Submitted:
13 June 2024
Posted:
14 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
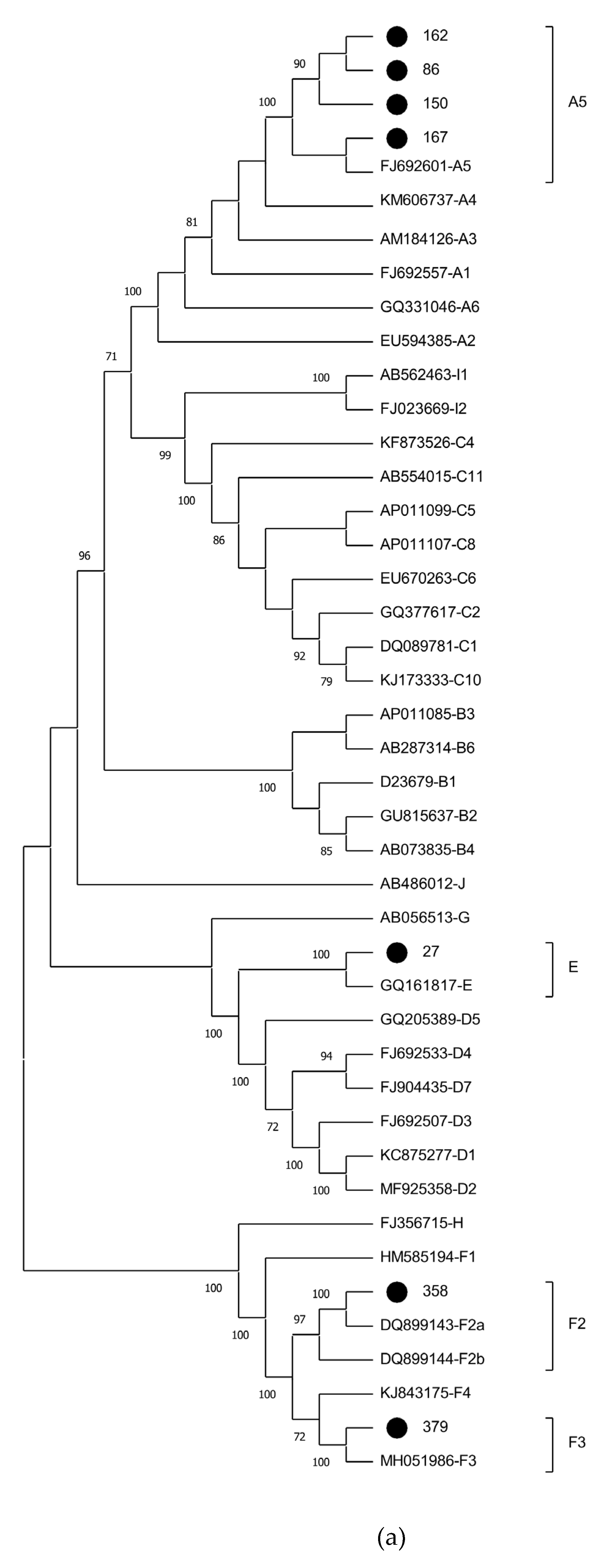
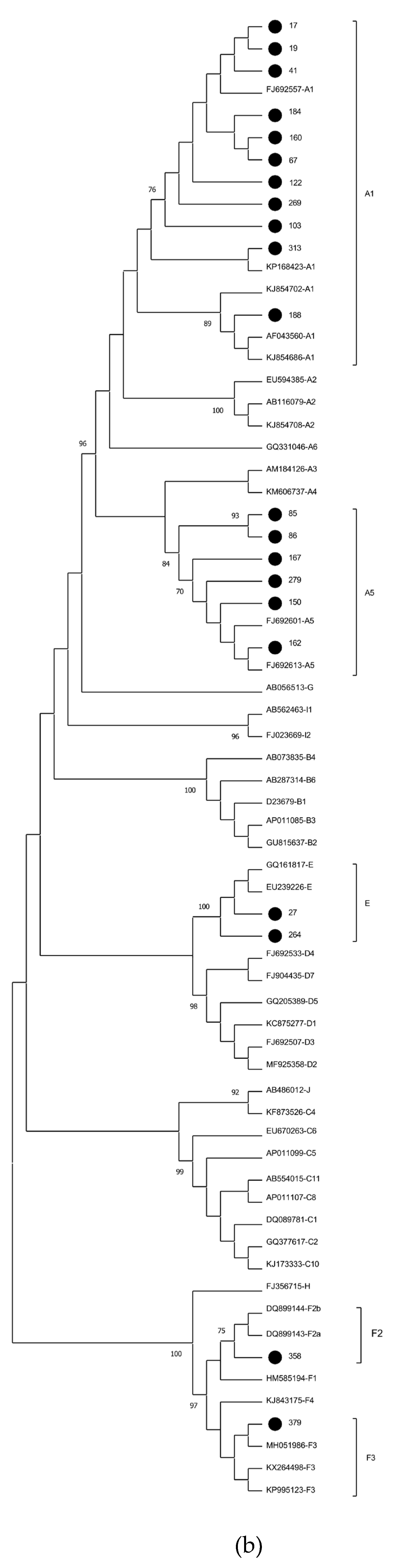
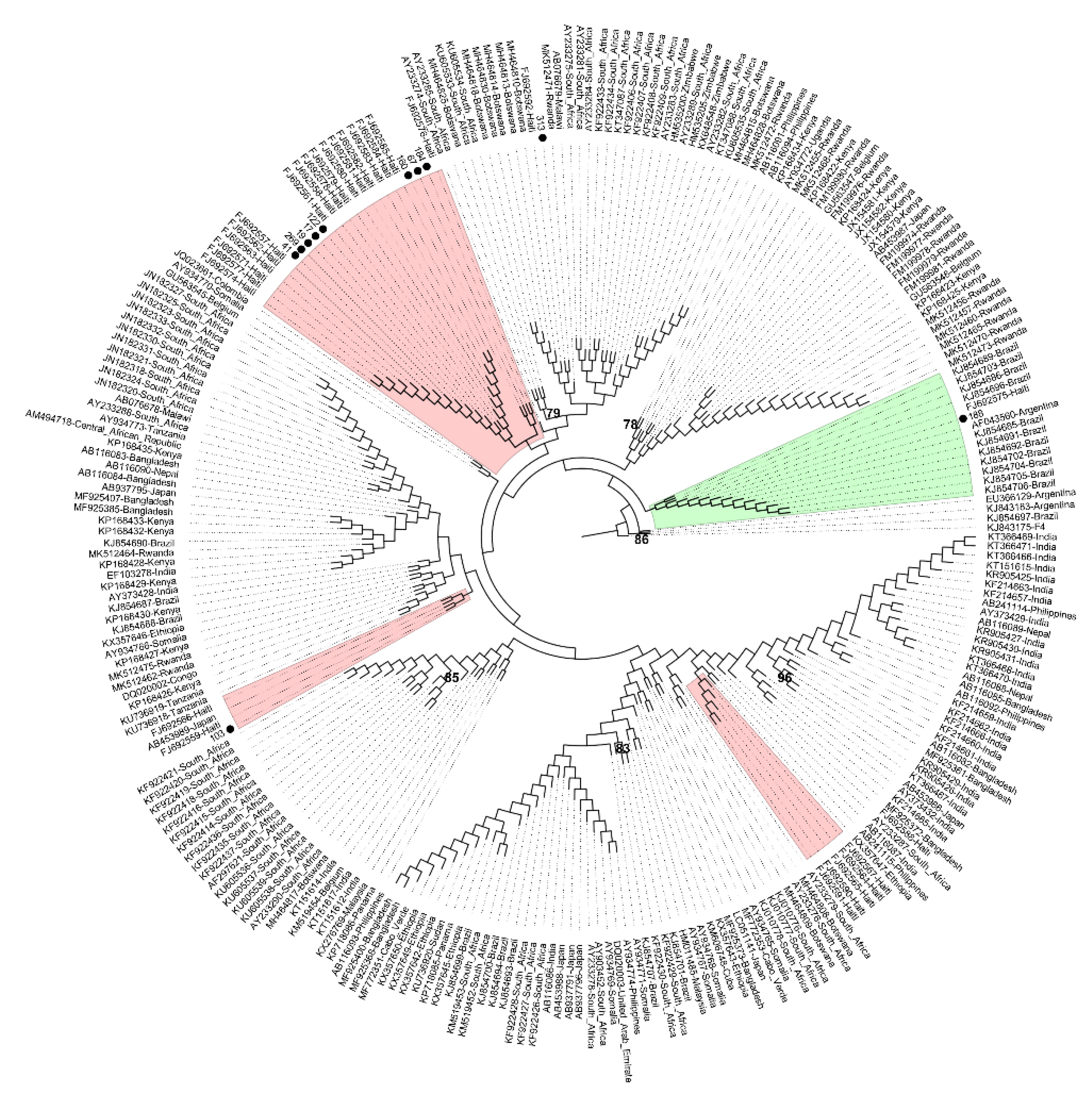
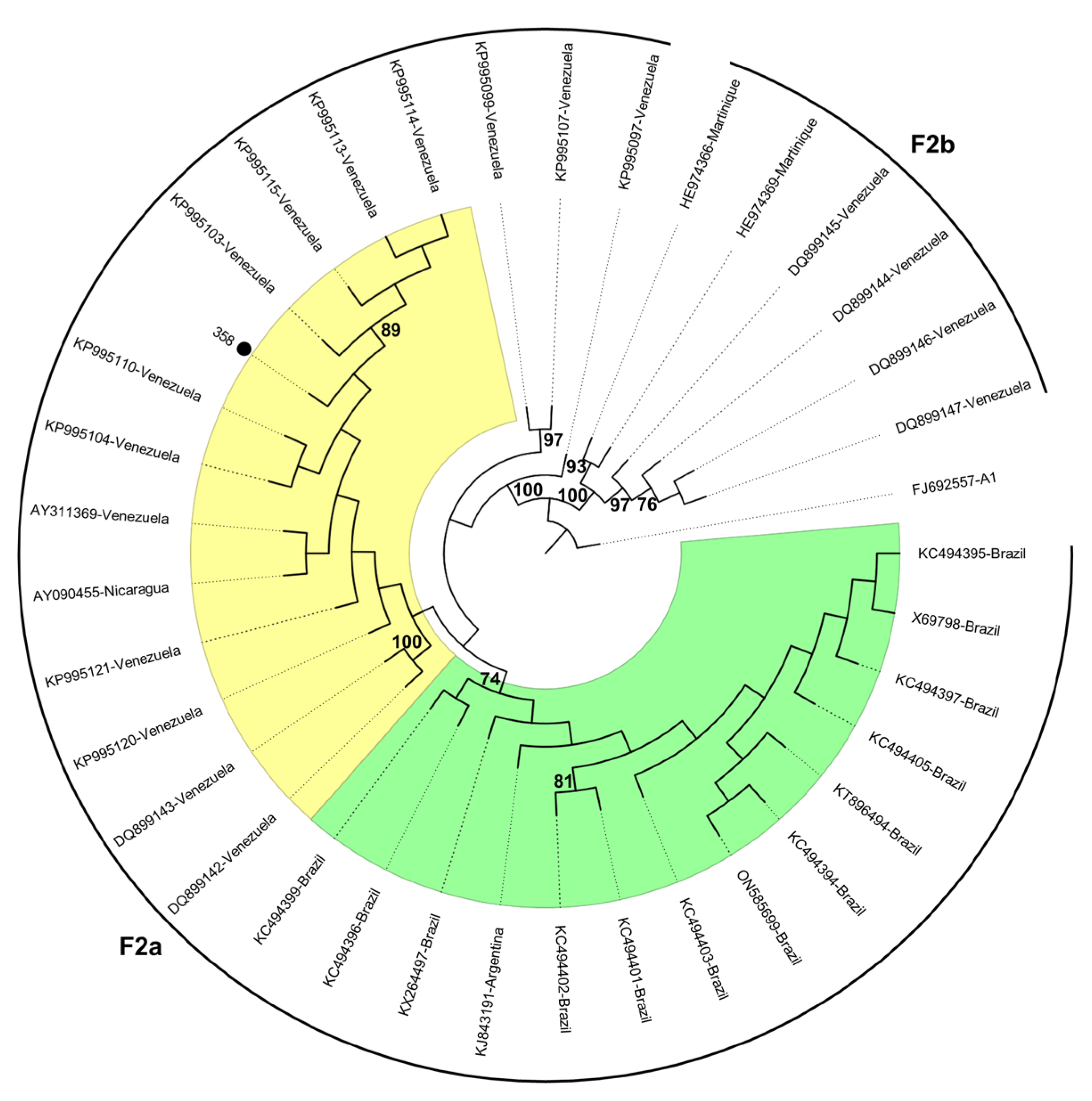
2. Results
3. Discussion
4. Materials and Methods
4.1. Study population
4.2. Viral DNA extraction and PCR amplification
4.3. Nucleotide sequencing
4.4. HBV genotyping and mutational analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO World Health Organization. Fact sheets. Hepatitis B. https://www.who.int/news-room/fact-sheets/detail/hepatitis-b (08 November 2023).
- Edmunds, W.J.; Medley, G.F.; Nokes, D.J.; O'Callaghan, C.J.; Whittle, H.C.; Hall, A.J. , Epidemiological patterns of hepatitis B virus (HBV) in highly endemic areas. Epidemiol Infect 1996, 117, 313–325. [Google Scholar] [CrossRef]
- Raimondo, G.; Locarnini, S.; Pollicino, T.; Levrero, M.; Zoulim, F.; Lok, A.S. , Update of the statements on biology and clinical impact of occult hepatitis B virus infection. J Hepatol 2019, 71, 397–408. [Google Scholar] [CrossRef]
- Karayiannis, P. , Hepatitis B virus: virology, molecular biology, life cycle and intrahepatic spread. Hepatol Int 2017, 11, 500–508. [Google Scholar] [CrossRef]
- Tsukuda, S.; Watashi, K. , Hepatitis B virus biology and life cycle. Antiviral Res 2020, 182, 104925. [Google Scholar] [CrossRef]
- Osiowy, C.; Giles, E.; Tanaka, Y.; Mizokami, M.; Minuk, G.Y. , Molecular evolution of hepatitis B virus over 25 years. J Virol 2006, 80, 10307–10314. [Google Scholar] [CrossRef]
- Revill, P.A.; Tu, T.; Netter, H.J.; Yuen, L.K.W.; Locarnini, S.A.; Littlejohn, M. , The evolution and clinical impact of hepatitis B virus genome diversity. Nat Rev Gastroenterol Hepatol 2020, 17, 618–634. [Google Scholar] [CrossRef]
- Kramvis, A. Genotypes and genetic variability of hepatitis B virus. Intervirology 2014, 57, 141–150. [Google Scholar] [CrossRef]
- Araujo, N.M.; Teles, S.A.; Spitz, N. , Comprehensive Analysis of Clinically Significant Hepatitis B Virus Mutations in Relation to Genotype, Subgenotype and Geographic Region. Front Microbiol 2020, 11, 616023. [Google Scholar] [CrossRef]
- Araujo, N.M.; Waizbort, R.; Kay, A. , Hepatitis B virus infection from an evolutionary point of view: how viral, host, and environmental factors shape genotypes and subgenotypes. Infect Genet Evol 2011, 11, 1199–1207. [Google Scholar] [CrossRef]
- Chen, J.; Li, L.; Yin, Q.; Shen, T. , A review of epidemiology and clinical relevance of Hepatitis B virus genotypes and subgenotypes. Clin Res Hepatol Gastroenterol 2023, 47, 102180. [Google Scholar] [CrossRef]
- Lin, C.L.; Kao, J.H. , The clinical implications of hepatitis B virus genotype: Recent advances. J Gastroenterol Hepatol 2011, 26 Suppl 1, 123–130. [Google Scholar] [CrossRef]
- Kocher, A.; Papac, L.; Barquera, R.; Key, F.M.; Spyrou, M.A.; Hubler, R.; Rohrlach, A.B.; Aron, F.; Stahl, R.; Wissgott, A.; van Bommel, F.; Pfefferkorn, M.; Mittnik, A.; Villalba-Mouco, V.; Neumann, G.U.; Rivollat, M.; van de Loosdrecht, M.S.; Majander, K.; Tukhbatova, R.I.; Musralina, L.; Ghalichi, A.; Penske, S.; Sabin, S.; Michel, M.; Gretzinger, J.; Nelson, E.A.; Ferraz, T.; Nagele, K.; Parker, C.; Keller, M.; Guevara, E.K.; Feldman, M.; Eisenmann, S.; Skourtanioti, E.; Giffin, K.; Gnecchi-Ruscone, G.A.; Friederich, S.; Schimmenti, V.; Khartanovich, V.; Karapetian, M.K.; Chaplygin, M.S.; Kufterin, V.V.; Khokhlov, A.A.; Chizhevsky, A.A.; Stashenkov, D.A.; Kochkina, A.F.; Tejedor-Rodriguez, C.; de Lagran, I.G.; Arcusa-Magallon, H.; Garrido-Pena, R.; Royo-Guillen, J.I.; Novacek, J.; Rottier, S.; Kacki, S.; Saintot, S.; Kaverzneva, E.; Belinskiy, A.B.; Veleminsky, P.; Limbursky, P.; Kostka, M.; Loe, L.; Popescu, E.; Clarke, R.; Lyons, A.; Mortimer, R.; Sajantila, A.; de Armas, Y.C.; Hernandez Godoy, S.T.; Hernandez-Zaragoza, D.I.; Pearson, J.; Binder, D.; Lefranc, P.; Kantorovich, A.R.; Maslov, V.E.; Lai, L.; Zoledziewska, M.; Beckett, J.F.; Langova, M.; Danielisova, A.; Ingman, T.; Atienzar, G.G.; de Miguel Ibanez, M.P.; Romero, A.; Sperduti, A.; Beckett, S.; Salter, S.J.; Zilivinskaya, E.D.; Vasil'ev, D.V.; von Heyking, K.; Burger, R.L.; Salazar, L.C.; Amkreutz, L.; Navruzbekov, M.; Rosenstock, E.; Alonso-Fernandez, C.; Slavchev, V.; Kalmykov, A.A.; Atabiev, B.C.; Batieva, E.; Calmet, M.A.; Llamas, B.; Schultz, M.; Krauss, R.; Jimenez-Echevarria, J.; Francken, M.; Shnaider, S.; de Knijff, P.; Altena, E.; Van de Vijver, K.; Fehren-Schmitz, L.; Tung, T.A.; Losch, S.; Dobrovolskaya, M.; Makarov, N.; Read, C.; Van Twest, M.; Sagona, C.; Ramsl, P.C.; Akar, M.; Yener, K.A.; Ballestero, E.C.; Cucca, F.; Mazzarello, V.; Utrilla, P.; Rademaker, K.; Fernandez-Dominguez, E.; Baird, D.; Semal, P.; Marquez-Morfin, L.; Roksandic, M.; Steiner, H.; Salazar-Garcia, D.C.; Shishlina, N.; Erdal, Y.S.; Hallgren, F.; Boyadzhiev, Y.; Boyadzhiev, K.; Kussner, M.; Sayer, D.; Onkamo, P.; Skeates, R.; Rojo-Guerra, M.; Buzhilova, A.; Khussainova, E.; Djansugurova, L.B.; Beisenov, A.Z.; Samashev, Z.; Massy, K.; Mannino, M.; Moiseyev, V.; Mannermaa, K.; Balanovsky, O.; Deguilloux, M.F.; Reinhold, S.; Hansen, S.; Kitov, E.P.; Dobes, M.; Ernee, M.; Meller, H.; Alt, K.W.; Prufer, K.; Warinner, C.; Schiffels, S.; Stockhammer, P.W.; Bos, K.; Posth, C.; Herbig, A.; Haak, W.; Krause, J.; Kuhnert, D. , Ten millennia of hepatitis B virus evolution. Science 2021, 374, 182–188. [Google Scholar] [CrossRef]
- Paraskevis, D.; Angelis, K.; Magiorkinis, G.; Kostaki, E.; Ho, S.Y.; Hatzakis, A. , Dating the origin of hepatitis B virus reveals higher substitution rate and adaptation on the branch leading to F/H genotypes. Mol Phylogenet Evol 2015, 93, 44–54. [Google Scholar] [CrossRef]
- Spitz, N.; Mello, F.C.A.; Moreira, A.S.; Gusatti, C.S.; Martins, R.M.B.; Gomes, S.A.; Bello, G.; Araujo, N.M. , Reconstruction of the spatial and temporal dynamics of hepatitis B virus genotype D in the Americas. PLoS One 2019, 14, e0220342. [Google Scholar] [CrossRef]
- Zehender, G.; Ebranati, E.; Gabanelli, E.; Sorrentino, C.; Lo Presti, A.; Tanzi, E.; Ciccozzi, M.; Galli, M. , Enigmatic origin of hepatitis B virus: an ancient travelling companion or a recent encounter? World J Gastroenterol 2014, 20, 7622–7634. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, Y.; Xu, M.; Li, X.; Zhang, Z. , Distribution of hepatitis B virus genotypes and subgenotypes: A meta-analysis. Medicine (Baltimore) 2021, 100, e27941. [Google Scholar] [CrossRef]
- Velkov, S.; Ott, J.J.; Protzer, U.; Michler, T. , The Global Hepatitis B Virus Genotype Distribution Approximated from Available Genotyping Data. Genes (Basel) 2018, 9. [Google Scholar] [CrossRef]
- WHO World Health Organization. Global hepatitis report. https://www.who.int/publications/i/item/9789241565455 (09 November 2023).
- Coppola, N.; Alessio, L.; Pisaturo, M.; Macera, M.; Sagnelli, C.; Zampino, R.; Sagnelli, E. , Hepatitis B virus infection in immigrant populations. World J Hepatol 2015, 7, 2955–2961. [Google Scholar] [CrossRef]
- Razavi-Shearer, D.; Gamkrelidze, I.; Pan, C.Q.; Razavi-Shearer, K.; Blach, S.; Estes, C.; Mooneyhan, E.; Razavi, H. , The impact of immigration on hepatitis B burden in the United States: a modelling study. Lancet Reg Health Am 2023, 22, 100516. [Google Scholar] [CrossRef]
- OBMigra Ministério da Justiça e Segurança Pública, Observatório das Migrações Internacionais (OBMigra) - Relatório Anual 2022 https://portaldeimigracao.mj.gov.br/pt/dados/relatorios-a (14 November 2023).
- Lago, B.V.; do Espirito-Santo, M.P.; Costa, V.D.; Marques, V.A.; Villar, L.M.; Lewis-Ximenez, L.L.; Lampe, E.; Mello, F.C.A. , Genetic Diversity of the Hepatitis B Virus Subgenotypes in Brazil. Viruses 2019, 11. [Google Scholar] [CrossRef]
- Lampe, E.; Mello, F.C.A.; do Espirito-Santo, M.P.; Oliveira, C.M.C.; Bertolini, D.A.; Goncales, N.S.L.; Moreira, R.C.; Fernandes, C.A.S.; Nascimento, H.C.L.; Grotto, R.M.T.; Pardini, M.; On Behalf Of The Brazilian Hepatitis, B.R.G. , Nationwide overview of the distribution of hepatitis B virus genotypes in Brazil: a 1000-sample multicentre study. J Gen Virol 2017, 98, 1389–1398. [Google Scholar] [CrossRef] [PubMed]
- Wolf, J.M.; Simon, D.; Lunge, V.R. , Hepatitis B virus genotypes in Brazil: Introduction and dissemination. Infect Genet Evol 2021, 93, 104936. [Google Scholar] [CrossRef] [PubMed]
- Andernach, I.E.; Nolte, C.; Pape, J.W.; Muller, C.P. , Slave trade and hepatitis B virus genotypes and subgenotypes in Haiti and Africa. Emerg Infect Dis 2009, 15, 1222–1228. [Google Scholar] [CrossRef] [PubMed]
- Lenders, S. Bolivians, Haitians and Venezuelans – three cases of immigration in Brazil (Bolivianos, haitianos e venezuelanos – três casos de imigração no Brasil). https://br.boell.org/pt-br/2019/04/15/bolivianos-haitianos-e-venezuelanos-tres-casos-de-imigracao-no-brasil#_ftn1 (23 November 2023.
- Motta-Castro, A.R.; Martins, R.M.; Araujo, N.M.; Niel, C.; Facholi, G.B.; Lago, B.V.; Mello, F.C.; Gomes, S.A. , Molecular epidemiology of hepatitis B virus in an isolated Afro-Brazilian community. Arch Virol 2008, 153, 2197–2205. [Google Scholar] [CrossRef] [PubMed]
- Sitnik, R.; Sette, H., Jr.; Santana, R.A.; Menezes, L.C.; Graca, C.H.; Dastoli, G.T.; Silbert, S.; Pinho, J.R. , Hepatitis B virus genotype E detected in Brazil in an African patient who is a frequent traveler. Braz J Med Biol Res 2007, 40, 1689–1692. [Google Scholar] [CrossRef] [PubMed]
- Honge, B.L.; Jespersen, S.; Medina, C.; Te Dda, S.; da Silva, Z.J.; Lewin, S.; Ostergaard, L.; Erikstrup, C.; Wejse, C.; Laursen, A.L.; Krarup, H.; Bissau, H.I.V. c. s. g. , Hepatitis B and Delta virus are prevalent but often subclinical co-infections among HIV infected patients in Guinea-Bissau, West Africa: a cross-sectional study. PLoS One 2014, 9, e99971. [Google Scholar] [CrossRef] [PubMed]
- Ingasia, L.A.O.; Wose Kinge, C.; Kramvis, A. , Genotype E: The neglected genotype of hepatitis B virus. World J Hepatol 2021, 13, 1875–1891. [Google Scholar] [CrossRef]
- Pujol, F.; Jaspe, R.C.; Loureiro, C.L.; Chemin, I. , Hepatitis B virus American genotypes: Pathogenic variants ? Clin Res Hepatol Gastroenterol 2020, 44, 825–835. [Google Scholar] [CrossRef] [PubMed]
- Kowalec, K.; Minuk, G.Y.; Borresen, M.L.; Koch, A.; McMahon, B.J.; Simons, B.; Osiowy, C. , Genetic diversity of hepatitis B virus genotypes B6, D and F among circumpolar indigenous individuals. J Viral Hepat 2013, 20, 122–130. [Google Scholar] [CrossRef]
- Mojsiejczuk, L.N.; Torres, C.; Pisano, M.B.; Re, V.; Campos, R.H.; Flichman, D.M. , New pieces on genetic diversity and evolutionary history of hepatitis B virus: Characterization of the novel subgenotype F6. Infect Genet Evol 2017, 47, 140–142. [Google Scholar] [CrossRef]
- Mello, F.C.; Araujo, O.C.; Lago, B.V.; Motta-Castro, A.R.; Moraes, M.T.; Gomes, S.A.; Bello, G.; Araujo, N.M. , Phylogeography and evolutionary history of hepatitis B virus genotype F in Brazil. Virol J 2013, 10, 236. [Google Scholar] [CrossRef]
- Sousa, D.D.; Silva, C.R.S.; Lima Junior, W.P.; Barros, J.A.; Nascimento, I.; Souza, V.C.; Naveca, F.G.; Granja, F. , Phylogenetic analysis and genotype distribution of Hepatitis B Virus (HBV) in Roraima, Brazil. Rev Inst Med Trop Sao Paulo 2018, 60, e35. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhang, H.; Gu, C.; Yin, J.; He, Y.; Xie, J.; Cao, G. , Associations between hepatitis B virus mutations and the risk of hepatocellular carcinoma: a meta-analysis. J Natl Cancer Inst 2009, 101, 1066–1082. [Google Scholar] [CrossRef]
- Nishiya, A.S.; Levi, J.E.; de Almeida-Neto, C.; Witkin, S.S.; Ferreira, S.C.; Bassit, L.; Sabino, E.C.; Di-Lorenzo-Oliveira, C.; Salles, N.A.; Coutinho, A.S.; Bellesa, M.A.; Rocha, V.; Mendrone-Jr, A. , Occult and active hepatitis B virus detection in donated blood in Sao Paulo, Brazil. Transfusion 2021, 61, 1495–1504. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, M.P.; Matos, M.A.; Silva, A.M.; Lopes, C.L.; Teles, S.A.; Matos, M.A.; Spitz, N.; Araujo, N.M.; Mota, R.M.; Kerr, L.R.; Martins, R.M. , Prevalence, Risk Behaviors, and Virological Characteristics of Hepatitis B Virus Infection in a Group of Men Who Have Sex with Men in Brazil: Results from a Respondent-Driven Sampling Survey. PLoS One 2016, 11, e0160916. [Google Scholar] [CrossRef]
- Oliveira, M.P.; Lemes, P.S.; Matos, M.A.; Del-Rios, N.H.; Carneiro, M.A.; Silva, A.M.; Lopes, C.L.; Teles, S.A.; Aires, R.S.; Lago, B.V.; Araujo, N.M.; Martins, R.M. , Overt and occult hepatitis B virus infection among treatment-naive HIV-infected patients in Brazil. J Med Virol 2016, 88, 1222–1229. [Google Scholar] [CrossRef]
- Matos, M.A.; Ferreira, R.C.; Rodrigues, F.P.; Marinho, T.A.; Lopes, C.L.; Novais, A.C.; Motta-Castro, A.R.; Teles, S.A.; Souto, F.J.; Martins, R.M. , Occult hepatitis B virus infection among injecting drug users in the Central-West Region of Brazil. Mem Inst Oswaldo Cruz 2013, 108, 386–389. [Google Scholar] [CrossRef]
- Martins, T.L.S.; Silva, G.; Silva, C.A.; Gomes, D.O.; Diniz, E.S.B.V.; Carneiro, M.; Pacheco, L.R.; Araujo, N.M.; Zanchetta, M.S.; Teles, S.A.; Caetano, K.A.A. , Hepatitis B and C in Immigrants and Refugees in Central Brazil: Prevalence, Associated Factors, and Immunization. Viruses 2022, 14. [Google Scholar] [CrossRef] [PubMed]
- Gunther, S.; Li, B.C.; Miska, S.; Kruger, D.H.; Meisel, H.; Will, H. , A novel method for efficient amplification of whole hepatitis B virus genomes permits rapid functional analysis and reveals deletion mutants in immunosuppressed patients. J Virol 1995, 69, 5437–5444. [Google Scholar] [CrossRef]
- McNaughton, A.L.; Revill, P.A.; Littlejohn, M.; Matthews, P.C.; Ansari, M.A. , Analysis of genomic-length HBV sequences to determine genotype and subgenotype reference sequences. J Gen Virol 2020, 101, 271–283. [Google Scholar] [CrossRef]

| Variables | n = 102 | (%) |
| Sex | ||
| Male | 70 | 68.6 |
| Female | 32 | 31.4 |
| Age | ||
| 2 – 11 | 0 | 0 |
| 12 – 17 | 0 | 0 |
| 18 – 30 | 38 | 37.3 |
| 31 – 50 | 58 | 56.9 |
| ≥51 | 6 | 5.9 |
| Country of Origin | ||
| Colombia | 1 | 1 |
| Guinea-Bissau | 12 | 11.8 |
| Haiti | 81 | 79.4 |
| Venezuela | 8 | 7.8 |
| Continent of Origin | ||
| Africa | 12 | 11.8 |
| Caribbean | 81 | 79.4 |
| South America | 9 | 8.8 |
| HBsAg | ||
| Positive | 24 | 23.5 |
| Negative | 78 | 76.5 |
| HBV DNA | ||
| Positive | 21 | 20.6 |
| Negative | 81 | 79.4 |
| ID | Country of origin | HBV serology | Amplified genomic region | Mutation sites | Subgenotype | ||||
|---|---|---|---|---|---|---|---|---|---|
| HBsAg | Anti-HBc | HBsAgb | pre-C/Cc | RTd | Other | ||||
| 17 | Haiti | Pos | Pos | S | None | ND | Nonee | A1 | |
| 19 | Haiti | Nega | Pos | S, pre-C/C | None | A1762T/G1764A | Nonee | A1 | |
| 41 | Haiti | Pos | Pos | S | None | ND | Nonee | A1 | |
| 67 | Guine Bissau | Nega | Pos | S | None | ND | Nonee | A1 | |
| 103 | Haiti | Pos | Pos | S | None | ND | Nonee | 12-nt insertion at S region | A1 |
| 122 | Haiti | Pos | Pos | S, pre-C/C | None | None | Nonee | A1 | |
| 160 | Haiti | Pos | Pos | S | None | ND | Nonee | A1 | |
| 184 | Haiti | Pos | Pos | S | None | ND | Nonee | A1 | |
| 188 | Haiti | Nega | Pos | S | None | ND | Nonee | A1 | |
| 269 | Haiti | Pos | Pos | S, pre-C/C | None | A1762T/G1764A | Nonee | A1 | |
| 313 | Haiti | Pos | Pos | S, pre-C/C | None | A1762T/G1764A | Nonee | A1 | |
| 85 | Haiti | Nega | Pos | S | None | ND | Nonee | A5 | |
| 86 | Haiti | Pos | Pos | complete genome | None | A1762T/G1764A, G1896A | None | A5 | |
| 150 | Haiti | Pos | Pos | complete genome | None | A1762T/G1764A | None | A5 | |
| 162 | Haiti | Pos | Pos | complete genome | None | A1762T/G1764A | None | A5 | |
| 167 | Haiti | Pos | Pos | complete genome | None | A1762T/G1764A | None | A5 | |
| 279 | Haiti | Pos | Pos | S, pre-C/C | None | None | Nonee | A5 | |
| 27 | Haiti | Pos | Pos | complete genome | None | G1896A | None | E | |
| 264 | Guine Bissau | Pos | Pos | S | None | ND | Nonee | 12-nt insertion at S region | E |
| 358 | Venezuela | Pos | Pos | complete genome | None | None | None | 21-nt deletion at pre-S2 region | F2 |
| 379 | Venezuela | Pos | Pos | complete genome | None | None | None | F3 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





