Submitted:
14 June 2024
Posted:
17 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
1.1. The Role of Phytochemicals in Traditional Medicine
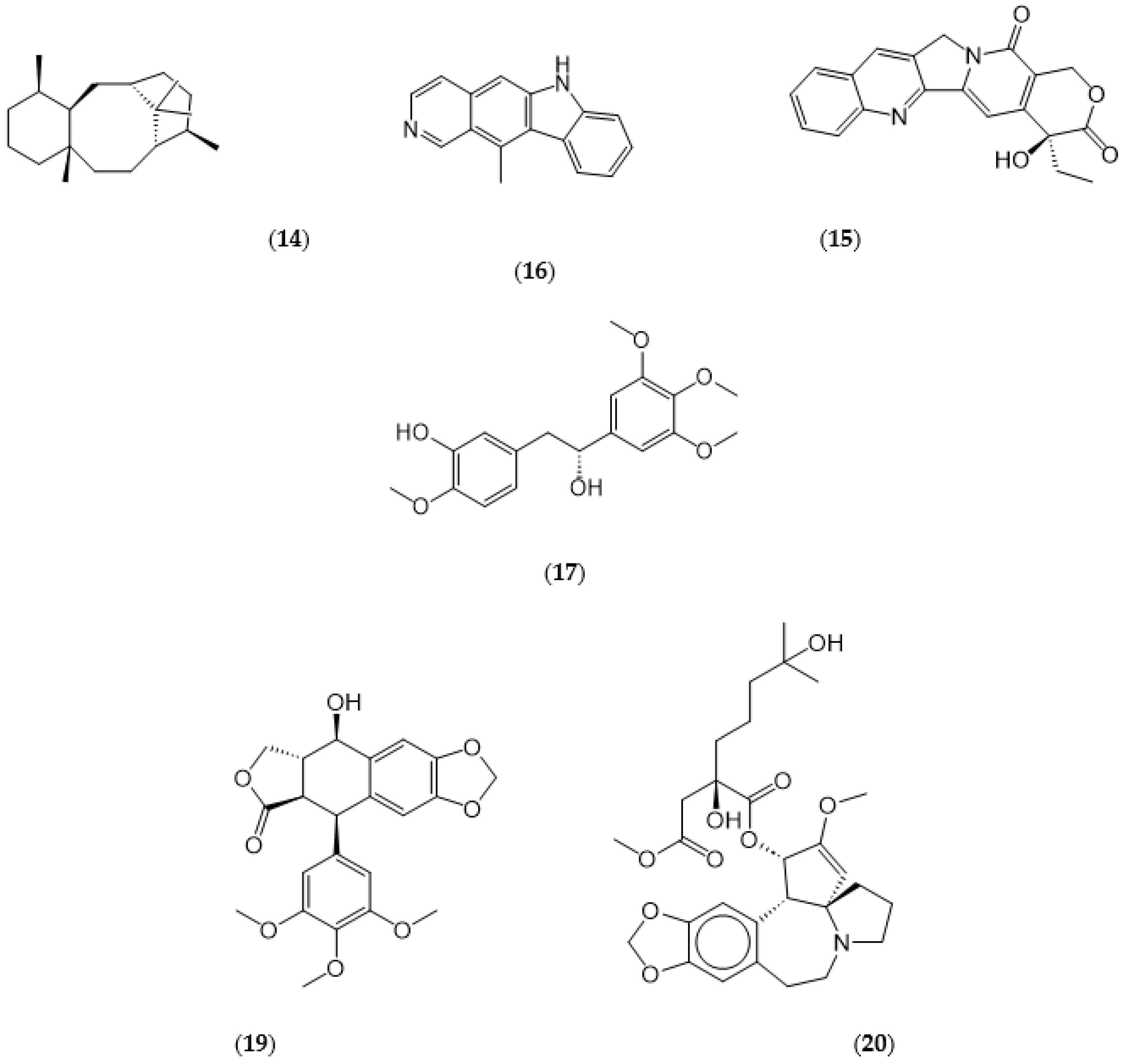
1.2. Examples of approved commercial phytochemical drugs
2. Phytochemicals and their modern-day applications
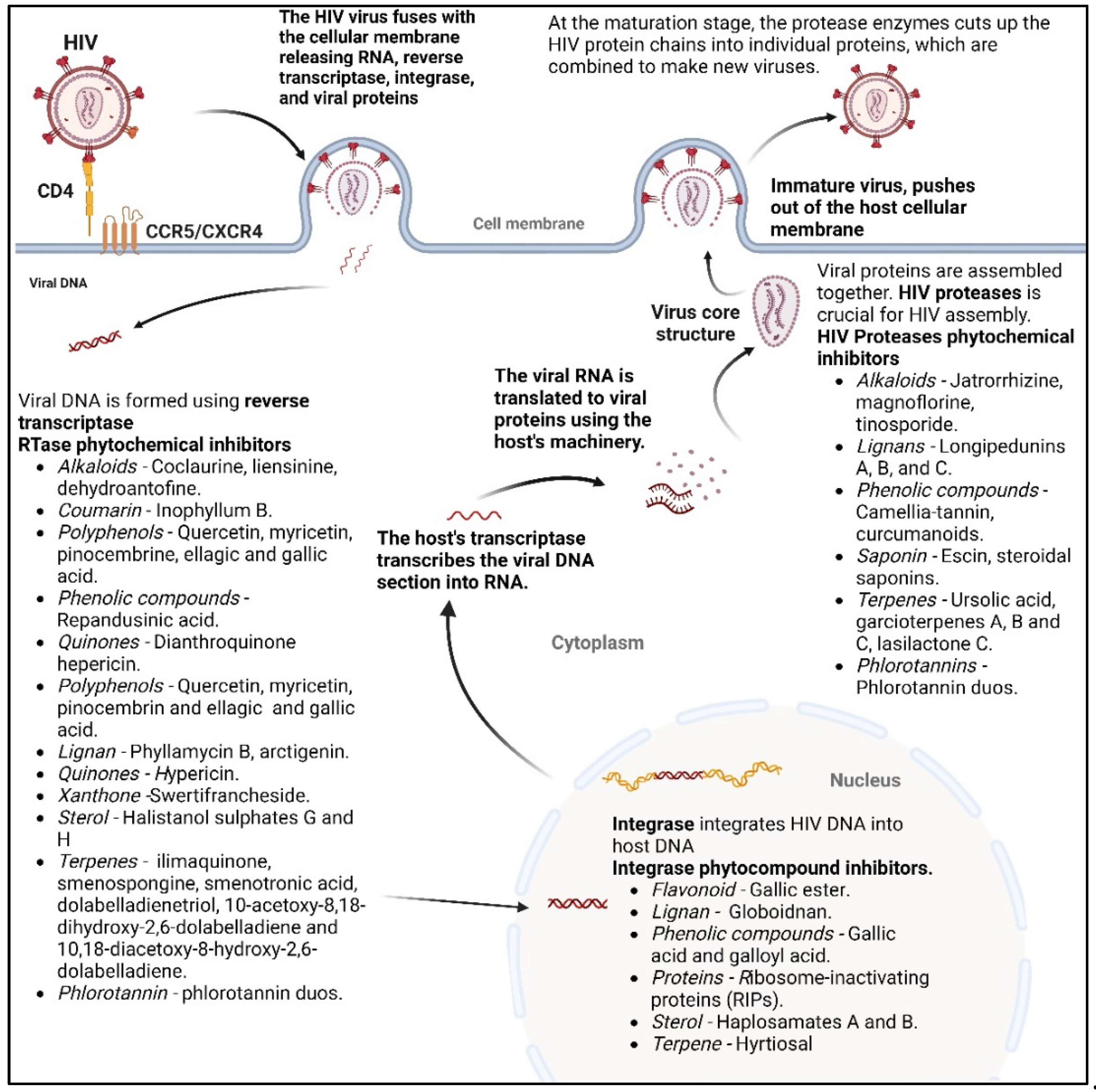
2.1. Phytochemicals as antivirals
2.2. Phytochemicals in Cancer Combination Therapies
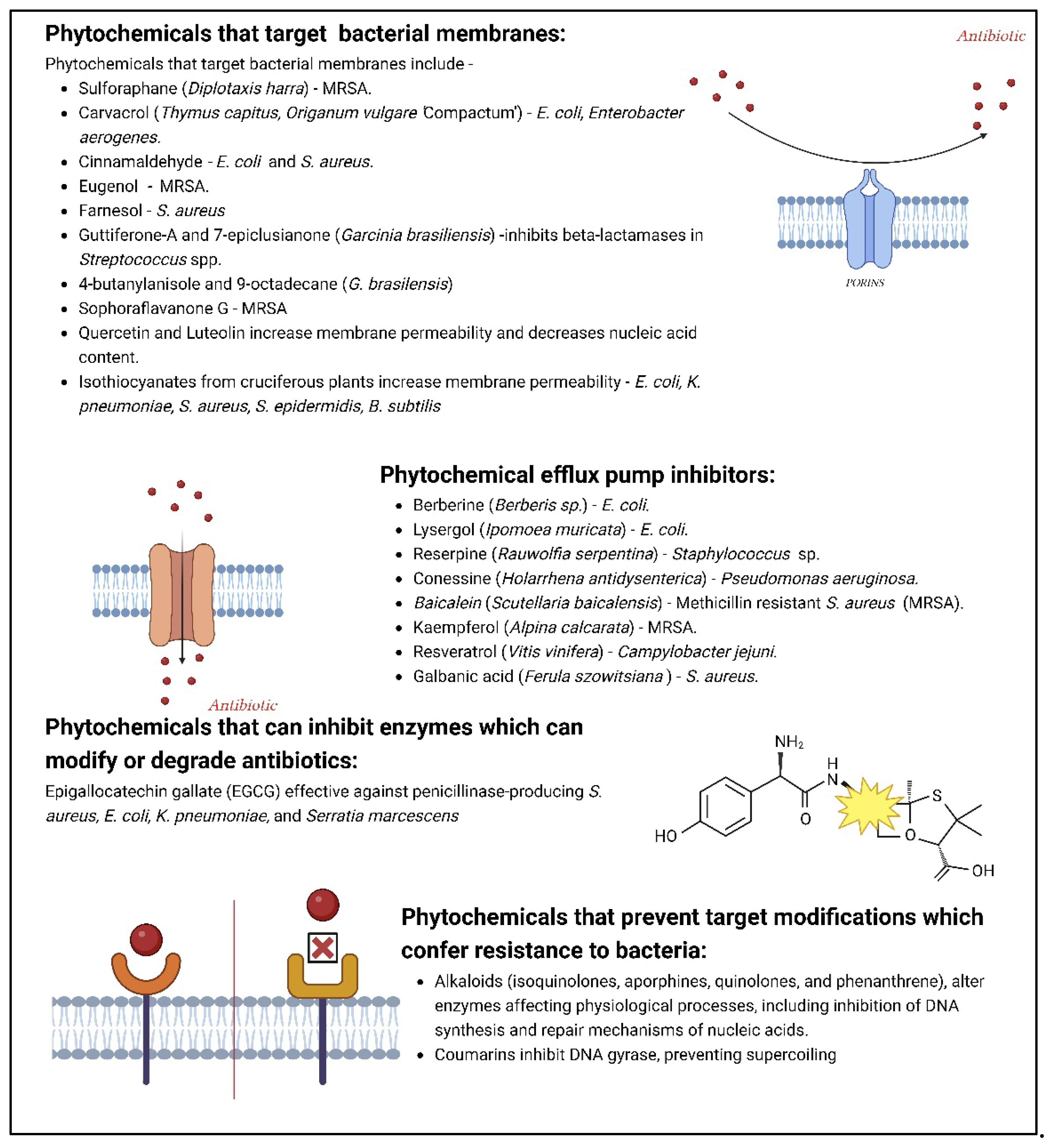
2.3. Phytochemicals as antimicrobials
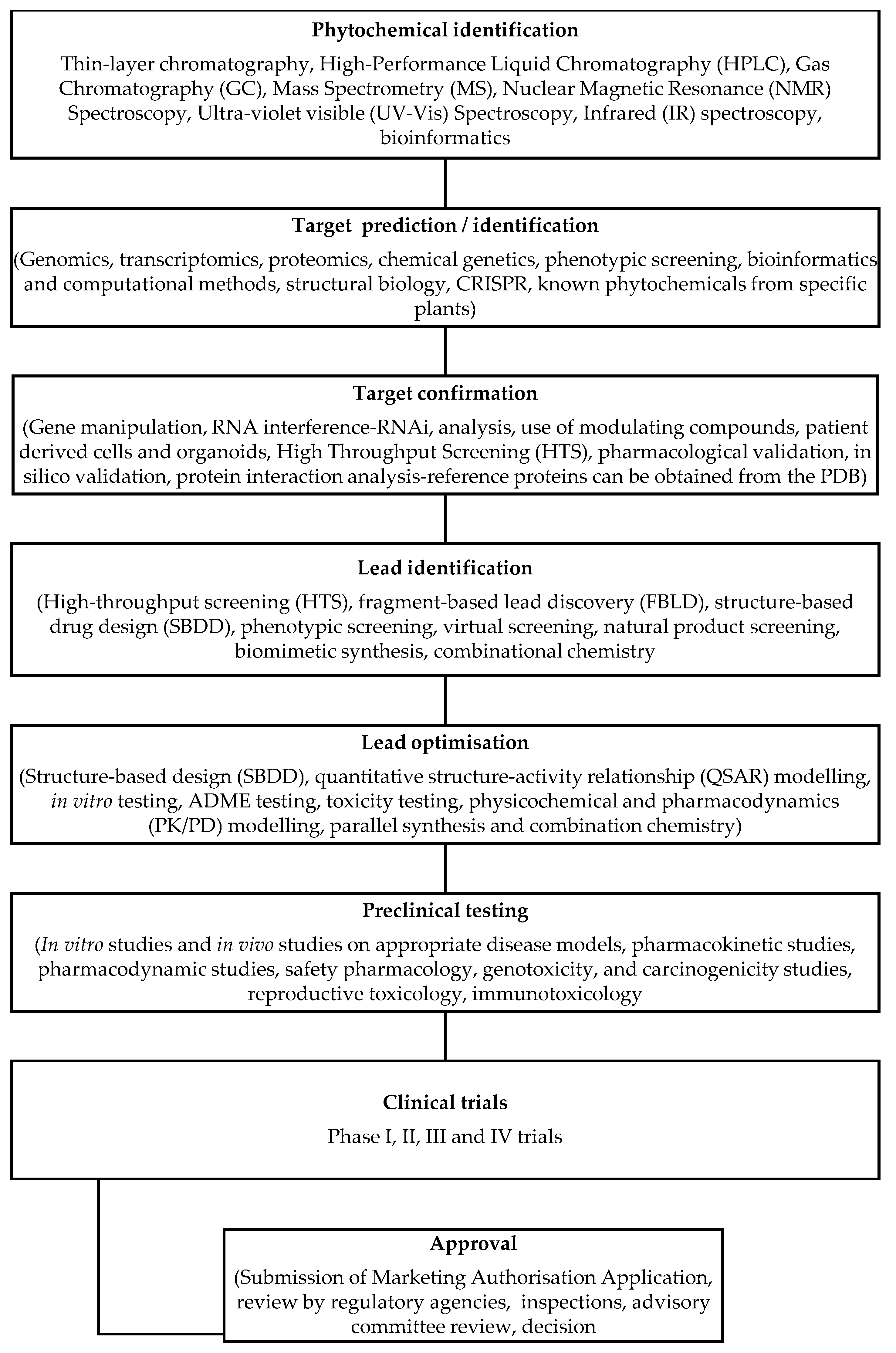
3. Drug discovery approaches using phytochemicals
3.1. Traditional versus modern drug discovery methods
3.1.1. Traditional Drug Discovery Methods
3.1.2. Modern Drug Discovery Methods
4. Computational approaches to identifying potential phytochemical drugs
4.1. Molecular Docking
4.2. Molecular Dynamics
4.3. Machine Learning and Artificial Intelligence
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Orhan, I.E.; Banach, M.; Rollinger, J.M.; et al. Natural products in drug discovery: advances and opportunities. Nat. Rev. Drug Discov 2021, 20, 200–216. [Google Scholar] [CrossRef] [PubMed]
- Ourmazd, A.; Moffat, K.; Lattman, E.E. Structural biology is solved — now what? Nat. Methods 2022, 19, 24–26. [Google Scholar] [CrossRef] [PubMed]
- Lemonnier, N.; Zhou, G.-B.; Prasher, B.; Mukerji, M.; Chen, Z.; Brahmachari, S.K.; et al. Traditional Knowledge-based Medicine: A Review of History, Principles, and Relevance in the Present Context of P4 Systems Medicine. Prog Prev Med 2017, 2, e0011. [Google Scholar] [CrossRef]
- Sheng-Ji, P. Ethnobotanical Approaches of Traditional Medicine Studies: Some Experiences From Asia. Pharm Biol 2001, 39, 74–79. [Google Scholar]
- Garcia, S. Pandemics and Traditional Plant-Based Remedies. A Historical-Botanical Review in the Era of COVID19. Front Plant Sci 2020, 11, 571042. [Google Scholar] [CrossRef] [PubMed]
- Kurhekar, J.V. Chapter 4 - Ancient and modern practices in phytomedicine. In Preparation of Phytopharmaceuticals for the Management of Disorders; Egbuna, C., Mishra, A.P., Goyal, M.R., Eds.; Eds.; Publisher: Academic Press, 2021; pp. 55–75. [Google Scholar]
- Mendelsohn, R.; Balick, M.J. The value of undiscovered pharmaceuticals in tropical forests. Econ Bot 1995, 49, 223–228. [Google Scholar] [CrossRef]
- Weng, J.K. Plant Solutions for the COVID-19 Pandemic and Beyond: Historical Reflections and Future Perspectives. Mol Plant 2020, 13, 803–807. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, S.S.; Sahoo, C.R.; Paidesetty, S.K.; Padhy, R.N. Role of phytocompounds as the potential anti-viral agent: an overview. Naunyn Schmiedebergs Arch Pharmacol 2023, 396, 2311–2329. [Google Scholar] [CrossRef] [PubMed]
- Farmanpour-Kalalagh, K.; Beyraghdar Kashkooli, A.; Babaei, A.; Rezaei, A. , van der Krol, A.R. Artemisinins in Combating Viral Infections Like SARS-CoV-2, Inflammation and Cancers and Options to Meet Increased Global Demand. Front Plant Sci 2022, 13. [Google Scholar] [CrossRef]
- Chang, Y.S.; Seo, E.K.; Gyllenhaal, C.; Block, K.I. Panax ginseng: a role in cancer therapy? Integr Cancer Ther 2003, 2, 13–33. [Google Scholar] [CrossRef]
- Akaberi, M.; Sahebkar, A.; Emami, S.A. Turmeric and Curcumin: From Traditional to Modern Medicine. Adv Exp Med Biol 2021, 1291, 15–39. [Google Scholar] [PubMed]
- Aucoin, M.; Cooley, K.; Saunders, P.R.; Carè, J.; Anheyer, D.; Medina, D.N.; et al. The effect of Echinacea spp. on the prevention or treatment of COVID-19 and other respiratory tract infections in humans: A rapid review. Adv Integr Med 2020, 7, 203–217. [Google Scholar] [CrossRef] [PubMed]
- Nagoor Meeran, M.F.; Javed, H.; Sharma, C.; Goyal, S.N.; Kumar, S.; Jha, N.K.; et al. Can Echinacea be a potential candidate to target immunity, inflammation, and infection - The trinity of coronavirus disease 2019. Heliyon 2021, 7. [Google Scholar] [CrossRef]
- Manayi, A.; Vazirian, M.; Saeidnia, S. Echinacea purpurea: Pharmacology, phytochemistry and analysis methods. Pharmacogn Rev 2015, 9, 63–72. [Google Scholar] [PubMed]
- Barnes, J.; Anderson, L.A.; Gibbons, S.; Phillipson, J.D. Echinacea species (Echinacea angustifolia (DC. ) Hell., Echinacea pallida (Nutt.) Nutt.,Echinacea purpurea (L.) Moench): a review of their chemistry, pharmacology and clinical properties. J Pharm Pharmacol 2005, 57, 929–954. [Google Scholar]
- Rawat, P.; Singh, P.K.; Kumar, V. Evidence based traditional anti-diarrheal medicinal plants and their phytocompounds. Biomed Pharmacother 2017, 96, 1453–1464. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, M.; Lee Teoh, H. Galanthamine from snowdrop--the development of a modern drug against Alzheimer's disease from local Caucasian knowledge. J Ethnopharmacol 2004, 92, 147–162. [Google Scholar] [CrossRef] [PubMed]
- Deleu, D.; Hanssens, Y.; Northway, M.G. Subcutaneous apomorphine: an evidence-based review of its use in Parkinson's disease. Drugs Aging 2004, 21, 687–709. [Google Scholar] [CrossRef]
- van Agtmael, M.A.; Eggelte, T.A. , van Boxtel, C.J. Artemisinin drugs in the treatment of malaria: from medicinal herb to registered medication. Trends Pharmacol Sci 1999, 20, 199–205. [Google Scholar] [CrossRef]
- Mundy, C.; Kirkpatrick, P. Tiotropium bromide. Nat Rev Drug Discov 2004, 3, 643–644. [Google Scholar] [CrossRef]
- Khameneh, B.; Eskin, N.A.M.; Iranshahy, M.; Fazly Bazzaz, B.S. Phytochemicals: A Promising Weapon in the Arsenal against Antibiotic-Resistant Bacteria. Antibiotics (Basel) 2021, 10, 10. [Google Scholar] [CrossRef] [PubMed]
- de la Torre, B.G.; Albericio, F. The Pharmaceutical Industry in 2023: An Analysis of FDA Drug Approvals from the Perspective of Molecules. Mol 2024, 29. [Google Scholar] [CrossRef] [PubMed]
- Choudhari, A.S.; Mandave, P.C.; Deshpande, M.; Ranjekar, P.; Prakash, O. Phytochemicals in Cancer Treatment: From Preclinical Studies to Clinical Practice. Front Pharmacol 2019, 10, 1614. [Google Scholar] [CrossRef] [PubMed]
- Gasparini, R.; Amicizia, D.; Lai, P.L.; Panatto, D. Clinical and socioeconomic impact of seasonal and pandemic influenza in adults and the elderly. Hum Vaccin Immunother 2012, 8, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Jassim, S.A.A.; Naji, M.A. Novel antiviral agents: a medicinal plant perspective. J Appl Microbiol 2003, 95, 412–427. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, R.; Dev, K.; Sourirajan, A. Antiviral activity of bioactive phytocompounds against coronavirus: An update. J Virol Methods 2021, 290, 114070. [Google Scholar] [CrossRef] [PubMed]
- Sankar, M.; Ramachandran, B.; Pandi, B.; Mutharasappan, N.; Ramasamy, V.; Prabu, P.G.; et al. In silico Screening of Natural Phytocompounds Towards Identification of Potential Lead Compounds to Treat COVID-19. Front Mol Biosci 2021, 8, 8. [Google Scholar] [CrossRef] [PubMed]
- Kar, B.; Dehury, B.; Singh, M.K.; Pati, S.; Bhattacharya, D. Identification of phytocompounds as newer antiviral drugs against COVID-19 through molecular docking and simulation based study. J Mol Graph Model 2022, 114, 108192. [Google Scholar] [CrossRef] [PubMed]
- Gul, I.; Hassan, A.; Haq, E.; Ahmad, S.M.; Shah, R.A.; Ganai, N.A.; et al. An Investigation of the Antiviral Potential of Phytocompounds against Avian Infectious Bronchitis Virus through Template-Based Molecular Docking and Molecular Dynamics Simulation Analysis. Viruses 2023, 15, 847. [Google Scholar] [CrossRef]
- Abd El Fadeel, M.R.; El-Dakhly, A.T.; Allam, A.M.; Farag, T.K.; El-Kholy, A.A. Preparation and efficacy of freeze-dried inactivated vaccine against bovine viral diarrhea virus genotypes 1 and 2, bovine herpes virus type 1.1, bovine parainfluenza-3 virus, and bovine respiratory syncytial virus. Clin Exp Vaccine Res.
- Reed, J.; Orme, A.; El-Demerdash, A.; Owen, C.; Martin, L.B.B.; Misra, R.C.; et al. Elucidation of the pathway for biosynthesis of saponin adjuvants from the soapbark tree. Science 2023, 379, 1252–1264. [Google Scholar] [CrossRef]
- Jamshidnia, M.; Sewell, R.D.E.; Rafieian-Kopaei, M. An Update on Promising Agents against COVID-19: Secondary Metabolites and Mechanistic Aspects. Curr Pharm Des 2022, 28, 2415–2425. [Google Scholar] [CrossRef] [PubMed]
- Musarra-Pizzo, M.; Pennisi, R.; Ben-Amor, I.; Mandalari, G.; Sciortino, M.T. Antiviral Activity Exerted by Natural Products against Human Viruses. Viruses 2021, 13. [Google Scholar] [CrossRef] [PubMed]
- Thakur, M.; Singh, M.; Kumar, S.; Dwivedi, V.P.; Dakal, T.C.; Yadav, V. A Reappraisal of the Antiviral Properties of and Immune Regulation through Dietary Phytochemicals. ACS Pharmacol Transl Sci 2023, 6, 1600–1615. [Google Scholar] [CrossRef] [PubMed]
- Loureiro, J.A.; Pereira, M.C. PLGA Based Drug Carrier and Pharmaceutical Applications: The Most Recent Advances. Pharmaceutics 2020, 12. [Google Scholar] [CrossRef]
- Mares, A.G.; Pacassoni, G.; Marti, J.S.; Pujals, S.; Albertazzi, L. Formulation of tunable size PLGA-PEG nanoparticles for drug delivery using microfluidic technology. PLoS One 2021, 16, e0251821. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Yao, L. Antiviral Effects of Plant-Derived Essential Oils and Their Components: An Updated Review. Mol 2020, 25. [Google Scholar] [CrossRef] [PubMed]
- Priyadarsani Mandhata, C.; Ranjan Sahoo, C.; Nath Padhy, R. A comprehensive overview on the role of phytocompounds in human immunodeficiency virus treatment. J Integr Med 2023, 21, 332–353. [Google Scholar] [CrossRef]
- Kaur, R.; Sharma, P.; Gupta, G.K.; Ntie-Kang, F.; Kumar, D. Structure-Activity-Relationship and Mechanistic Insights for Anti-HIV Natural Products. Mol 2020, 25, 2070. [Google Scholar] [CrossRef]
- Zhang, H.-J.; Rumschlag-Booms, E.; Guan, Y.-F.; Wang, D.-Y.; Liu, K.-L.; Li, W.-F.; et al. Potent Inhibitor of Drug-Resistant HIV-1 Strains Identified from the Medicinal Plant Justicia gendarussa. J Nat Prod 2017, 80, 1798–1807. [Google Scholar] [CrossRef]
- Oladele, J.O.; Ajayi, E.I.; Oyeleke, O.M.; Oladele, O.T.; Olowookere, B.D.; Adeniyi, B.M.; et al. A systematic review on COVID-19 pandemic with special emphasis on curative potentials of Nigeria based medicinal plants. Heliyon 2020, 6, e04897. [Google Scholar] [CrossRef]
- da Silva, A.M.; Horsth, A.L.; Timóteo, É.d.S.; Faria, R.J.; Bazoni, P.S.; Meira, E.F.; et al. Use of medicinal plants during COVID-19 pandemic in Brazil. Sci Rep 2023, 13, 16558. [Google Scholar] [CrossRef] [PubMed]
- Khadka, D.; Dhamala, M.K.; Li, F.; Aryal, P.C.; Magar, P.R.; Bhatta, S.; et al. The use of medicinal plants to prevent COVID-19 in Nepal. J Ethnobiology Ethnomed 2021, 172, 6. [Google Scholar] [CrossRef] [PubMed]
- Bellik, Y. ; M. Hammoudi, S.; Abdellah, F.; Iguer-Ouada, M.; Boukraa, L. Phytochemicals to Prevent Inflammation and Allergy. Recent Pat on Inflam Allergy Drug Discov 2012, 6, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Wahedi, H.M.; Ahmad, S.; Abbasi, S.W. Stilbene-based natural compounds as promising drug candidates against COVID-19. J Biomol Struct Dyn 2021, 39, 3225–3234. [Google Scholar] [CrossRef] [PubMed]
- Ahmad Hafidul, A.; Feri Eko, H.; Adzral, A.; Iva Himmatul, A.; Fatchiyah, F. Virtual prediction of antiviral potential of ginger (Zingiber officinale) bioactive compounds against spike and MPro of SARS-CoV2 protein. Berk Penelit Hayati 2020, 25. [Google Scholar]
- Choudhary, N.; Bawari, S.; Burcher, J.T.; Sinha, D.; Tewari, D.; Bishayee, A. Targeting Cell Signaling Pathways in Lung Cancer by Bioactive Phytocompounds. Cancers 2023, 15, 3980. [Google Scholar] [CrossRef]
- Ansari, P.; Akther, S.; Hannan, J.M.A.; Seidel, V.; Nujat, N.J.; Abdel-Wahab, Y.H.A. Pharmacologically Active Phytomolecules Isolated from Traditional Antidiabetic Plants and Their Therapeutic Role for the Management of Diabetes Mellitus. Mol 2022, 27. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, S.; Matsumori, N. Chemical diversity and mode of action of natural products targeting lipids in the eukaryotic cell membrane. Nat Prod Rep 2020, 37, 677–702. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, A. Potential Role of Bioactive Phytochemicals in Combination Therapies against Antimicrobial Activity. J Pharmacopuncture 2022, 25, 79–87. [Google Scholar] [CrossRef]
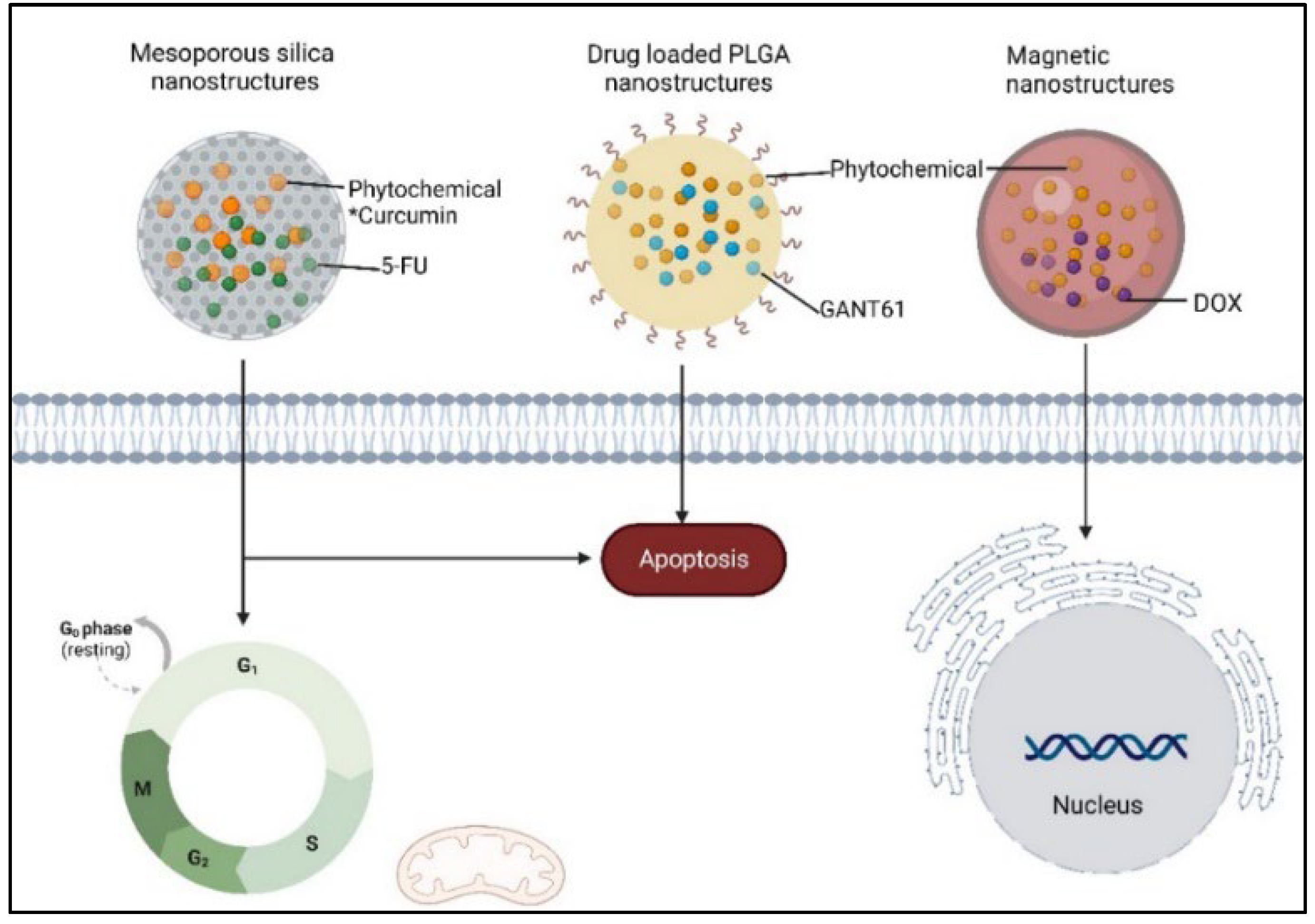
- Zandieh, M.A.; Farahani, M.H.; Daryab, M.; Motahari, A.; Gholami, S.; Salmani, F.; et al. Stimuli-responsive (nano)architectures for phytochemical delivery in cancer therapy. Biomedicine & Pharmacotherapy 2023, 166, 115283. [Google Scholar]
- Rossiter, S.E.; Fletcher, M.H.; Wuest, W.M. Natural Products as Platforms To Overcome Antibiotic Resistance. Chem Rev 2017, 117, 12415–12474. [Google Scholar] [CrossRef] [PubMed]
- Talib, W.H.; Alsayed, A.R.; Barakat, M.; Abu-Taha, M.I.; Mahmod, A.I. Targeting Drug Chemo-Resistance in Cancer Using Natural Products. Biomedicines 2021, 9. [Google Scholar] [CrossRef] [PubMed]
- Reichel, A.; Lienau, P. Pharmacokinetics in Drug Discovery: An Exposure-Centred Approach to Optimising and Predicting Drug Efficacy and Safety. Handb Exp Pharmacol 2016, 232, 235–260. [Google Scholar] [PubMed]
- Irurzun-Arana, I.; McDonald, T.O.; Trocóniz, I.F.; Michor, F. Pharmacokinetic Profiles Determine Optimal Combination Treatment Schedules in Computational Models of Drug Resistance. Cancer Res 2020, 80, 3372–3382. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Warden, A.R.; Ding, X. The optimization of combinatorial drug therapies: Strategies and laboratorial platforms. Drug Discov Today 2021, 26, 2646–2659. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, V.M.; Zhang, H.; Dalby, P.A.; Aylott, J.W. Advancements in the co-formulation of biologic therapeutics. J Control Release 2020, 327, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Strohbehn, G.W.; Kacew, A.J.; Goldstein, D.A.; Feldman, R.C.; Ratain, M.J. Combination therapy patents: a new front in evergreening. Nat Biotechnol 2021, 39, 1504–1510. [Google Scholar] [CrossRef] [PubMed]
- Plana, D.; Palmer, A.C.; Sorger, P.K. Independent Drug Action in Combination Therapy: Implications for Precision Oncology. Cancer Discov 2022, 12, 606–624. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, L.; Lin, Z.; Yu, D.; Jin, M.; Zhou, P.; et al. Targeting DCLK1 overcomes 5-fluorouracil resistance in colorectal cancer through inhibiting CCAR1/beta-catenin pathway-mediated cancer stemness. Clin Transl Med 2022, 12. [Google Scholar]
- Wang, D.; Yu, D.; Liu, X.; Wang, Q.; Chen, X.; Hu, X.; et al. Targeting laryngeal cancer cells with 5-fluorouracil and curcumin using mesoporous silica nanoparticles. Technol Cancer Res T 2020, 19, 1533033820962114. [Google Scholar] [CrossRef]
- Harada, K.; Ohashi, R.; Naito, K.; Kanki, K. Hedgehog Signal Inhibitor GANT61 Inhibits the Malignant Behavior of Undifferentiated Hepatocellular Carcinoma Cells by Targeting Non-Canonical GLI Signaling. Int J Mol Sci 2020, 21, 21. [Google Scholar] [CrossRef] [PubMed]
- Borah, A.; Pillai, S.C.; Rochani, A.K.; Palaninathan, V.; Nakajima, Y.; Maekawa, T.; et al. GANT61 and curcumin-loaded PLGA nanoparticles for GLI1 and PI3K/Akt-mediated inhibition in breast adenocarcinoma. Nanotechnology 2020, 31. [Google Scholar] [CrossRef] [PubMed]
- Ak, G.; Karakayalı, T.; Cin, A.N.; Özel, B.; Şanlıer, Ş.H. One pot green synthesis of doxorubicin and curcumin loaded magnetic nanoparticles and cytotoxicity studies. Anticancer Agents Med Chem 2021, 21, 2563–2571. [Google Scholar] [CrossRef]
- Mazumder, K.; Aktar, A.; Roy, P.; Biswas, B.; Hossain, M.E.; Sarkar, K.K.; et al. A Review on Mechanistic Insight of Plant Derived Anticancer Bioactive Phytocompounds and Their Structure Activity Relationship. Mol 2022, 27. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Guo, X.; Han, F.; He, Z.; Wang, Y. Emerging role of natural products in cancer immunotherapy. Acta Pharmaceutica Sinica B 2022, 12, 1163–1185. [Google Scholar] [CrossRef] [PubMed]
- Mignani, S.; Rodrigues, J.; Tomas, H.; Zablocka, M.; Shi, X.; Caminade, A.M.; et al. Dendrimers in combination with natural products and analogues as anticancer agents. Chem Soc Reviews 2018, 47, 514–532. [Google Scholar] [CrossRef] [PubMed]
- Braicu, C.; Mehterov, N.; Vladimirov, B.; Sarafian, V.; Nabavi, S.M.; Atanasov, A.G.; et al. Nutrigenomics in cancer: Revisiting the effects of natural compounds. Semin Cancer Biol 2017, 46, 84–106. [Google Scholar] [CrossRef] [PubMed]
- Efferth, T.; Saeed, M.E.M.; Kadioglu, O.; Seo, E.-J.; Shirooie, S.; Mbaveng, A.T.; et al. Collateral sensitivity of natural products in drug-resistant cancer cells. Biotechnol Adv 2020, 38, 107342. [Google Scholar] [CrossRef] [PubMed]
- Fontana, F.; Raimondi, M.; Di Domizio, A.; Moretti, R.M.; Montagnani Marelli, M.; Limonta, P. Unraveling the molecular mechanisms and the potential chemopreventive/therapeutic properties of natural compounds in melanoma. Sem Cancer Biol 2019, 59, 266–282. [Google Scholar] [CrossRef]
- Ye, J.; Zhang, R.; Wu, F.; Zhai, L.; Wang, K.; Xiao, M.; et al. Non-apoptotic cell death in malignant tumor cells and natural compounds. Cancer Lett 2018, 420, 210–227. [Google Scholar] [CrossRef]
- Fontana, F.; Raimondi, M.; Marzagalli, M.; Di Domizio, A.; Limonta, P. The emerging role of paraptosis in tumor cell biology: Perspectives for cancer prevention and therapy with natural compounds. Biochim Biophys Acta - Rev Cancer, 1873. [Google Scholar]
- Chhabra, G.; Singh, C.K.; Ndiaye, M.A.; Fedorowicz, S.; Molot, A.; Ahmad, N. Prostate cancer chemoprevention by natural agents: Clinical evidence and potential implications. Cancer Lett 2018, 422, 9–18. [Google Scholar] [CrossRef]
- Garg, N.; Luzzatto-Knaan, T.; Melnik, A.V.; Caraballo-Rodríguez, A.M.; Floros, D.J.; Petras, D.; et al. Natural products as mediators of disease. Nat Prod Rep 2017, 34, 194–219. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.D.; Liu, X.P.; Zhao, W.J.; Dong, Q.; Li, F.N.; Wang, H.B.; et al. Curcumin induces apoptosis in breast cancer cells and inhibits tumor growth in vitro and in vivo. Int J Clin Exp Pathol 2014, 7, 2818–2824. [Google Scholar] [PubMed]
- Gallardo, M.; Calaf, G.M. Curcumin inhibits invasive capabilities through epithelial mesenchymal transition in breast cancer cell lines. Int J Oncol 2016, 49, 1019–1027. [Google Scholar] [CrossRef] [PubMed]
- Varinska, L.; Gal, P.; Mojzisova, G.; Mirossay, L.; Mojzis, J. Soy and Breast Cancer: Focus on Angiogenesis. Int J Mol Sci 2015, 16, 11728–11749. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.-J.; Xie, M.-Y.; Kluxen, F.M.; Diel, P. Genistein modulates the anti-tumor activity of cisplatin in MCF-7 breast and HT-29 colon cancer cells. Arch Toxicol 2014, 88, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J Nat Prod 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed]
- Khare, T.; Anand, U.; Dey, A.; Assaraf, Y.G.; Chen, Z.S.; Liu, Z.; et al. Exploring Phytochemicals for Combating Antibiotic Resistance in Microbial Pathogens. Front Pharmacol 2021, 12, 720726. [Google Scholar] [CrossRef] [PubMed]
- Mak, S.; Xu, Y.; Nodwell, J.R. The expression of antibiotic resistance genes in antibiotic-producing bacteria. Mol Microbiol. 2014, 93, 391–402. [Google Scholar] [CrossRef]
- Perumal Samy, R.; Gopalakrishnakone, P. Therapeutic Potential of Plants as Anti-microbials for Drug Discovery. Evid Based Complement Alternat Med 2010, 7, 283–294. [Google Scholar] [CrossRef]
- Moiketsi, B.N.; Makale, K.P.P.; Rantong, G.; Rahube, T.O.; Makhzoum, A. Potential of Selected African Medicinal Plants as Alternative Therapeutics against Multi-Drug-Resistant Bacteria. Biomedicines 2023, 11, 11. [Google Scholar] [CrossRef]
- Suganya, T.; Packiavathy, I.A.S.V.; Aseervatham, G.S.B.; Carmona, A.; Rashmi, V.; Mariappan, S.; et al. Tackling Multiple-Drug-Resistant Bacteria With Conventional and Complex Phytochemicals. Front Cell Infect Microbiol 2022, 12. [Google Scholar] [CrossRef]
- Scherlach, K.; Hertweck, C. Mining and unearthing hidden biosynthetic potential. Nat Comm 2021, 12, 3864. [Google Scholar] [CrossRef]
- Campos, K.R.; Coleman, P.J.; Alvarez, J.C.; Dreher, S.D.; Garbaccio, R.M.; Terrett, N.K.; et al. The importance of synthetic chemistry in the pharmaceutical industry. Science 2019, 363, eaat0805. [Google Scholar] [CrossRef]
- Holzmeyer, L.; Hartig, A.-K.; Franke, K.; Brandt, W.; Muellner-Riehl, A.N.; Wessjohann, L.A.; et al. Evaluation of plant sources for antiinfective lead compound discovery by correlating phylogenetic, spatial, and bioactivity data. Proc Nat Acad Sci 2020, 117, 12444–12451. [Google Scholar] [CrossRef]
- Yu, W.; MacKerell, A.D.; Jr. Computer-Aided Drug Design Methods. Methods Mol Biol 2017, 1520, 85–106. [Google Scholar]
- Mithun, R.; Shubham, J.K.; Anil, G.J. Drug Repurposing (DR): An Emerging Approach in Drug Discovery. In IntechOpen, Farid, A.B., editor. Drug Repurposing. Rijeka, 2020.
- Vijayakumar, S.; Prabhu, S.; Rajalakhsmi, S.; Manogar, P. Review on potential phytocompounds in drug development for Parkinson disease: A pharmacoinformatic approach. Inform Med Unlocked 2016, 5, 15–25. [Google Scholar] [CrossRef]
- Han, H.S.; Koo, S.Y.; Choi, K.Y. Emerging nanoformulation strategies for phytocompounds and applications from drug delivery to phototherapy to imaging. Bioact Mater 2022, 14, 182–205. [Google Scholar] [CrossRef]
- Li, J.W.; Vederas, J.C. Drug discovery and natural products: end of an era or an endless frontier? Science. 2009, 325, 161–165. [Google Scholar] [CrossRef]
- Zhou, S.F.; Zhong, W.Z. Drug Design and Discovery: Principles and Applications. Mol 2017, 22, 22. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, S.; Kleywegt, G.J. A paradigm shift in structural biology. Nat Methods 2022, 19, 20–23. [Google Scholar] [CrossRef]
- Tan, S.Y.; Tatsumura, Y. Alexander Fleming (1881-1955): Discoverer of penicillin. Singapore Med J 2015, 56, 366–367. [Google Scholar] [CrossRef]
- Beutler, J.A. Natural Products as a Foundation for Drug Discovery. Curr Protoc Pharmacol 2019, 86, e67. [Google Scholar] [CrossRef]
- Bárcena, M.; Barnes, C.O.; Beck, M.; Bjorkman, P.J.; Canard, B.; Gao, G.F.; et al. Structural biology in the fight against COVID-19. Nat Struct Mol Biol 2021, 28, 2–7. [Google Scholar] [CrossRef]
- Masrati, G.; Landau, M.; Ben-Tal, N.; Lupas, A.; Kosloff, M.; Kosinski, J. Integrative Structural Biology in the Era of Accurate Structure Prediction. J Mol Biol 2021, 433, 167127. [Google Scholar] [CrossRef]
- Ziegler, S.J.; Mallinson, S.J.B.; St John, P.C.; Bomble, Y.J. Advances in integrative structural biology: Towards understanding protein complexes in their cellular context. Comput Struct Biotechnol J 2021, 19, 214–225. [Google Scholar] [CrossRef]
- Mahapatra, M.K.; Karuppasamy, M. Fundamental considerations in drug design. Computer Aided Drug Design (CADD): From Ligand-Based Methods to Structure-Based Approaches. 2022, 17-55.
- Dreiman, G.H.S.; Bictash, M.; Fish, P.V.; Griffin, L.; Svensson, F. Changing the HTS Paradigm: AI-Driven Iterative Screening for Hit Finding. SLAS Discov 2021, 26, 257–262. [Google Scholar] [CrossRef]
- Prudent, R.; Annis, D.A.; Dandliker, P.J.; Ortholand, J.-Y.; Roche, D. Exploring new targets and chemical space with affinity selection-mass spectrometry. Nat Rev Chem 2021, 5, 62–71. [Google Scholar] [CrossRef]
- Bender, B.J.; Gahbauer, S.; Luttens, A.; Lyu, J.; Webb, C.M.; Stein, R.M.; et al. A practical guide to large-scale docking. Nat Protoc 2021, 16, 4799–4832. [Google Scholar] [CrossRef]
- Stanzione, F.; Giangreco, I.; Cole, J.C. Use of molecular docking computational tools in drug discovery. Prog Med Chem 2021, 60, 273–343. [Google Scholar]
- Ou-Yang, S.S.; Lu, J.Y.; Kong, X.Q.; Liang, Z.J.; Luo, C.; Jiang, H. Computational drug discovery. Acta Pharmacol Sin 2012, 33, 1131–1140. [Google Scholar] [CrossRef]
- Leelananda, S.P.; Lindert, S. Computational methods in drug discovery. Beilstein J Org Chem 2016, 12, 2694–2718. [Google Scholar] [CrossRef]
- Sadybekov, A.V.; Katritch, V. Computational approaches streamlining drug discovery. Nature 2023, 616, 673–685. [Google Scholar] [CrossRef]
- Rallabandi, H.R.; Mekapogu, M.; Natesan, K.; Saindane, M.; Dhupal, M.; Swamy, M.K.; et al. Computational Methods Used in Phytocompound-Based Drug Discovery. In Plant-derived Bioactives: Chemistry and Mode of Action; Swamy, M.K., editor. Springer Singapore, 2020. pp. 549–573.
- Das, S.K.; Mahanta, S.; Tanti, B.; Tag, H.; Hui, P.K. Identification of phytocompounds from Houttuynia cordata Thunb. as potential inhibitors for SARS-CoV-2 replication proteins through GC-MS/LC-MS characterization, molecular docking and molecular dynamics simulation. Mol Divers 2022, 26, 365–388. [Google Scholar] [CrossRef]
- Imaduwage, K.P.; Lakbub, J.; Go, E.P.; Desaire, H. Rapid LC-MS Based High-Throughput Screening Method, Affording No False Positives or False Negatives, Identifies a New Inhibitor for Carbonic Anhydrase. Sci Rep 2017, 7, 10324. [Google Scholar] [CrossRef]
- Wills, L.P. The use of high-throughput screening techniques to evaluate mitochondrial toxicity. Toxicology 2017, 391, 34–41. [Google Scholar] [CrossRef]
- Roy, A. High-Throughput Screening (HTS) Technology. In Encyclopedia of Molecular Pharmacology; Offermanns, S., Rosenthal, W., Eds, *!!! REPLACE !!!*, Eds.; Cham: Springer International Publishing, 2021; pp. 787–799. [Google Scholar]
- Li, G.; Peng, X.; Guo, Y.; Gong, S.; Cao, S.; Qiu, F. Currently Available Strategies for Target Identification of Bioactive Natural Products. Front Chem 2021, 9, 761609. [Google Scholar] [CrossRef]
- Asiamah, I.; Obiri, S.A.; Tamekloe, W.; Armah, F.A.; Borquaye, L.S. Applications of molecular docking in natural products-based drug discovery. Sci Afr 2023, 20, e01593. [Google Scholar] [CrossRef]
- Reddy, A.S.; Pati, S.P.; Kumar, P.P.; Pradeep, H.N.; Sastry, G.N. Virtual screening in drug discovery -- a computational perspective. Curr Protein Pept Sci 2007, 8, 329–351. [Google Scholar] [CrossRef]
- Lavecchia, A.; Di Giovanni, C. Virtual screening strategies in drug discovery: a critical review. Curr Med Chem 2013, 20, 2839–2860. [Google Scholar] [CrossRef]
- Kontoyianni, M. Docking and Virtual Screening in Drug Discovery. Methods Mol Biol 2017, 1647, 255–266. [Google Scholar]
- Hert, J.; Willett, P.; Wilton, D.J.; Acklin, P.; Azzaoui, K.; Jacoby, E.; et al. Comparison of fingerprint-based methods for virtual screening using multiple bioactive reference structures. J Chem Inf Comput Sci 2004, 44, 1177–1185. [Google Scholar] [CrossRef]
- Qing, X.; Yin Lee, X.; De Raeymaeker, J.; Rh Tame, J.; Yj Zhang, K.; De Maeyer, M.; et al. Pharmacophore modeling: advances, limitations, and current utility in drug discovery. J Receptor Ligand Channel Res 2014, 7, 81–92. [Google Scholar]
- Muegge, I.; Mukherjee, P. An overview of molecular fingerprint similarity search in virtual screening. Expert Opin Drug Discov 2016, 11, 137–148. [Google Scholar] [CrossRef]
- Santana, K. , do Nascimento, L.D.; Lima, E.L.A.; Damasceno, V.; Nahum, C.; Braga, R.C.; et al. Applications of Virtual Screening in Bioprospecting: Facts, Shifts, and Perspectives to Explore the Chemo-Structural Diversity of Natural Products. Front Chem 2021, 9, 662688. [Google Scholar] [CrossRef]
- Das, A.P.; Agarwal, S.M. Recent advances in the area of plant-based anticancer drug discovery using computational approaches. Mol Divers 2023, 1–25. [Google Scholar]
- Padhi, A.K.; Janežič, M.; Zhang, K.Y.J. Chapter 26 - Molecular dynamics simulations: Principles, methods, and applications in protein conformational dynamics. In Advances in Protein Molecular and Structural Biology Methods; Tripathi, T.; Dubey, V.K.; Eds.; Academic Press, 2022, pp. 439–454.
- Zhang, W.; Chien, J.; Yong, J.; Kuang, R. Network-based machine learning and graph theory algorithms for precision oncology. Precis Oncol 2017, 1, 25. [Google Scholar] [CrossRef]
- Kumar, S.; Kumar, S. Chapter 6 - Molecular Docking: A Structure-Based Approach for Drug Repurposing. In: Roy, K., editor. In Silico Drug Design: Academic Press; 2019. pp. 161–189.
- Silvestrini, L.; Belhaj, N.; Comez, L.; Gerelli, Y.; Lauria, A.; Libera, V.; et al. The dimer-monomer equilibrium of SARS-CoV-2 main protease is affected by small molecule inhibitors. Sci Rep 2021, 11, 9283. [Google Scholar] [CrossRef]
- Chugh, A.; Sehgal, I.; Khurana, N.; Verma, K.; Rolta, R.; Vats, P.; et al. Comparative docking studies of drugs and phytocompounds for emerging variants of SARS-CoV-2. 3 Biotech 2023, 13, 36. [Google Scholar] [CrossRef]
- Jamiu, A.T.; Pohl, C.H.; Bello, S.; Adedoja, T.; Sabiu, S. A review on molecular docking analysis of phytocompounds against SARS-CoV-2 druggable targets. All Life 2021, 14, 1100–1128. [Google Scholar] [CrossRef]
- Swargiary, G.; Mani, S. ER and PGR targeting ability of phytocompounds derived from Centella asiatica and Andrographis paniculata: An in-silico approach. J Herb Med 2022, 32, 100541. [Google Scholar] [CrossRef]
- Gayathiri, E.; Prakash, P.; Ahamed, M.; Pandiaraj, S.; Venkidasamy, B.; Dayalan, H.; et al. Multitargeted pharmacokinetics, molecular docking and network pharmacology-based identification of effective phytocompounds from Sauropus androgynus (L.) Merr for inflammation and cancer treatment. J Biomol Struct Dyn.
- Mao, J.; Akhtar, J.; Zhang, X.; Sun, L.; Guan, S.; Li, X.; et al. Comprehensive strategies of machine-learning-based quantitative structure-activity relationship models. iScience 2021, 24, 103052. [Google Scholar] [CrossRef]
- Paul, D.; Sanap, G.; Shenoy, S.; Kalyane, D.; Kalia, K.; Tekade, R.K. Artificial intelligence in drug discovery and development. Drug Discov Today 2021, 26, 80–93. [Google Scholar] [CrossRef]
- Shin, S.H.; Hur, G.; Kim, N.R.; Park, J.H.Y.; Lee, K.W.; Yang, H. A machine learning-integrated stepwise method to discover novel anti-obesity phytochemicals that antagonize the glucocorticoid receptor. Food Func 2023, 14, 1869–1883. [Google Scholar] [CrossRef]
- Yabuuchi, H.; Hayashi, K.; Shigemoto, A.; Fujiwara, M.; Nomura, Y.; Nakashima, M.; et al. Virtual screening of antimicrobial plant extracts by machine-learning classification of chemical compounds in semantic space. PLoS One 2023, 18, e0285716. [Google Scholar] [CrossRef]
- García-Pérez, P.; Lozano-Milo, E.; Landín, M.; Gallego, P.P. Combining Medicinal Plant In Vitro Culture with Machine Learning Technologies for Maximizing the Production of Phenolic Compounds. Antioxidants (Basel) 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Idowu, S.O.; Fatokun, A.A. Artificial Intelligence (AI) to the Rescue: Deploying Machine Learning to Bridge the Biorelevance Gap in Antioxidant Assays. SLAS Technol 2021, 26, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Orhan, I.E.; Banach, M.; Rollinger, J.M.; et al. Natural products in drug discovery: advances and opportunities. Nat. Rev. Drug Discov. 2021, 202, 00–216. [Google Scholar] [CrossRef] [PubMed]
- Ourmazd, A.; Moffat, K.; Lattman, E.E. Structural biology is solved — now what? Nature Methods 2022, 192, 4–26. [Google Scholar] [CrossRef] [PubMed]
- Lemonnier, N.; Zhou, G.-B.; Prasher, B.; Mukerji, M.; Chen, Z.; Brahmachari, S.K.; et al. Traditional Knowledge-based Medicine: A Review of History, Principles, and Relevance in the Present Context of P4 Systems Medicine. Prog. Prev. Med. 2017, 2, e0011. [Google Scholar] [CrossRef]
- Sheng-Ji, P. Ethnobotanical Approaches of Traditional Medicine Studies: Some Experiences From Asia. Pharm. Biol. 2001, 39, 74–79. [Google Scholar] [PubMed]
- Garcia, S. Pandemics and Traditional Plant-Based Remedies. A Historical-Botanical Review in the Era of COVID19. Front Plant Sci 2020, 115, 71042. [Google Scholar] [CrossRef] [PubMed]
- Kurhekar, J.V. Chapter 4 - Ancient and modern practices in phytomedicine. In: Egbuna, C.; Mishra, A.P.; Goyal, M.R., editors. Preparation of Phytopharmaceuticals for the Management of Disorders: Academic Press; 2021. p. 55-75.
- Mendelsohn, R.; Balick, M.J. The value of undiscovered pharmaceuticals in tropical forests. Econ. Bot. 1995, 492, 23–228. [Google Scholar] [CrossRef]
- Weng, J.K. Plant Solutions for the COVID-19 Pandemic and Beyond: Historical Reflections and Future Perspectives. Mol. Plant 2020, 138, 03–807. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, S.S.; Sahoo, C.R.; Paidesetty, S.K.; Padhy, R.N. Role of phytocompounds as the potential anti-viral agent: an overview. Naunyn Schmiedebergs Arch. Pharmacol. 2023, 3962, 311–2329. [Google Scholar] [CrossRef] [PubMed]
- Farmanpour-Kalalagh, K.; Beyraghdar Kashkooli, A.; Babaei, A.; Rezaei, A. , van der Krol, A.R. Artemisinins in Combating Viral Infections Like SARS-CoV-2, Inflammation and Cancers and Options to Meet Increased Global Demand. Frontiers in Plant Science, 2022; 12. [Google Scholar]
- Chang, Y.S.; Seo, E.K.; Gyllenhaal, C.; Block, K.I. Panax ginseng: a role in cancer therapy? Integr Cancer Ther 2003, 21, 3–33. [Google Scholar] [CrossRef]
- Akaberi, M.; Sahebkar, A.; Emami, S.A. Turmeric and Curcumin: From Traditional to Modern Medicine. Adv. Exp. Med. Biol. 2021, 12911, 5–39. [Google Scholar]
- Aucoin, M.; Cooley, K.; Saunders, P.R.; Carè, J.; Anheyer, D.; Medina, D.N.; et al. The effect of Echinacea spp. on the prevention or treatment of COVID-19 and other respiratory tract infections in humans: A rapid review. Adv. Integr. Med. 2020, 72, 03–217. [Google Scholar] [CrossRef] [PubMed]
- Nagoor Meeran, M.F.; Javed, H.; Sharma, C.; Goyal, S.N.; Kumar, S.; Jha, N.K.; et al. Can Echinacea be a potential candidate to target immunity, inflammation, and infection - The trinity of coronavirus disease 2019. Heliyon 2021, 7, e05990. [Google Scholar] [CrossRef] [PubMed]
- Manayi, A.; Vazirian, M.; Saeidnia, S. Echinacea purpurea: Pharmacology, phytochemistry and analysis methods. Pharmacogn. Rev. 2015, 96, 3–72. [Google Scholar]
- Barnes, J.; Anderson, L.A.; Gibbons, S.; Phillipson, J.D. Echinacea species (Echinacea angustifolia (DC.) Hell., Echinacea pallida (Nutt.) Nutt.,Echinacea purpurea (L.) Moench): a review of their chemistry, pharmacology and clinical properties. J. Pharm. Pharmacol. 2005, 579, 29–954. [Google Scholar]
- Rawat, P.; Singh, P.K.; Kumar, V. Evidence based traditional anti-diarrheal medicinal plants and their phytocompounds. Biomed. Pharmacother. 2017, 961, 453–1464. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, M.; Lee Teoh, H. Galanthamine from snowdrop--the development of a modern drug against Alzheimer's disease from local Caucasian knowledge. J. Ethnopharmacol. 2004, 92, 147–162. [Google Scholar] [CrossRef] [PubMed]
- Deleu, D.; Hanssens, Y.; Northway, M.G. Subcutaneous apomorphine : an evidence-based review of its use in Parkinson's disease. Drugs Aging 2004, 216, 87–709. [Google Scholar] [CrossRef] [PubMed]
- van Agtmael, M.A.; Eggelte, T.A. , van Boxtel, C.J. Artemisinin drugs in the treatment of malaria: from medicinal herb to registered medication. Trends Pharmacol. Sci. 1999, 201, 99–205. [Google Scholar]
- Mundy, C.; Kirkpatrick, P. Tiotropium bromide. Nat. Rev. Drug Discov. 2004, 36, 43–644. [Google Scholar] [CrossRef]
- Khameneh, B.; Eskin, N.A.M.; Iranshahy, M.; Fazly Bazzaz, B.S. Phytochemicals: A Promising Weapon in the Arsenal against Antibiotic-Resistant Bacteria. Antibiotics 2021, 10. [Google Scholar] [CrossRef]
- de la Torre, B.G.; Albericio, F. The Pharmaceutical Industry in 2023: An Analysis of FDA Drug Approvals from the Perspective of Molecules. Molecules 2024, 29. [Google Scholar] [CrossRef]
- Gasparini, R.; Amicizia, D.; Lai, P.L.; Panatto, D. Clinical and socioeconomic impact of seasonal and pandemic influenza in adults and the elderly. Hum. Vaccin. Immunother. 2012, 82, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Jassim, S.A.A.; Naji, M.A. Novel antiviral agents: a medicinal plant perspective. J. Appl. Microbiol. 2003, 954, 12–427. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, R.; Dev, K.; Sourirajan, A. Antiviral activity of bioactive phytocompounds against coronavirus: An update. J. Virol. Methods 2021, 2901, 14070. [Google Scholar] [CrossRef] [PubMed]
- Sankar, M.; Ramachandran, B.; Pandi, B.; Mutharasappan, N.; Ramasamy, V.; Prabu, P.G.; et al. In silico Screening of Natural Phytocompounds Towards Identification of Potential Lead Compounds to Treat COVID-19. Frontiers in Molecular Biosciences 2021, 8. [Google Scholar] [CrossRef]
- Kar, B.; Dehury, B.; Singh, M.K.; Pati, S.; Bhattacharya, D. Identification of phytocompounds as newer antiviral drugs against COVID-19 through molecular docking and simulation based study. J. Mol. Graph. Model. 2022, 1141, 08192. [Google Scholar] [CrossRef] [PubMed]
- Gul, I.; Hassan, A.; Haq, E.; Ahmad, S.M.; Shah, R.A.; Ganai, N.A.; et al. An Investigation of the Antiviral Potential of Phytocompounds against Avian Infectious Bronchitis Virus through Template-Based Molecular Docking and Molecular Dynamics Simulation Analysis. Viruses 2023, 158, 47. [Google Scholar] [CrossRef]
- Abd El Fadeel, M.R.; El-Dakhly, A.T.; Allam, A.M.; Farag, T.K.; El-Kholy, A.A. Preparation and efficacy of freeze-dried inactivated vaccine against bovine viral diarrhea virus genotypes 1 and 2, bovine herpes virus type 1.1, bovine parainfluenza-3 virus, and bovine respiratory syncytial virus. Clin. Exp. Vaccine Res. 2020, 91, 19–125. [Google Scholar] [CrossRef] [PubMed]
- Reed, J.; Orme, A.; El-Demerdash, A.; Owen, C.; Martin, L.B.B.; Misra, R.C.; et al. Elucidation of the pathway for biosynthesis of saponin adjuvants from the soapbark tree. Science 2023, 3791, 252–1264. [Google Scholar] [CrossRef]
- Jamshidnia, M.; Sewell, R.D.E.; Rafieian-Kopaei, M. An Update on Promising Agents against COVID-19: Secondary Metabolites and Mechanistic Aspects. Curr. Pharm. Des. 2022, 282, 415–2425. [Google Scholar] [CrossRef]
- Musarra-Pizzo, M.; Pennisi, R.; Ben-Amor, I.; Mandalari, G.; Sciortino, M.T. Antiviral Activity Exerted by Natural Products against Human Viruses. Viruses 2021, 13. [Google Scholar] [CrossRef] [PubMed]
- Thakur, M.; Singh, M.; Kumar, S.; Dwivedi, V.P.; Dakal, T.C.; Yadav, V. A Reappraisal of the Antiviral Properties of and Immune Regulation through Dietary Phytochemicals. ACS Pharmacol. Transl. Sci. 2023, 61, 600–1615. [Google Scholar] [CrossRef] [PubMed]
- Loureiro, J.A.; Pereira, M.C. PLGA Based Drug Carrier and Pharmaceutical Applications: The Most Recent Advances. Pharmaceutics 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Mares, A.G.; Pacassoni, G.; Marti, J.S.; Pujals, S.; Albertazzi, L. Formulation of tunable size PLGA-PEG nanoparticles for drug delivery using microfluidic technology. PLoS One, 2021; 16, e0251821. [Google Scholar]
- Ma, L.; Yao, L. Antiviral Effects of Plant-Derived Essential Oils and Their Components: An Updated Review. Molecules 2020, 25. [Google Scholar] [CrossRef] [PubMed]
- Priyadarsani Mandhata, C.; Ranjan Sahoo, C.; Nath Padhy, R. A comprehensive overview on the role of phytocompounds in human immunodeficiency virus treatment. J. Integr. Med. 2023, 213, 32–353. [Google Scholar] [CrossRef] [PubMed]
- Kaur, R.; Sharma, P.; Gupta, G.K.; Ntie-Kang, F.; Kumar, D. Structure-Activity-Relationship and Mechanistic Insights for Anti-HIV Natural Products. Molecules 2020, 252, 070. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-J.; Rumschlag-Booms, E.; Guan, Y.-F.; Wang, D.-Y.; Liu, K.-L.; Li, W.-F.; et al. Potent Inhibitor of Drug-Resistant HIV-1 Strains Identified from the Medicinal Plant Justicia gendarussa. J. Nat. Prod. 2017, 801, 798–1807. [Google Scholar]
- Oladele, J.O.; Ajayi, E.I.; Oyeleke, O.M.; Oladele, O.T.; Olowookere, B.D.; Adeniyi, B.M.; et al. A systematic review on COVID-19 pandemic with special emphasis on curative potentials of Nigeria based medicinal plants. Heliyon, 2020; 6, e04897. [Google Scholar]
- Villena-Tejada, M.; Vera-Ferchau, I.; Cardona-Rivero, A.; Zamalloa-Cornejo, R.; Quispe-Florez, M.; Frisancho-Triveño, Z.; et al. Use of medicinal plants for COVID-19 prevention and respiratory symptom treatment during the pandemic in Cusco, Peru: A cross-sectional survey. PLoS One, 2021; 16, e0257165. [Google Scholar]
- da Silva, A.M.; Horsth, A.L.; Timóteo, É.d.S.; Faria, R.J.; Bazoni, P.S.; Meira, E.F.; et al. Use of medicinal plants during COVID-19 pandemic in Brazil. Sci. Rep. 2023, 131, 6558. [Google Scholar] [CrossRef]
- Khadka, D.; Dhamala, M.K.; Li, F.; Aryal, P.C.; Magar, P.R.; Bhatta, S.; et al. The use of medicinal plants to prevent COVID-19 in Nepal. J. Ethnobiol. Ethnomedicine 2021, 172, 6. [Google Scholar] [CrossRef]
- Bellik, Y.; M. Hammoudi, S.; Abdellah, F.; Iguer-Ouada, M.; Boukraa, L. Phytochemicals to Prevent Inflammation and Allergy. Recent. Pat. Inflamm. Allergy Drug Discov. 2012, 61, 47–158. [Google Scholar]
- Wahedi, H.M.; Ahmad, S.; Abbasi, S.W. Stilbene-based natural compounds as promising drug candidates against COVID-19. J. Biomol. Struct. Dyn. 2021, 393, 225–3234. [Google Scholar] [CrossRef] [PubMed]
- Ahmad Hafidul, A.; Feri Eko, H.; Adzral, A.; Iva Himmatul, A.; Fatchiyah, F. Virtual prediction of antiviral potential of ginger (Zingiber officinale) bioactive compounds against spike and MPro of SARS-CoV2 protein. Berkala Penelitian Hayati 2020, 25. [Google Scholar]
- Choudhary, N.; Bawari, S.; Burcher, J.T.; Sinha, D.; Tewari, D.; Bishayee, A. Targeting Cell Signaling Pathways in Lung Cancer by Bioactive Phytocompounds. Cancers 2023, 153, 980. [Google Scholar] [CrossRef]
- Ansari, P.; Akther, S.; Hannan, J.M.A.; Seidel, V.; Nujat, N.J.; Abdel-Wahab, Y.H.A. Pharmacologically Active Phytomolecules Isolated from Traditional Antidiabetic Plants and Their Therapeutic Role for the Management of Diabetes Mellitus. Molecules 2022, 27. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, S.; Matsumori, N. Chemical diversity and mode of action of natural products targeting lipids in the eukaryotic cell membrane. Nat. Prod. Rep. 2020, 376, 77–702. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, A. Potential Role of Bioactive Phytochemicals in Combination Therapies against Antimicrobial Activity. J. Pharmacopunct. 2022, 257, 9–87. [Google Scholar] [CrossRef] [PubMed]
- Zandieh, M.A.; Farahani, M.H.; Daryab, M.; Motahari, A.; Gholami, S.; Salmani, F.; et al. Stimuli-responsive (nano)architectures for phytochemical delivery in cancer therapy. Biomed. Pharmacother. 2023, 1661, 15283. [Google Scholar] [CrossRef] [PubMed]
- Rossiter, S.E.; Fletcher, M.H.; Wuest, W.M. Natural Products as Platforms To Overcome Antibiotic Resistance. Chem. Rev. 2017, 1171, 2415–12474. [Google Scholar] [CrossRef]
- Talib, W.H.; Alsayed, A.R.; Barakat, M.; Abu-Taha, M.I.; Mahmod, A.I. Targeting Drug Chemo-Resistance in Cancer Using Natural Products. Biomedicines 2021, 9. [Google Scholar] [CrossRef]
- Reichel, A.; Lienau, P. Pharmacokinetics in Drug Discovery: An Exposure-Centred Approach to Optimising and Predicting Drug Efficacy and Safety. Handb. Exp. Pharmacol. 2016, 2322, 35–260. [Google Scholar]
- Irurzun-Arana, I.; McDonald, T.O.; Trocóniz, I.F.; Michor, F. Pharmacokinetic Profiles Determine Optimal Combination Treatment Schedules in Computational Models of Drug Resistance. Cancer Res. 2020, 803, 372–3382. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Warden, A.R.; Ding, X. The optimization of combinatorial drug therapies: Strategies and laboratorial platforms. Drug Discov. Today 2021, 262, 646–2659. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, V.M.; Zhang, H.; Dalby, P.A.; Aylott, J.W. Advancements in the co-formulation of biologic therapeutics. J. Control. Release 2020, 3273, 97–405. [Google Scholar] [CrossRef] [PubMed]
- Strohbehn, G.W.; Kacew, A.J.; Goldstein, D.A.; Feldman, R.C.; Ratain, M.J. Combination therapy patents: a new front in evergreening. Nat. Biotechnol. 2021, 391, 504–1510. [Google Scholar] [CrossRef] [PubMed]
- Plana, D.; Palmer, A.C.; Sorger, P.K. Independent Drug Action in Combination Therapy: Implications for Precision Oncology. Cancer Discov. 2022, 126, 06–624. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhao, L.; Lin, Z.; Yu, D.; Jin, M.; Zhou, P.; et al. Targeting DCLK1 overcomes 5-fluorouracil resistance in colorectal cancer through inhibiting CCAR1/beta-catenin pathway-mediated cancer stemness. Clin. Transl. Med. 2022, 12. [Google Scholar]
- Wang, D.; Yu, D.; Liu, X.; Wang, Q.; Chen, X.; Hu, X.; et al. Targeting laryngeal cancer cells with 5-fluorouracil and curcumin using mesoporous silica nanoparticles. Technol. Cancer Res. Treat. 2020, 191, 533033820962114. [Google Scholar] [CrossRef] [PubMed]
- Harada, K.; Ohashi, R.; Naito, K.; Kanki, K. Hedgehog Signal Inhibitor GANT61 Inhibits the Malignant Behavior of Undifferentiated Hepatocellular Carcinoma Cells by Targeting Non-Canonical GLI Signaling. Int. J. Mol. Sci. 2020, 21. [Google Scholar] [CrossRef] [PubMed]
- Borah, A.; Pillai, S.C.; Rochani, A.K.; Palaninathan, V.; Nakajima, Y.; Maekawa, T.; et al. GANT61 and curcumin-loaded PLGA nanoparticles for GLI1 and PI3K/Akt-mediated inhibition in breast adenocarcinoma. Nanotechnology 2020, 31. [Google Scholar] [CrossRef]
- Ak, G.; Karakayalı, T.; Cin, A.N.; Özel, B.; Şanlıer, Ş.H. One pot green synthesis of doxorubicin and curcumin loaded magnetic nanoparticles and cytotoxicity studies. Anti-Cancer Agents Med. Chem. 2021, 212, 563–2571. [Google Scholar] [CrossRef]
- Mazumder, K.; Aktar, A.; Roy, P.; Biswas, B.; Hossain, M.E.; Sarkar, K.K.; et al. A Review on Mechanistic Insight of Plant Derived Anticancer Bioactive Phytocompounds and Their Structure Activity Relationship. Molecules 2022, 27. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Guo, X.; Han, F.; He, Z.; Wang, Y. Emerging role of natural products in cancer immunotherapy. Acta Pharm. Sin. B 2022, 121, 163–1185. [Google Scholar] [CrossRef] [PubMed]
- Mignani, S.; Rodrigues, J.; Tomas, H.; Zablocka, M.; Shi, X.; Caminade, A.M.; et al. Dendrimers in combination with natural products and analogues as anti-cancer agents. Chem. Soc. Rev. 2018, 475, 14–532. [Google Scholar] [CrossRef] [PubMed]
- Braicu, C.; Mehterov, N.; Vladimirov, B.; Sarafian, V.; Nabavi, S.M.; Atanasov, A.G.; et al. Nutrigenomics in cancer: Revisiting the effects of natural compounds. Semin. Cancer Biol. 2017, 468, 4–106. [Google Scholar] [CrossRef] [PubMed]
- Efferth, T.; Saeed, M.E.M.; Kadioglu, O.; Seo, E.-J.; Shirooie, S.; Mbaveng, A.T.; et al. Collateral sensitivity of natural products in drug-resistant cancer cells. Biotechnol. Adv. 2020, 381, 07342. [Google Scholar] [CrossRef] [PubMed]
- Fontana, F.; Raimondi, M.; Di Domizio, A.; Moretti, R.M.; Montagnani Marelli, M.; Limonta, P. Unraveling the molecular mechanisms and the potential chemopreventive/therapeutic properties of natural compounds in melanoma. Semin. Cancer Biol. 2019, 592, 66–282. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Zhang, R.; Wu, F.; Zhai, L.; Wang, K.; Xiao, M.; et al. Non-apoptotic cell death in malignant tumor cells and natural compounds. Cancer Lett. 2018, 4202, 10–227. [Google Scholar] [CrossRef] [PubMed]
- Fontana, F.; Raimondi, M.; Marzagalli, M.; Di Domizio, A.; Limonta, P. The emerging role of paraptosis in tumor cell biology: Perspectives for cancer prevention and therapy with natural compounds. Biochimica et Biophysica Acta - Reviews on Cancer, 2020; 1873. [Google Scholar]
- Chhabra, G.; Singh, C.K.; Ndiaye, M.A.; Fedorowicz, S.; Molot, A.; Ahmad, N. Prostate cancer chemoprevention by natural agents: Clinical evidence and potential implications. Cancer Letters 2018, 4229. [Google Scholar] [CrossRef] [PubMed]
- Garg, N.; Luzzatto-Knaan, T.; Melnik, A.V.; Caraballo-Rodríguez, A.M.; Floros, D.J.; Petras, D.; et al. Natural products as mediators of disease. Nat. Prod. Rep. 2017, 341, 94–219. [Google Scholar] [CrossRef]
- Lv, Z.D.; Liu, X.P.; Zhao, W.J.; Dong, Q.; Li, F.N.; Wang, H.B.; et al. Curcumin induces apoptosis in breast cancer cells and inhibits tumor growth in vitro and in vivo. Int. J. Clin. Exp. Pathol. 2014, 72, 818–2824. [Google Scholar]
- Gallardo, M.; Calaf, G.M. Curcumin inhibits invasive capabilities through epithelial mesenchymal transition in breast cancer cell lines. Int. J. Oncol. 2016, 491, 019–1027. [Google Scholar] [CrossRef]
- Varinska, L.; Gal, P.; Mojzisova, G.; Mirossay, L.; Mojzis, J. Soy and Breast Cancer: Focus on Angiogenesis. Int. J. Mol. Sci. 2015, 161, 1728–11749. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.-J.; Xie, M.-Y.; Kluxen, F.M.; Diel, P. Genistein modulates the anti-tumor activity of cisplatin in MCF-7 breast and HT-29 colon cancer cells. Arch. Toxicol. 2014, 886, 25–635. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 837, 70–803. [Google Scholar] [CrossRef]
- Khare, T.; Anand, U.; Dey, A.; Assaraf, Y.G.; Chen, Z.S.; Liu, Z.; et al. Exploring Phytochemicals for Combating Antibiotic Resistance in Microbial Pathogens. Front. Pharmacol. 2021, 127, 20726. [Google Scholar] [CrossRef]
- Mak, S.; Xu, Y.; Nodwell, J.R. The expression of antibiotic resistance genes in antibiotic-producing bacteria. Mol. Microbiol. 2014, 933, 91–402. [Google Scholar] [CrossRef]
- Perumal Samy, R.; Gopalakrishnakone, P. Therapeutic Potential of Plants as Anti-microbials for Drug Discovery. Evid. Based Complement. Altern. Med. 2010, 72, 83–294. [Google Scholar] [CrossRef] [PubMed]
- Moiketsi, B.N.; Makale, K.P.P.; Rantong, G.; Rahube, T.O.; Makhzoum, A. Potential of Selected African Medicinal Plants as Alternative Therapeutics against Multi-Drug-Resistant Bacteria. Biomedicines 2023, 11. [Google Scholar] [CrossRef] [PubMed]
- Suganya, T.; Packiavathy, I.A.S.V.; Aseervatham, G.S.B.; Carmona, A.; Rashmi, V.; Mariappan, S.; et al. Tackling Multiple-Drug-Resistant Bacteria With Conventional and Complex Phytochemicals. Frontiers in Cellular and Infection Microbiology 2022, 12. [Google Scholar] [CrossRef]
- Scherlach, K.; Hertweck, C. Mining and unearthing hidden biosynthetic potential. Nat. Commun. 2021, 123, 864. [Google Scholar] [CrossRef]
- Campos, K.R.; Coleman, P.J.; Alvarez, J.C.; Dreher, S.D.; Garbaccio, R.M.; Terrett, N.K.; et al. The importance of synthetic chemistry in the pharmaceutical industry. Science 2019, 363, eaat0805. [Google Scholar] [CrossRef]
- Holzmeyer, L.; Hartig, A.-K.; Franke, K.; Brandt, W.; Muellner-Riehl, A.N.; Wessjohann, L.A.; et al. Evaluation of plant sources for antiinfective lead compound discovery by correlating phylogenetic, spatial, and bioactivity data. Proc. Natl. Acad. Sci. 2020, 1171, 2444–12451. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; MacKerell, A.D.; Jr. Computer-Aided Drug Design Methods. Methods Mol. Biol. 2017, 15208, 5–106. [Google Scholar]
- Mithun, R.; Shubham, J.K.; Anil, G.J. Drug Repurposing (DR): An Emerging Approach in Drug Discovery. In: Farid, A.B., editor. Drug Repurposing. Rijeka: IntechOpen; 2020. p. Ch. 1.
- Vijayakumar, S.; Prabhu, S.; Rajalakhsmi, S.; Manogar, P. Review on potential phytocompounds in drug development for Parkinson disease: A pharmacoinformatic approach. Inform. Med. Unlocked 2016, 51, 5–25. [Google Scholar] [CrossRef]
- Han, H.S.; Koo, S.Y.; Choi, K.Y. Emerging nanoformulation strategies for phytocompounds and applications from drug delivery to phototherapy to imaging. Bioact. Mater. 2022, 141, 82–205. [Google Scholar] [CrossRef] [PubMed]
- Li, J.W.; Vederas, J.C. Drug discovery and natural products: end of an era or an endless frontier? Science 2009, 3251, 61–165. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.F.; Zhong, W.Z. Drug Design and Discovery: Principles and Applications. Molecules 2017, 22. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, S.; Kleywegt, G.J. A paradigm shift in structural biology. Nat. Methods 2022, 192, 0–23. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.Y.; Tatsumura, Y. Alexander Fleming (1881-1955): Discoverer of penicillin. Singap. Med. J. 2015, 563, 66–367. [Google Scholar] [CrossRef]
- Beutler, J.A. Natural Products as a Foundation for Drug Discovery. Curr. Protoc. Pharmacol. 2019; 86, e67. [Google Scholar]
- Bárcena, M.; Barnes, C.O.; Beck, M.; Bjorkman, P.J.; Canard, B.; Gao, G.F.; et al. Structural biology in the fight against COVID-19. Nat. Struct. Mol. Biol. 2021, 282. [Google Scholar] [CrossRef]
- Masrati, G.; Landau, M.; Ben-Tal, N.; Lupas, A.; Kosloff, M.; Kosinski, J. Integrative Structural Biology in the Era of Accurate Structure Prediction. J. Mol. Biol. 2021, 4331, 67127. [Google Scholar] [CrossRef]
- Ziegler, S.J.; Mallinson, S.J.B.; St John, P.C.; Bomble, Y.J. Advances in integrative structural biology: Towards understanding protein complexes in their cellular context. Comput. Struct. Biotechnol. J. 2021, 192, 14–225. [Google Scholar] [CrossRef]
- Mahapatra, M.K.; Karuppasamy, M. Fundamental considerations in drug design. Comput. Aided Drug Des. (CADD): Ligand-Based Methods Struct. -Based Approaches 2022, 1, 7–55. [Google Scholar]
- Dreiman, G.H.S.; Bictash, M.; Fish, P.V.; Griffin, L.; Svensson, F. Changing the HTS Paradigm: AI-Driven Iterative Screening for Hit Finding. SLAS Discov. 2021, 262, 57–262. [Google Scholar] [CrossRef] [PubMed]
- Prudent, R.; Annis, D.A.; Dandliker, P.J.; Ortholand, J.-Y.; Roche, D. Exploring new targets and chemical space with affinity selection-mass spectrometry. Nat. Rev. Chem. 2021, 56, 2–71. [Google Scholar] [CrossRef]
- Bender, B.J.; Gahbauer, S.; Luttens, A.; Lyu, J.; Webb, C.M.; Stein, R.M.; et al. A practical guide to large-scale docking. Nat. Protoc. 2021, 164, 799–4832. [Google Scholar] [CrossRef]
- Stanzione, F.; Giangreco, I.; Cole, J.C. Use of molecular docking computational tools in drug discovery. Prog. Med. Chem. 2021, 602, 73–343. [Google Scholar]
- Ou-Yang, S.S.; Lu, J.Y.; Kong, X.Q.; Liang, Z.J.; Luo, C.; Jiang, H. Computational drug discovery. Acta Pharmacol. Sin. 2012, 331, 131–1140. [Google Scholar] [CrossRef]
- Leelananda, S.P.; Lindert, S. Computational methods in drug discovery. Beilstein J. Org. Chem. 2016, 122, 694–2718. [Google Scholar] [CrossRef] [PubMed]
- Sadybekov, A.V.; Katritch, V. Computational approaches streamlining drug discovery. Nature 2023, 6166, 73–685. [Google Scholar] [CrossRef]
- Rallabandi, H.R.; Mekapogu, M.; Natesan, K.; Saindane, M.; Dhupal, M.; Swamy, M.K.; et al. Computational Methods Used in Phytocompound-Based Drug Discovery. In: Swamy, M.K., editor. Plant-derived Bioactives: Chemistry and Mode of Action. Singapore: Springer Singapore; 2020. p. 549-573.
- Das, S.K.; Mahanta, S.; Tanti, B.; Tag, H.; Hui, P.K. Identification of phytocompounds from Houttuynia cordata Thunb. as potential inhibitors for SARS-CoV-2 replication proteins through GC-MS/LC-MS characterization, molecular docking and molecular dynamics simulation. Mol. Divers. 2022, 263, 65–388. [Google Scholar] [CrossRef] [PubMed]
- Imaduwage, K.P.; Lakbub, J.; Go, E.P.; Desaire, H. Rapid LC-MS Based High-Throughput Screening Method, Affording No False Positives or False Negatives, Identifies a New Inhibitor for Carbonic Anhydrase. Sci. Rep. 2017, 71, 0324. [Google Scholar] [CrossRef] [PubMed]
- Wills, L.P. The use of high-throughput screening techniques to evaluate mitochondrial toxicity. Toxicology 2017, 3913, 4–41. [Google Scholar] [CrossRef] [PubMed]
- Roy, A. High-Throughput Screening (HTS) Technology. In: Offermanns, S.; Rosenthal, W., editors. Encyclopedia of Molecular Pharmacology. Cham: Springer International Publishing; 2021. p. 787-799.
- Li, G.; Peng, X.; Guo, Y.; Gong, S.; Cao, S.; Qiu, F. Currently Available Strategies for Target Identification of Bioactive Natural Products. Front. Chem. 2021, 97, 61609. [Google Scholar] [CrossRef] [PubMed]
- Asiamah, I.; Obiri, S.A.; Tamekloe, W.; Armah, F.A.; Borquaye, L.S. Applications of molecular docking in natural products-based drug discovery. Sci. Afr. 2023, 20, e01593. [Google Scholar] [CrossRef]
- Reddy, A.S.; Pati, S.P.; Kumar, P.P.; Pradeep, H.N.; Sastry, G.N. Virtual screening in drug discovery -- a computational perspective. Curr. Protein Pept. Sci. 2007, 83, 29–351. [Google Scholar]
- Lavecchia, A.; Di Giovanni, C. Virtual screening strategies in drug discovery: a critical review. Curr. Med. Chem. 2013, 202, 839–2860. [Google Scholar] [CrossRef]
- Kontoyianni, M. Docking and Virtual Screening in Drug Discovery. Methods Mol. Biol. 2017, 16472, 55–266. [Google Scholar]
- Hert, J.; Willett, P.; Wilton, D.J.; Acklin, P.; Azzaoui, K.; Jacoby, E.; et al. Comparison of fingerprint-based methods for virtual screening using multiple bioactive reference structures. J. Chem. Inf. Comput. Sci. 2004, 441, 177–1185. [Google Scholar] [CrossRef]
- Qing, X.; Yin Lee, X.; De Raeymaeker, J.; Rh Tame, J.; Yj Zhang, K.; De Maeyer, M.; et al. Pharmacophore modeling: advances, limitations, and current utility in drug discovery. Journal of Receptor, Ligand Channel Res. 2014, 7, 81–92. [Google Scholar]
- Muegge, I.; Mukherjee, P. An overview of molecular fingerprint similarity search in virtual screening. Expert. Opin. Drug Discov. 2016, 111, 37–148. [Google Scholar] [CrossRef] [PubMed]
- Santana, K. , do Nascimento, L.D.; Lima, E.L.A.; Damasceno, V.; Nahum, C.; Braga, R.C.; et al. Applications of Virtual Screening in Bioprospecting: Facts, Shifts, and Perspectives to Explore the Chemo-Structural Diversity of Natural Products. Front. Chem. 2021, 96, 62688. [Google Scholar]
- Das, A.P.; Agarwal, S.M. Recent advances in the area of plant-based anti-cancer drug discovery using computational approaches. Mol. Divers. 2023, 1, 25. [Google Scholar] [CrossRef]
- Padhi, A.K.; Janežič, M.; Zhang, K.Y.J. Chapter 26 - Molecular dynamics simulations: Principles, methods, and applications in protein conformational dynamics. In: Tripathi, T.; Dubey, V.K., editors. Advances in Protein Molecular and Structural Biology Methods: Academic Press; 2022. p. 439-454.
- Zhang, W.; Chien, J.; Yong, J.; Kuang, R. Network-based machine learning and graph theory algorithms for precision oncology. npj Precis. Oncol. 2017, 12, 5. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Kumar, S. Chapter 6 - Molecular Docking: A Structure-Based Approach for Drug Repurposing. In: Roy, K., editor. In Silico Drug Design: Academic Press; 2019. p. 161-189.
- Silvestrini, L.; Belhaj, N.; Comez, L.; Gerelli, Y.; Lauria, A.; Libera, V.; et al. The dimer-monomer equilibrium of SARS-CoV-2 main protease is affected by small molecule inhibitors. Sci. Rep. 2021, 119, 283. [Google Scholar] [CrossRef]
- Chugh, A.; Sehgal, I.; Khurana, N.; Verma, K.; Rolta, R.; Vats, P.; et al. Comparative docking studies of drugs and phytocompounds for emerging variants of SARS-CoV-2. Biotech. 2023, 133, 6. [Google Scholar] [CrossRef]
- Jamiu, A.T.; Pohl, C.H.; Bello, S.; Adedoja, T.; Sabiu, S. A review on molecular docking analysis of phytocompounds against SARS-CoV-2 druggable targets. All. Life 2021, 141, 100–1128. [Google Scholar] [CrossRef]
- Swargiary, G.; Mani, S. ER and PGR targeting ability of phytocompounds derived from Centella asiatica and Andrographis paniculata: An in-silico approach. J. Herb. Med. 2022, 321, 00541. [Google Scholar] [CrossRef]
- Gayathiri, E.; Prakash, P.; Ahamed, M.; Pandiaraj, S.; Venkidasamy, B.; Dayalan, H.; et al. Multitargeted pharmacokinetics, molecular docking and network pharmacology-based identification of effective phytocompounds from Sauropus androgynus (L.) Merr for inflammation and cancer treatment. J. Biomol. Struct. Dyn. 2023, 1, 14. [Google Scholar] [CrossRef]
- Mao, J.; Akhtar, J.; Zhang, X.; Sun, L.; Guan, S.; Li, X.; et al. Comprehensive strategies of machine-learning-based quantitative structure-activity relationship models. iScience 2021, 241, 03052. [Google Scholar] [CrossRef]
- Paul, D.; Sanap, G.; Shenoy, S.; Kalyane, D.; Kalia, K.; Tekade, R.K. Artificial intelligence in drug discovery and development. Drug Discov. Today 2021, 268, 0–93. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.H.; Hur, G.; Kim, N.R.; Park, J.H.Y.; Lee, K.W.; Yang, H. A machine learning-integrated stepwise method to discover novel anti-obesity phytochemicals that antagonize the glucocorticoid receptor. Food Funct. 2023, 141, 869–1883. [Google Scholar] [CrossRef] [PubMed]
- Yabuuchi, H.; Hayashi, K.; Shigemoto, A.; Fujiwara, M.; Nomura, Y.; Nakashima, M.; et al. Virtual screening of antimicrobial plant extracts by machine-learning classification of chemical compounds in semantic space. PLoS One 2023, 18, e0285716. [Google Scholar] [CrossRef] [PubMed]
- García-Pérez, P.; Lozano-Milo, E.; Landín, M.; Gallego, P.P. Combining Medicinal Plant In Vitro Culture with Machine Learning Technologies for Maximizing the Production of Phenolic Compounds. Antioxidants 2020, 9. [Google Scholar] [CrossRef]
- Idowu, S.O.; Fatokun, A.A. Artificial Intelligence (AI) to the Rescue: Deploying Machine Learning to Bridge the Biorelevance Gap in Antioxidant Assays. SLAS Technol. 2021, 261, 6–25. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




