1. Introduction
One of the primary functions of the skin, the largest organ in the human body, is to act as a physical barrier to pathogenic microbes and toxic substances thus making it one of the first lines of defence against infection [
1]. The quality of an individual’s skin is influenced by a multitude of internal and external factors [
1,
2]. External factors include the environment (sun exposure, wind, humidity, air conditioning), physical abrasions, microbes (bacteria, fungi, viruses), and exposure to toxins. Internal factors include genetics, medications (immunosuppressant drugs, birth control), antibiotics, and diet. Other factors which directly exert an effect on the skin are the types of cosmetics and skin care products, washing detergent, soap, and perfume an individual may use [
3].
A combination of these factors alongside switches in microbial diversity can lead to dysbiosis [
4]; a state of microbial imbalance within areas of the body sometimes impacting the vitality and health status of those areas [
1,
2]. Many human skin disorders and diseases have been linked to changes within the skin microbiome, including acne, atopic dermatitis, and psoriasis [
3,
5,
6,
7,
8,
9]. Unsurprisingly this has led to a new area of research looking at how to mediate the skin microbiome in both dermatological and cosmetic fields for common conditions such as acne, body odour, and atopic dermatitis [
3,
5,
6,
7,
8,
9].
The recent advancement of technologies has allowed researchers to evaluate the human microbiome in greater depth to better understand the connections between specific microbes and a variety of disease presentations, leading to the development of many novel and emerging therapies to treat commonly occurring skin issues [
10]. The influx of information relating to the skin microbiome and its use in pharmaceuticals has stimulated the conversation and development around probiotics.
It is known that probiotics interfere with the activity of pathogenic bacteria by producing antimicrobial substances including metabolites, peptides and, bacteriocins [
10,
11]. Further to the direct inhibition of the microorganism, they can also create an unfavourable environment for the pathogen as they can alter pH, compete for nutrients, and inhibit pathogen adhesion to epithelial cells by adhering to these cells themselves [
12].
Recently, a live probiotic hydration serum containing
M. luteus Q24 was introduced in New Zealand.
M. luteus Q24 is a skin commensal probiotic bacterium that resides on the skin of healthy human adults and has been identified to produce an interesting antimicrobial spectrum that is inhibitory towards pathogenic bacteria associated with many skin diseases and found to be safe for topical applications in humans [
12].
The primary aim of this study was to carry out a cosmetic trial to measure changes in (1) skin quality parameters using an advanced handheld skin analyzer and (2) align this to any changes in microbial diversity on the skin, using whole genome sequencing platform, following twice daily application of live M. luteus Q24 probiotic serum for 25 days in healthy adult participants.
2. Materials and Methods
Live probiotic hydration serum was supplied by Unconditional Skincare Co, Blis Technologies Limited, Dunedin, New Zealand. The product packaging is composed of two chambers: Chamber A containing Serum: Medium chain triglyceride, Silica Dimethyl Silylate, Polysorbate 80 (T), BLIS Q24TM (M. luteus Q24 - DSMZ 17,172) and Chamber B: Hydrator: Aqua, Coco-caprylate, Glycerin, Cetearyl alcohol, Glyceryl stearate SE, Cetearyl wheat straw glycosides, Sodium stearoyl glutamate, Xanthan gum, Phenoxyethanol, Ethylhexylglycerin. Skin analyser device (dpViso, Dermobella app) was purchased from Chowis Co. Ltd, Gyeonggi-do, South Korea. Samsung Tablet 10.1 was purchased from PB Technologies, Auckland, New Zealand. Skin swabs were collected using a DNA/RNA Shield Collection Tube kit (Zymo Research, Irvine, CA, USA).
2.1. Cosmetic and Colonisation Efficacy Trial
A cosmetic trial involving the topical application of a live probiotic hydration serum product was conducted in healthy adult participants. M. luteus was previously assessed for safety as a probiotic [REF]. This product is safe and was marketed in New Zealand and contains skin commensal probiotic M. luteus Q24. All subjects gave their informed consent for inclusion before they participated in the study. "All subjects gave their informed consent for inclusion before they participated in the study. The study was conducted in accordance with the Declaration of Helsinki, and the protocol was reviewed by the Health and Disability Ethics Committee (HDEC) (New Zealand). This committee advised that since the scope of these currently proposed pilot trials is cosmetic-based rather than directed at health-related outcomes they were outside of the scope of HDEC review. Therefore, due to cosmetic product testing, ethics committee approval was not required.
2.2. Trial Design
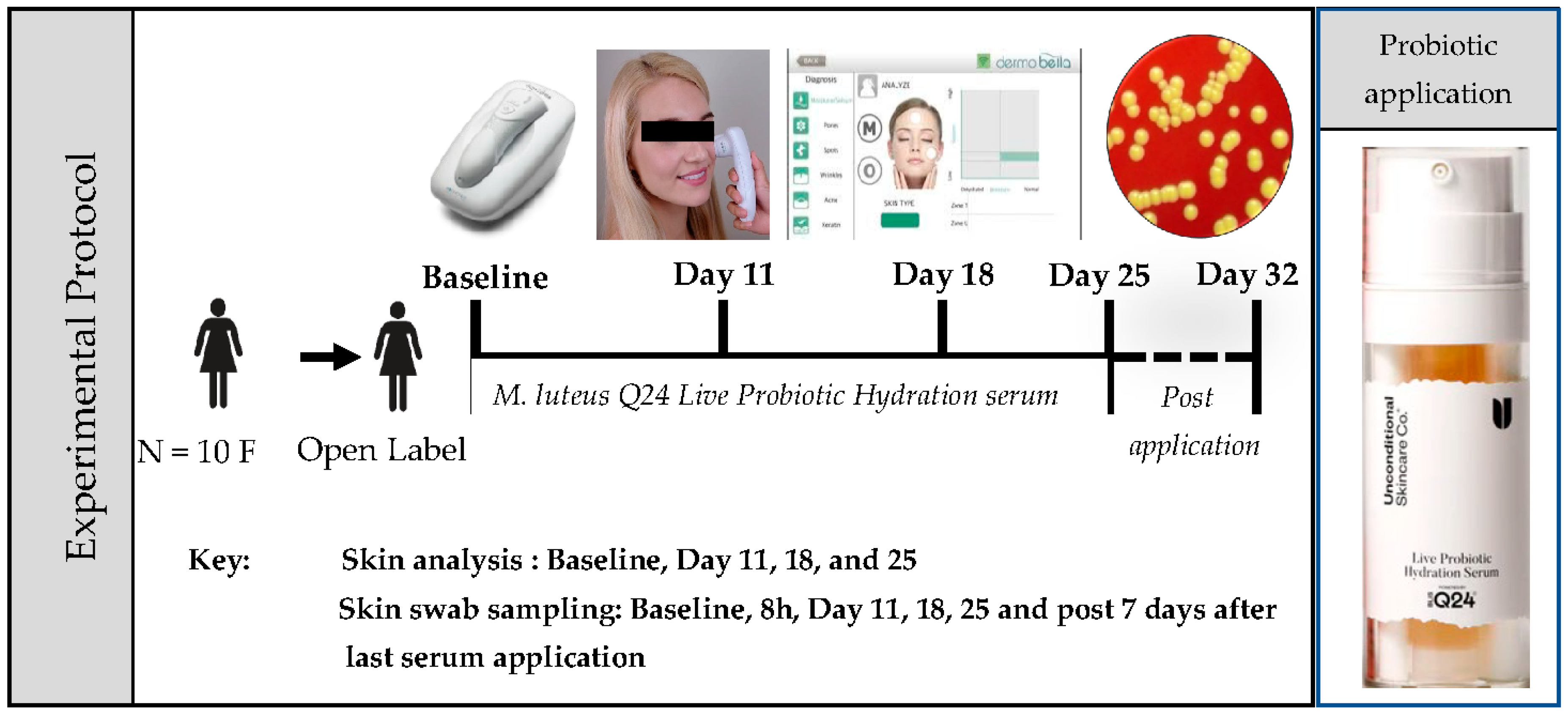
Ten females participated in a single-site, open-label, non – randomized baseline-controlled trial. The study design is shown in
Figure 1. Potential participants were enrolled if they met the following inclusion criteria: healthy adult females, 18y – 60y of age, having generally healthy skin, with mild to moderate breakouts, fine lines, wrinkles, spots, dull skin, redness, and seeking improvement in facial appearance. Participants were excluded if they had cut or wounded skin, had an active infection, had a history of autoimmune disease, or were currently being treated with antibiotics or topical steroids, or anti-inflammatories. Participants were asked to apply two pumps equating to >1E+8 cfu/dose (at the date of manufacturing) of the serum product on the face twice daily (morning and night) for 25 days.
Measurement of skin parameters from the face was taken at Baseline, Days 11, 18, and 25, using a novel skin analyser device. Skin swab samples were also collected from the same site post-skin quality analysis at each sample point. Additional swab samples were also collected at 8h post-first application and Day 7 post-last application of the serum. Participants were instructed not to change their skincare routine or try out new products during the trial except to replace their current moisturiser with a probiotic serum product. Participants were also asked to report adverse events, if any.
2.3. Measurement of Skin Quality
Quantitative measurement of changes in various skin parameters such as hydration /moisture, pores, spots (pigmentation), wrinkles, and impurities (porphyrins) were carried out using a handheld skin analyser (dP/Viso, Chowis, South Korea). The device utilises advanced optic technology with interchangeable lenses and AI-powered analysis to allow for an in-depth measurement of different skin quality parameters. At each time point, the device camera/sensor was placed on the left cheek in line with the corner of the eye and the tip of the nose. This method was kept consistent so that the triplicate readings were taken approximately from the same area on the cheek. The data was captured as scores or images and analysed in the related app “DermoBella” Skin app.
2.4. Statistical Analysis of Skin Analyser Data
Statistical analysis and graphing were performed using Prism 9.4.0 (GraphPad Software). Data were analysed by one-way repeated measures analysis of variance (ANOVA).
2.5. Microbial Analysis
2.5.1. Sample Collection
At each time point, post skin quality parameter measurement, skin swab samples were collected. Briefly, a sterile swab from the DNA/RNA Shield Collection Tube kit (Zymo Research, USA) was used to collect the sample by swabbing the cheek in a zig-zag manner covering about 4 cm x 4 cm area. The tip of the swab was then cut and placed into the collection tube prefilled with the DNA/RNA Shield reagent. The tube was then capped and inverted several times to ensure homogeneity and stored at -20 oC until analysed. Following samples: A1: Pre-sample Day 0 (n= 10), and then post M. luteus Q24 serum application A2: 8h (n = 6), A3: 11 days (n = 10), A4: 25 days (n = 10) and A5: post-trial 7 days (n= 6) were sent to an external independent laboratory COSMOS ID, USA for Whole Genome Sequencing (WGS) and bioinformatic analysis.
2.5.2. DNA Extraction
DNA from the skin swab samples was isolated using the Zymo BIOMICS Micro Prep according to the manufacturer’s protocol. Extracted DNA samples were quantified using Qubit 4 fluorometer and Qubit™ dsDNA HS Assay Kit (Thermofisher Scientific, USA). DNA libraries were prepared using the Nextera XT DNA Library Preparation Kit (Illumina) and IDT Unique Dual Indexes with total DNA input of 1ng. Genomic DNA was fragmented using a proportional amount of Illumina Nextera XT fragmentation enzyme. Unique dual indexes were added to each sample followed by 12 cycles of PCR to construct libraries. DNA libraries were purified using AMpure magnetic Beads (Beckman Coulter) and eluted in QIAGEN EB buffer. DNA libraries were quantified using Qubit 4 fluorometer and Qubit™ dsDNA HS Assay Kit.
2.5.3. Library Preparation and Sequencing
DNA libraries were prepared using the Nextera XT DNA Library Preparation Kit (Illumina) and IDT Unique Dual Indexes with a total DNA input of 1ng. Genomic DNA was fragmented using a proportional amount of Illumina Nextera XT fragmentation enzyme. Unique dual indexes were added to each sample followed by 12 cycles of PCR to construct libraries. DNA libraries were purified using AMpure magnetic Beads (Beckman Coulter) and eluted in QIAGEN EB buffer. DNA libraries were quantified using Qubit 4 fluorometer and Qubit™ dsDNA HS Assay Kit. Libraries were then sequenced on an Illumina HiSeq X platform 2x150 bp.
2.5.4. Bioinformatics Analysis
Unassembled sequencing reads were directly analyzed by CosmosID-HUB Microbiome Platform (CosmosID Inc., Germantown, MD) [
13,
14,
15,
16] for multi-kingdom microbiome analysis. Briefly, the system utilizes curated genome databases and a high-performance data-mining algorithm that rapidly disambiguates hundreds of millions of metagenomic sequence reads into the discrete microorganisms engendering the particular sequences.
2.5.5. Statistical Analysis of Bioinformatic Analysis Data
Statistical analysis was performed using Prism 9.4.0 (GraphPad Software). Data were analysed by one-way analysis of variance (ANOVA).
3. Results
3.1. Cosmetic Efficacy Trial
3.1.1. Skin Quality Measurement
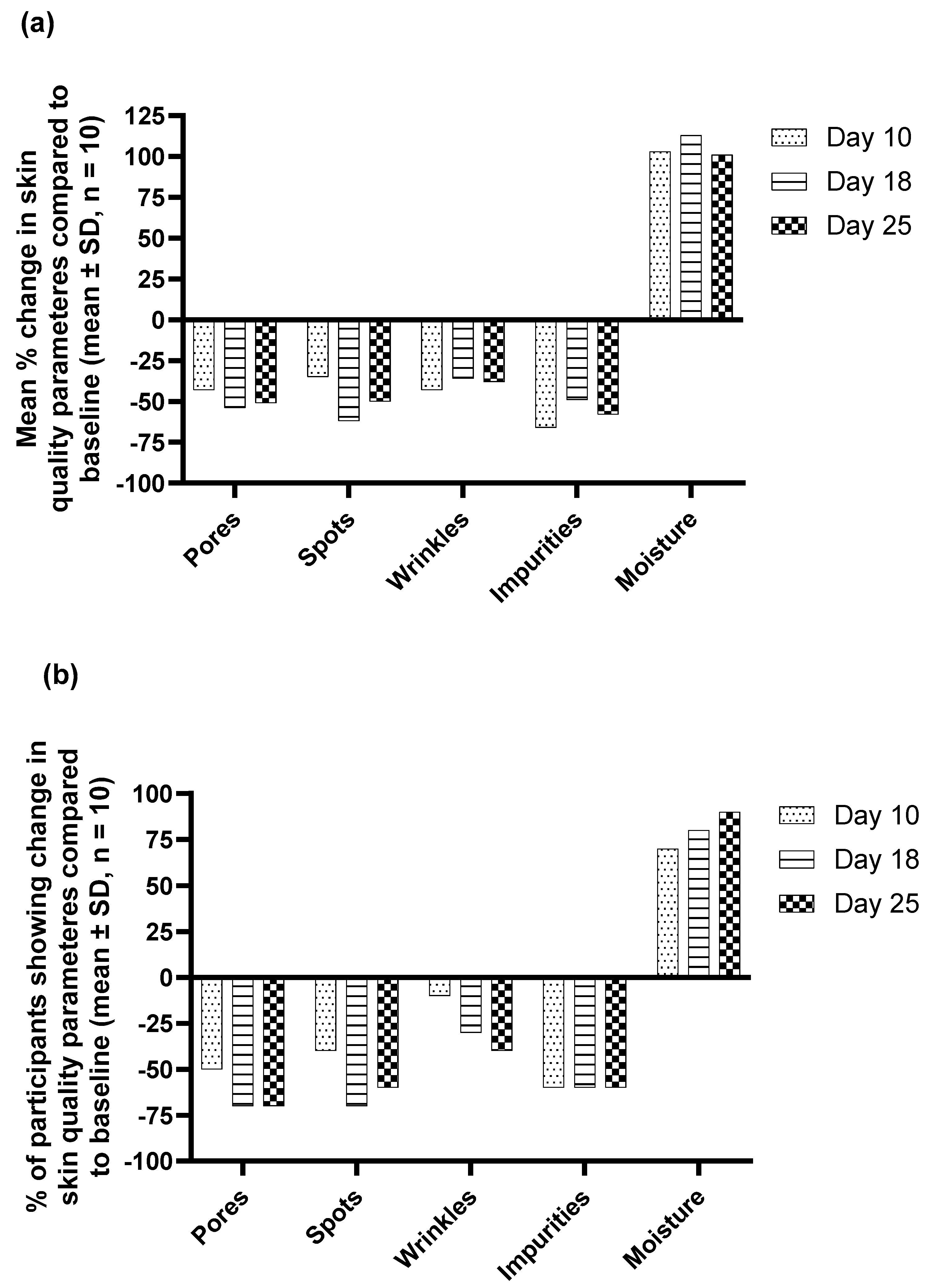
All participants completed the trial with no adverse events related to the product were reported. Compared to the baseline (
Figure 2), a significant (p ≤ 0.05) reduction in the mean scores for pores, spots, and wrinkles at all test points and on day 18 and day 25 for impurities (porphyrins) was observed. On the other hand, a significant increase in the hydration scores was observed after 18 and 25 days demonstrating the efficacy of the
M. luteus Q24 probiotic serum.
Further, compared to baseline, mean % reduction in the pore, spots, wrinkles, and impurities scores of 51%, 50%, 46%, and 56% respectively, and an increase of 101% in the hydration scores was observed after only 25-day application of the probiotic serum (
Figure 3a). Additionally, 45-80% of participants showed a decrease in skin quality parameters (
Figure 3b). The increase in hydration levels was observed with almost 60% of participants showed enhanced hydration in as early as 10 days and about 90% participants showed an increase in hydration after 25 days of probiotic serum application (
Figure 3b).
3.1.2. Microbial Analysis
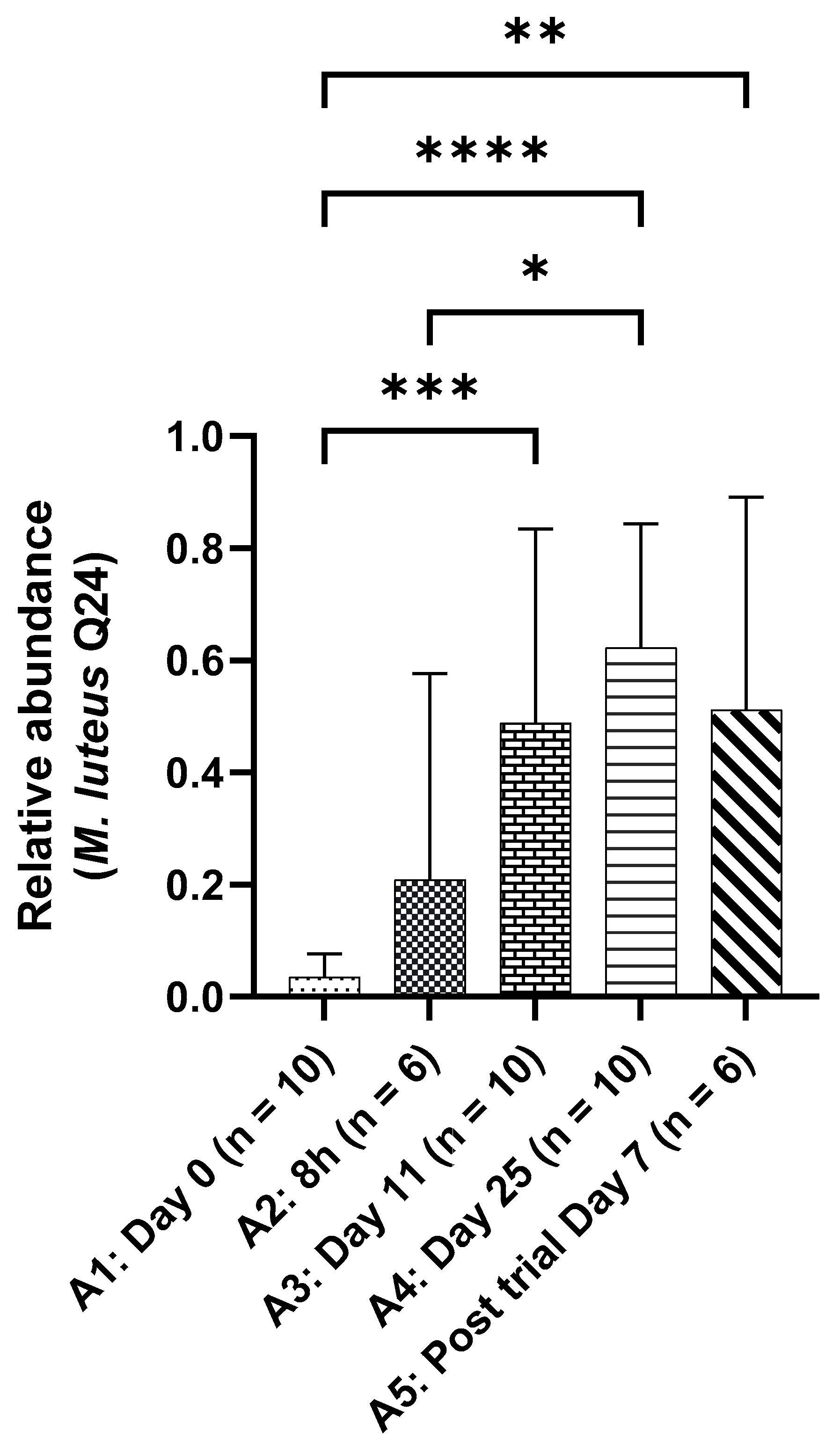
The WGS analysis of the skin swab samples showed a significant increase in the relative abundance of
M. luteus Q24 (
Figure 4). This increase was observed as early as post 8h after the first application and post-day 7 samples collected after the participants stopped using the product, showed persistence of
M. luteus Q24.
Under the conditions tested in this study involving healthy individuals, application of the serum seemed to maintain homeostasis of the microbiome as there was no significant difference observed in the levels of specific organisms commonly found on skin surface and sometimes implicated in diseases such as
Cutibacterium acnes, Staphylococcus aureus, Pseudomonas aeruginosa,
Corynebacterium or skin commensal such as
Staphylococcus epidermidis across the samples tested before and after the study (
Figure 5). This further highlight that the overall microbiome diversity is not impacted by the application of the
M. luteus Q24 serum in healthy individuals.
4. Discussion
The community of micro-organisms that inhabit the human skin is commonly referred to as the skin microbiome and includes pathogenic and commensal bacteria, fungi, viruses, and skin mites [
3,
17,
18,
19]. The colonisation of the human microbiome is initiated at birth, where the composition of microbial communities is strongly influenced by the route of delivery e.g., vaginal or caesarean (c) section [
1,
4]. A vaginal birth results in the newborn being colonised with microbes from the vaginal and faecal tract whereas a child born via a c-section immediately encounters the mother’s skin microbiota as well as microbes shed by the hospital staff or those present in the operating theatre [
20,
21]. From this point on the composition of an individual’s microbiota is determined by a multitude of internal and external host factors generating inter- and intrapersonal variations.
Several commensal and pathogenic bacteria co-exist among different niches on the surface of the skin [
5]. Commensal microbes inhabit different regions of the skin and contribute to symbiosis in various ways [
5]. The switch from a homeostatic state to a diseased state can be affected by microorganisms and their metabolites. It is not completely understood what initiates this process and if it is microbe-dependent or environmentally triggered or a combination of both. Potential theories include physical damage to the mucosa, disruption of the microbiota with antimicrobials, expression of specific virulence factors, and access to the site by several pathogens [
21,
22,
23]. However recent evidence supports the claim that a synergistic relationship between the species of bacteria that inhabit the skin influences the pathological progression of skin diseases [
19,
22].
A specific strain of probiotic bacteria
M. luteus Q24 that resides on the skin of healthy human adults has been identified to produce a unique antimicrobial spectrum that is inhibitory towards pathogenic bacteria associated with many skin diseases [
23]. It is thought that microbes not naturally found on the skin surface are less likely to survive or colonize efficiently on the skin surface in comparison with common skin commensals. Reasons for this include that the skin presents a continually shedding environment, is more suited for salt-tolerant microbes, and is exposed to many environmental factors [
3]. This means that any beneficial activity associated with the topical application of gut probiotics is probably akin to the relatively shorter-term benefits mediated by postbiotics or prebiotics and thus is unlikely to be maintained in the manner associated with the use of a colonising probiotic. Hence the importance of identifying and using live skin-specific microbes [
3,
12].
M. luteus Q24 was found to interact with the skin to enhance skin moisture and reduce pores, spots, wrinkles, and impurities [
12]. Unhealthy undernourished skin with poor skin quality is a result of a lack of hydration (moisture) that leads to dryness, loss in skin elasticity, and smoothness. One of the main reasons for dry skin is the damaged skin barrier. The skin barrier functions not only by maintaining the water content but also by protecting against the penetration of microorganisms, allergens, irritants, etc [
24]. In this study, a significant improvement in skin hydration was observed following the probiotic serum. This can be attributed to the interaction of the probiotic with the skin layers by hydrating them for repair and improvement of the skin barrier function [
25]. Further, the probiotic serum, due to its nature forms a protective occlusive layer on the skin to create a hydrophobic barrier over the skin and block the epidermal water loss thereby maintaining the integrity of the skin [
24,
25]. It is possible that the active metabolites produced by
M. luteus Q24 penetrate deeper into the skin tissues to positively influence the structure and function of the skin barrier, avoid desiccation of the underlying skin layers, and thus also help in reducing the pore size and coarse and fine wrinkles, the key signs of skin ageing [
24,
26]. Hyperpigmentation or spots is also a common condition of ageing skin that makes some areas of skin darker than others. The common exogenous factor responsible for skin spots is the photodamage caused by the sun, other factors could include injury to the skin, acne, and cuts. A significant improvement in the skin spots following the application of probiotic serum observed in this study is not surprising as
Micrococcus luteus has been reported to be UV resistant, with antioxidant, and ultraviolet protective properties [
27].
The impact of probiotic serum on the skin quality parameters was captured using a skin analyser that measures these parameters using advanced optics, sensors, and artificial intelligence in conjunction with advanced software and a complex algorithm to generate quantitative scores that can compare the skin quality before and after the application of the cosmetic product containing active. The skin analyzer measures the moisture level using a sensor that detects the changes in capacitance which differ depending on the moisture distribution. Moisture measurement indicates the hydration of the skin. When the level of hydration experienced by the skin is low it alters confounding issues associated with aging (dark, sunken, and dry skin) [
28]. Pores are small openings on the skin surface where sebaceous glands open to the external environment [
28]. The relative size of a pore may appear larger in areas where excess sebum is produced, where the pores are blocked with impurities or there is a loss of elasticity due to aging. Skin analyzer measures pore size by using the differences in brightness between the skin surface and the pore itself. Using brightness, morphological changes such as size, shape, depth, and elasticity of the pore are measured and used by the device software to calculate the pore index. Spots are markers of hyperpigmentation. This is due to ultraviolet rays, skin infections, or areas of scarring [
29]. Melanin, the pigment responsible for skin colour is excessively produced with hyperpigmentation, therefore the difference in brightness allows detection. The numerical value for pigmentation is computed by the analyser using the brightness value. Wrinkles are the folds and creases or ridges in the skin due to loss of skin elasticity, collagen, and moisture, UV radiation exposure, or a result of aging. Similarly, to the pores, the skin analyser uses light to detect wrinkles on the skin’s surface. The difference in brightness allows the length and depth of the wrinkle to be measured and quantified into a fixed value using AI. Finally, impurities (porphyrins) are an indicator of the amount of porphyrin on the skin, a substance produced by
Cutibacterium acnes [
30]. As a result, the number of impurities identified relates to the skin disease acne. This is identified as an orange colour which can be detected by light response using specific wavelength range detection.
There was a significant enhancement in moisture score observed as early as 10-day time point with enhanced score obtained throughout the application of the serum. The enhanced moisture level was complimented with a significant reduction in wrinkles, pores, and impurities again from the 10-day time point onwards showing the potential of the probiotic in providing cosmetic benefits of the probiotic in reducing signs of skin fatigue and ageing.
The skin samples were analysed by an independent laboratory using whole genome sequencing on the swab samples highlighting the microbiome modulation mediated by the application of
M. luteus Q24 serum. It was interesting but unsurprising to see the prominent relative abundance of
M. luteus Q24 in the skin swab samples with a significant increase throughout the application. The change in the relative abundance of
M. luteus Q24 coincided with the improvement in hydration level and reduction in the skin parameters associated with skin quality suggesting that the presence of
M. luteus Q24 may have been responsible for underlying mechanisms. Lastly, the preservation of microbial diversity on healthy skin suggests that topical application of
M. luteus Q24 probiotic may not only improves skin quality but also be considered microbiome-friendly. This is supported by its natural presence on human skin (commensal origin) [
3], its established safety profile [
31], and the positive outcomes observed in this study.
5. Conclusions
The topical application of probiotic M. luteus Q24 containing serum was evaluated for the potential of enhancing the cosmetic parameter in healthy adult female participants. Microbiome analysis of the skin swab samples was also carried out. The results support the efficacy of topical application of M. luteus Q24, with good colonisation efficacy and improvement in hydration level and reduction in parameters such as pores, spots, and wrinkles associated with ageing thereby highlighting the potential beneficial effects in cosmetics.
6. Patents
Some of the work included in this paper has been part of the provisional patent “Topical composition and use thereof”— PCT/IB2022/051860, Mar 2021.
Author Contributions
Conceptualization, I.J.M., R.J., A.L.V. and J.D.F.H.; methodology, R.J., A.L.V, and J.D.F.H.; formal analysis, A.L.V.; investigation, A.L.V., and R.J.; writing—original draft preparation, R.J, and I.M.; writing—review and editing, R.J., A.L.V., J.D.F.H.; project administration, J.D.F.H., and R.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and the protocol was reviewed by the Health and Disability Ethics Committee (New Zealand). “Ethical review and approval were waived for this study as the review board advised that since the scope of pilot trials were cosmetic-based rather than directed at health-related outcomes they were outside of the scope of HDEC review.
Informed Consent Statement
“Informed consent was obtained from all subjects involved in the study”, and “Written informed consent has been obtained from the participants(s) to publish this paper”.
Conflicts of Interest
All authors are past or current employees of Blis Technologies Limited, the manufacturer of Micrococcus luteus Q24TM probiotics.
References
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The Human Skin Microbiome. Nature Reviews Microbiology 2018 16:3 2018, 16, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Nakatsuji, T.; Gallo, R.L. The Role of the Skin Microbiome in Atopic Dermatitis. Ann Allergy Asthma Immunol 2019, 122, 263–269. [Google Scholar] [CrossRef] [PubMed]
- McLoughlin, I.J.; Wright, E.M.; Tagg, J.R.; Jain, R.; Hale, J.D.F. Skin Microbiome-The Next Frontier for Probiotic Intervention. Probiotics Antimicrob Proteins 2022, 14. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.; Byrd, A.L.; Park, M.; Kong, H.H.; Segre, J.A. Temporal Stability of the Human Skin Microbiome. Cell 2016, 165, 854–866. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Li, H. Acne, the Skin Microbiome, and Antibiotic Treatment. Am J Clin Dermatol 2019, 20, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Schommer, N.N.; Gallo, R.L. Structure and Function of the Human Skin Microbiome. Trends Microbiol 2013, 21, 660–668. [Google Scholar] [CrossRef] [PubMed]
- Egert, M.; Simmering, R.; Riedel, C.U. The Association of the Skin Microbiota With Health, Immunity, and Disease. Clin Pharmacol Ther 2017, 102, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Rather, I.A.; Bajpai, V.K.; Kumar, S.; Lim, J.; Paek, W.K.; Park, Y.-H. Probiotics and Atopic Dermatitis: An Overview. Front Microbiol 2016, 7, 507. [Google Scholar] [CrossRef] [PubMed]
- Clausen, M.L.; Edslev, S.M.; Andersen, P.S.; Clemmensen, K.; Krogfelt, K.A.; Agner, T. Staphylococcus Aureus Colonization in Atopic Eczema and Its Association with Filaggrin Gene Mutations. British Journal of Dermatology 2017, 177, 1394–1400. [Google Scholar] [CrossRef]
- Hale, J.D.F.; Wescombe, P.A.; Tagg, J.R.; Heng, N.C.K. Streptococcal Bacteriocin-Producing Strains as Oral Probiotic Agents. The Bacteriocins: Current Knowledge and Future Prospects 2016, 103–126. [Google Scholar] [CrossRef]
- Riley, M.A.; Wertz, J.E. Bacteriocins: Evolution, Ecology, and Application. Annu Rev Microbiol 2002, 56, 117–137. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.; Voss, A.L.; Tagg, J.R.; Hale, J.D.F. Evaluation of the Preliminary Safety, Tolerability and Colonisation Efficacy of Topical Probiotic Formulations Containing Micrococcus Luteus Q24 in Healthy Human Adults. Cosmetics 2022, 9, 121. [Google Scholar] [CrossRef]
- Ottesen, A.; Ramachandran, P.; Reed, E.; White, J.R.; Hasan, N.; Subramanian, P.; Ryan, G.; Jarvis, K.; Grim, C.; Daquiqan, N.; et al. Enrichment Dynamics of Listeria Monocytogenes and the Associated Microbiome from Naturally Contaminated Ice Cream Linked to a Listeriosis Outbreak. BMC Microbiol 2016, 16. [Google Scholar] [CrossRef] [PubMed]
- Ponnusamy, D.; Kozlova, E. v.; Sha, J.; Erova, T.E.; Azar, S.R.; Fitts, E.C.; Kirtley, M.L.; Tiner, B.L.; Andersson, J.A.; Grim, C.J.; et al. Cross-Talk among Flesh-Eating Aeromonas Hydrophila Strains in Mixed Infection Leading to Necrotizing Fasciitis. Proc Natl Acad Sci U S A 2016, 113, 722–727. [Google Scholar] [CrossRef] [PubMed]
- Hasan, N.A.; Young, B.A.; Minard-Smith, A.T.; Saeed, K.; Li, H.; Heizer, E.M.; McMillan, N.J.; Isom, R.; Abdullah, A.S.; Bornman, D.M.; et al. Microbial Community Profiling of Human Saliva Using Shotgun Metagenomic Sequencing. PLoS One 2014, 9, e97699. [Google Scholar] [CrossRef] [PubMed]
- Lax, S.; Smith, D.P.; Hampton-Marcell, J.; Owens, S.M.; Handley, K.M.; Scott, N.M.; Gibbons, S.M.; Larsen, P.; Shogan, B.D.; Weiss, S.; et al. Longitudinal Analysis of Microbial Interaction between Humans and the Indoor Environment. Science 2014, 345, 1048–1052. [Google Scholar] [CrossRef] [PubMed]
- Dréno, B.; Araviiskaia, E.; Berardesca, E.; Gontijo, G.; Sanchez Viera, M.; Xiang, L.F.; Martin, R.; Bieber, T. Microbiome in Healthy Skin, Update for Dermatologists. Journal of the European Academy of Dermatology and Venereology 2016, 30, 2038–2047. [Google Scholar] [CrossRef]
- Kong, H.H. Skin Microbiome: Genomics-Based Insights into the Diversity and Role of Skin Microbes. Trends Mol Med 2011, 17, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Cooper, A.J.; Weyrich, L.S.; Dixit, S.; Farrer, A.G. The Skin Microbiome: Associations between Altered Microbial Communities and Disease. Australasian Journal of Dermatology 2015, 56, 268–274. [Google Scholar]
- Maguire, M.; Maguire, G. The Role of Microbiota, and Probiotics and Prebiotics in Skin Health. Arch Dermatol Res 2017, 309, 411–421. [Google Scholar] [CrossRef]
- Milani, C.; Duranti, S.; Bottacini, F.; Casey, E.; Turroni, F.; Mahony, J.; Belzer, C.; Delgado Palacio, S.; Arboleya Montes, S.; Mancabelli, L.; et al. The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota. Microbiol Mol Biol Rev 2017, 81. [Google Scholar] [CrossRef] [PubMed]
- Huttenhower, C.; Gevers, D.; Knight, R.; Abubucker, S.; Badger, J.H.; Chinwalla, A.T.; Creasy, H.H.; Earl, A.M.; Fitzgerald, M.G.; Fulton, R.S.; et al. Structure, Function and Diversity of the Healthy Human Microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [PubMed]
- WO2006104403A1 - Skin Treatment Compositions - Google Patents. Available online: https://patents.google.com/patent/WO2006104403A1/en (accessed on 28 July 2021).
- Draelos, Z.D. Proper Skin Hydration and Barrier Function. Nutritional Cosmetics: Beauty from Within 2009, 355–363. [Google Scholar] [CrossRef]
- Spada, F.; Barnes, T.M.; Greive, K.A. Skin Hydration Is Significantly Increased by a Cream Formulated to Mimic the Skin’s Own Natural Moisturizing Systems. Clin Cosmet Investig Dermatol 2018, 11, 491–497. [Google Scholar] [CrossRef]
- Purnamawati, S.; Indrastuti, N.; Danarti, R.; Saefudin, T. The Role of Moisturizers in Addressing Various Kinds of Dermatitis: A Review. Clin Med Res 2017, 15, 75. [Google Scholar] [CrossRef]
- Mohana, D.; Thippeswamy, S.; Abhishek, R. Antioxidant, Antibacterial, and Ultraviolet-Protective Properties of Carotenoids Isolated from Micrococcus Spp. Radiation Protection and Environment 2013, 36, 168–174. [Google Scholar] [CrossRef]
- Roh, M.; Han, M.; Kim, D.; Chung, K. Sebum Output as a Factor Contributing to the Size of Facial Pores. Br J Dermatol 2006, 155, 890–894. [Google Scholar] [CrossRef] [PubMed]
- Desai, S.R. Hyperpigmentation Therapy: A Review. J Clin Aesthet Dermatol 2014, 7, 13. [Google Scholar]
- Mayslich, C.; Grange, P.A.; Dupin, N. Cutibacterium Acnes as an Opportunistic Pathogen: An Update of Its Virulence-Associated Factors. Microorganisms 2021, Vol. 9, Page 303 2021, 9, 303. [Google Scholar] [CrossRef]
- Jain, R.; Voss, A.L.; Tagg, J.R.; Hale, J.D.F. Evaluation of the Preliminary Safety, Tolerability and Colonisation Efficacy of Topical Probiotic Formulations Containing Micrococcus Luteus Q24 in Healthy Human Adults. Cosmetics 2022, 9. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).