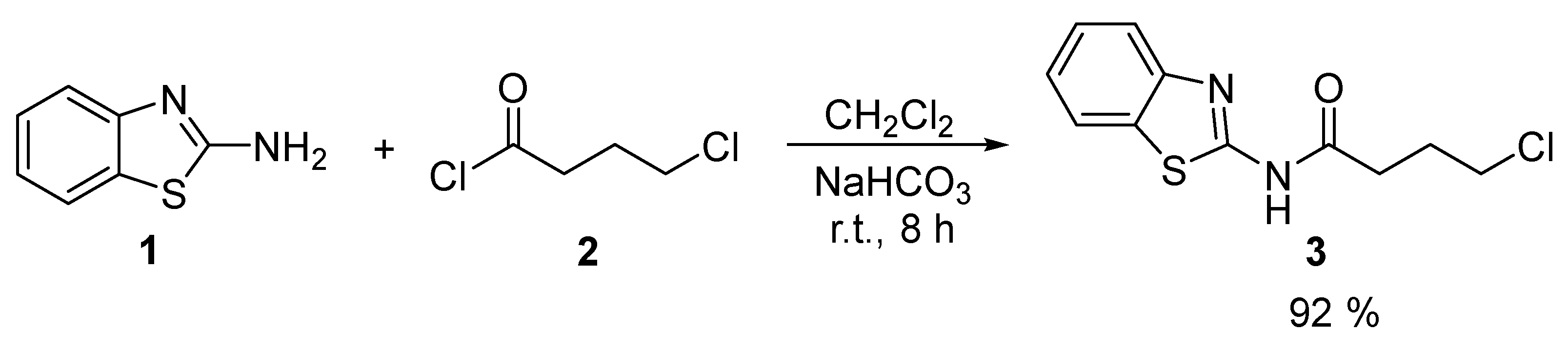1. Introduction
Benzazoles, including benzoxazoles and benzothiazoles, are aromatic compounds with good chemical stability [
1]. These compounds consist of a benzene ring attached to either oxazole or thiazole. They are important raw materials because they are heterocycles with fascinating physicochemical properties [
2]. In addition to its reactivity, several reports in the literature mentioned very varied pharmacological properties of this type of compound, such as antidiabetic [
3], anti-inflammatory [
4], neuroprotective [
5] and antibiotic [
6,
7] effects.
Diabetes mellitus type II is a widespread disease that affects many people worldwide. One of the most common treatments for this disease is inhibiting the alpha-glucosidase enzyme, which metabolizes carbohydrates [
8]. Acarbose is an example of a drug that works through this mechanism of action [
9]. Therefore, we are interested in synthesizing compounds with antidiabetic activity, particularly of the alpha-glucosidase inhibitor type. Considering the antidiabetic properties of benzoxazole, we decided to synthesize a compound that contains both a benzoxazole unit and a benzothiazole unit in its structure.
2. Results
2.1. Synthesis
We were able to synthesize butanamide
5 using a simple and inexpensive two-step methodology. The first step involved an
N-acylation reaction of 2-aminobenzothiazole
1 with 4-chlorobutanoyl chloride
2 in CH
2Cl
2 with NaHCO
3 as a base at room temperature for 8 h. The resultant 4-chlorobutanamide
3 was purified through crystallization in cold water and obtained as a white solid with a yield of 92 % [
10].
Scheme 1
The
1H spectrum confirmed the formation of butanamide
5. This spectrum shows the characteristic signals for the H of the three CH
2, which appear at 2.05 ppm(t), 2.65 ppm(m), and 3.69 ppm (t). The amide’s NH signal appears at 12.45 ppm. On the other hand, in the
13C spectrum, the signal corresponding to the carbonyl group can be observed at 171.4 ppm (please refer to
Figures S1 and S2 in the supplementary material).
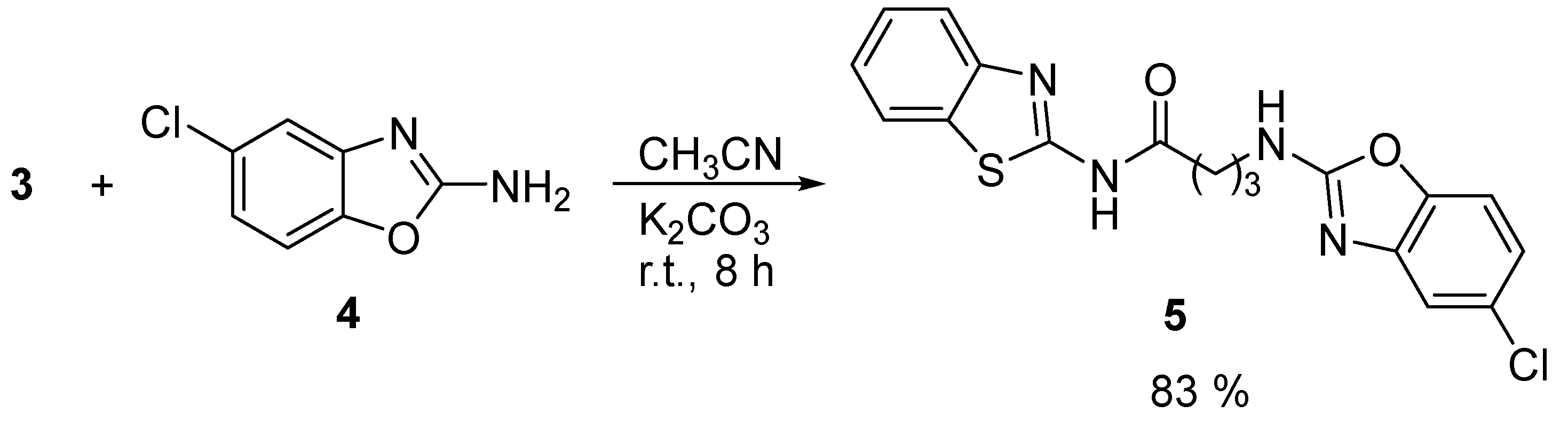
In the second step, the 4-chlorobutanamide
3 underwent a nucleophilic substitution reaction with 5-chloro-2-aminobenzoxazole
4 in CH
3CN, a non-protic polar solvent, with K
2CO
3 as base at room temperature for 8 h. The resulting compound
5 was also purified by recrystallization in cold water and obtained as a yellow solid with a yield of 83 %.
Scheme 2.
Compound
5 was successfully confirmed in the
1H NMR spectrum. The spectrum shows observable signals from the aromatic ring of both 5-chlorobenzoxazole and benzothiazole from 6.96 to 7.98 ppm. Additionally, a wide signal that integrates for two hydrogens NH was observed at 7.60 ppm (please refer to
Figure S3 in the supplementary material). The two-step synthesis resulted in an overall yield of 76 % of
N-(benzothiazol-2-yl)-2-((5-chlorobenzoxazol-2-yl)amino)butanamide
5.
2.2. Molecular Docking Validation
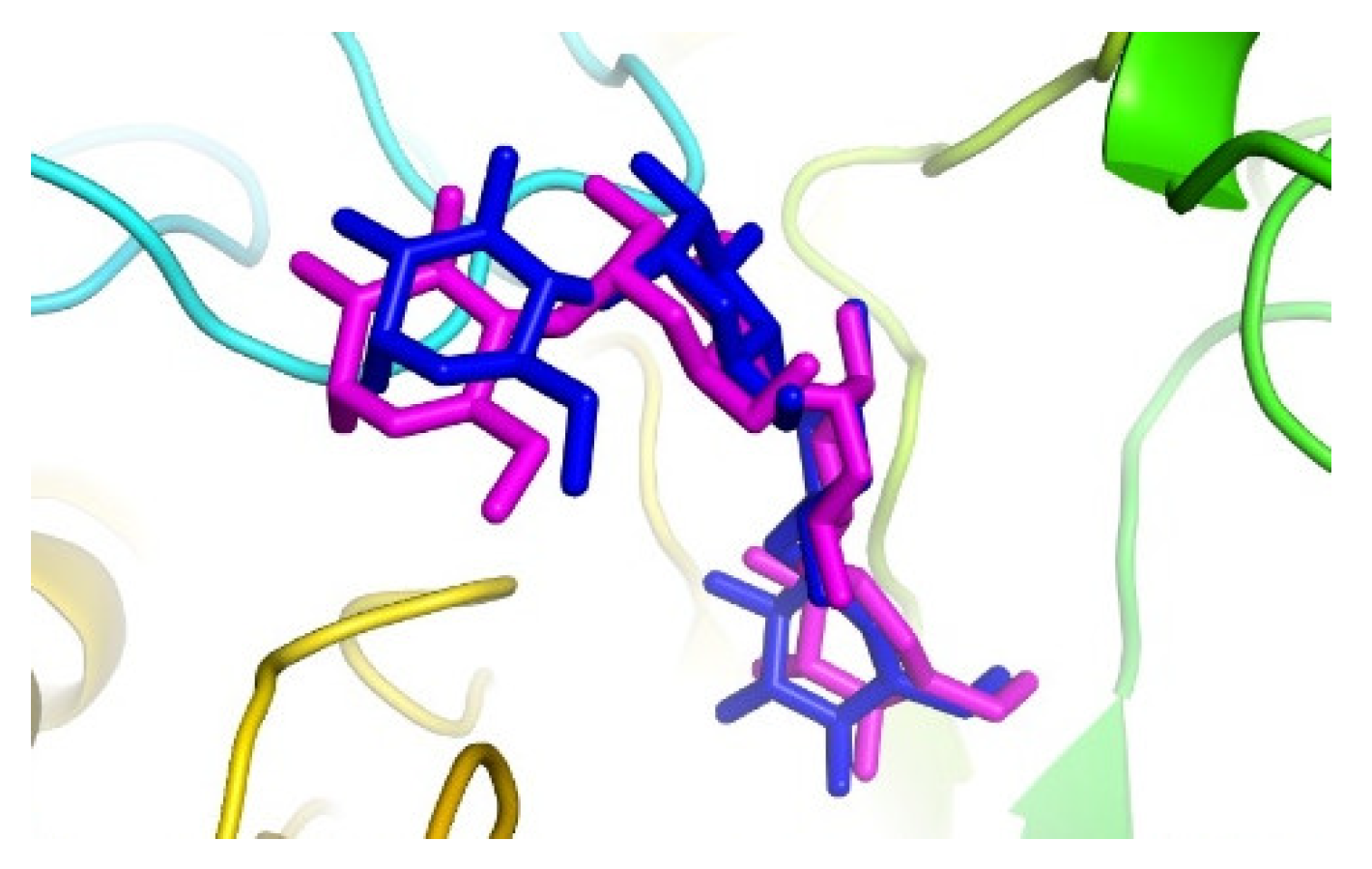
We performed computational analyses by docking, confirming the hypothesis that this compound can act as an inhibitor of the alpha-glucosidase enzyme. In this sense, the active site of 3-TOP protein was validated with the redock co-crystallized native ligand acarbose. The protein 3-TOP is a human maltase-glucoamylase, and its function is to hydrolyze linear alpha-1,4-linked oligosaccharide substrates. Comparison of the poses obtained by the AutoDock Vina program against those of the crystallized protein yielded root mean square deviation (RMSD) = 1.27 Å [
11,
12]. (
Figure 1).
2.3. Molecular Docking Studies
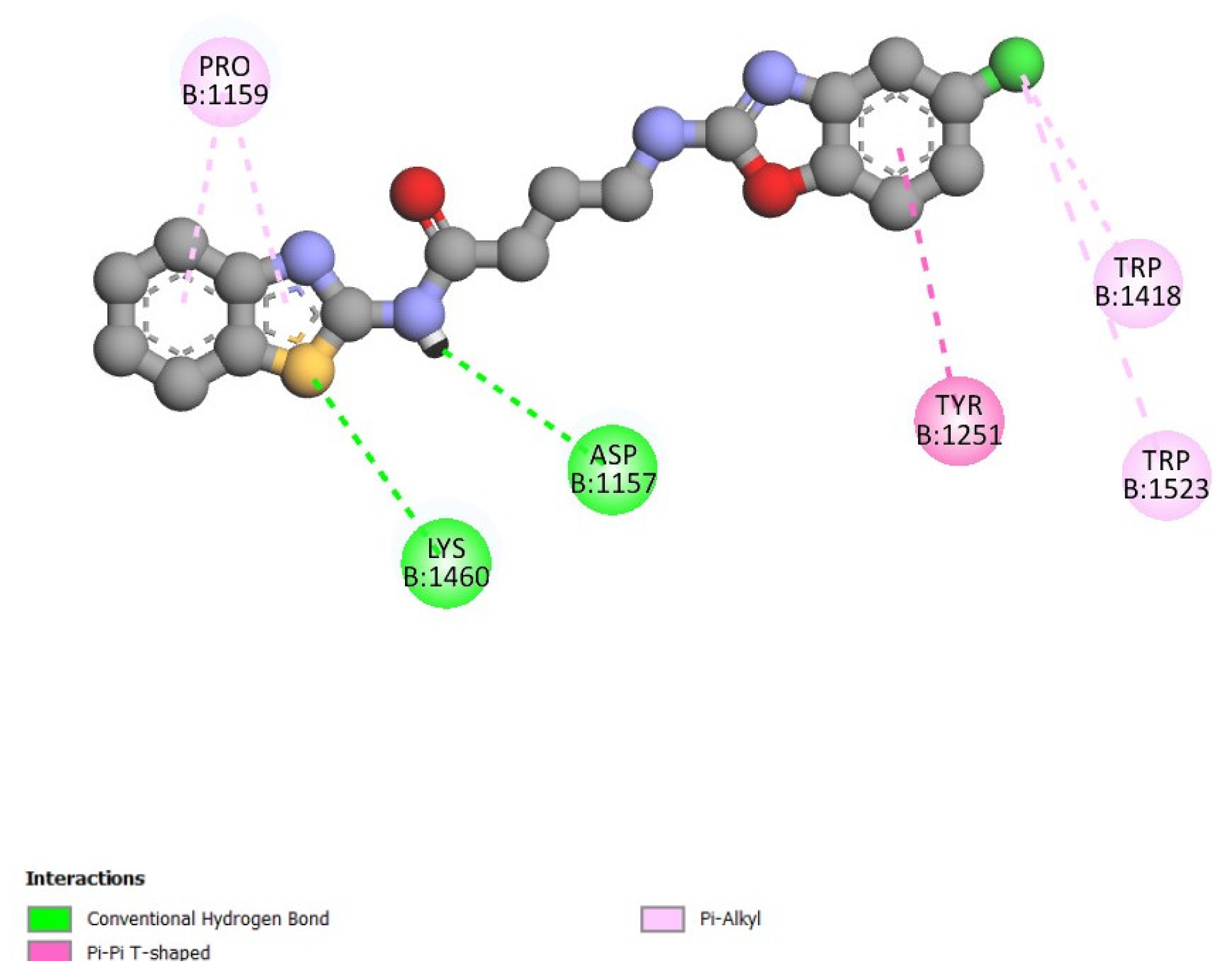
The AutoDock Vina open-source program was used to model the docking of butanamide 5 with 3-TOP protein. The optimized structure of butanamide 5 is shown in
Figure 2. The docking analysis revealed that butanamide 5 had high binding affinities with the 3-TOP protein, as evident from the docking score of -8.4 kcal/mol. According to the results, it is worth highlighting that the benzothiazole unit presents more interaction than the benzoxazole unit with some of the amino acids of the 3-TOP protein. It is relevant to note that benzothiazole presents a pi-alkyl interaction with proline 1159, both in the benzene ring and with the thiazole fragment, aside from the sulfur itself having a hydrogen bond interaction with Lysine 1460. Finally, amidic N also presents a hydrogen bond, where appropriate, with aspartate 1157. Additionally, the benzoxazole unit has a pi-pi interaction between the benzene ring and the tyrosine 1251 unit. However, neither the oxazole nor the oxygen atom presents any interaction. Chlorine has two pi-alkyl interactions, with tryptophan’s 1418 and 1523.
3. Discussion
This research involved a two-step linear synthesis process to obtain the desired product butanamide 5 with good chemical yields and an overall good yield of the reaction. The synthesis was completed without complications, and no by-products were observed using a simple reaction methodology. Furthermore, the two synthesized compounds were easily purified through a crystallization process using cold water.
In the computational studies, validation comparison of the poses obtained by the AutoDock Vina program against those of the crystallized protein indicates an appropriate optimization score. These values are small and support binding at the simulation site with the original orientation of the co-crystallized molecule. The interactions among butanamide 5 and specific amino acids of 3-TOP protein involve hydrogen bonds, pi-pi interactions, and pi-alkyl interactions. The docking analysis used showed that butanamide 5 exhibited docking poses with high binding affinities (in terms of affinity energy), and therefore, it might have antidiabetic activity.
4. Materials and Methods
4.1. General
All commercial reagents and solvents were used without any further purification. 1H and 13C NMR spectra were recorded on a 600 MHz Varian AR spectrometer, with DMSO-d6 as solvent. Infrared spectra were obtained using a Thermo Scientific Nicolet. Mass spectra were recorded on a GC-MS Agilent Technologies. The reactions were TLC monitored on silica gel 60 F254 (Merck).
4.2. Synthesis of N-(benzothiazol-2-yl)-4-chlorobutanamide (3).
Departing on a solution of 2-aminobenzothiazole 1 (500 mg, 3.33 mmol) in CH
2Cl
2 (10 mL), NaHCO
3 (419 mg, 4.99 mmol) was added. This mixture was stirred in a cold-water bath for 20 minutes. Then, 4-chlorobutanoyl chloride (448 µL, 4.00 mmol) was added dropwise. The reaction was stirred for 8 h at room temperature and monitored by TLC. After the reaction concluded, the resulting mixture was concentrated under reduced pressure. The obtained product was dissolved in cold water for 10 min. Finally, it was filtered and dried in a desiccator for 24 h. The yield reached 92 % after purification [
10].
1H NMR (600 MHz, DMSO-d
6) δ ppm 2.05 (m, 2H-CH
2), 2.65 (t,
J = 7.1 Hz, 2H-CH
2), 3.69 (t,
J = 6.6 Hz, 2H-CH
2), 7.28 (t,
J = 7.6 Hz, 1H-CH), 7.41 (t,
J = 7.7 Hz, 1H-CH), 7.72 (d,
J = 8.0 Hz, 1H-CH), 7.95 (d,
J = 7.9 Hz, 1H-CH), 12.45 (s, 1H-NH).
Figure S1.
13C NMR (150 MHz, DMSO-d
6) δ ppm 27.3, 32.4, 44.8, 120.6, 121.7, 123.6, 126.1, 131.5, 148.6, 157.8, 171.4.
Figure S2.
4.3. Synthesis of N-(benzothiazol-2-yl)-4-((5-chlorobenzoxazol-2-yl)amino)butanamide (5).
To a solution of N-(benzothiazol-2-yl)-4-chlorobutanoamide (279 mg, 1.10 mmol) in CH3CN (5 mL) was added K2CO3 (304.8 mg, 2.21 mmol), then was stirred at cold water bath over 20 min. After, a solution of 2-amino-5-chlorobenzoxazole (396 mg, 1.10 mmol) was added dropwise in CH3CN (5 mL). The reaction was TLC monitored. When the reaction ended, it was concentrated under reduced pressure. The compound obtained was dissolved in cold water for 10 min. Finally, it was filtered and dried in a desiccator for 24 h. The yield reached 76 % after purification.
1H NMR (600 MHz, DMSO-d
6) δ ppm 2.17 (m, 2H-CH
2), 2.66 (t,
J = 8.0 Hz, 2H-CH
2), 4.13 (t,
J = 7.2 Hz, 2H-CH
2), 6.96 (dd,
J = 2.2, 8.4 Hz, 1H-CH), 7.22 (d,
J = 2.2 Hz, 1H-CH), 7.31 (m, 2H-CH), 7.43 (t,
J = 7.6 Hz, 1H-CH), 7.60 (s, 2H-NH), 7.79 (d,
J = 8.0 Hz, 1H-CH), 7.98 (d,
J = 7.9 Hz, 1H-CH).
Figure S3.
13C NMR (150 MHz, DMSO-d
6) δ ppm 18.0, 31.9, 48.5, 109.9, 115.3, 119.9, 121.3, 122.3, 124.2, 126.6, 128.1, 132.1, 145.7, 147.2, 148.8, 157.1, 164.3, 175.2.
Figure S4. FAB-(+) MS. 391 m/z (5H+ high protonate). Fragment Molecular Formula: C
11H
15N
3OS
2+ 237 m/z. Fragment Molecular Formula: C
11H
15N
3OS
2+ 219 m/z.
Figure S5.
4.4. Validation of Active Site
The active site of 3-TOP was validated using acarbose as a native ligand. Autodock Vina generated an RMSD value of 1.27 Å. The validation was carried out with 1000 modes and exhaustiveness of 1000, selecting the lowest energy value. Visualization and overlay of the co-crystalized ligand and the validation ligand were performed with symbol 2.5.
4.5. Molecular Docking
The docking of 3-TOP protein with butanamide
5 was simulated using AutoDock Vina, which has been used to estimate the conformation of protein-ligand complexes [35] and significantly improves the average accuracy of the binding mode predictions. The ligand and protein were prepared and saved in PDBQT format to carry out molecular docking. The x,y,z box size was set to 20 Å with grid spacing of 1.00 Å and centered at x = -51.08, y = 8.075, and z = -62.481. Autodock Vina was configured for 1000 modes and a exhaustiveness of 1000. The lowest energy mode was aligned to the receiver structure for analysis. Both pymol 2.5 (
https://pymol.org) and Discovery Studio 2021 (
https://discover.3ds.com/discovery-studio-visualizer-download) were used to visualize the protein-ligand interaction.
5. Conclusions
With a straightforward methodology, this two-step linear synthesis allowed us to obtain the compound of interest in an overall yield of 70%. Based on the results of the docking studies carried out, this compound has the potential to be an inhibitor of the alpha-glucosidase enzyme and, thus, an antidiabetic drug.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, 1H and 13C NMR spectra of the compounds 3 and 5 are available online.
Author Contributions
Conceptualization, H.P.X and E.H.N; performing synthesis, H.P.X.; computational analysis, E.H.N. and G.C.N.; investigation, R.O.A.; resources, E.H.N. and G.N.V.; writing—original draft preparation, H.P.X.; review and editing, E.H.N and G.N.V.; supervision, E.H.N and G.N.V. All authors have read and agreed to the published version of the manuscript.
Funding
The authors thank the National Council for Science and Technology (CONACYT) for its financial support for project 254321.
Data Availability Statement
Not applicable.
Acknowledgments
The authors thank Patricia Quintana for providing access to LANNBIO facilities. GCN and HPX also thank CONACYT for Graduate Scholarship 832462 and Postdoctoral Scholarship 592119, respectively.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Nguyen, T. B.; Ermolenko, L.; Dean, W. A.; Al-Mourabit, A. Benzazoles from aliphatic amines and o-amino/mercaptan/hidroxyanilines: elemental sulfur as a highly efficient and traceless oxidizing agent. Org. Lett. 2012, 14, 5948-5951. [CrossRef]. [CrossRef]
- Shrivas, P.; Zodape, S.; Wankhade, A.; Pratap, U. Facile synthesis of benzazoles through biocatalytic cyclization and dehydrogenation employing catalase in water. Enzyme Microb. Technol. 2020, 138, 109562. [CrossRef]. [CrossRef]
- Wang, M. Y.; Cheng, X. C.; Chen, X. B.; Li, Y.; Zang, L. L.; Duan, Y. Q.; Chen, M. Z.; Yu, P.; Sun, H.; Wang, R. L. Synthesis and biological evaluation of novel N-aryl-ω-(benzoazol-2-yl)-sulfanylalkanamides as dual inhibitors of α-glucosidase and protein tyrosine phosphatase 1B. Chemical Biology and Drug Design. 2018, 92(3), 1647–1656. [CrossRef]
- Papadopoulou, C.; Geronikaki, A.; Hadjipavlou-Litina, D. Synthesis and biological evaluation of new thiazolyl/benzothiazolyl-amides, derivatives of 4-phenyl-piperazine. Il Farmaco. 2005, 60(11-12), 969-973. [CrossRef]
- Azbill, R. D.; Mu, X.; Springer, J. E. Riluzole increases high-affinity glutamate uptake in rat spinal cord synaptosomes. Brain research. 2000, 871(2), 175-180. [CrossRef]
- Prabhat, P.; Das, S. K.; Shafaat, K.; Anand, J. P. Evaluation of N-(6-chlorobenzothiazol-2-yl)-2-(substitutedamino)acetamide for its Anti-bacterial Activity. Int J Pharm Sci Res. 2012, 3(8), 2669-2674. [CrossRef]
- Bhausaheb, D. K.; Madhukar, G. V.; Balkrishna, S. S. Jippr.Human. Synthesis and Evaluation of Antifungal Activity of Benzothiazole Derivatives. 2015, 3(1), 112-124.
- Sun, H.; Saedi, P.; Karuranga, S.; Pinkepank, M.; Orgutsova, K.; Dunca, B. B.; Stein, C.; Basit, A.; Chan, J. C.; Mbanya, J. C. IDF Diabetes Atlas: global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. J. Diabetes Res. 2022, 183, 109119. [CrossRef]
- Zang, W.; Kim, D.; Philip, E.; Miyan, Z.; Barykina, I.; Schmidt.; Stein, H. A multitational, observational study to investigate the efficacy, safety and tolerability of acarbose as add-on or monotherapy in a range of patiens: The glucoVIP study. Clinical Drug Investigation. 2013, 33(4), 263-274. [CrossRef]
- El-Subbagh, H. I.; Hassan, G. S.; El-Azab, A. S.; Abdel-Aziz, A. A. M.; Kadi, A. A.; Al-Obaid, A. M.; Al-Shabanah, O. A.; Sayed-Ahmed, M. M. Synthesis and anticonvulsant activity of some new thiazolo [3,2-a][1,3] diazepine, benzo[d]thiazolo[5,2-a][12,6]diazepine and benzo[d]oxazolo[5,2-a][12, 6]diazepine analogues. Eur. J. Med. Chem. 2011, 46(11), 5567–5572. [CrossRef]
- Trott, O.; Olson, A. J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31(2), 455–461. [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A. F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61(8), 3891–3898. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).