Submitted:
11 June 2024
Posted:
12 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results and Discussion
2.1. Green Synthesis of CuO Nanoparticles from Macroalgae
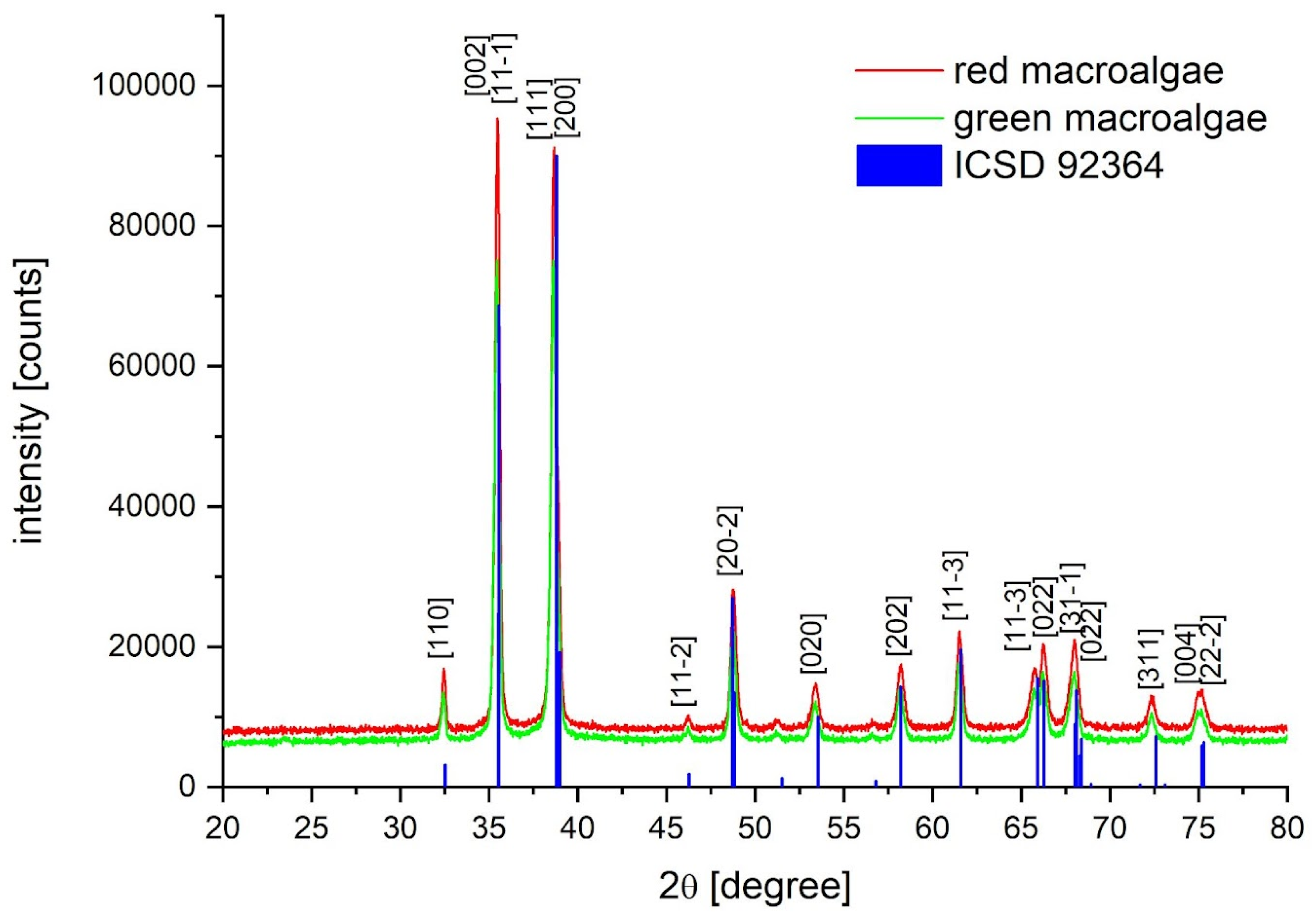
2.2. XRD Nanoparticle Characterization
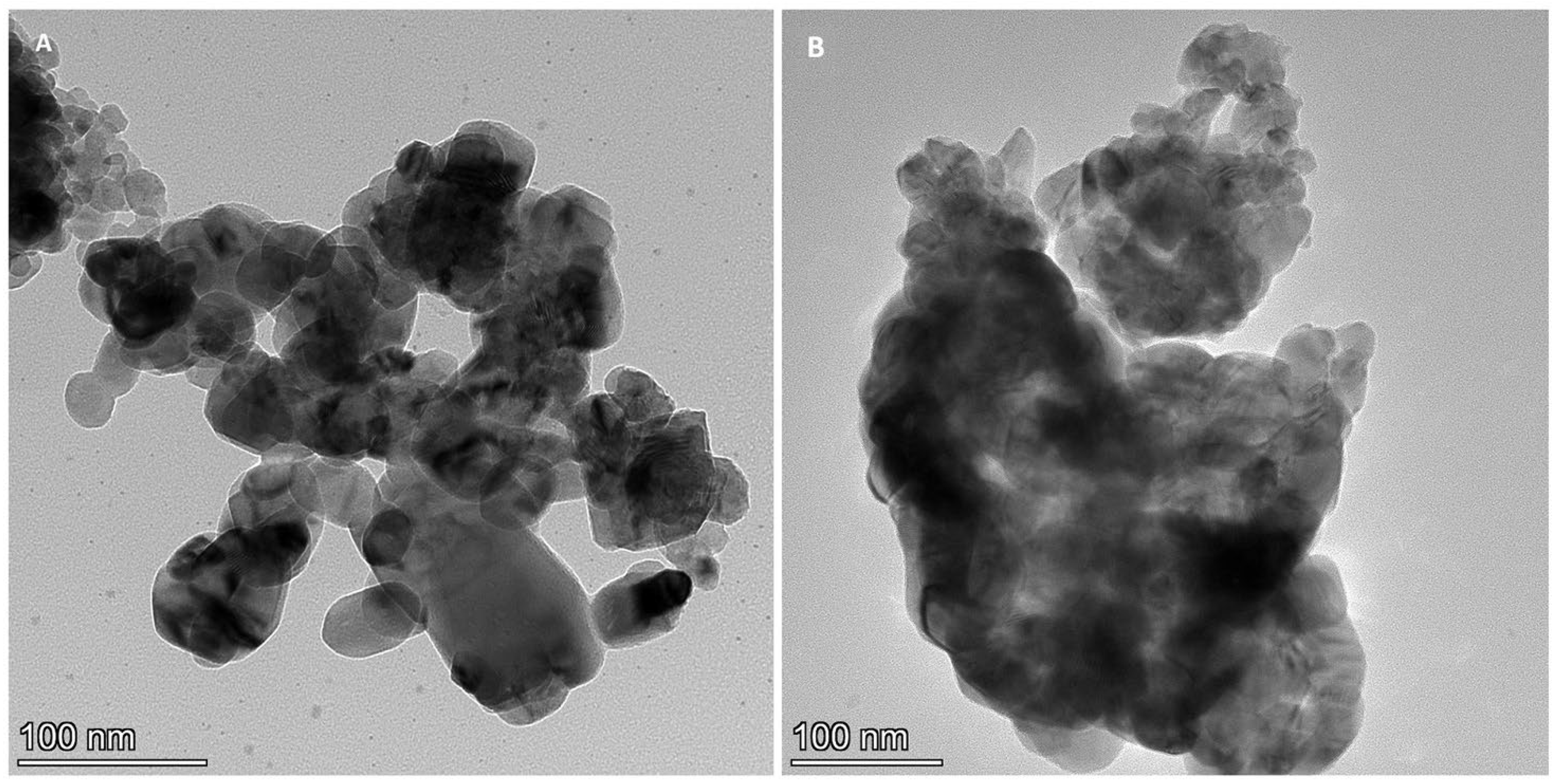
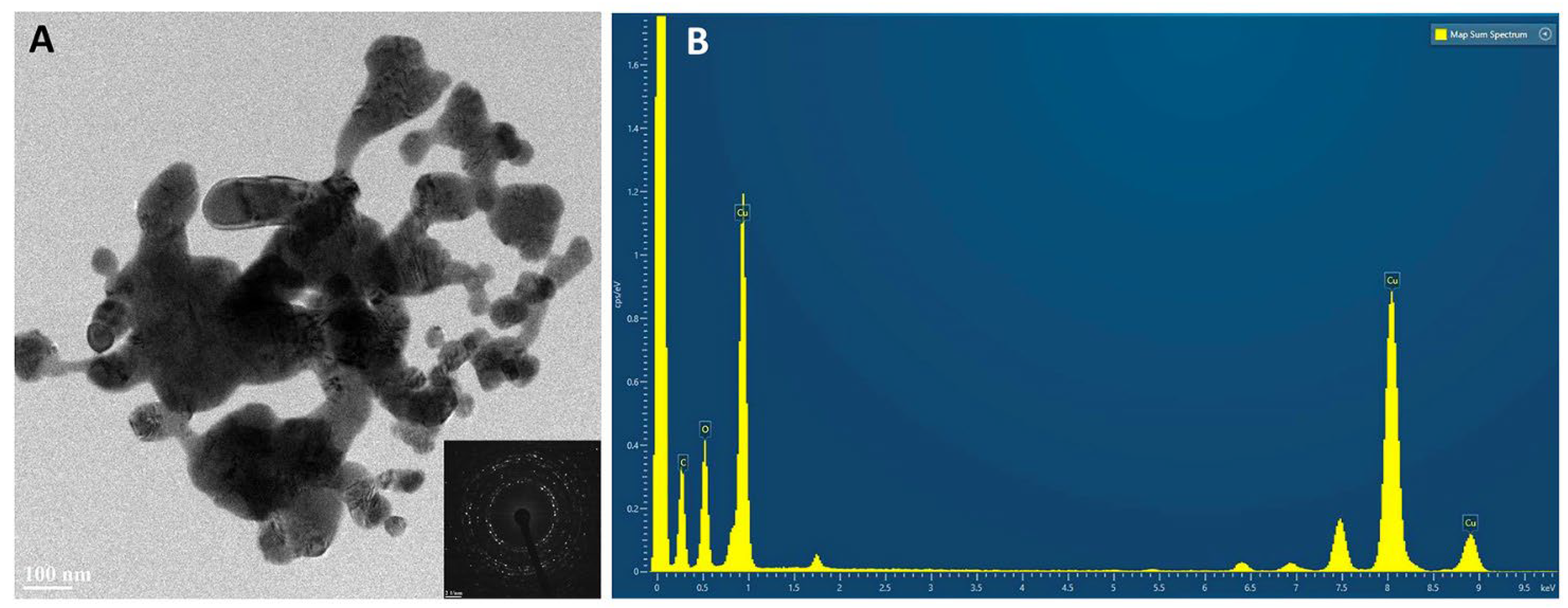
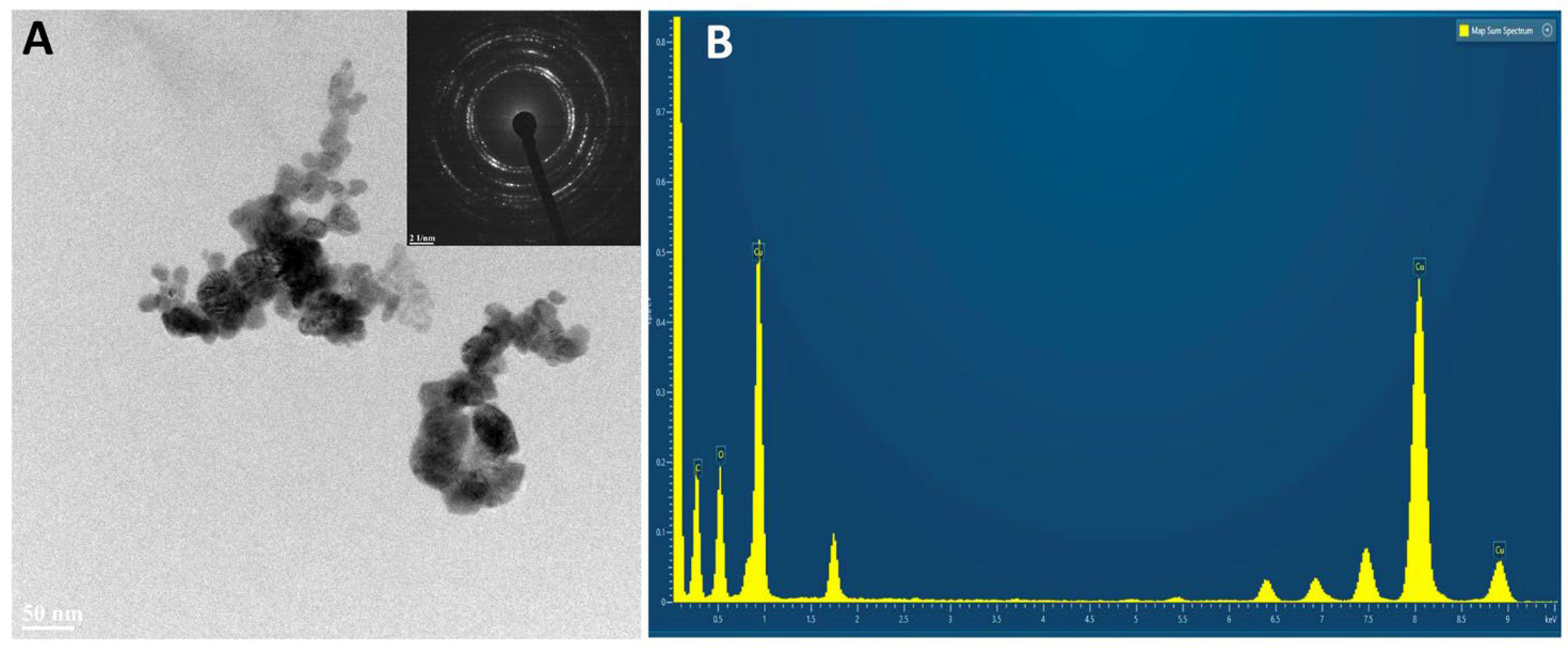
2.3. HR TEM/EDX
2.4. FTIR Analyses
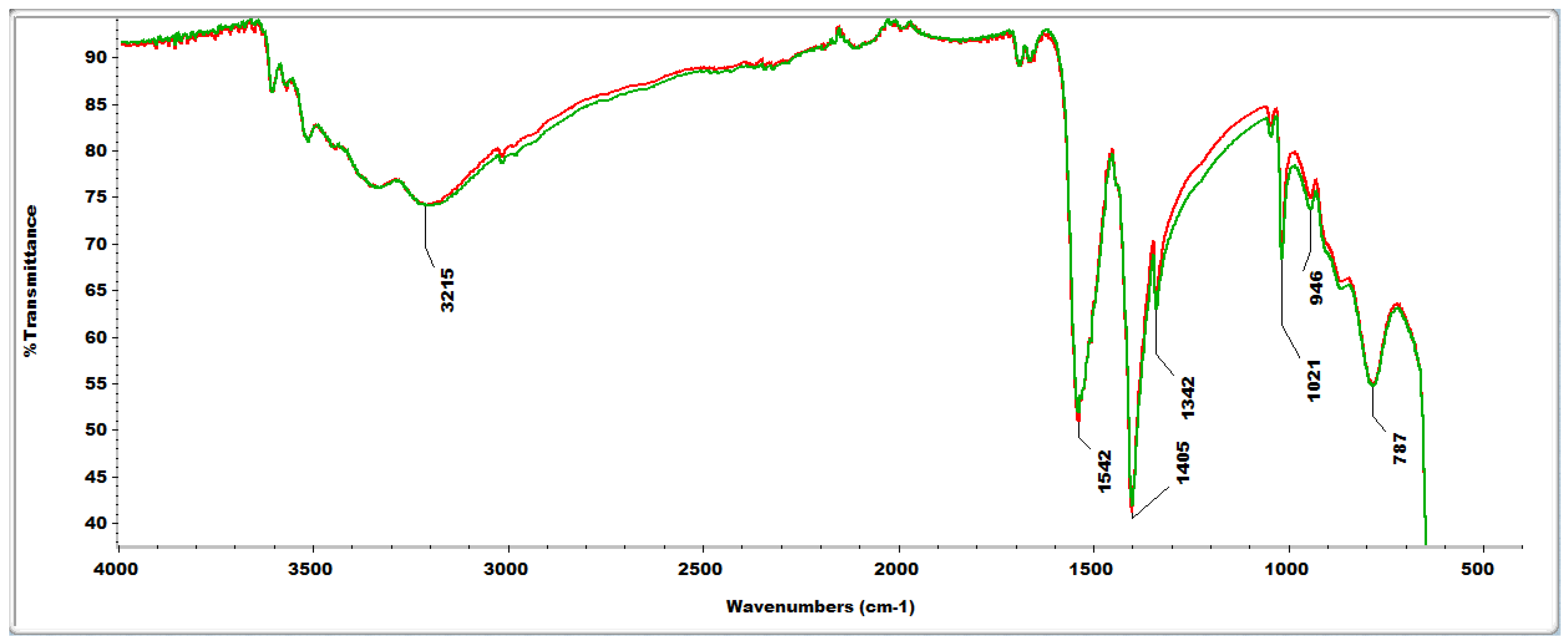
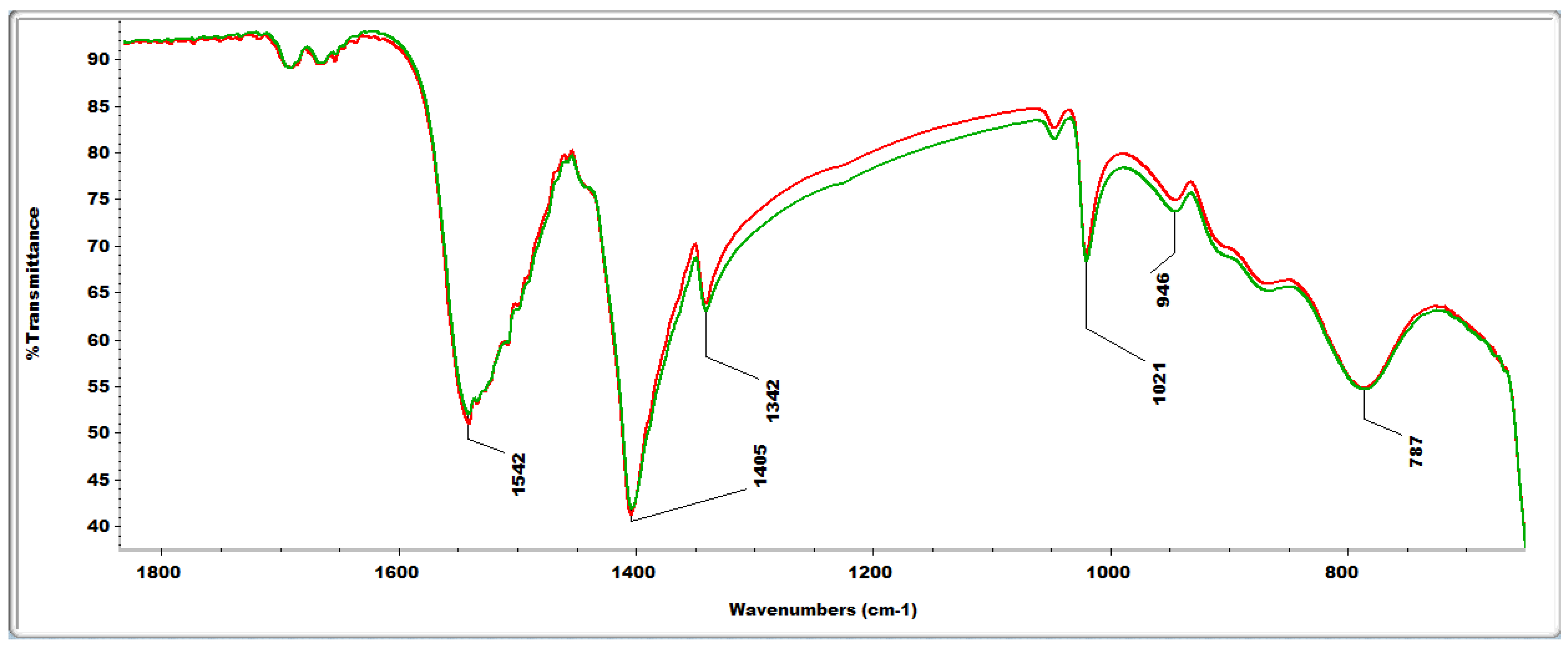
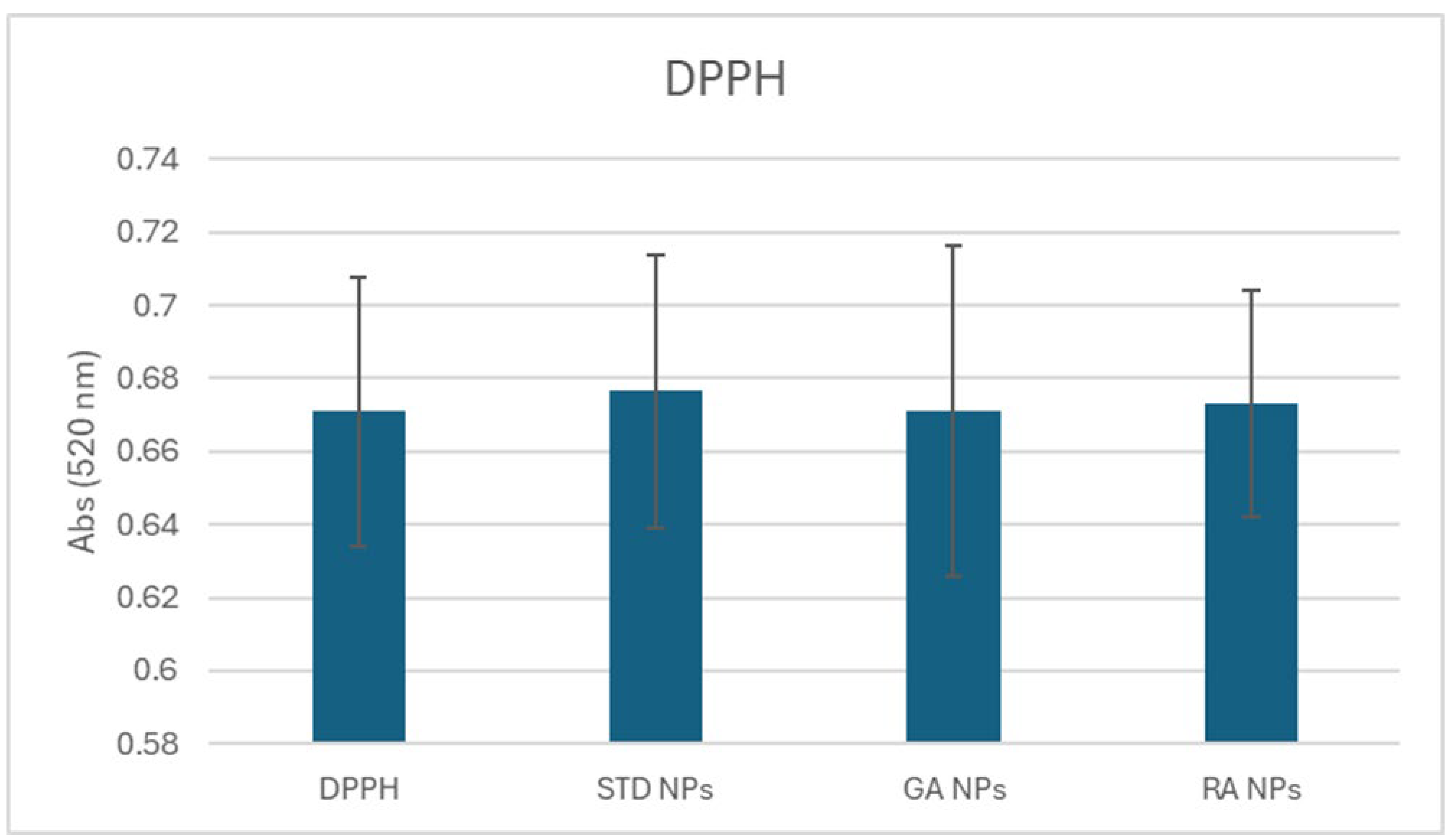
2.4. Reactive Oxygen Species (ROS) Scavenging Capacity
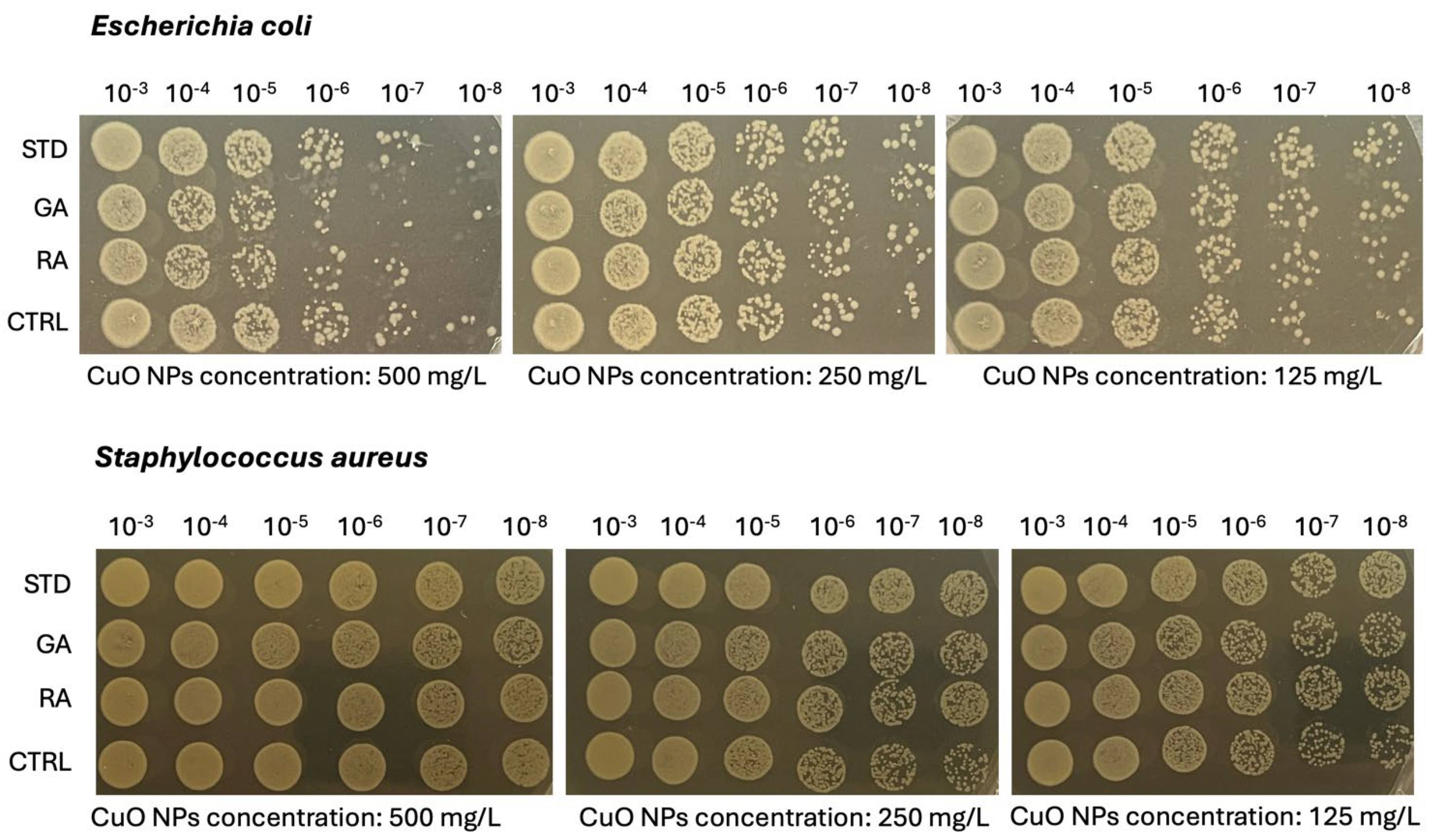
2.5. Microbiological Analysis on Copper Oxide (CuO) NPs
3. Materials and Methods
3.1. Macroalgae Biomass Collection and Extraction Process
3.2. NPs Synthesis
3.3. Nanoparticles Characterization
3.3.1. XRD
3.3.2. HR TEM/EDX
3.3.3. Zeta Potential, Particle Size and Dissolution
3.3.4. FTIR
3.3.5. DPPH Assay for Free Radical Scavenging Capacity
3.3.6. Microbiological Analysis on Copper Oxide (CuO) Nanoparticles (NPs)
3.3.7. Flow Cytometry on Yeasts
4. Conclusions
Supplementary Materials
Acknowledgments
References
- Abid, N.; Khan, A.M.; Shujait, S.; Chaudhary, K.; Ikram, M.; Imran, M.; Haider, J.; Khan, M.; Khan, Q.; et al. Synthesis of nanomaterials using various top-down and bottom-up approaches, influencing factors, advantages, and disadvantages: A review. Advances in Colloid and Interface. 2022, 18, 156–166. [Google Scholar] [CrossRef]
- Abidi, N. FTIR Microspectroscopy Selected Emerging Applications, Springer Nature Switzerland AG 2021, ISBN 978-3-030-84424-0.
- Barciela, P.; Carpena, M.; Li, N.Y.; Liu, C.; Jafari, S.M.; Simal-Gandara, J.; Prieto, M.A. Macroalgae as biofactories of metal nanoparticles; biosynthesis and food applications. Advances in Colloid and Interface Science 2023, 311, 102829. [Google Scholar] [CrossRef]
- Castro, L.; Blázquez, M.L.; Muñoz, J.A.; González, F.; Ballester, A. Biological synthesis of metallic nanoparticles using algae. IET Nanobiotechnology 2013, 7, 109–116. [Google Scholar] [CrossRef]
- Chatterjee, A.; Kwatra, N.; Abraham, J. Chapter 8—Challenges and Prospects Micro and Nano Technologies 2020, 143-157.
- Chellapandian, C.; Ramkumar, B.; Puja, P.; Shanmuganathan, R.; Pugazhendhi, A.; Kumar, P. Gold nanoparticles using red seaweed Gracilaria verrucosa: Green synthesis, characterization and biocompatibility studies. Process Biochem. 2019, 80, 58–63. [Google Scholar] [CrossRef]
- Coates, J. Interpretation of Infrared Spectra, A Practical Approach, in Encyclopedia of Analytical Chemistry, RA. Meyers (Ed.), 10815–10837, 2000, John Wiley & Sons Ltd., Chichester.
- De Almeida, C.L.F.; Falc˜ao, H.d.S.; Lima, G.R.d.M.; Montenegro, C.d.A.; Lira, N.S.; de Athayde-Filho, P.F.; Rodrigues, L.C.; De Souza, M.d.F.V.; Barbosa-Filho, J.M.; Batista, L.M. Bioactivities from Marine Algae of the Genus Gracilaria. Int. J. Mol. Sci. 2011, 12, 4550–4573. [Google Scholar] [CrossRef]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef]
- Dominguez, H.; Loret, E.P. Ulva lactuca, A Source of Troubles and Potential Riches. Mar. Drugs 2019, 17, 357. [Google Scholar] [CrossRef]
- EClinica Medicine. Antimicrobial Resistance: A Top Ten Global Public Health Threat. EClinical,Medicine 2021, 41, 101221. [Google Scholar]
- El-Seedi, H.R.; El-Shabasy, R.M.; Shaden Khalifa, A.M.; et al. Metal nanoparticles fabricated by green chemistry using natural extracts: biosynthesis, mechanisms,and applications. RSC Adv. 2019. 9, 24539. [CrossRef]
- Fatima, R.; Priya, M.; Indurthi, L.; Radhakrishnan, V.; Sudhakaran, R. Biosynthesis of silver nanoparticles using red algae Portieria hornemannii and its antibacterial activity against fish pathogens. Microbial Pathogenesis 2020, 138, 103780. [Google Scholar] [CrossRef]
- Ficko-Blean, E.; Hervé, C.; Michel, G. Sweet and sour sugars from the sea: the biosynthesis and remodeling of sulfated cell wall polysaccharides from marine macroalgae. Perspectives in Phycology 2015, 000, 251–264. [Google Scholar] [CrossRef]
- Fredericq, S.; Hommersand, M.H. Proposal of the Gracilariales ord. nov. (Rhodophyta) based on an analysis of the reproductive development of Gracilaria verrucosa. Journal of Phycology 1989, 25, 213–227, 57. [Google Scholar]
- Gnanajobitha, G. Fruit-mediated synthesis of silver nanoparticles using Vitis vinifera and evaluation of their antimicrobial efficacy. Journal of Nanostructure in Chemistry 2013, 3, 1–6. [Google Scholar] [CrossRef]
- Hu, J.; Wang, Z.; Li, J. Gold nanoparticles with special shapes: controlled synthesis, surface-enhanced Raman scattering, and the application in biodetection. Sensors 2007, 7, 3299–3311. [Google Scholar] [CrossRef]
- Jayarambabu, N.; Akshaykranth, A.; Venkatappa Rao, T.; Venkateswara Rao, K.; Rakesh Kumar, R. Green synthesis of Cu nanoparticles using Curcuma longa extract and their application in antimicrobial activity. Materials Letters 2020, 259, 126813. [Google Scholar] [CrossRef]
- Jin, R. Nanotechnology Reviews. The impacts of nanotechnology on catalysis by precious metal nanoparticles. 2012, 1, 31–56. [Google Scholar]
- Kasemets, K.; Ivask, A.; Dubourguier, H.C.; Kahru, A. Toxicity of nanoparticles of ZnO, CuO and TiO2 to yeast Saccharomyces cerevisiae. Toxicology in Vitro 2009, 23, 1116–1122. [Google Scholar] [CrossRef]
- Khalil, I.; Yehye, W.A.; Etxeberria, A.E.; Alhadi, A.A.; Dezfooli, S.M.; Julkapli, N.B.M.; Basirun, W.J.; Seyfoddin, A. Nanoantioxidants: Recent Trends in Antioxidant Delivery Applications. Antioxidants 2020, 9, 24. [Google Scholar]
- Khan, I.; Saeed, K.; Khan, I. Nanoparticles: Properties, applications and toxicities. Arabian Journal of Chemistry. 2019, 12, 908–931. [Google Scholar] [CrossRef]
- Kloareg, B.; Badis, Y.; Cock, J.M.; Michel, G. Role and Evolution of the Extracellular Matrix in the Acquisition of Complex Multicellularity in Eukaryotes: A Macroalgal Perspective. Genes 2021, 12, 1059–167. [Google Scholar] [CrossRef]
- Kumar, D.A.; Palanichamy, V.; Roopan, S.M. Green synthesis of silver nanoparticles using Alternanthera dentata leaf extract at room temperature and their antimicrobial activity. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy 2014, 127, 168–171. [Google Scholar] [CrossRef]
- Makarov, M.V.; Ryzhik, I.V.; Voskoboinikov, G.M. The Effect of Fucus vesiculosus L. (Phaeophyceae) Depth of Vegetation in the Barents Sea (Russia) on Its Morphophysiological Parameters. International Journal on Algae 2013, 15, 77–90. [Google Scholar]
- Mantri, V.A.; Kavale, M.G.; Kazi, M.A. Seaweed Biodiversity of India: Reviewing Current Knowledge to Identify Gaps, Challenges, and Opportunities. Diversity 2019, 12, 13. [Google Scholar] [CrossRef]
- Mariselvam, R.; Ranjitsingh AJ, A.; Usha Raja Nanthini, A.; Kalirajan, K.; Padmalatha, C.; Mosae Selvakumar, P. Green synthesis of silver nanoparticles from the extract of the inflorescence of Cocos nucifera (Family: Arecaceae) for enhanced antibacterial activity. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy 2014, 129, 537–541. [Google Scholar] [CrossRef]
- Marmiroli, M.; Pagano, L.; Rossi, R.; De La Torre-Roche, R.; Lepore, G.O.; Ruotolo, R.; Gariani, G.; Bonanni, V.; Pollastri, S.; Puri, A.; Gianoncelli, A.; Aquilanti, G.; d’Acapito, F.; White, J.C.; Marmiroli, N. Copper Oxide nanomaterial fate in plant tissue: nanoscale impacts on reproductive tissues. Environ. Sci. Technol. 2021, 55, 10769–10783. [Google Scholar] [CrossRef]
- Mourdikoudis, S.; Pallares, R.M.; Thanh, N.T.K. Characterization techniques for nanoparticles: comparison and complementarity upon studying nanoparticle properties. Nanoscale 2018, 10, 12871. [Google Scholar] [CrossRef]
- Mussin, J.; Giusiano, G. Biogenic silver nanoparticles as antifungal agents. Front. Chem. 2022, 10, 1023542. [Google Scholar] [CrossRef]
- Nath, D.; Banerjee, P. Green nanotechnology–a new hope for medical biology. Toxicology and Pharmacology. 2013, 36, 997–1014. [Google Scholar] [CrossRef]
- Pagano, L.; Marmiroli, M.; Villani, M.; Magnani, J.; Rossi, R.; Zappettini, A.; White, J.C.; Marmiroli, N. Engineered nanomaterial exposure affects organelle genetic material replication in Arabidopsis thaliana. ACS Nano 2022, 16, 2249–2260. [Google Scholar] [CrossRef]
- Pagano, L.; Maestri, E.; Caldara, M.; White, J.C.; Marmiroli, N.; Marmiroli, M. Engineered nanomaterial activity at the organelle level: Impacts on the chloroplasts and mitochondria. ACS Sustain. Chem. Eng. 2018, 6, 12562–12579. [Google Scholar] [CrossRef]
- Pappou, S.; Dardavila, M.M.; Savvidou, M.G.; Louli, V.; Magoulas, K.; Voutsas, E. Extraction of Bioactive Compounds from Ulva lactuca. Appl. Sci. 2022, 12, 2117. [Google Scholar] [CrossRef]
- Pearce, A.K.; Wilks, T.R.; Arno, M.C.; et al. Synthesis and applications of anisotropic nanoparticles with precisely defined dimensions. Nat Rev Chem. 2021, 5, 21–45. [Google Scholar] [CrossRef]
- Pereira, A.M.; Costa, A.D.; Dias, S.C.; Casal, M.; Machado, R. Production and Purification of Two Bioactive Antimicrobial Peptides Using a Two-Step Approach Involving an Elastin-Like Fusion Tag. Pharmaceuticals 2021, 14, 956. [Google Scholar] [CrossRef]
- Rajeshkumar, S.; Aboelfetoh, E.F.; Balusamy, S.R.; Ali, D.; Almarzoug, M.H.A.; Tesfaye, J.L.; Krishnaraj, R. Anticancer, Enhanced Antibacterial, and Free Radical Scavenging Potential of Fucoidan- (Fucus vesiculosus Source) Mediated Silver Nanoparticles. Oxid Med Cell Longev. 2021, 2021, 8511576. [Google Scholar] [CrossRef]
- Ray, B.; Lahaye, M. Cell-wall polysaccharides from the marine green alga Ulva “rigida” (Ulvales, Chlorophyta). Chemical structure of ulvan. Carbohydrate Research 1995, 274, 251–261. [Google Scholar] [CrossRef]
- Sangeetha, N.; Saravanan, K. Biogenic silver nanoparticles using marine seaweed (Ulva lactuca) and evaluation of its antibacterial activity. Journal of Nanoscience and Nanotechnology. 2014, 2, 99–102. [Google Scholar]
- Sivakumar, S.; Jin, D.X.; Rathod, R.; Ross, J.; Cantley, L.C.; Scaltriti, M.; Chen, J.W.; Hutchinson, K.E.; Wilson, T.R.; Sokol, E.S.; Vasan, N. Genetic Heterogeneity and Tissue-specific Patterns of Tumors with Multiple PIK3CA Mutations. Clin Cancer Res. 2023, 29, 1125–1136. [Google Scholar] [CrossRef]
- Singh, A.; Jain, D.; Upadhyay, M.K.; et al. Green synthesis of silver nanoparticles using Argemone mexicana leaf extract and evaluation of their antimicrobial activities. Digest Journal of Nanomaterials and Biostructures 2010, 2, 483–489. [Google Scholar]
- Singaravelu, G.; Arockiamary, J.; Kumar, V.G.; Govindaraju, K. A novel extracellular synthesis of monodisperse gold nanoparticles using marine alga, Sargassum wightii Greville. Colloids Surf. B Biointerfaces 2007, 57, 97–101. [Google Scholar] [CrossRef]
- Suresh, A.K.; Pelletier, D.A.; Wang, W.; et al. Ciprofloxacin functionalized biogenic gold nanoflowers as nanoantibiotics against pathogenic bacterial strains. Acta Biomaterialia 2011, 7, 2148–2152. [Google Scholar] [CrossRef]
- Thakkar, K.N.; Mhatre, S.S.; Parikh, R.Y. Biological synthesis of metallic nanoparticles. Nanomedicine: Nanotechnology, Biology and Medicine. 2010, 6, 257–262. [Google Scholar] [CrossRef]
- Türk, M.; Erkey, C. Synthesis of supported nanoparticles in supercritical fluids by supercritical fluid reactive deposition: Current state, further perspectives and needs. The Journal of Supercritical Fluids 2018. 134, 176–183. [CrossRef]
- Vestby, L.K.; Grønseth, T.; Simm, R.; Nesse, L.L. Bacterial Biofilm and its Role in the Pathogenesis of Disease. Antibiotics 2020, 9, 59. [Google Scholar] [CrossRef]
- Viaroli, P.; Azzoni, R.; Bartoli, M.; Giordani, G.; Tajé, L. Evolution of the trophic conditions and dystrophic outbreaks in the Sacca di Goro lagoon; In: Northern Adri-atic Sea. In Structure and processes in the Mediterranean ecosystems; Faranda, F.M., Guglielmo, L., Spezie, G., Eds.; Springer: Berlin Heidelberg New York; p. 443.
- World Health Organitation. Antimicrobial Resistance; WHO: Geneva, Switzerland, 2021. [Google Scholar]
- World Health Organization. Fungal Priority Pathogens List to Guide Research, Development and Public Health Action; WHO: Geneva, Switzerland, 2022. [Google Scholar]
- Zhao, J.; Stenzel, M.H. Entry of nanoparticles into cells: The importance of nanoparticle properties. Polym. Chem. 2018, 9, 259–272. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, L.; Xu, X.; Li, F.; Wu, Q. Combined systems of different antibiotics with nano-CuO against Escherichia coli and the mechanisms involved. Nanomedicine (Lond). 2018, 13, 339–351. [Google Scholar] [CrossRef]
- Zaldívar, J.M.; Cattaneo, E.; Plus, M.; Murray, C.N.; Giordani, G.; Viaroli, P. Long-term simulation of main biogeochemical events in a coastal lagoon: Sacca Di Goro (Northern Adriatic Coast, Italy). Continental Shelf Research 2003, 23, 1847–1875. [Google Scholar] [CrossRef]
- Zulfiqar, H.; Zafar, A.; Rasheed, M.N.; Ali, Z.; Mehmood, K.; Mazher, A.; Hasan, M.; Mahmood, N. Synthesis of silver nanoparticles using Fagonia cretica and their antimicrobial activities. Nanoscale Adv. 2018, 1, 1707–1713. [Google Scholar] [CrossRef]
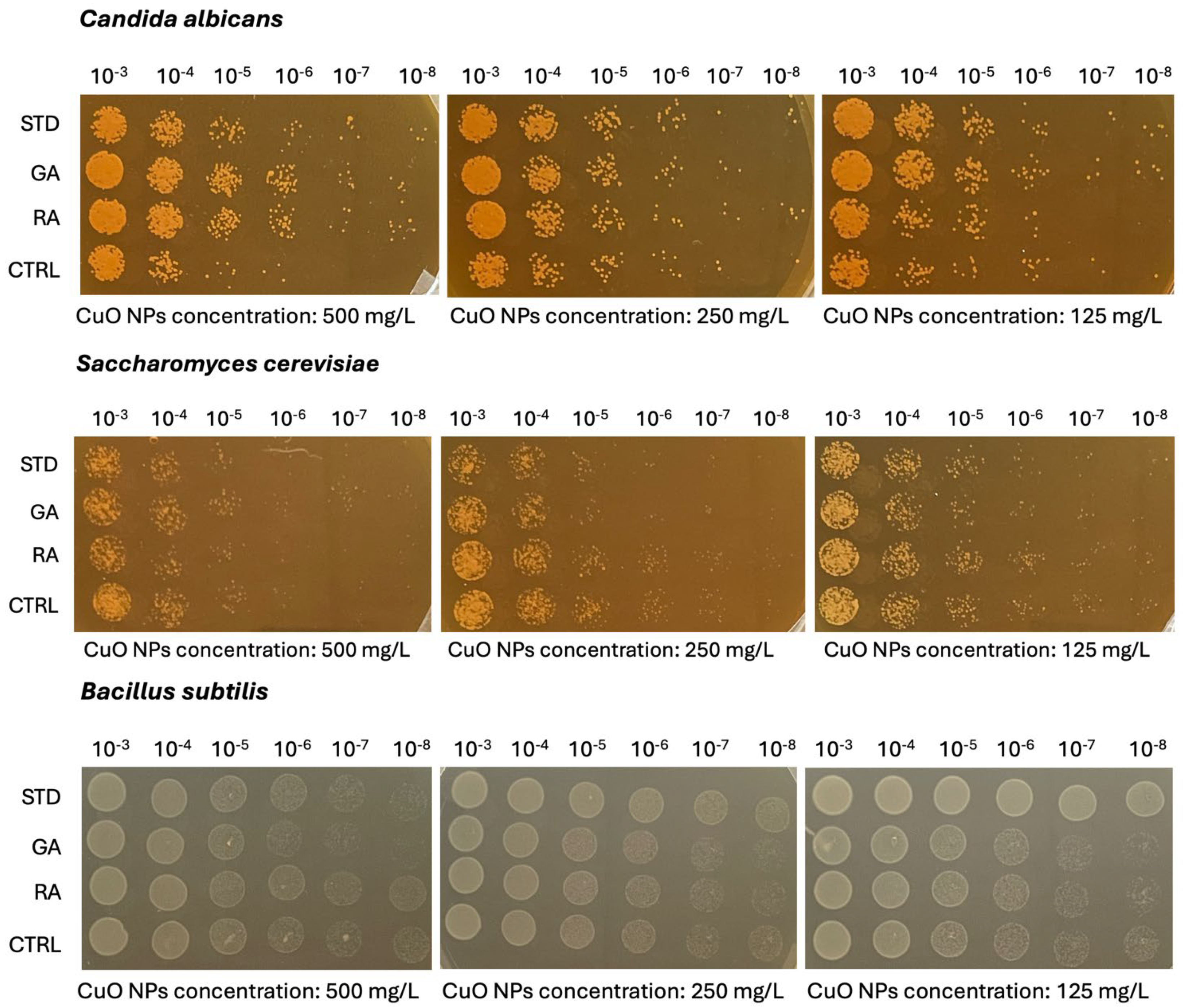
| Candida albicans | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NPs concentration (mg L-1) | ||||||||||||
| 500 | 250 | 125 | 62.5 | 31.3 | 15.6 | 7.8 | 3.9 | 2.0 | 1.0 | 0.5 | CTRL | |
| STD NPs | 1.098 | 1.233 | 1.31 | 1.342 | 1.349 | 1.335 | 1.333 | 1.361 | 1.364 | 1.377 | 1.372 | 1.341 |
| GA NPs | 1.092 | 1.259 | 1.326 | 1.363 | 1.362 | 1.367 | 1.386 | 1.393 | 1.382 | 1.383 | 1.391 | 1.343 |
| RA NPs | 1.11 | 1.304 | 1.363 | 1.383 | 1.374 | 1.398 | 1.407 | 1.387 | 1.399 | 1.393 | 1.396 | 1.407 |
| Saccharomyces cerevisiae | ||||||||||||
| NPs concentration (mg L-1) | ||||||||||||
| 500 | 250 | 125 | 62.5 | 31.3 | 15.6 | 7.8 | 3.9 | 2.0 | 1.0 | 0.5 | CTRL | |
| STD NPs | 0.915 | 1.021 | 1.111 | 1.13 | 1.218 | 1.141 | 1.246 | 1.249 | 1.248 | 1.249 | 1.206 | 1.114 |
| GA NPs | 0.863 | 0.982 | 1.079 | 1.177 | 1.158 | 1.191 | 1.226 | 1.253 | 1.235 | 1.283 | 1.314 | 1.231 |
| RA NPs | 0.831 | 1.013 | 1.129 | 1.127 | 1.2 | 1.233 | 1.248 | 1.304 | 1.304 | 1.334 | 1.375 | 1.354 |
| Bacillus subtilis | ||||||||||||
| NPs concentration (mg L-1) | ||||||||||||
| 500 | 250 | 125 | 62.5 | 31.3 | 15.6 | 7.8 | 3.9 | 2.0 | 1.0 | 0.5 | CTRL | |
| STD NPs | 0.05 | 0.05 | 0.05 | 0.11 | 0.634 | 0.921 | 0.965 | 0.996 | 0.965 | 1.025 | 0.995 | 0.997 |
| GA NPs | 0.566 | 0.856 | 0.921 | 0.985 | 1.005 | 1.025 | 1.008 | 1.013 | 1.007 | 0.966 | 0.975 | 0.999 |
| RA NPs | 0.613 | 0.811 | 0.892 | 0.964 | 0.984 | 1.014 | 1.037 | 0.998 | 1.029 | 1.028 | 1.022 | 1.053 |
| Escherichia coli | ||||||||||||
| NPs concentration (mg L-1) | ||||||||||||
| 500 | 250 | 125 | 62.5 | 31.3 | 15.6 | 7.8 | 3.9 | 2.0 | 1.0 | 0.5 | CTRL | |
| STD NPs | 0.605 | 0.711 | 0.72 | 0.723 | 0.725 | 0.722 | 0.716 | 0.712 | 0.703 | 0.716 | 0.721 | 0.662 |
| GA NPs | 0.402 | 0.564 | 0.644 | 0.673 | 0.713 | 0.703 | 0.673 | 0.672 | 0.657 | 0.678 | 0.666 | 0.66 |
| RA NPs | 0.05 | 0.074 | 0.52 | 0.706 | 0.737 | 0.732 | 0.695 | 0.698 | 0.675 | 0.695 | 0.7 | 0.705 |
| Staphylococcus aureus | ||||||||||||
| NPs concentration (mg L-1) | ||||||||||||
| 500 | 250 | 125 | 62.5 | 31.3 | 15.6 | 7.8 | 3.9 | 2.0 | 1.0 | 0.5 | CTRL | |
| STD NPs | 0.619 | 0.549 | 0.618 | 0.596 | 0.536 | 0.561 | 0.563 | 0.529 | 0.55 | 0.588 | 0.6 | 0.675 |
| GA NPs | 0.481 | 0.473 | 0.555 | 0.523 | 0.528 | 0.542 | 0.534 | 0.523 | 0.54 | 0.552 | 0.538 | 0.666 |
| RA NPs | 0.455 | 0.371 | 0.542 | 0.452 | 0.531 | 0.607 | 0.506 | 0.549 | 0.465 | 0.58 | 0.52 | 0.69 |
| Growth inhibition | |||
| (OD600 = 0.05) | |||
| NPs | |||
| concentrations (mg L-1) | |||
| STD NPs | GA NPs | RA NPs | |
| Candida albicans | 0 | 0 | 0 |
| Saccharomyces cerevisiae | 0 | 0 | 0 |
| Bacillus subtilis | 125.0 | 0 | 0 |
| Escherichia coli | 0 | 0 | 500.0 |
| Staphylococcus aureus | 0 | 0 | 0 |
| 50% growth reduction compared to the | |||
| untreated control | |||
| NPs | |||
| concentrations (mg L-1) | |||
| STD NPs | GA NPs | RA NPs | |
| Candida albicans | 0 | 0 | 0 |
| Saccharomyces cerevisiae | 0 | 0 | 0 |
| Bacillus subtilis | 62.5 | 0 | 0 |
| Escherichia coli | 0 | 0 | 250.0 |
| Staphylococcus aureus | 0 | 0 | 0 |
| 25% growth reduction compared to the | |||
| untreated control | |||
| NPs | |||
| concentrations (mg L-1) | |||
| STD NPs | GA NPs | RA NPs | |
| Candida albicans | 0 | 0 | 0 |
| Saccharomyces cerevisiae | 0 | 500 | 250.0 |
| Bacillus subtilis | 31.25 | 500 | 500.0 |
| Escherichia coli | 0 | 500 | 125.0 |
| Staphylococcus aureus | 0 | 500 | 250.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





