A.Y.; Qiu, J. 2010. Parvovirus infection-induced cell death and cell cycle arrest. Future Virology 5, 731-743. [CrossRef]
- Söderlund-Venermo, M. 2019. Emerging human parvoviruses: The rocky road to fame. The Annual Review of Virology 6, 71-91. [CrossRef]
- Kailasan, S.; Agbandje-McKenna, M.; Parrish, C.R. 2015. Parvovirus family conundrum: What makes a killer? Annu Rev Virol. 2, 425-450. [CrossRef]
- Deng, X.; Yan, Z.; Cheng, F.; Engelhardt, J.F.; Qiu, J. 2016. Replication of an autonomous human parvovirus in non-dividing human airway epithelium is facilitated through the DNA damage and repair pathways. PLOS Pathogens 12, e1005399. [CrossRef]
- Deng, X.; Xu, P.; Zou, W.; Shen, W.; Peng, J.; Liu, K.; et al. 2017. DNA damage signaling is required for replication of human bocavirus 1 DNA in dividing HEK293 cells. Journal of Virology 91, e01831-16. [CrossRef]
- Bucci, C.; Francoeur, M.; McGreal, J.; Smolowitz, R.; Zazueta-Novoa, V.; Wessel, G.M.; Gomez-Chiarri, M. 2017. Sea Star Wasting Disease in Asterias forbesi along the Atlantic Coast of North America. PLoS One. 12(12):e0188523. [CrossRef]
- Flegel, T.W. 2006. Detection of major penaeid shrimp viruses in Asia, a historical perspective with emphasis on Thailand. Aquaculture 258, 1-33. [CrossRef]
- Spann, K.M.; Aldard, R.D.; Hudson, D.A.; Dyecroft, S.B.; Jones, T.C.; Voigt, M.O.C. 1997. Hepatopancreatic parvo-like virus (HPV) of Penaeus japonicus cultured in Australia. Dis. Aquat. Org. 31, 239-241. [CrossRef]
- Manivannan, S.; Otta, S.K.; Karunasagar, I.; Karunasagar, I. 2002. Multiple viral infections in Penaeus monodon prawn post larvae in an Indian hatchery. Diseases of Aquatic Organisms 48, 233-236. [CrossRef]
- Pénzes, J.J.; de Souza, W.M.; Agbandje-McKenna, M.; Gifford, R.J. 2019. An ancient lineage of highly divergent parvoviruses infects both vertebrate and invertebrate hosts. Viruses 11, 525. [CrossRef]
- Vega-Heredia, S.; Mendoza-Cano, F.; Sanchez-Paz, A. ; 2012. The infectious hypodermal and haematopoietic necrosis virus: A brief review of what we do and do not know. Transboundary and Emerging Diseases 59, 95–105. [CrossRef]
- Hsieh, C.Y.; Chuang, P.C.; Chen, L.C.; Tu, C.; Chien, M.S.; Huang, K.C.; Kao, H.F.; Tung, M.C.; Tsai, S.S. ; 2006. Infectious hypodermal and haematopoietic necrosis virus (IHHNV) infections in giant freshwater prawn, Macrobrachium rosenbergii. Aquaculture 258, 73-79. [CrossRef]
- WOAH (World Organization for Animal Health). 2021. Infection with infectious hypodermal and haematopoietic necrosis virus. Chapter 2.2.4. In Manual of Diagnostic Tests for Aquatic Animals. pp 138-153. Available at: https://www.woah.org/fileadmin/Home/eng/Health_standards/aahm/current/2.2.04_IHHN.
- Miller, K.M.; Trudel, M.; Patterson, D.A.; Schulze, A.; Kaukinen, K.; Li, S.; Ginther, N.; Ming, T.; Tabata, A. ; 2013. Are smolts healthier in years of good ocean productivity? North Pacific Anadromous Fish Commission, Technical Report No. 9, 165-168. https://www.npafc.org/wp-content/uploads/TechReport9.
- Van Eynde, B.; Christiaens, O.; Delbare, D.; Shi, C.; Vanhulle, E.; Yinda, C.K.; Matthijnssens, J.; Smagghe, G. 2020. Exploration of the virome of the European brown shrimp (Crangon crangon). Journal of General Virology 101, 651-666. [CrossRef]
- Owens, L.; McElnea, C.; Snape, N.; Harris, L.; Smith, M. 2003. Prevalence and effect of spawner-isolated mortality virus on the hatchery phases of Penaeus monodon and P. merguiensis in Australia. Diseases of Aquatic Organisms 53, 101-106. [CrossRef]
- NCBI Taxonomy Browser. Available at: https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi.
- Cotmore, S.F.; Agbandje-McKenna, M.; Chiorini, J.A.; Mukha, D.V.; Pintel, D.J.; Qiu, J.; Soderlund-Venermo, M.; Tattersall, P.; Tijssen, P.; Gatherer, D. 2014. The family Parvoviridae. Archives of Virology 159, 1239-1247. [CrossRef]
- Edgerton, B.; Webb, R.; Wingfield, M. 1997. A systematic parvo-like virus in the freshwater crayfish Cherax destructor. Diseases of Aquatic Organisms 29:73-78. [CrossRef]
- Bochow, S. 2016. Characterisation of Cherax quadricarinatus densovirus: the first virus characterised from Australian freshwater crayfish. PhD thesis, James Cook University. Available at: https://researchonline.jcu.edu.au/48889/.
- Bochow, S.; Condon, K.; Elliman, J.; Owens, L. 2015. First complete genome of an Ambidensovirus; Cherax quadricarinatus densovirus, from freshwater crayfish Cherax quadricarinatus. Mar Genomics 3:305-312. [CrossRef]
- Gudenkauf, B.M.; Eaglesham, J.B.; Aragundi, W.M.; Hewson, I. 2014. Discovery of urchin-associated densoviruses (family Parvoviridae) in coastal waters of the Big Island, Hawaii. Journal of General Virology 95, 652-658. [CrossRef]
- Hewson, I.; Bistolas, K.S.I.; Quijano Cardé, E.M.; Button, J.B.; Foster, P.J.; Flanzenbaum, J.M.; Kocian, J.; Lewis, C.K. ; 2018. Investigating the complex association between viral ecology, environment, and northeast Pacific sea star wasting. Frontiers in Marine Science 5, 1-14. [CrossRef]
- Aquino, C.A.; Besemer, R.M.; DeRito, C.M.; Kocian, J.; Porter, I.R.; Raimondi, P.T.; Rede, J.E.; Schiebelhut, L.M.; Sparks, J.P.; Wares, J.P.; Hewson, I. 2021. Evidence that microorganisms at the animal-water interface drive sea star wasting disease. Frontiers in Microbiology 11, 610009. [CrossRef]
- Jackson, E.W.; Wilhelm, R.C.; Johnson, M.R.; Lutz, H.L.; Danforth, I.; Gaydos, J.K.; Hart, M.W.; Hewson, I. 2021. Diversity of sea star-associated densoviruses and transcribed endogenous viral elements of densovirus origin. Journal of Virology 95, e01594-20. [CrossRef]
- Eisenlord, M.E.; Groner, M.L.; Yoshioka, R.M.; Elliott, J.; Maynard, J.; Fradkin, S.; Turner, M.; Pyne, K.; Rivlin, N.; van Hooidonk, R.; Harvell, C.D. 2016. Ochre star mortality during the 2014 wasting disease epizootic: role of population size structure and temperature. Philosophical Transactions Royal Society London Series B Biological Science 371, 20150212. [CrossRef]
- La Fauce, K.A.; Elliman, J.; Owens, L. 2007. Molecular characterisation of hepatopancreatic parvovirus (PmergDNV) from Australian Penaeus merguiensis. Virology 5, 397-403. [CrossRef]
- Tang, K.F.J.; Pantoja, C.; Lightner, D.V. 2008. Nucleotide sequence of a Madagascar hepatopancreatic parvovirus (HPV) and comparison of genetic variation among geographic isolates. Diseases of Aquatic Organisms 80, 105-112. [CrossRef]
- Jeeva, S.; Kang, S.W.; Lee, Y.S.; Jang, I.K.; Seo, H.C.; Choi, T.J. 2012. Complete nucleotide sequence analysis of a Korean strain of hepatopancreatic parvovirus (HPV) from Fenneropenaeus chinensis. Virus Genes 44, 89-97. [CrossRef]
- Safeena, M.P.; Rai, P.; Karunasagar, I. 2012. Molecular biology and epidemiology of Hepatopancreatic parvovirus of Penaeid Shrimp. Indian Journal of Virology 23, 191-202. [CrossRef]
- Gangnonngiw, W.; Kiatpathomchai, W.; Sriurairatana, S.; Laisutisan, K.; Chuchird, N.; Limsuwan, C.; Flegel, T.W. 2009. A parvo-like virus in the hepatopancreas of freshwater prawns Macrobrachium rosenbergii cultivated in Thailand. Dis. Aquat. Org. 85, 167-173. [CrossRef]
- Srisala, J.; Thaiue, D.; Sanguanrut, P.; Aldama-Cano, D.J.; Flegel, T.W.; Sritunyalucksana, K. 2021. Potential universal PCR method to detect decapod hepanhamaparvovirus (DHPV) in crustaceans. Aquaculture 541, 736782. [CrossRef]
- Anderson, I.G.; Law, A.T.; Shariff, M.; Nash, G. 1990. A parvo-like virus in the giant freshwater prawn, Macrobrachium rosenbergii. Journal of Invertebrate Pathology 55, 447-449. [CrossRef]
- Lightner, D.V.; Redman, R.M.; Poulos, B.T.; Mari, J.L.; Bonami, J.R.; Shariff, M. 1994. Distinction of HPV-type virus in Penaeus chinensis and Macrobrachium rosenbergii using a DNA probe. Asian Fisheries Science 7, 267-272. [CrossRef]
- Lee, C.-F.; Chang, Y.-C.; Chiou, H.-Y.; Chang, H.-W. 2022. Concurrent infection of a novel genotype of hepatopancreatic parvovirus and Enterocytozoon hepatopenaei in Penaeus vannamei in Taiwan. Journal of Fish Diseases, 45, 1201–1210. [CrossRef]
- Chong, Y.C.; Loh, H. 1984. Hepatopancreas chlamydial and parvoviral infections of farmed marine prawns in Singapore. Singapore Veterinary Journal (Singapore) 9, 51-56.
- Lightner, D.V. ; 1996. A Handbook of Pathology and Diagnostic Procedures for Diseases of Penaeid Shrimp. World Aquaculture Society, Baton Rouge, LA. https://lib.ugent. 0021. [Google Scholar]
- Thitamadee, S.; Prachumwat, A.; Srisala, J.; Jaroenlak, P.; Salachan, P.V.; Sritunyalucksana, K.; Flegel, T.W.; Itsathitphaisarn, O. 2016. Review of current disease threats for cultivated penaeid shrimp in Asia. Aquaculture 452, 69-87. [CrossRef]
- Singaravel, V.; Gopalakrishnan, A.; Martin, G.G. ; 2021. Multiple infections of Enterocytozoon hepatopenaei and Hepatopancreatic parvovirus in pond-reared Penaeus vannamei in India. Aquaculture 545, 737232. [CrossRef]
- Dhar, A.K.; Cruz-Flores, R.; Caro, L.F.A.; Siewiora, H.M.; Jory, D. 2019. Diversity of single-stranded DNA containing viruses in shrimp. Virus Dis. 30, 43-57. [CrossRef]
- Flegel, T.; Thamavit, V.; Pasharawipas, T.; Alday-Sanz, V. 1999. Statistical correlation between severity of hepatopancreatic parvovirus infection and stunting of farmed black tiger shrimp (Penaeus monodon). Aquaculture 174, 197-206. [CrossRef]
- Chayaburakul, K.; Nash, G.; Pratanpipat, P.; Sriurairatana, S.; Withyachumnarnkul, B. 2004. Multiple pathogens found in growth-retarded black tiger shrimp Penaeus monodon cultivated in Thailand. Diseases of Aquatic Organisms 60, 89-96. [CrossRef]
- Chantanachookin, C.; Boonyaratanapalin, S.; Kasornchandra, J.; Direkbusarakom, S.; Ekpanithanpong, U.; Supamataya, K.; Siurairatana, S.; Flegel, T.W. 1993. Histology and ultrastructure reveal a new granulosis-like virus in Penaeus monodon affected by ‘‘yellowhead’’ disease. Diseases of Aquatic Organisms 17, 145-157. [CrossRef]
- Sukhumsirichart, W.; Attasart, P.; Boonsaeng, V.; Panyim, S. 2006. Complete nucleotide sequence and genomic organization of hepatopancreatic parvovirus (HPV) of Penaeus monodon. Virology 346, 266-277. [CrossRef]
- Cervellione, F.; McGurk, C.; Berger Eriksen, T.; Van Den Broeck, W. 2017. Use of computer-assisted image analysis for semi-quantitative histology of the hepatopancreas in whiteleg shrimp Penaeus vannamei (Boone). Journal of Fish Diseases 40, 1223-1234. [CrossRef]
- Phromjai, J.; Boonsaeng, V.; Withyachumnarnkul, B.; Flegel, T.W. 2002. Detection of hepatopancreatic parvovirus in Thai shrimp Penaeus monodon by in situ hybridization, dot blot hybridization and PCR amplification. Diseases of Aquatic Organisms 51, 227-232. [CrossRef]
- Attasart, P.; Kaewkhaw, R.; Chimwai, C.; Kongphom, U.; Panyim, S. 2011. Clearance of Penaeus monodon densovirus in naturally pre-infected shrimp by combined ns1 and vp dsRNAs. Virus Res. 159, 79-82. [CrossRef]
- Dhar, A.K.; Robles-Sikisaka, R.; Saksmerprome, V.; Lakshman, D.K. 2014. Biology, genome organization, and evolution of parvoviruses in marine shrimp. Advances in Virus Research 89, 85-139. [CrossRef]
- Lightner, D.V.; Redman, R.M. 1998. Shrimp diseases and current diagnostic methods. Aquaculture 164,201-220. [CrossRef]
- Tang, K.F.J.; Poulos, B.T.; Wang, J.; Redman, R.M.; Shih, H.-H.; Lightner, D.V. 2003. Geographic variations among infectious hypodermal and hematopoietic necrosis virus (IHHNV) isolates and characteristics of their infection. Diseases of Aquatic Organisms 53:91-99. [CrossRef]
- Yu, J.Y.; Yang, N.; Hou, Z.H.; Wang, J.J.; Li, T.; Chang, L.R.; Fang, Y.; Yan, D.C. 2021. Research progress on hosts and carriers, prevalence, virulence of infectious hypodermal and hematopoietic necrosis virus (IHHNV). Journal of Invertebrate Pathology 183, 107556. [CrossRef]
- Tang, K.F.J.; Lightner, D.V. 2006. Infectious hypodermal and hematopoietic necrosis virus (IHHNV)-related sequences in the genome of the black tiger prawn Penaeus monodon from Africa and Australia. Virus Res 118:185–191. [CrossRef]
- Rai, P.; Pradeep, B.; Safeena, M.P.; Karunasagar, I.; Karunasagar, I. 2009. Simultaneous presence of infectious hypodermal and hematopoietic necrosis virus (IHHNV) and type A virus-related sequence in Penaeus monodon from India. Aquaculture 295, 168-174. [CrossRef]
- Saksmerprome, V.; Jitrakorn, S.; Chayaburakul, K.; Laiphrom, S.; Boonsua, K.; Flegel, T.W. 2011. Additional random, single to multiple genome fragments of Penaeus stylirostris densovirus in the giant tiger shrimp genome have implications for viral disease diagnosis. Virus Research 160, 180-190. [CrossRef]
- Rusaini, La Fauce, K.A.; Elliman, J.; Bowater, R.O.; Owens, L. 2013. Endogenous Brevidensovirus-like elements in Cherax quadricarinatus: Friend or foe? Fish & Shellfish Immunology 396-399, 136-145. [CrossRef]
- Alcivar-Warren, A.; Bao, W.; Tang, K.F. 2022. Endogenous viral elements (EVE) of <italic>Decapod penstylhamaparvovirus 1</italic> (Infectious hypodermal and hematopoietic necrosis virus, IHHNV) – Implications for shrimp diagnosis. Aquaculture 2022, February 28 - March 4, 2022, San Diego, California. Abstract available at this link: https://www.was.org/Meeting/Program/PaperDetail/158479. (Accessed 7 June 2022).
- Salcedo-Mejía, L.A.; Durán-Ramirez, Y.; Velazco-Peña, R.Z.; Pinto, J.A.; Rebaza-Caballero, A. 2021. Near-complete genome sequences of 12 Peruvian strains of infectious hypodermal and hematopoietic necrosis virus infecting the shrimp Penaeus vannamei. Microbiol Resour Announc 10:e00169-21. [CrossRef]
- CEFAS (Centre for Environment Fisheries and Aquaculture Science). 2020. Infectious hypodermal and hematopoietic necrosis virus. Available at: https://www.cefas.co.uk/international-database-on-aquatic-animal-diseases/disease-data/?id=39 (accessed 29 June 2022).
- Bajaña, L.; Betancourt, I.; Bayot, B. 2022. Complete coding genome sequence of infectious hypodermal and hematopoietic necrosis virus isolated from Penaeus (Litopenaeus) vannamei shrimp in Ecuador. Microbiology Resource Announcements 11, 4. [CrossRef]
- Bell, T.A.; Lightner, D.V. 1984. IHHN virus: infectivity and pathogenicity studies in Penaeus stylirostris and Penaeus vannamei. Aquaculture 38, 185-194. [CrossRef]
- Chai, C.; Liu, Y.; Xia, X.; Wang, H.; Pan, Y.; Yan, S.; Wang, Y. 2016. Prevalence and genomic analysis of infectious hypodermal and hematopoietic necrosis virus (IHHNV) in Litopenaeus vannamei shrimp farmed in Shanghai, China. Arch Virol 161:3189–3201. [CrossRef]
- Lightner, D.V.; Redman, R.M.; Poulos, B.T.; Nunan, L.M.; Mari, J.L.; Hasson, K.W. 1997. Risk of spread of penaeid shrimp viruses in the Americas by the international movement of live and frozen shrimp. Revue Scientifique et Technique 16, 146-160. [CrossRef]
- Hazreen-Nita, M.K.; Kua, B.C.; Bhassu, S.; Othman, R.Y. ; 2012. Detection and genetic profiling of infectious hypodermal and haematopoietic necrosis virus (IHHNV) infections in wild berried freshwater prawn, Macrobrachium rosenbergii collected for hatchery production. Mol. Biol. Rep. 39, 3785-3790. [CrossRef]
- Chen, B.K.; Dong, Z.; Liu, D.P.; Yan, Y.B.; Pang, N.Y.; Nian, Y.Y.; Yan, D.C. ; 2017. Infectious hypodermal and haematopoietic necrosis virus (IHHNV) infection in freshwater crayfish Procambarus clarkii. Aquaculture 477, 76-79. [CrossRef]
- Chen, B.K.; Dong, Z.; Pang, N.Y.; Nian, Y.Y.; Yan, D.C. ; 2018. A novel real-time PCR approach for detection of infectious hypodermal and haematopoietic necrosis virus (IHHNV) in the freshwater crayfish Procambarus clarkii. J. Invertebr. Pathol. 157, 100-103. [CrossRef]
- Lee, C.; Choi, S.-K.; Jeon, H.J.; Lee, S.H.; Kim, Y.K.; Park, S.; Han, S.-Y.; Bae, S.; Kim, J.H.; Han, J.E. 2021. Detection of infectious hypodermal and hematopoietic necrosis virus (IHHNV, Decapod Penstylhamaparvovirus 1) in commodity Red Claw crayfish (Cherax quadricarinatus) imported into South Korea. Journal of Marine Science and Engineering 9, 856. [CrossRef]
- Yang, B.; Song, X.L.; Huang, J.; Shi, C.Y.; Liu, L. 2007. Evidence of existence of infectious hypodermal and hematopoietic necrosis virus in penaeid shrimp cultured in China. Vet. Microbiol. 120, 63-70. [CrossRef]
- Cavalli, L.S.; Batista, C.R.; Nornberg, B.F.S.; Mayer, F.Q.; Seixas, F.K.; Romano, L.A.; Marins, L.F.; Abre, P.C. 2013. Natural occurrence of White spot syndrome virus and Infectious hypodermal and hematopoietic necrosis virus in Neohelice granulata crab. J. Invertebr. Pathol. 114, 86-88. [CrossRef]
- Macías-Rodríguez, N.A.; Manon-Ríos, N.; Romero-Romero, J.L.; Camacho-Beltrán, E.; Magallanes-Tapia, M.A.; Leyva-López, N.E.; Hernández-López, J.; Magallón-Barajas, F.J.; Perez-Enriquez, R.; Sánchez-González, S.; Méndez-Lozano, J. 2014. Prevalence of viral pathogens WSSV and IHHNV in wild organisms at the Pacific Coast of Mexico. J. Invertebr. Pathol. 116, 8-12. [CrossRef]
- Wei, Y.W.; Fan, D.D.; Chen, J. 2017. The mussel Mytilus edulis L. as an important reservoir of infectious hypodermal and hematopoietic necrosis virus (IHHNV). Aquaculture Research 48, 758-759. [CrossRef]
- Rai, P.; Safeena, M.P.; Krabsetsve, K.; La-Fauce, K.; Owens, L.; Karunasagar, I. ; 2012. Genomics, molecular epidemiology and diagnostics of infectious hypodermal and hematopoietic necrosis virus. Indian Journal of Virology 23, 203-214. [CrossRef]
- Sellars, M.J.; Cowley, J.A.; Musson, D.; Rao, M.; Menzies, M.L.; Coman, G.J.; Murphy, B.S. 2019. Reduced growth performance of black tiger shrimp (Penaeus monodon) infected with infectious hypodermal and hematopoietic necrosis virus. Aquaculture 499:160-166. [CrossRef]
- DAWE (Australian Government Department of Agriculture, Water and the Environment). 2020. Infection with infectious hypodermal and haematopoietic necrosis virus (IHHNV). Available at: https://www.agriculture.gov.au/sites/default/files/documents/infection-infectious-hypodermal-haematopoietic-necrosis-virus.pdf.
- Lightner, D.V. ; 2011. Virus diseases of farmed shrimp in the Western Hemisphere (the Americas): A review. Journal of Invertebrate Pathology 106, 110-130. [CrossRef]
- Jagadeesan, V.; Praveena, P.E.; Otta, S.K.; Jithendran, K.P. 2019. Classical runt deformity syndrome cases in farmed Penaeus vannamei along the east coast of India. Journal of Coastal Research, Special Issue No. 86, 107-111. [CrossRef]
- Zhang, C.; Yuan, J.F.; Shi, Z.L. ; 2007. Molecular epidemiological investigation of infectious hypodermal and hematopoietic necrosis virus and Taura syndrome virus in Penaeus vannamei cultured in China. Virologica Sinica 22, 380-388. [CrossRef]
- Nian, Y.-Y.; Chen, B.-K.; Wang, J.-J.; Zhong, W.-T.; Fang, Y.; Li, Z.; Zhang, Q.-S.; Yan, D.-C. 2020. Transcriptome analysis of Procambarus clarkii infected with infectious hypodermal and haematopoietic necrosis virus. Fish and Shellfish Immunology 98, 766-772. [CrossRef]
- Lee, D.; Yu, Y.-B.; Choi, J.-H.; Jo, A.-H.; Hong, S.-M.; Kang, J.-C.; Kim, J.-H. 2022. Viral shrimp diseases listed by the OIE: A review. Viruses 14, 585. [CrossRef]
- Bonnichon, V.; Lightner, D.V.; Bonami, J.R. 2006. Viral interference between infectious hypodermal and hematopoietic necrosis virus and white spot syndrome virus in Litopenaeus vannamei. Diseases of Aquatic Organisms 72, 179-184. [CrossRef]
- Molthathong, S.; Jitrakorn, S.; Joyjinda, Y.; Boonchird, C.; Witchayachamnarnkul, B.; Pongtippatee, P.; Flegel, T.; Saksmerprome, V. 2013. Persistence of Penaeus stylirostris densovirus delays mortality caused by white spot syndrome virus infection in black tiger shrimp (Penaeus monodon). BMC Veterinary Research 9, 33. [CrossRef]
- Robles-Sikisaka, R.; Bohonak, A.J.; McClenaghan, L.R., Jr.; Dhar, A.K. 2010. Genetic signature of rapid IHHNV (Infectious hypodermal and hematopoietic necrosis virus) expansion in wild penaeus shrimp populations. PLoS ONE 5, e11799. [CrossRef]
- Tang, K.F.; Navarro, S.A.; Lightner, D.V. 2007. PCR assay for discriminating between infectious hypodermal and hematopoietic necrosis virus (IHHNV) and virus-related sequences in the genome of Penaeus monodon. Diseases of Aquatic Organisms 74, 165-170. [CrossRef]
- Dhar, A.K.; Roux, M.M.; Klimpel, K.R. 2001. Detection and quantification of infectious hypodermal and hematopoietic necrosis virus and white spot virus in shrimp using real-time quantitative PCR and SYBR Green chemistry. Journal of Clinical Microbiology 39, 2835-2845. [CrossRef]
- Silva, D.C.D.; Nunes, A.R.D.; Teixeira, D.I.A.; Lima, J.P.M.S.; Lanza, D.C.F. 2014. Infectious hypodermal and hematopoietic necrosis virus from Brazil: sequencing, comparative analysis and PCR detection. Virus Research 189, 136-146. [CrossRef]
- Bower, S.M. 1996. Synopsis of infectious diseases and parasites of commercially exploited shellfish: Infectious hypodermal and haematopoietic necrosis virus (IHHNV) of penaeid shrimp. Available at: https://www.dfo-mpo.gc.ca/science/aah-saa/diseases-maladies/ihnvsp-eng.html (accessed 29 June 2022).
- Dong, H.T.; Sangpo, P.; Dien, L.T.; Mai, T.T.; Linh, N.V.; Del-Pozo, J.; Salin, K.R.; Senapin, S. 2022. Usefulness of the pancreas as a prime target for histopathological diagnosis of Tilapia parvovirus (TiPV) infection in Nile tilapia, Oreochromis niloticus. Journal of Fish Diseases, 45, 1323-1331. [CrossRef]
- Rajendran, K.V.; Sood, N.; Rao, B.M.; Valsalam, A.; Bedekar, M.K.; Jeena, K.; Pradhan, P.K.; Paria, A.; Swaminathan, T.R.; Verma, D.K.; Sood, N.K. 2023. Widespread occurrence of Tilapia parvovirus in farmed Nile tilapia Oreochromis niloticus from India. Journal of Fish Diseases, 00, 1–11. [CrossRef]
- Hargitai, R.; Pankovics, P.; Boros, Á.; Mátics, R.; Altan, E.; Delwart, E.; Reuter, G. 2021. Novel picornavirus (family Picornaviridae) from freshwater fishes (Perca fluviatilis, Sander lucioperca and Ameiurus melas) in Hungary. Archives of Virology 166, 2627-2632. [CrossRef]
- Suchard, M.A.; Lemey, P.; Baele, G.; Ayres, D.L.; Drummond, A.J.; Rambaut, A. 2018. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol 4, vey016. [CrossRef]
- Fraser, C.A.; Owens, L. 1996. Spawner-isolated mortality virus from Australia Penaeus monodon. Diseases of Aquatic Organisms 27, 141-148. [CrossRef]
- Owens, L.; Haqshenas, G.; McElnea, C.; Coelen, R. 1998. Putative spawner-isolated mortality virus associated with mid-crop mortality syndrome in farmed Penaeus monodon from northern Australia. Diseases of Aquatic Organisms 34, 177-185. [CrossRef]
- Owens, L.; McElnea, C. 2000. Natural infection of the redclaw crayfish Cherax quadricarinatus with presumptive spawner-isolated mortality virus. Diseases of Aquatic Organisms 40, 219-223. [CrossRef]
| Subfamily and Genus | Species1 | Virus common name (abbreviation) | Sampled host | Geographical area | Clinical disease | Reference |
| Densovirinae, Aquambidensovirus | Decapod aquambidensovirus 1 | Cherax quadricarinatus densovirus [previously known as Cherax quadricarinatus parvo-like virus (CqPlV)]; Cherax destructor systemic parvo-like virus (CdSPV) | redclaw crayfish Cherax quadricarinatus and Cherax destructor | Australia | Chronic mortality or mass mortality | [7,8] |
| Asteroid aquambidensovirus 1 | Sea star-associated densovirus (SSaDV) | sea stars and sea urchins | West coast of USA and Atlantic coast of North America | Originally associated with sea star wasting syndrome (SSWS) | [9,29] | |
| Clinch densovirus 1 | Freshwater mussels | Clinch River, Virginia, and Tennessee, USA | Episodic mass mortality | [10] | ||
| Hamaparvovirinae, Hepanhamaparvovirus | Decapod hepanhamaparvovirus 1 | Fenneropenaeus chinensis hepatopancreatic densovirus [previously known as Hepatopancreatic parvovirus (HPV)] | Prawns & Shrimp | Widespread | Reduced growth in juvenile shrimp; Mortalities during the larval stages. | [30-32] |
| Hamaparvovirinae, Ichtahamaparvovirus | Syngnathid ichthamaparvovirus 1 | Syngnathus scovelli chapparvovirus |
gulf pipefish | Gulf of Mexico and parts of South America | Not known | [33] |
| Tilapia parvovirus (TiPV) | Nile tilapia | China and Thailand | Mass morbidity and 60-70% mortality in adult Nile tilapia (500-600 g); mass mortality of 50-75% in juvenile red hybrid tilapia (10-30 g to 300-800 g). | [15-17] | ||
| unclassified Ichthamaparvovirus | ||||||
| Hamaparvovirinae, Penstylhamaparvovirus | Decapod penstylhamaparvovirus 1 | Penaeus stylirostris penstyldensovirus (PstDV 1&2); Penaeus monodon penstyldensovirus (PmoPDV 1&2) [previously known as Infectious hypodermal & hematopoietic necrosis virus (IHHNV)] | Prawns & Shrimp | Widespread | IHHN; 80-100% mortality in postlarvae and juveniles of Penaeus (Litopenaeus) stylirostris and postlarvae of Macrobrachium rosenbergii; runt-deformity syndrome (RDS) in juvenile of P. vannamei and P. monodon |
[34-36] |
| Unassigned | Novel salmon parvovirus | Sockeye salmon | BC-Canada | Not known | [37] | |
| Unassigned in subfamily Parvovirinae | Novel zander parvovirus (zander/M5/2015/HUN, OK236393) | Zander or pikeperch (Sander lucioperca) | Hungary | Not known | [5] | |
| Unassigned | Crangon crangon parvo-like virus 1 (CcPaLV 1) | Molluscs | Europe | Not known | [38] | |
| Unassigned | Spawner-isolated mortality virus (SMV) | Freshwater crayfish | Northern Australia | Mortalities in broodstock of Penaeus monodon with mid-crop mortality syndrome on grow-out farms | [39] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





