Submitted:
11 June 2024
Posted:
12 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Kefir Production
2.3. Analysis of Fatty Acids by GC-FID
2.4. Analysis of Volatile Compounds by Headspace GC-QqQ-MS
2.5. Analysis of Non-Volatile Compounds by GC-ToF-MS
2.6. Analysis of Non-Volatile Compounds by UHPLC-QToF-MS
2.7. NIRS Analysis
2.8. Data and Statistical Analysis
3. Results
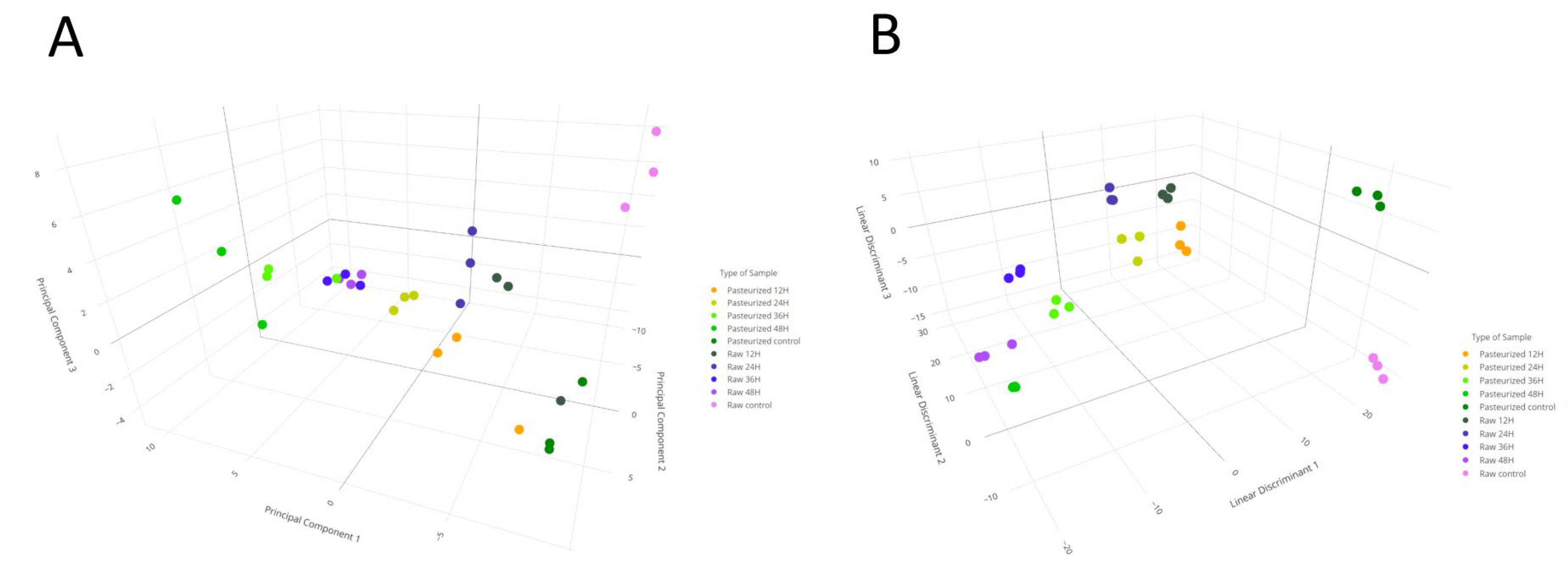
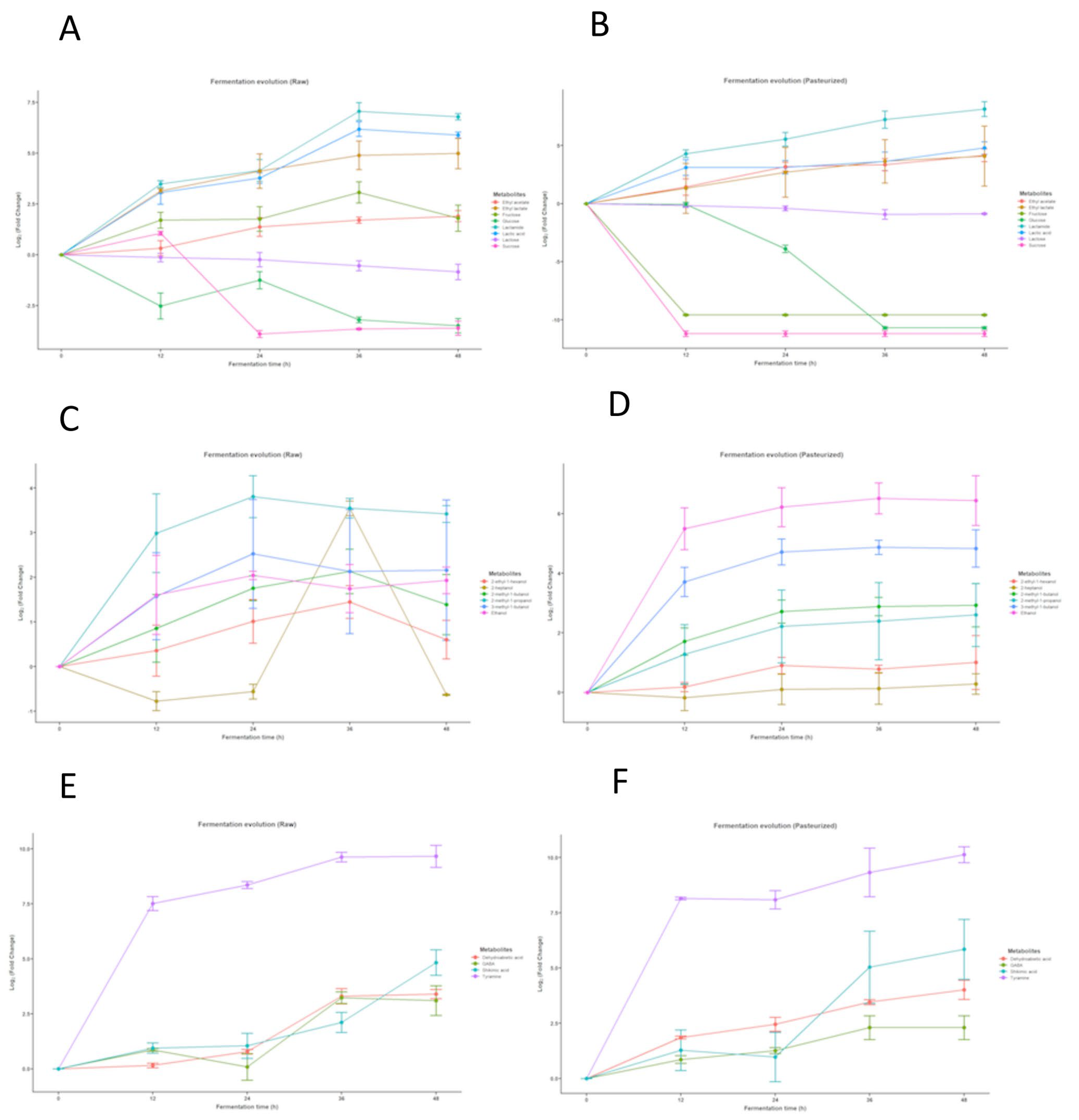
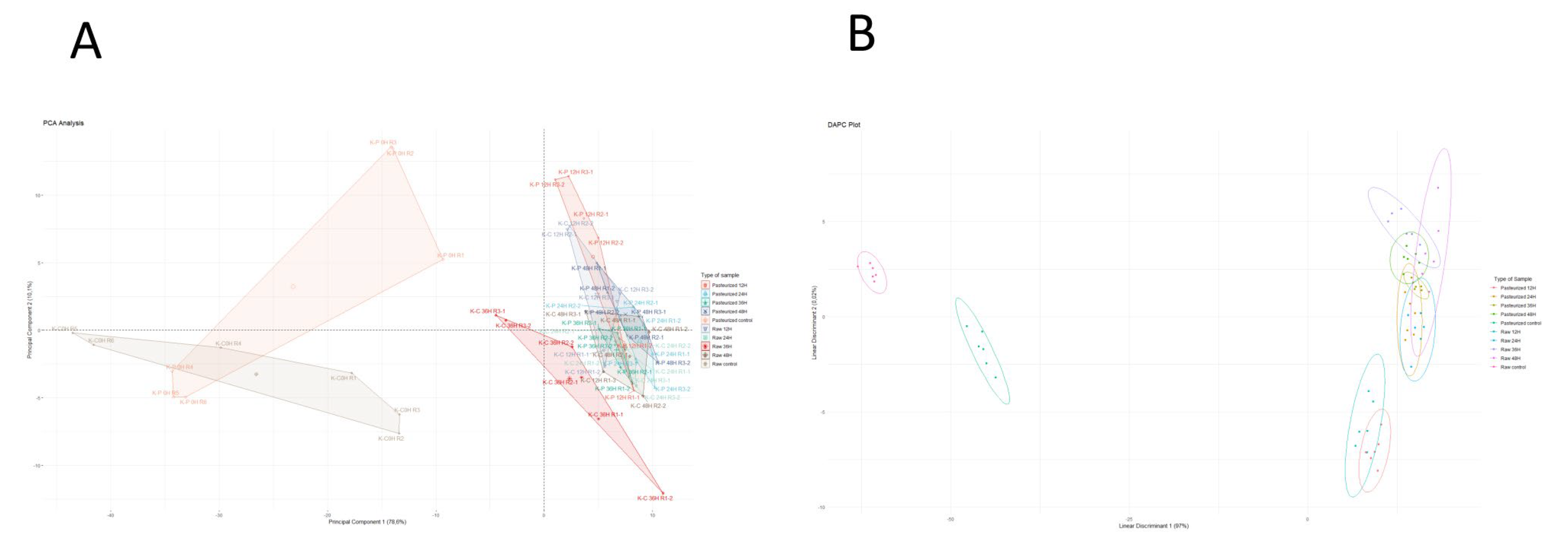
3.1. Metabolomic Analysis of Goat Milk Kefir
3.2. NIRS Analysis of Goat Milk Kefir
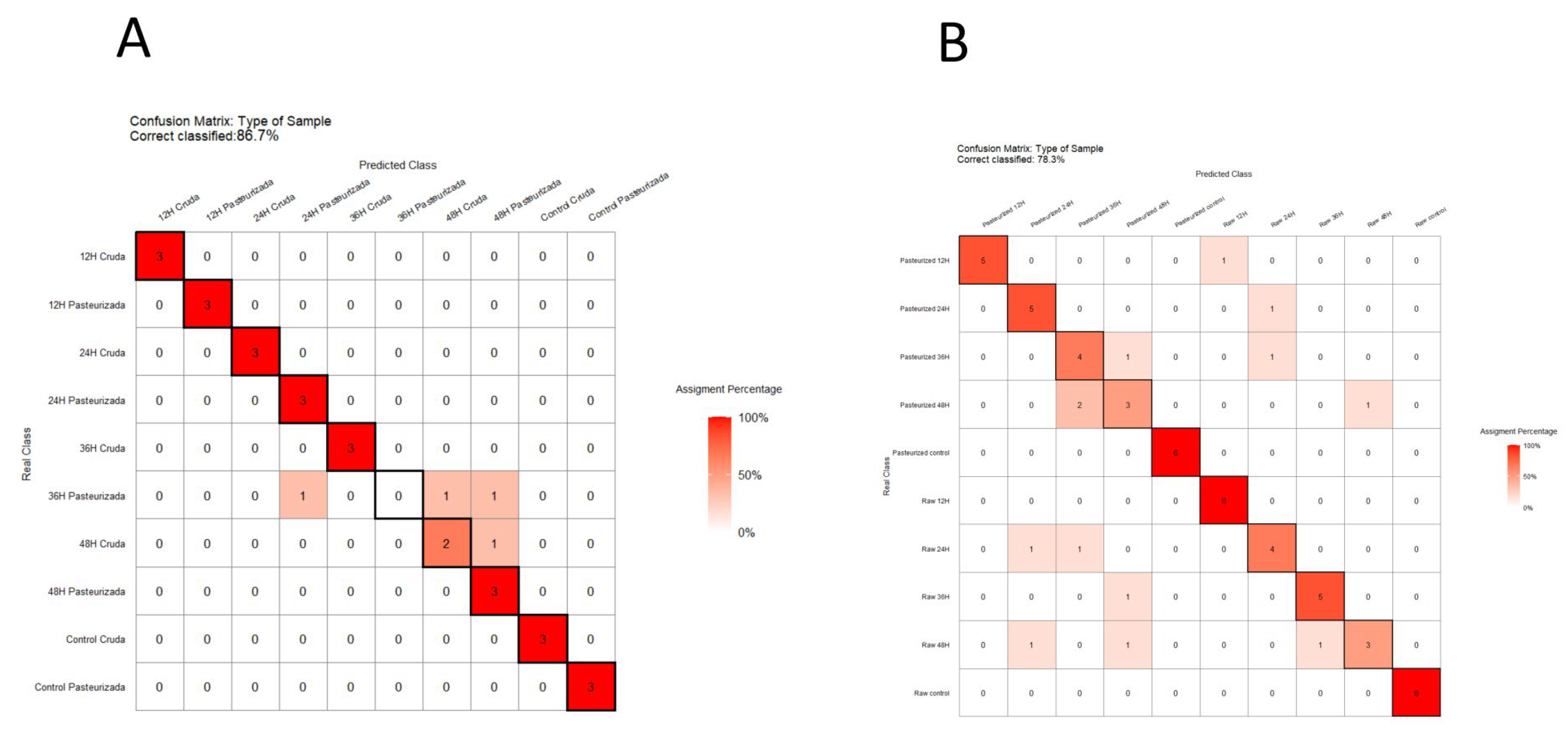
3.3. Predictive Capacity of Metabolomics and NIRS to Discriminate Kefir Samples
4. Discussion
Limitations and Future Research
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Escareño, L.; Salinas-Gonzalez, H.; Wurzinger, M.; Iñiguez, L.; Sölkner, J.; Meza-Herrera, C. Dairy Goat Production Systems: Status Quo, Perspectives and Challenges. Trop. Anim. Health Prod. 2012, 45, 17–34. [Google Scholar] [CrossRef]
- Ministerio de Agricultura, Pesca y Alimentación Caracterización del sector ovino y caprino de leche en España; Madrid, Spain, 2022; https://www.mapa.gob.es/es/ganaderia/temas/produccion-y-mercados-ganaderos/caracterizacionovinoycaprinolechedatos2021_tcm30-562416.pdf.
- Prado, M.R.; Blandón, L.M.; Vandenberghe, L.P.S.; Rodrigues, C.; Castro, G.R.; Thomaz-Soccol, V.; Soccol, C.R. Milk Kefir: Composition, Microbial Cultures, Biological Activities, and Related Products. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Bourrie, B.C.T.; Willing, B.P.; Cotter, P.D. The Microbiota and Health Promoting Characteristics of the Fermented Beverage Kefir. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef]
- Farag, M.A.; Jomaa, S.A.; El-Wahed, A.A.; El-Seedi, A.H.R. The Many Faces of Kefir Fermented Dairy Products: Quality Characteristics, Flavour Chemistry, Nutritional Value, Health Benefits, and Safety. Nutrients 2020, 12, 346. [Google Scholar] [CrossRef] [PubMed]
- Azizi, N.F.; Kumar, M.R.; Yeap, S.K.; Abdullah, J.O.; Khalid, M.; Omar, A.R.; Osman, M.A.; Mortadza, S.A.S.; Alitheen, N.B. Kefir and Its Biological Activities. Foods Basel Switz. 2021, 10, 1210. [Google Scholar] [CrossRef] [PubMed]
- Clark, S.; Mora García, M.B. A 100-Year Review: Advances in Goat Milk Research. J. Dairy Sci. 2017, 100, 10026–10044. [Google Scholar] [CrossRef]
- Prosser, C.G. Compositional and Functional Characteristics of Goat Milk and Relevance as a Base for Infant Formula. J. Food Sci. 2021, 86, 257–265. [Google Scholar] [CrossRef]
- Ebner, J.; Aşçı Arslan, A.; Fedorova, M.; Hoffmann, R.; Küçükçetin, A.; Pischetsrieder, M. Peptide Profiling of Bovine Kefir Reveals 236 Unique Peptides Released from Caseins during Its Production by Starter Culture or Kefir Grains. J. Proteomics 2015, 117, 41–57. [Google Scholar] [CrossRef]
- Dallas, D.C.; Citerne, F.; Tian, T.; Silva, V.L.M.; Kalanetra, K.M.; Frese, S.A.; Robinson, R.C.; Mills, D.A.; Barile, D. Peptidomic Analysis Reveals Proteolytic Activity of Kefir Microorganisms on Bovine Milk Proteins. Food Chem. 2016, 197, 273–284. [Google Scholar] [CrossRef]
- Amorim, F.G.; Coitinho, L.B.; Dias, A.T.; Friques, A.G.F.; Monteiro, B.L.; Rezende, L.C.D.D.; Pereira, T.D.M.C.; Campagnaro, B.P.; De Pauw, E.; Vasquez, E.C.; et al. Identification of New Bioactive Peptides from Kefir Milk through Proteopeptidomics: Bioprospection of Antihypertensive Molecules. Food Chem. 2019, 282, 109–119. [Google Scholar] [CrossRef]
- Wang, H.; Zhou, X.; Sun, Y.; Sun, X.; Guo, M. Differences in Protein Profiles of Kefir Grains from Different Origins When Subcultured in Goat Milk. J. Agric. Food Chem. 2022, 70, 7515–7524. [Google Scholar] [CrossRef]
- Izquierdo-González, J.J.; Amil-Ruiz, F.; Zazzu, S.; Sánchez-Lucas, R.; Fuentes-Almagro, C.A.; Rodríguez-Ortega, M.J. Proteomic Analysis of Goat Milk Kefir: Profiling the Fermentation-Time Dependent Protein Digestion and Identification of Potential Peptides with Biological Activity. Food Chem. 2019, 295, 456–465. [Google Scholar] [CrossRef]
- Dalabasmaz, S.; De La Torre, E.P.; Gensberger-Reigl, S.; Pischetsrieder, M.; Rodríguez-Ortega, M.J. Identification of Potential Bioactive Peptides in Sheep Milk Kefir through Peptidomic Analysis at Different Fermentation Times. Foods 2023, 12, 2974. [Google Scholar] [CrossRef]
- Cui, Y.; Wang, X.; Yue, Y.; Du, G.; Chen, H.; Ning, M.; Yuan, Y.; Yue, T. Metagenomic Features of Tibetan Kefir Grains and Its Metabolomics Analysis during Fermentation. LWT 2023, 175, 114502. [Google Scholar] [CrossRef]
- Qin, C.; Liu, L.; Wang, Y.; Leng, T.; Zhu, M.; Gan, B.; Xie, J.; Yu, Q.; Chen, Y. Advancement of Omics Techniques for Chemical Profile Analysis and Authentication of Milk. Trends Food Sci. Technol. 2022, 127, 114–128. [Google Scholar] [CrossRef]
- Folch, J.; Lees, M.; Stanley, G.H.S. A SIMPLE METHOD FOR THE ISOLATION AND PURIFICATION OF TOTAL LIPIDES FROM ANIMAL TISSUES. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef]
- Osorio, S.; Do, P.T.; Fernie, A.R. Profiling Primary Metabolites of Tomato Fruit with Gas Chromatography/Mass Spectrometry. In Plant Metabolomics; Hardy, N.W., Hall, R.D., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, 2011; ISBN 978-1-61779-593-0. [Google Scholar]
- Settachaimongkon, S.; Van Valenberg, H.J.F.; Winata, V.; Wang, X.; Nout, M.J.R.; Van Hooijdonk, T.C.M.; Zwietering, M.H.; Smid, E.J. Effect of Sublethal Preculturing on the Survival of Probiotics and Metabolite Formation in Set-Yoghurt. Food Microbiol. 2015, 49, 104–115. [Google Scholar] [CrossRef]
- Batista, L.L.; Malta, S.M.; Guerra Silva, H.C.; Borges, L.D.F.; Rocha, L.O.; Da Silva, J.R.; Rodrigues, T.S.; Venturini, G.; Padilha, K.; Da Costa Pereira, A.; et al. Kefir Metabolites in a Fly Model for Alzheimer’s Disease. Sci. Rep. 2021, 11, 11262. [Google Scholar] [CrossRef]
- Walsh, A.M.; Crispie, F.; Kilcawley, K.; O’Sullivan, O.; O’Sullivan, M.G.; Claesson, M.J.; Cotter, P.D. Microbial Succession and Flavor Production in the Fermented Dairy Beverage Kefir. mSystems 2016, 1, e00052-16. [Google Scholar] [CrossRef] [PubMed]
- Pisano, M.B.; Scano, P.; Murgia, A.; Cosentino, S.; Caboni, P. Metabolomics and Microbiological Profile of Italian Mozzarella Cheese Produced with Buffalo and Cow Milk. Food Chem. 2016, 192, 618–624. [Google Scholar] [CrossRef] [PubMed]
- Caboni, P.; Maxia, D.; Scano, P.; Addis, M.; Dedola, A.; Pes, M.; Murgia, A.; Casula, M.; Profumo, A.; Pirisi, A. A Gas Chromatography-Mass Spectrometry Untargeted Metabolomics Approach to Discriminate Fiore Sardo Cheese Produced from Raw or Thermized Ovine Milk. J. Dairy Sci. 2019, 102, 5005–5018. [Google Scholar] [CrossRef] [PubMed]
- Murgia, A.; Scano, P.; Cacciabue, R.; Dessì, D.; Caboni, P. GC-MS Metabolomics Comparison of Yoghurts from Sheep’s and Goats’ Milk. Int. Dairy J. 2019, 96, 44–49. [Google Scholar] [CrossRef]
- Scano, P.; Cagliani, L.R.; Consonni, R. 1H NMR Characterisation of the Lipid Fraction and the Metabolite Profiles of Fossa (Pit) Cheese. Int. Dairy J. 2019, 90, 39–44. [Google Scholar] [CrossRef]
- Settachaimongkon, S.; Nout, M.J.R.; Antunes Fernandes, E.C.; Hettinga, K.A.; Vervoort, J.M.; Van Hooijdonk, T.C.M.; Zwietering, M.H.; Smid, E.J.; Van Valenberg, H.J.F. Influence of Different Proteolytic Strains of Streptococcus Thermophilus in Co-Culture with Lactobacillus Delbrueckii Subsp. Bulgaricus on the Metabolite Profile of Set-Yoghurt. Int. J. Food Microbiol. 2014, 177, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Beshkova, D.M.; Simova, E.D.; Frengova, G.I.; Simov, Z.I.; Dimitrov, Zh.P. Production of Volatile Aroma Compounds by Kefir Starter Cultures. Int. Dairy J. 2003, 13, 529–535. [Google Scholar] [CrossRef]
- Bourrie, B.C.T.; Diether, N.; Dias, R.P.; Nam, S.L.; De La Mata, A.P.; Forgie, A.J.; Gaur, G.; Harynuk, J.J.; Gänzle, M.; Cotter, P.D.; et al. Use of Reconstituted Kefir Consortia to Determine the Impact of Microbial Composition on Kefir Metabolite Profiles. Food Res. Int. 2023, 173, 113467. [Google Scholar] [CrossRef] [PubMed]
- Cais-Sokolińska, D.; Wójtowski, J.; Pikul, J.; Danków, R.; Majcher, M.; Teichert, J.; Bagnicka, E. Formation of Volatile Compounds in Kefir Made of Goat and Sheep Milk with High Polyunsaturated Fatty Acid Content. J. Dairy Sci. 2015, 98, 6692–6705. [Google Scholar] [CrossRef]
- Guzel-Seydim, Z.; Seydim, A.C.; Greene, A.K. Organic Acids and Volatile Flavor Components Evolved During Refrigerated Storage of Kefir. J. Dairy Sci. 2000, 83, 275–277. [Google Scholar] [CrossRef]
- Zhao, M.; Ma, H.; Hou, Y.; Li, J.; Zou, T.; Zhang, D.; Wen, R.; Li, H.; Song, H. Characterization of Key Odor-Active Off-Flavor Compounds in Aged Pasteurized Yogurt by Sensory-Directed Flavor Analysis. J. Agric. Food Chem. 2022, 70, 14439–14447. [Google Scholar] [CrossRef]
- Silva, T.; Pires, A.; Gomes, D.; Viegas, J.; Pereira-Dias, S.; Pintado, M.E.; Henriques, M.; Pereira, C.D. Sheep’s Butter and Correspondent Buttermilk Produced with Sweet Cream and Cream Fermented by Aromatic Starter, Kefir and Probiotic Culture. Foods 2023, 12, 331. [Google Scholar] [CrossRef]
- Bai, J.; Wu, Y.; Liu, X.; Zhong, K.; Huang, Y.; Gao, H. Antibacterial Activity of Shikimic Acid from Pine Needles of Cedrus Deodara against Staphylococcus Aureus through Damage to Cell Membrane. Int. J. Mol. Sci. 2015, 16, 27145–27155. [Google Scholar] [CrossRef]
- Cui, Y.; Miao, K.; Niyaphorn, S.; Qu, X. Production of Gamma-Aminobutyric Acid from Lactic Acid Bacteria: A Systematic Review. Int. J. Mol. Sci. 2020, 21, 995. [Google Scholar] [CrossRef]
- Hao, M.; Xu, J.; Wen, H.; Du, J.; Zhang, S.; Lv, M.; Xu, H. Recent Advances on Biological Activities and Structural Modifications of Dehydroabietic Acid. Toxins 2022, 14, 632. [Google Scholar] [CrossRef]
- Gillman, P.K. A Reassessment of the Safety Profile of Monoamine Oxidase Inhibitors: Elucidating Tired Old Tyramine Myths. J. Neural Transm. Vienna Austria 1996 2018, 125, 1707–1717. [Google Scholar] [CrossRef]
- Salter, M.; Kenney, A. Myocardial Injury from Tranylcypromine-Induced Hypertensive Crisis Secondary to Excessive Tyramine Intake. Cardiovasc. Toxicol. 2018, 18, 583–586. [Google Scholar] [CrossRef]
- Windarsih, A.; Rohman, A.; Irnawati; Riyanto, S. The Combination of Vibrational Spectroscopy and Chemometrics for Analysis of Milk Products Adulteration. Int. J. Food Sci. 2021, 2021, 1–15. [Google Scholar] [CrossRef]
- Xu, L.; Yan, S.-M.; Cai, C.-B.; Wang, Z.-J.; Yu, X.-P. The Feasibility of Using Near-Infrared Spectroscopy and Chemometrics for Untargeted Detection of Protein Adulteration in Yogurt: Removing Unwanted Variations in Pure Yogurt. J. Anal. Methods Chem. 2013, 2013, 1–9. [Google Scholar] [CrossRef]
- Gorla, G.; Fumagalli, S.; Jansen, J.J.; Giussani, B. Acquisition Strategies for Fermentation Processes with a Low-Cost Miniaturized NIR-Spectrometer from Scratch: Issues and Challenges. Microchem. J. 2022, 183, 108035. [Google Scholar] [CrossRef]
- Grassi, S.; Alamprese, C.; Bono, V.; Picozzi, C.; Foschino, R.; Casiraghi, E. Monitoring of Lactic Acid Fermentation Process Using Fourier Transform near Infrared Spectroscopy. J. Infrared Spectrosc. 2013, 21, 417–425. [Google Scholar] [CrossRef]
- Ntsame Affane, A.L.; Fox, G.P.; Sigge, G.O.; Manley, M.; Britz, T.J. Quantitative Analysis of DL-Lactic Acid and Acetic Acid in Kefir Using near Infrared Reflectance Spectroscopy. J. Infrared Spectrosc. 2009, 17, 255–264. [Google Scholar] [CrossRef]
- Ntsame Affane, A.L.; Fox, G.P.; Sigge, G.O.; Manley, M.; Britz, T.J. Simultaneous Prediction of Acidity Parameters (pH and Titratable Acidity) in Kefir Using near Infrared Reflectance Spectroscopy. Int. Dairy J. 2011, 21, 896–900. [Google Scholar] [CrossRef]
- Shao, Y.; He, Y. Measurement of Soluble Solids Content and pH of Yogurt Using Visible/Near Infrared Spectroscopy and Chemometrics. Food Bioprocess Technol. 2009, 2, 229–233. [Google Scholar] [CrossRef]
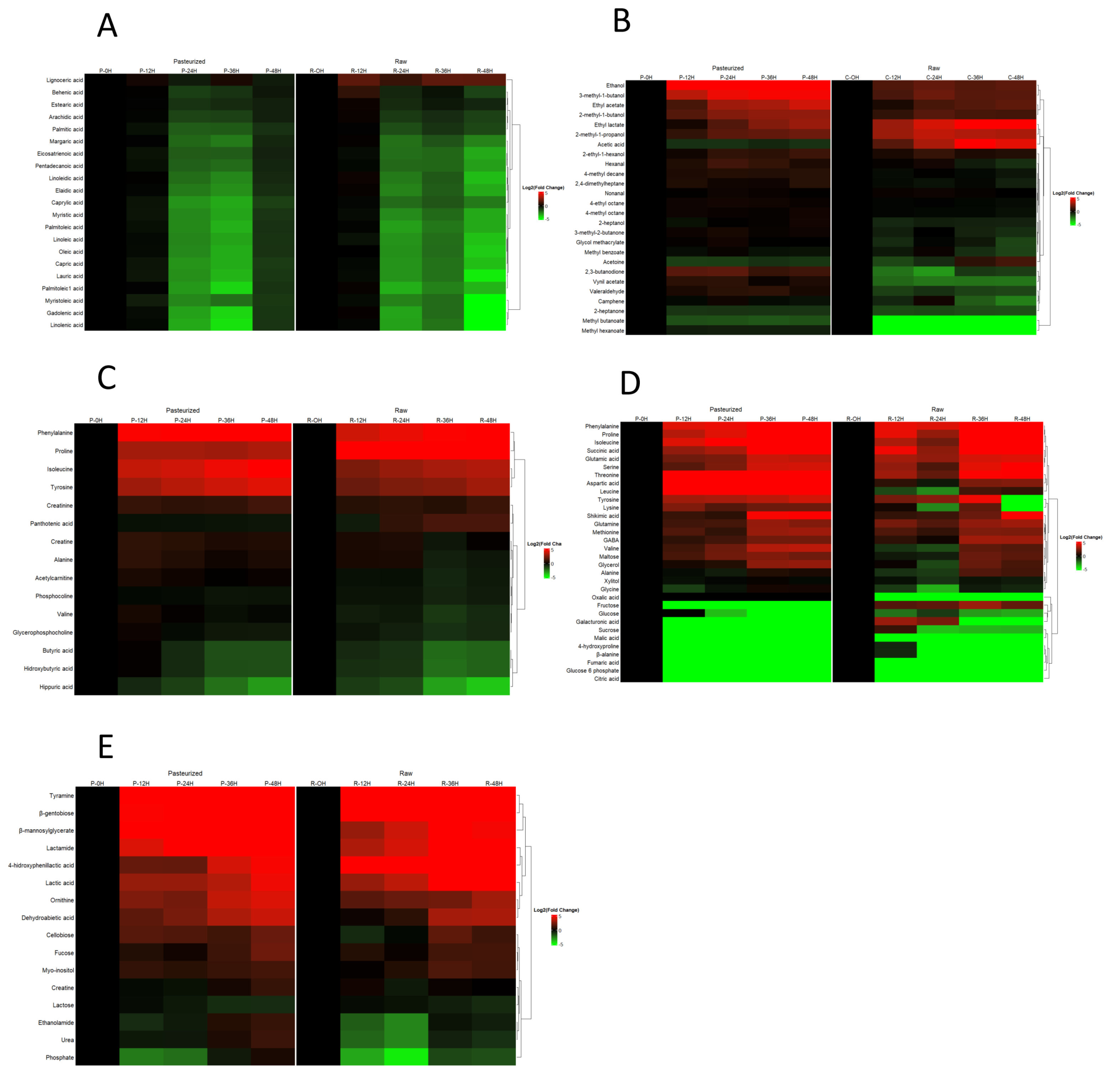
| Technique | Metabolites detected | Name |
|---|---|---|
| GC-FID | 21 | Arachidic acid, behenic acid, capric acid, caprylic acid, eicosatrienoic acid, elaidic acid, estearic acid, gadolenic acid, lauric acid, lignoceric acid, linoleic acid, linoleidic acid, linolenic acid, margaric acid, myristic acid, myristoleic acid, oleic acid, palmitic acid, palmitoleic acid, palmitoleic1 acid, pentadecanoic acid |
| UHPLC-MS (QToF) | 15 | Acetylcarnitine, alanine, butyric acid, creatine, creatinine, Glycerophosphocholine, hippuric acid, hidroxybutyric acid, isoleucine, panthotenic acid, fenilalanine, tyrosine, proline, phosphocoline, valine |
| GC-MS (QqQ) | 26 | 2-methyl-1-butanol,2-methyl-1-propanol, 3-methyl-1-butanol, 2,3-butanodione, 2,4-dimethylheptane, 2-ethyl-1-hexanol, 2-heptanol, 2-heptanone, 3-methyl-2-butanone, 4-ethyl octane, 4-methyl decane, 4-methyl octane, ethanol, ethyl acetate, vynil acetate, acetoine, acetic acid, methyl benzoate, methyl butanoate, camphene, glycol methacrylate, hexanal, mehtyl hexanoate, ethyl lactate, nonanal, valeraldehyde |
| GC-MS (Tof) | 32 | 4-hidroxyproline, aspartic acid, citric acid, fumaric acid, galacturonic acid, glutamic acid, malic acid, oxalic acid, shikimic acid, succinic acid, alanine, phenylalanine, fructose, GABA, glycerol, glycine, glucose, glucose-6-phosphate, glutamine, isoleucine, leucine, lysine, maltose, methionine, proline, sucrose, serine, tyrosine, threonine, valine, xylitol, β-alanine |
| 16 | 4-hidroxyphenillactic acid, dehidroabietic acid, lactic acid, celobiose, creatine, ethanolamide, phosphate, fucose, lactamide, lactose, myo-inositol, ornithine, tyramine, urea, β-gentobiose, β-mannosylglycerate |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





