Submitted:
14 June 2024
Posted:
17 June 2024
Read the latest preprint version here
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Hybridization Method
2.3. Plant Genomic DNA Isolation
2.4. Multiplex PCR Analysis
2.5. Gel Electrophoresis
2.6. Phytopathological Screening
2.7. Analysis of Structural Elements of Yield of Hybrid Rice Lines
2.8. Statistical Analyses
3. Results and Discussion
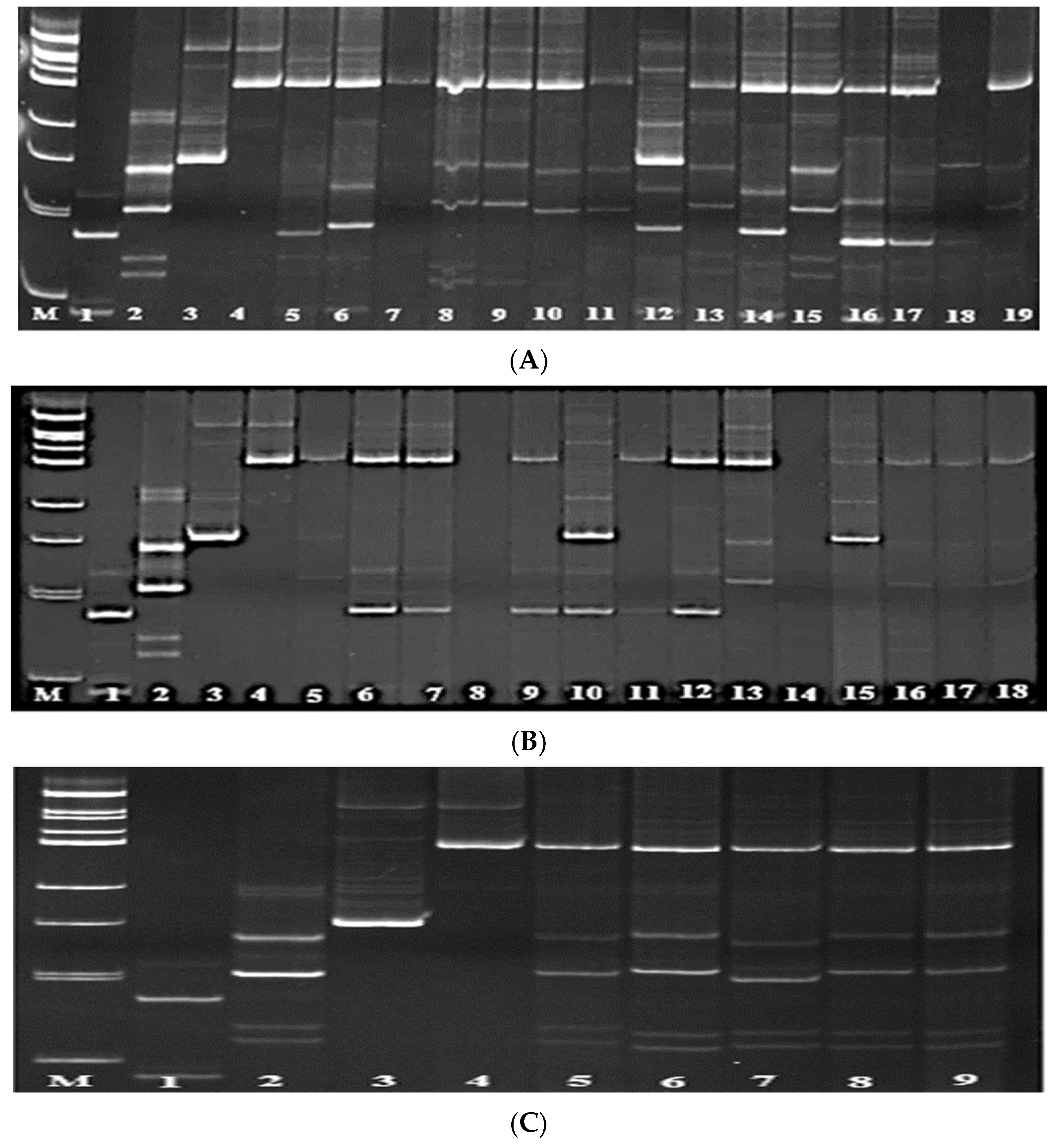
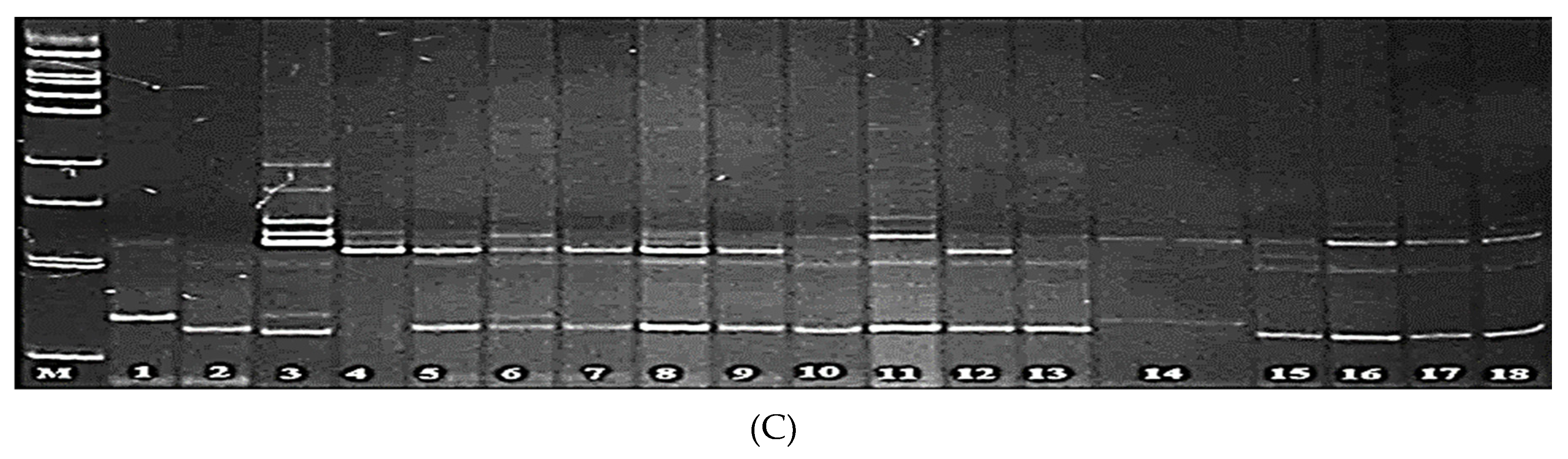
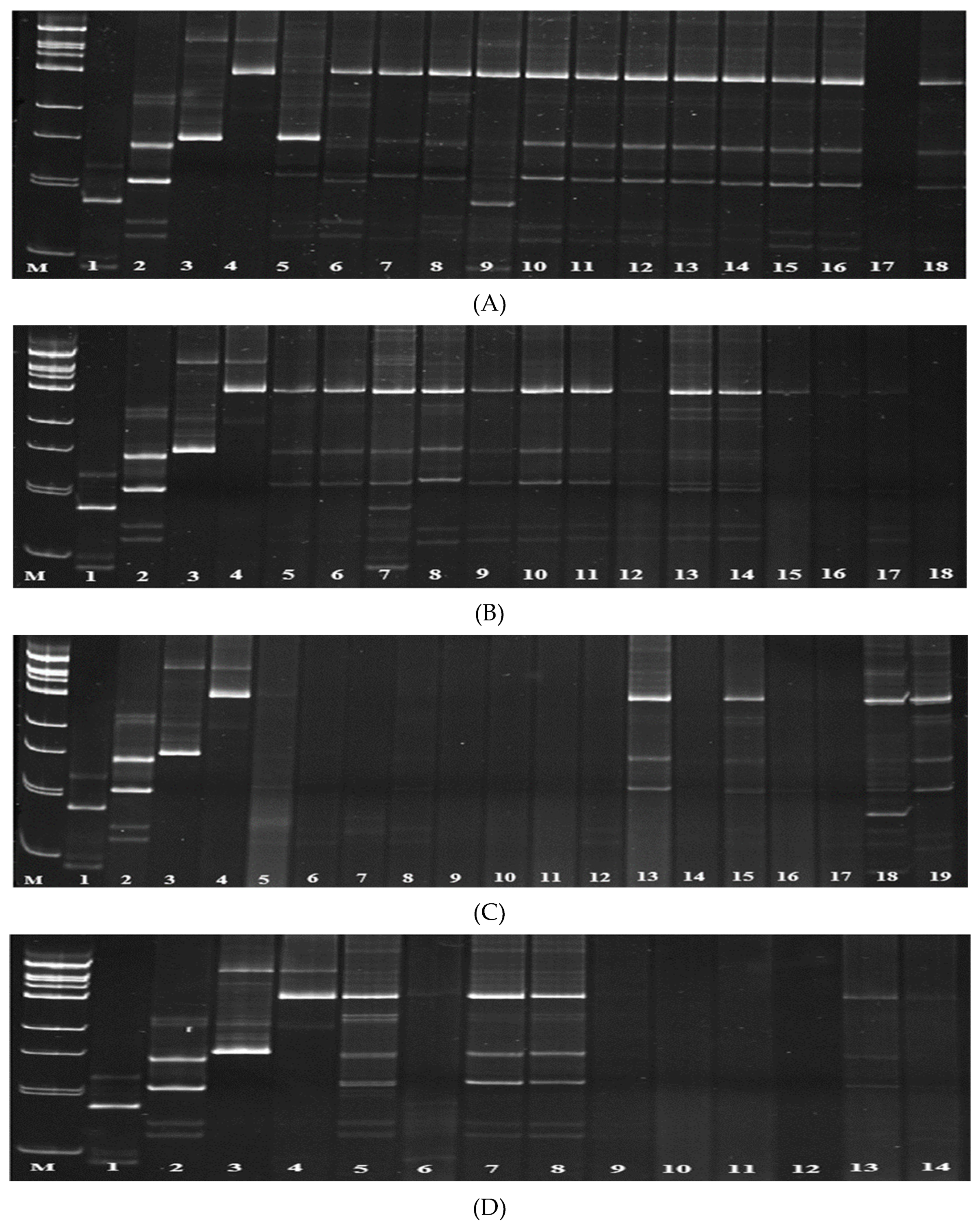
3.1. Screening of Parental Rice Genotypes for Blast Resistance Genes Pi-1 and Pi-2
3.2. Screening of Parental Genotypes for the Presence of Pi-33 and Pi-ta Genes
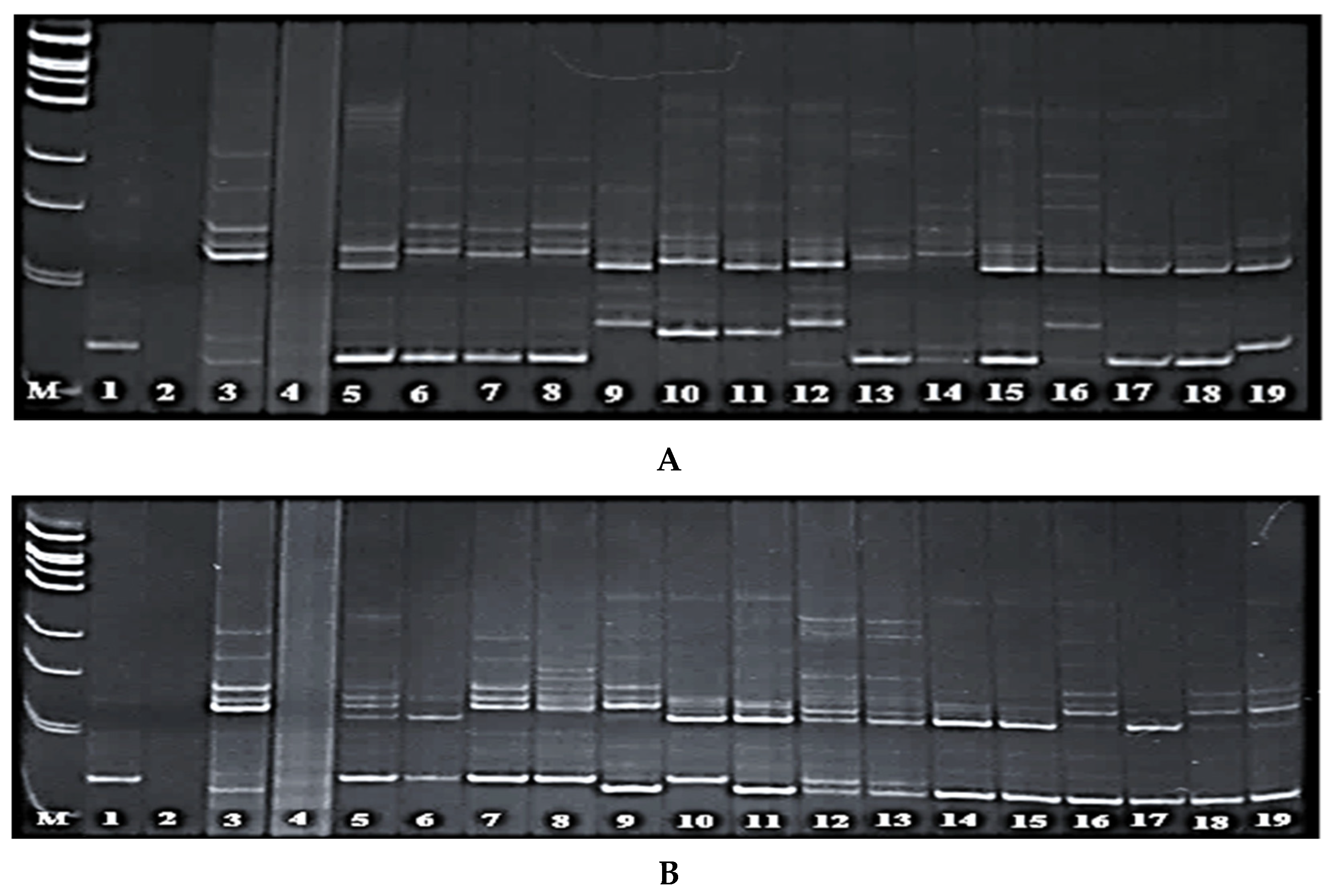
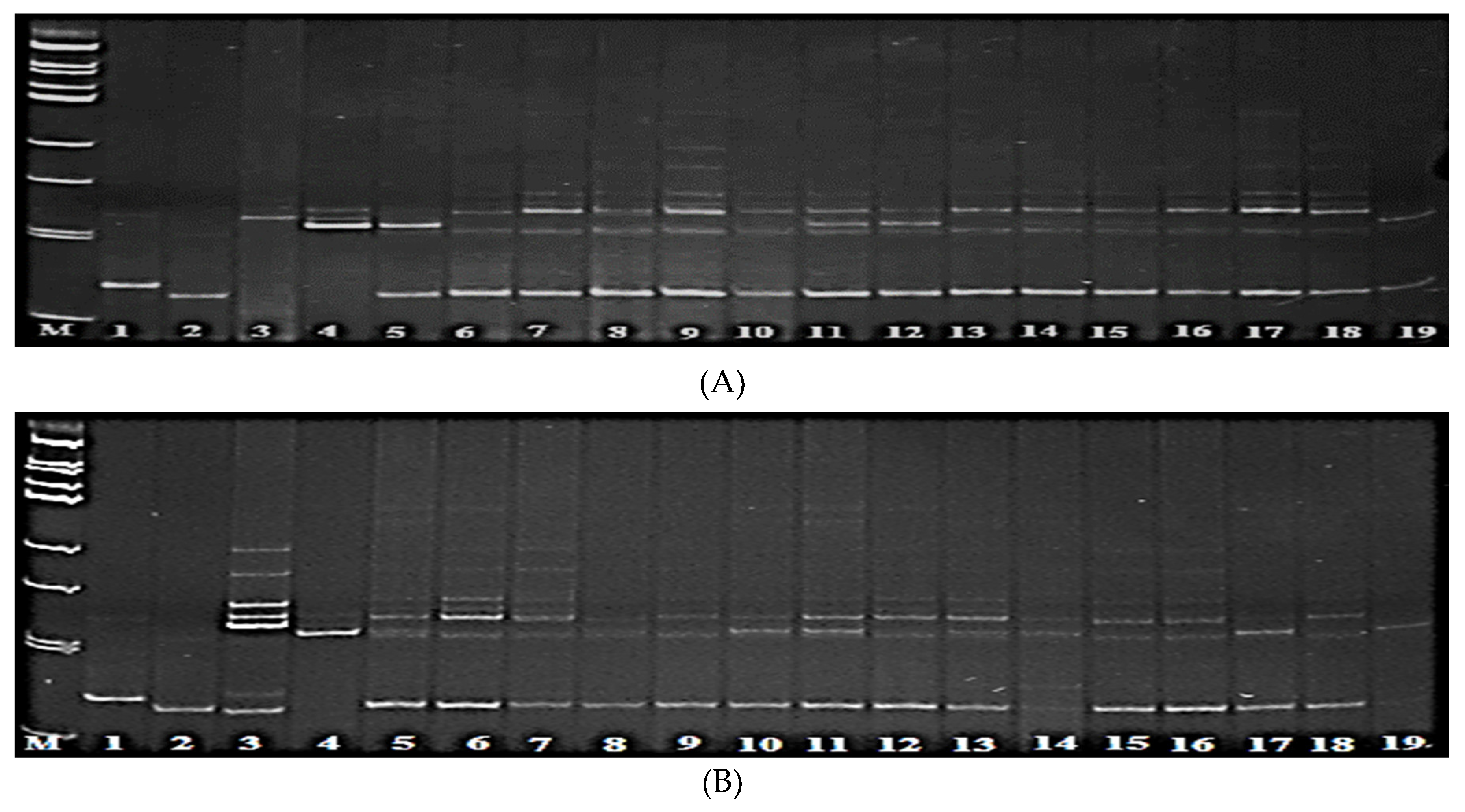
3.3. Screening of Rice Hybrids for the Presence of Pi-1 and Pi-2 Genes
3.4. Screening of Rice Hybrids for the Presence of Pi-33 and Pi-ta Genes
3.5. Phytopathological Test of Cultivars and Pyramided Lines for Resistance to Blast Disease
3.6. Assessment of the Productivity of Pyramided Rice Lines
4. Discussion
5. Conclusions
Author Contributions
Data Availability Statement
Conflicts of Interest
References
- Khush, G.S.; Jena, K.K. Current status and future prospects for research on blast resistance in rice (Oryza sativa L.). In Wang G.L., Valent B. (Eds.). Advances in genetics, genomics and control of rice blast disease. Springer, Dordrecht., 2009, 1-10. [CrossRef]
- Liu, J.; Wang, X.; Mitchell, T.; Hu, Y.; Liu, X.; Dai, L.; Wang, G.L. Recent progress and understanding of the molecular mechanisms of the rice-Magnaporthe oryzae interaction. Molecular Plant Pathology. 2010, 11, 419–427. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.B.; Wu, C.J.; Jiang, G.H.; Wang, L.Q.; He, Y.Q. Dynamic analyses of rice blast resistance for the assessment of genetic and environmental effects. Plant Breeding. 2007, 126, 541–547. [Google Scholar] [CrossRef]
- Zhang, T.; Huang, L.; Wang, Y.; Wang, W.; Zhao, X.; Zhang, S.; Zhang, J.; Hu, F.; Fu, B.; Li, Z. Differential transcriptome profiling of chilling stress response between shoots and rhizomes of Oryza longistaminata using RNA sequencing. PLoS One 2017, 12, e0188625. [Google Scholar] [CrossRef]
- Xiao, W.; Yang, Q.; Huang, M.; Guo, T.; Liu, Y.; Wang, J.; Yang, G.; Zhou, J.; Yang, J.; Zhu, X.; Chen, Zh.; Wang, H. Improvement of rice blast resistance by developing monogenic lines, two-gene pyramids and three-gene pyramid through MAS. Rice 2019, 12. [Google Scholar] [CrossRef]
- Liu, Z.; Zhu, Y.; Shi, H.; Qiu, J.; Ding, X.; Kou, Y. Recent Progress in Rice Broad-Spectrum Disease Resistance. Int. J. Mol. Sci. 2021, 22, 11658. [Google Scholar] [CrossRef] [PubMed]
- Miah, G.; Rafii, M.Y.; Ismail, M.R.; Puteh, A.B.; Rahim, H.A.; Latif, M.A. Marker-assisted introgression of broad-spectrum blast resistance genes into the cultivated MR219 rice variety. J. Sci. Food Agric. 2017, 97, 2810–2818. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Li, Z.; Liu, J.; Shen, Z.; Gao, G.; Zhang, Q.; He, Y. Development and evaluation of improved lines with broad-spectrum resistance to rice blast using nine resistance genes. Rice. 2019; 12. [Google Scholar] [CrossRef]
- Jain, P.; Dubey, H.; Singh, P.K.; Solanke, A.U.; Singh, A.K.; Sharma, T.R. Deciphering signaling network in broad spectrum near isogenic lines of rice resistant to Magnaporthe oryzae. Sci. Rep. 2019, 9, 16939. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Xiao, N.; Chen, Y.; Yu, L.; Pan, C.; Li, Y.; Zhang, X.; Huang, N.; Ji, H.; Dai, Z.; Chen, X.; Li, A. Comprehensive evaluation of resistance effects of pyramiding lines with different broad-spectrum resistance genes against Magnaporthe oryzae in rice (Oryza sativa L.). Rice. 2019, 12, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Tang, J.; Yang, D.; Chen, Y.; Ali, J.; Mou, T. Improving rice blast resistance of Feng39S through molecular marker-assisted backcrossing. Rice. 2019, 12, 1–16. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, L.; Zhang, X.; Zhang, Q.; Jia, Y.; Wang, G.; Li, S.; Tian, D.; Li, W.H.; Yang, S. Large-scale identification and functional analysis of NLR genes in blast resistance in the Tetep rice genome sequence. Proc. Natl. Acad. Sci. USA. 2019, 116, 18479–18487. [Google Scholar] [CrossRef]
- Ramalingam, J.; Palanisamy, S.; Alagarasan, G.; Renganathan, V.G.; Ramanathan, A.; Saraswathi, R. Improvement of stable restorer lines for blast resistance through functional marker in rice (Oryza sativa L.). Genes. 2020, 11, 1266. [Google Scholar] [CrossRef]
- Peng, M.; Lin, X.; Xiang, X.; Ren, H.; Fan, X.; Chen, K. Characterization and evaluation of transgenic rice pyramided with the Pi Genes Pib, Pi25 and Pi54. Rice. 2021, 14, 1–14. [Google Scholar] [CrossRef]
- Devanna, N.B.; Vijayan, J.; Sharma, T.R. The blast resistance gene Pi54 of cloned from Oryza officinalis interacts with Avr-Pi54 through its novel non-LRR domains. PLoS One. 2014, 9, 104–40. [Google Scholar] [CrossRef]
- Deng, Y.; Zhai, K.; Xie, Z.; Yang, D.; Zhu, X.; Liu, J.; Wang, X.; Qin, P.; Yang, Y.; Zhang, G.; Li, Q.; Zhang, J.; Wu, S.; Milazzo, J.; Mao, B.; Wang, E.; Xie, H.; Tharreau, D.; He, Z. Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science. 2017, 355, 962–965. [Google Scholar] [CrossRef]
- Xie, Z.; Yan, B.; Shou, J.; Tang, J.; Wang, X.; Zhai, K.; Liu, J.; Li, Q.; Luo, M.; Deng, Y.; He, Z. A nucleotide-binding site-leucine-rich repeat receptor pair confers broad-spe;ctrum disease resistance through physical association in rice. Philosophical Transactions of the Royal Society B. 2019, 374, 20180308. [Google Scholar] [CrossRef]
- Zhao, H.; Wang, X.; Jia, Y.; Minkenberg, B.; Wheatley, M.; Fan, J.; Jia, M.H.; Famoso, A.; Edwards, J.D.; Wamishe, Y.; Valent, B.; Wang, G.L.; Yang, Y. The rice blast resistance gene Ptr encodes an atypical protein required for broad-spectrum disease resistance. Nat. Commun. 2018, 9, 2039. [Google Scholar] [CrossRef]
- Meng, X.; Xiao, G.; Telebanco-Yanoria, M.J.; Siazon, P.M.; Padilla, J.; Opulencia, R.; Bigirimana, J.; Habarugira, G.; Wu, J.; Li, M.; Wang, B.; Lu, G.; Zhou, B. The broad-spectrum rice blast resistance (R) gene Pita2 encodes a novel R protein unique from Pita. Rice. 2020, 13, 1–15. [Google Scholar] [CrossRef]
- Chen, D.H.; Zeigler, R.S.; Ahn, S.W.; Nelson, R.J. Phenotypic characterization of the rice blast resistance gene Pi-2(t). Plant Disease. 1996, 80, 52–56. [Google Scholar] [CrossRef]
- Amante-Bordeos, A.; Sitch, L.A.; Nelson, R.; Dalmacio, R.D.; Oliva, N.P.; Aswidinnoor, H.; Leung, H. Transfer of bacterial blight and blast resistance from the tetraploid wild rice Oryza minuta to cultivated rice Oryza sativa. Theor. Appl. Genet. 1992, 84, 345–354. [Google Scholar] [CrossRef]
- Jia, Y.; Wang, Z.; Singh, P. Development of Dominant Rice Blast Pi-ta Resistance Gene Markers. Crop Sci. 2002, 42, 2145–2149. [Google Scholar] [CrossRef]
- Mukhina, Zh.M. The use of DNA markers to study the genetic diversity of plant resources, Krasnodar: Prosveshenie-South, 2008, 98 p.
- Xu, X.; Chen, H.; Fujimura, T.; Kawasaki, S. Fine mapping of a strong QTL of field resistance against rice blast, Pikahei-1(t), from upland rice Kahei, utilizing a novel resistance evaluation system in the greenhouse. Theoretical and applied genetics. 2008, 117, 997–1008. [Google Scholar] [CrossRef]
- Ashkani, S.; Rafii, M.Y.; Rahim, H.A.; Latif, M.A. Genetic dissection of rice blast resistance by QTL mapping approach using an F3 population. Molecular Biology Reports. 2013, 40, 2503–2515. [Google Scholar] [CrossRef] [PubMed]
- Ballini, E.; Morel, J.B.; Droc, G.; Price, A.; Courtois, B.; Notteghem, J.L.; Tharreau, D. A genome-wide meta-analysis of rice blast resistance genes and quantitative trait loci provides new insights into partial and complete resistance. Molecular plant-microbe interactions: MPMI 2008, 21, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Padmavathi, G.; Ram, T.; Satyanarayana, K.; Mishra, B. Identification of blast (Magnaporthe grisea) resistance genes in rice. Current Science. 2005, 88, 28–630. [Google Scholar]
- Sharma, R.C.; Shrestha, S.M.; Pandey, M.P. Inheritance of blast resistance and associated microsatellite markers in rice cultivar ‘Laxmi’. Journal of Phytopathology. 2007, 155, 749–753. [Google Scholar] [CrossRef]
- Mohanty, C.R.; Gangopadhyay, S. Testing of blast resistance in F2 rice seedlings in different doses of nitrogen and seasons. Annals of the Phytopathological Society of Japan. 1982, 48(5), 648–658. [Google Scholar] [CrossRef]
- Flores-Gaxiola, J.A.; Nuque, F.L.; Crill, J.P.; Khush, G.S. Inheritance of blast Pyricularia oryzae resistance in rice. International Rice Research Newsletter 1983, 8, 5–6. [Google Scholar]
- Mackill, D.J.; Bonman, J.M.; Suh, H.S.; Srilingam, R. Genes for resistance to Philippine isolates of the rice blast pathogen. Rice Genetics Newsletter. 1985, 2, 80–81. [Google Scholar]
- Bonman, J.M.; Khush, G.S.; Nelson, R.J. Breeding rice for resistance to pests. Annual Review of Phytopathology. 1992, 30, 507–528. [Google Scholar] [CrossRef]
- Lang, N.T.; Luy, T.T.; Ha, P.T.T.; Buu, B.C. Monogenic lines resistance to blast disease in rice (Oryza sativa L.) in Vietnam. Int J Genet Mol Biol 2009, 1, 127–136. [Google Scholar]
- Hasan, N.; Choudhary, S.; Naaz, N.; Sharma, N.; Laskar, R.A. Recent advancements in molecular marker assisted selection and applications in plant breeding programmes. Journal of Genetic Engineering and Biotechnology. 2021, 19, 128. [Google Scholar] [CrossRef] [PubMed]
- Zelensky, G.L. Rice: biological foundations of breeding and agricultural technology. Monography. Krasnodar. KubSAU 2016, 177. [Google Scholar]
- Los, G.D. Rice hybridization technique. Rice growing [Los' G. D. Metodika gibridizacii risa. G. D. Los'. Risovodstvo. 2007, 10, 42–51. [Google Scholar]
- Williams, C.E.; Ronald, P.C. PCR template-DNA isolated quickly from monocot and dicot leaves without tissue homogenization. Nucleic Acids Research. 1994, 22, 1917–1918. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lu, Y.G.; Shi, Y.; Wu, L.; Xu, Y.J.; Huang, F.; Guo, X.Y.; Zhang, Y.; Fan, J.; Zhao, J.Q.; Zhang, H.Y.; Xu, P.Z.; Zhou, J.M. , Wu, X.J., Wang, P.R.; Wang, W.M. Multiple rice microRNAs are involved in immunity against the blast fungus Magnaporthe oryzae. PlantPhysiol. 2013, 164, 1077–92. [Google Scholar] [CrossRef]
- 39. Guidelines for the Conduct of Test for Distinctiveness, Uniformity and Stability On (Oryza sativa L.). Plant Variety Journal of India 2007, 1, 25.
- Li, W.; Lei, C.L.; Cheng, Z.J.; Jia, Y.L.; Huang, D.Y.; Wang, J.L.; Wang, J.K.; Zhang, X.; Su, N.; Guo, X.P.; Zhai, H.Q.; Wan, J.M. Identification of SSR markers for a broad-spectrum blast resistance gene Pi20(t) for marker-assisted breeding. Mol.Breed. 2008, 22, 141149. [Google Scholar] [CrossRef]
- Peng, P.; Jiang, H.; Luo, L.; Ye, C.; Xiao, Y. Pyramiding of Multiple Genes to Improve Rice Blast Resistance of Photo-Thermo Sensitive Male Sterile Line, without Yield Penalty in Hybrid Rice Production. Plants. 2023, 12, 1389. [Google Scholar] [CrossRef]
- Ospanova, A.; Mynbayeva, D.; Turganova, Ch.; Usenbekov, B.; Amirova, A.; Berkimbay, Kh.; Zhunusbayeva, Zh.; Utepbergenov, E. Molecular screening for the presence of Magnaporthe oryzae resistance Pi-b gene in rice hybrids. International Scientific Forum “Modern Trends in Sustainable Development of Biological Sciences”. BIO Web of Conferences 2024, 100, 03012. [Google Scholar] [CrossRef]
- Amirova, A.; Usenbekov, B.; Berkimbay, Kh.; Mynbayeva, D.; Atabayeva, S.; Baiseitova, G.; Meldebekova, A.; Zhunusbayeva, Zh.; Kenzhebayeva, S.; Mukhambetzhanov, S. Selection of rice breeding lines for resistance to biotic and abiotic stresses. Brazilian Journal of Biology. 2024, 84, e282495. [Google Scholar] [CrossRef]
| Gene | Localization on chromosome | Name of a marker |
Forward primer (5′-3′) Reverse sequence (5′-3′) |
| Pi-1 | 11 | Rm 224 | F - аtсgаtсgаtсttсасgаgg R - tgctataaaaggcattcggg |
| Rm144 | F - tgccctggcgcaaatttgatcc R - gctagaggagatcagatggtagtgcatg |
||
| Pi-2 | 6 | Rm 527 | F - ggctcgatctagaaaatccg R - ttgcacaggttgcgatagag |
| SSR 140 | F - aaggtgtgaaacaagctagcaa R - ttctaggggaggggtgtgaa |
||
| Pi-33 | 8 | Rm 310 | F - ссggсgаtаааасааtgаg R - gсаtсggtссtаасtааggg |
| Rm 72 | F - ссggсgаtаааасааtgаg R - gсаtсggtссtаасtааggg |
||
| Pi-ta | 12 | Pita | F1 - gссgtggсttсtаtсtttасatg R1 - аtссааgtgttаgggссаасаttс |
| F2 - ttgасасtсtсаааggасtgggаt R2 - tсааgtсаggttgааgаtgсаtсgа |
| SS | df | MS | F | P-value | F crit | |
| Between | 367,1166333 | 6 | 61,18610556* | 27,07338195 | 0,000000000000421.97 | 3,254124603 |
| Within | 97,18041667 | 43 | 2,26000969 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




