1. Introduction
Tuberculosis (TB) is caused by
Mycobacterium tuberculosis which is spread from person to person through the air, which makes it highly contagious. According to this, the World Health Organization (WHO) maintains that TB is among in the top ten causes of death worldwide. Moreover, until 2020, it was the leading cause of death caused by a single infectious agent, now overshadowed by COVID-19 [
1,
2]. In developing countries, the situation is worse due to almost
of the people with active TB are concentrated in 30 of these countries each year. Different efforts led by WHO, such as the End TB initiative, have been proposed, but it has been difficult to reduce the incidence rates (newly diagnosed cases), which had an increment of
between 2020 and 2021, due to the impact of the COVID-19 pandemic. In addition, it was estimated that 10.6 million people fell ill with TB in 2021, representing
more than in 2020 [
3].
One out of four people is estimated to be infected with latent TB worldwide. This means that the mycobacterium lives in the host without developing and transmitting the disease. Furthermore, people with this latent TB can develop active TB with a
more risk associated with compromised immune systems such as HIV, diabetes, malnutrition, tobacco, or homeless conditions [
4,
5]. TB can affect any organ of the body but mainly attacks the lungs, known as pulmonary TB (PTB), with symptoms such as severe cough lasting more than three weeks, chest pain, and coughing up blood or sputum [
6,
7]. Other forms of TB as recognized as extra-pulmonary TB, where the most common is pleural TB, and the more lethal is meningeal TB.
The present work analyzed a case with information from a developing country such as Colombia. This country has reported an incidence rate of
in 2021 and an increase to
in 2022 per 100000 population, registering 17341 new reported cases in the last year for PTB [
3,
8]. There, it is straightforward to identify some problems related to the inequality of resources regarding public health. There are regions or states in the national territory with higher PTB incidence rates, such as Amazonas or Antioquia, where the rural conditions established values of
and
, respectively. This last state had
of the total PTB cases in Colombia [
1,
8,
9]. These farming areas with precarious health systems are far from the main cities, as the capital, where the incidence rate reached 16.02. It is necessary to contribute to proposing alternative strategies that allow better disease management and identification and begin the antiPTB treatment as soon as possible.
In most cases, PTB can be treated aggressively using four drugs for six months or more in cases of drug-resistant TB [
10,
11]. The Colombian health system has a protocol for these treatments [
12], but in some regions, the diagnostic methods are not fully accessible, presenting fatal consequences. The same standard has PTB detection and reporting guidelines in the national health system [
12]. Besides, the diagnosis must be elaborated by microbiological confirmation of the presence of the mycobacteria in the sputum of suspicious patients. For this, there are three types of tests: smear microscopy, molecular tests, and culture. However, the protocol established that health professionals can initiate the treatment based on clinical analysis of the patient, even without bacteriological confirmation, to prevent the disease from spreading and to avoid its progress in the patient.
Smear microscopy is the simplest and least expensive test, with results that can take short periods. Despite this, its sensitivity is low, between
and
, depending on the quality of the sample. Molecular tests have a higher sensitivity, more than
, and results can be delivered within hours. The disadvantage of this method is the specialized requirements related to equipment and professionals, which can become more expensive than smear microscopy. Finally, cultures are the most potent methods, with high sensitivity and specificity, demanding skilled personnel and costly infrastructure, and the duration of the results can take two to three weeks at best [
12,
13]. Each test has its advantages and disadvantages. Depending on the availability of the tests, a patient may have one, two, or all three tests performed. Nevertheless, the time and costs associated with each test constitute a gap in its accessibility in some places, so it is necessary to create new low-cost and rapid technologies that can be used to support health professionals in diagnosing the disease [
14].
Despite the current technologies and the protocol to diagnose the PTB, some locations in the country need more conditions to develop this procedure. Sometimes health professionals hold traditional instruments to do their work and laboratories or sophisticated supplies can only be performed on time. Based on previous strategies reported by the same team of researchers, a search for alternatives for this scenario is related in [
15]. In the present case, an extension was based on a reported text that medical staff accomplished in the patient consultation, introducing a description with medical findings and terminology, which holds in the traditional Colombian health systems and can be analyzed as an additional data source in the diagnosis process.
In recent years, Artificial Intelligence (AI) has been used in medicine to support decisions [
16,
17]. The findings are supported by computer systems on which health professionals can rely to do a better job. These systems can process large volumes of data and use that learning for a specific task [
18,
19]. Among the advantages of these tools is that they are low-cost, and front-line personnel can use them, making them useful in situations where conventional methods are unavailable [
20]. Different applications of using AI in health problems can be seen in support of cancer detection [
21], COVID-19 diagnosis and treatment, and drug discovery through deep neural networks implementation [
22,
23,
24]. In the case of TB, [
25] and [
26] show how an artificial neural network (ANN) can be trained to diagnose TB using clinical data (CD), and in [
26] and [
27] variants of ANN for clustering are used to determine three risk groups (high, medium, and low risk) of the population concerning TB, showing good results.
Natural Language Processing (NLP) is a branch of AI that allows the analysis of texts that are not necessarily written in a structured language and can be used to design support tools. For example, NLP has been used to build AI systems that help in tasks such as searching for relevant information [
28], determining eligibility and tracking patients [
29,
30], or diagnosing diseases [
31], generally using information contained in the electronic medical records (EMR) such as clinical, laboratory or image data. For constructing these NLP systems, it has been found that the best-performing models are those that learn the rules of the data, i.e., those that use machine learning (ML) techniques [
32]. From 2021 to now, the popularity of large language models (LLM) has increased, motivating the use of generative models and proposals for text processing. Difficulties presented by these approaches are associated with the specific dataset employed to develop the model and the manual annotations made by expert medical professionals [
33,
34]. Examples of this can be seen in diabetes disease problem for evaluating the risks related to food intake [
35], a specific TB case with a comparison between NLP labels based on image and the TB experts screening on chest RX [
36], among other applications for data fusion in the medical context [
37]. An important difference from those previous works, we explored the use of the NLP representation according to available information for training the models in a limited scenario with little data. This represents the context of some developing countries, where there are challenges related to systematic health data acquisition, available data, and the health human resources component included in model development.
The present work is framed within that project and seeks to develop models based on computational intelligence to allow health personnel to make better decisions about suspected PTB patients. The data was collected from 151 patients, consisting of clinical data such as HIV status, location inside the city, and sex. In addition, physicians report (PR) at the moment of patient consultation as a routine practice. With these two sources of information, four schemes were proposed to create models that predict PTB status from data. This data fusion proposal explores information provided by CD, PR, or both sources in the PTB diagnosis support process. For this, two schemes merge the information from CD and PR, and the other two use them separately. In all of them, the use of five ML algorithms was explored: Support Vector Machine (SVM), K-Nearest Neighbors (KNN), Logistic Regression (LR), Random Forest (RF), and ANN. In the case of PR, it is necessary to employ NLP techniques to represent the texts in numerical form so that the ML algorithms can learn from this representation. The proposed scenario can contribute to training models employed in tasks associated with diagnosing PTB without the main cities’ evolved infrastructure.
2. Materials and Methods
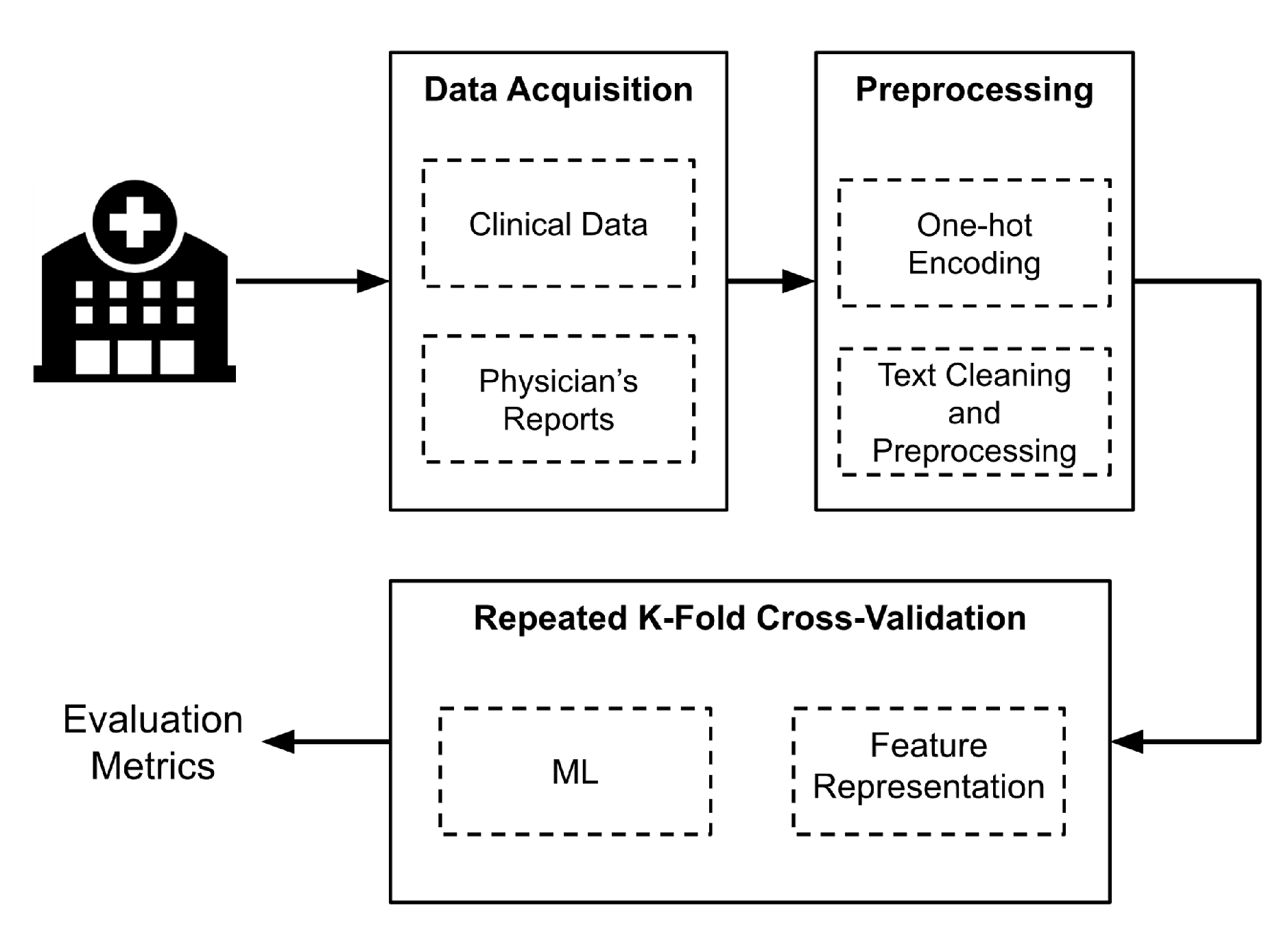
Figure 1 shows a general scheme of the methodology implemented. First, data were acquired at the Hospital Santa Clara in Bogota D.C., Colombia. Then, a preprocessing and cleaning process was implemented before extracting the features representing the data, which is the input of the algorithms. For the ML models, four types of schemes that use five ML algorithms were proposed, and a repeated through stratified
k folds cross-validation technique was performed to evaluate the performance of the models. Each step of
Figure 1 will be explained in detail in this section.
Dataset
The data were acquired during the period from 2017 to 2019 at the Hospital Santa Clara in Bogota D.C., Colombia, associated with the Middle-East Integrated Subnetwork of Health Services of the city (from Spanish Subred Integrada de Servicios de Salud Centro Oriente). The subnetwork ethics committee approved the study by act No 316 of May 24th, 2021. Approval was established based on anonymization of data with only population-related variables that posed no risks to subjects. Informed consent was not required because all data were retrospective and anonymous.
Data variables associated with CD were extracted from registers from the institutional PTB program. PR was obtained from EMR, which is standardized according to the record acquisition system of the hospital. Data belong to clinically suspected subjects of having PTB, and microbiological tests were performed on these patients to confirm the disease. These tests allowed us to determine 116 confirmed PTB cases and 35 patients without the disease from an initial dataset with 233 individual registers. The difference was not considered because of unavailable PTB confirmation or PR information.
For PR extraction, when usable, it was decided to employ dates related to diagnostic tests and treatment beginning. Temporal locating of the PR was taken into account due to the information holding that suggests PTB. Thus, five PR were extracted 30 days before to these dates and analyzed and summarized because not all the information in the EMR can be associated with PTB, but the patient’s entire medical history.
In the case of the CD, data were collected by the physicians involved in the PTB institutional program and members of the researchers’ team, including patient’s information such as
i) Bogota’s inside location,
ii) sex,
iii) human immunodeficiency (HIV) status,
iv) antiretroviral treatment (ART) status,
v) type of population associated with risk groups such as homeless, migrants, displaced persons, or belonging to indigenous people. These variables were considered according to suggestions from physicians and their essential codification by the binary representation [
38].
Table 1 visualizes the variables and possible values.
Preprocessing
In the preprocessing and data cleaning, an exploratory analysis was performed to determine any errors in acquiring the data and to fix them if possible. The database of 151 patients was constructed considering this analysis, because there were cases of patients in which, for external reasons, the EMR was not available, or the dates recorded for the performance of the diagnostic tests did not correspond to those appearing in the EMR. It was also decided to organize the population of Bogota into five zones based on where they live in the north, south, east, west, and outside Bogota so that there is a significant number of patients per zone. The localities in each area correspond to those assigned by the city to each one of the four integrated subnetworks health services. This allows including indirect sociodemographic information.
On the other hand, the preprocessing of the PRs consists of cleaning up the text and removing Spanish stop words to reduce noise and ensure that the text representation (feature extraction) and the ML focus more on words and concepts that can provide relevant information regarding PTB. The stop words are meaningless words within a language, such as articles, pronouns, and prepositions, and in this case, units of physiological variables were included. Removing stop words is relevant in this work because these meaningless words do not provide information for the classification task.
Other words in the clinical reports indicate information and events unrelated to PTB [
39]. However, the effect of these last terms is intended to be reduced with post-processing. Additionally, characters that were not letters of the Spanish alphabet were eliminated, including accents, punctuation marks, numbers, and other strange symbols. Also, double spaces and line breaks were eliminated. All this was done to remove from the text elements that are not useful and may confuse the algorithms. More information about the construction of the dataset from the EMR and a preliminary analysis of the content of the texts can be found in [
39]. For practical issues, the patient document is called the union of the five PR preprocessed and cleaned for each patient.
Feature Representation
The CD source data was established from nominal variables represented by numerical vectors. For this, the variables were one-hot encoded, employing binary unique values associated to the original variable value. For example, the variable HIV has three values: positive, negative, and unknown. Therefore, three columns were created, representing each value, assigning one (1) binary value for the corresponding relation of existence or zero (0) in the absence of the value variable. This numerical representation allows ML algorithms to be trained from the categorical variables of the CD without a specific order or preference.
For NLP applications, computers receive language in the form of text coming from the patients’ documents. These texts need to be encoded into a numerical representation so that ML algorithms can learn from the data. For this, two modes of representing the text of the patients’ documents were explored, in which each document has a vector representing its content.
Term Frequency - Inverse Document Frequency: The first method uses the measure Term Frequency- Inverse Document Frequency (TF-IDF) [
40]. This metric helps highlight words that appear in a document but penalizes them if they appear in several documents. The information of the word is related inversely to the number of times that appears in the documents, due to the low relevance when the classifying of the documents is done. Equation (
1) shows how IDF was calculated for each term
, where
is the number of documents, and
is the number of documents in the document set that contain the term
. The set of all the terms that appear in the documents is known as the vocabulary.
After obtaining the IDF for all the terms, the TF-IDF is computed by multiplying each item (TF) occurrence in a document with the corresponding IDF. Thus, each document has a vector with the TF-IDF value for each term in the set. In addition, an L2 normalization was applied for each vector, so the sum of squares of the vector elements is 1.
On the other hand, the terms for TF-IDF do not necessarily have to be single words, the use of n-grams can be implemented. N-grams are sub-sequences of elements, for example, a 2-gram is a sequence of two consecutive words that appear in a document. This strategy can capture the language structure and add context to the words. Different combinations of n-grams were explored to build the vocabulary: i) 1-grams (a single term), ii) 1-grams and 2-grams combination, iii) 2-grams, iv) 1-grams, 2-grams, and 3-grams combination, v) 3-grams. As the number of terms in the vocabulary can be large, the size of the vectors was limited to a range of , the maximum and minimum values of that range were explored during the process, and the vocabulary size was also limited to 1000 terms at most. However, the length of the vectors remains large to apply ML with the number of samples available, so a dimensionality reduction was employed using truncated single value decomposition (SVD), where the number of components was also explored.
Version 1.0 of the
Scikit Learn library was used to obtain the TF-IDF vector for each document [
41]. The vector computation was based on parameters explained above, such as analyzer by words,
n-grams from one to three grams, and a maximum number of features of 1000. According to the previous subsection, stop-words were not considered when a preprocessing was implemented based on the Spanish language.
Embeddings Representation: The second method employed embeddings representation. In the embeddings, each term is represented by a vector so that words with similar meanings, or used in a similar context, have a close vector representation. There are different ways to obtain embeddings, they can be obtained from a layer of an ML model by training in a specific task, or they can also be obtained in an unsupervised way from document statistics [
42]. In the present work, Word2Vec model was used to build the embeddings [
43]. There, two algorithms were applied, continuous bag-of-words (COBW), which predicts a word in the middle of a window using words around it. The second algorithm is skip-grams, the reverse of COBW one, which indicates the context word for a given target word based on unsupervised learning techniques. Both methods employ neural networks, layer, and weights concepts to predict the words and to obtain the embeddings.
Finally, there is a vector that represents the terms in the document, but a vector that represents each document is needed to use it to train classic ML algorithms. For this reason, a pooling step was added using two different operations, the first one by calculating the maximum between the elements of the vectors of each word and the other one by calculating the mean. Therefore, each document was represented by a vector containing either the maximum or the mean of the set of embeddings that represents each word.
Machine Learning Models
Before discussing the experimental design, it was necessary to understand the ML algorithms used briefly. As mentioned above, the five algorithms used are KNN, LR, SVM, ANN, and RF. These models were trained to do a binary classification between patients with confirmed TB and patients without TB. KNN is an algorithm that does not attempt to construct a general internal model, it simply stores instances of the training data, so it is an instance-based learning classifier. The classification was computed using the information of the
k (a specified number) nearest neighbors. In this approach, the discrimination is based on the voting of the majority of the samples with the nearest neighbors or assigning weights according to the inverse proportional distance from the point to be classified [
44].
The regression task developed by the LR model consists of optimizing a cost function. There, the problem is to find the probability of doing the best job based on a logistic regression function, modeling the probability for a binary output, which in the present case is the TB detection [
41].
In the case of the ANN, different architectures can be employed, however, the current one consists of one hidden layer with a set of neurons. Each neuron performs a weighted sum of its inputs and applies an activation function on the output to add nonlinearity to the data. The weights of the weighted sum are learned from the training set and generalized for new inputs [
45].
Another approach for classification can be implemented through the use of SVM. The objective here is to maximize the margin, defined as the distance between the separation hyperplane and the training labeled samples closest to this hyperplane [
46]. These samples are called support vectors and contribute to the classification of two main classes: TB-detected and non-detected.
Finally, the RF model assembles a set of decision trees that are trained separately like a real forest. This training has the objective of maximizing the information. However, one of these trees usually needs to be more capable of solving a classification problem independently. What RF achieves is to put together a set of weak classifiers to improve the classification task [
47]. The
Scikit Learnlibrary was used to implement all algorithms except ANN, which was implemented with Keras library and its version 2.7.0rc0 [
48]. For each case, the hyperparameters or each approach were established through a grid search strategy, where different values were tested and those with better results were chosen.
Experimental Design
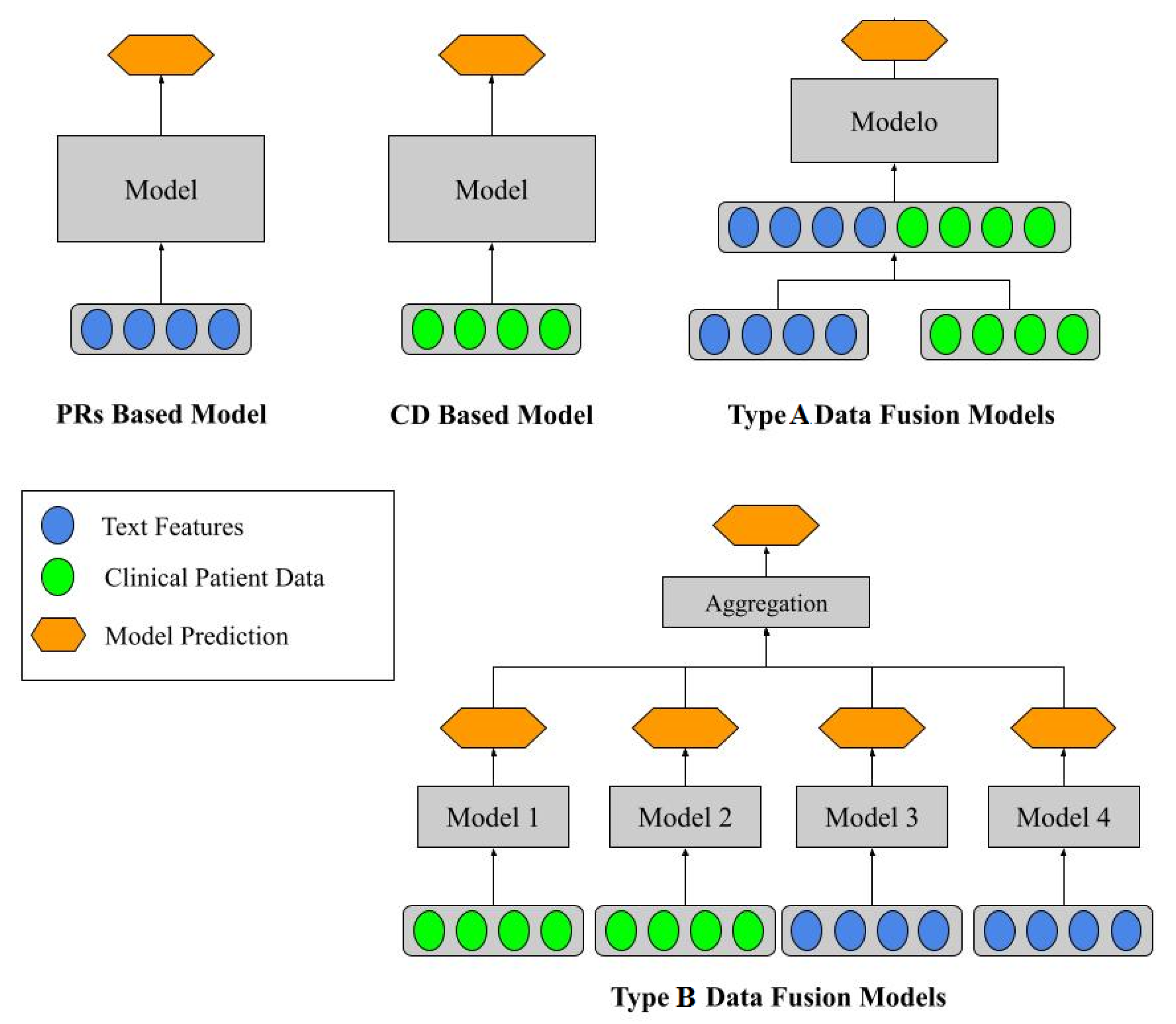
As mentioned above, five ML schemes were proposed to perform the classification.
Figure 2 shows these schemes. Two approaches use PR and CD isolated and two fusion schemes that merge the two sources of information: type A and type B data fusions. The ML approaches were employed for the PR and CD-based and type A data fusion models. By contrast, in the type B data fusion models, where the ML-employed models were selected based on the results of PR and CD-based isolated approaches. This was because it is possible to know which are the best ML algorithms for the problem.
In the type B data fusion model, there is an aggregation layer with two different strategies. The first one is a simple majority vote of the four classifiers. In cases of a tie, the classifier selects the class by giving the highest priority to model one and the lowest priority to model four. The second strategy is called stacking classifier, where an ML algorithm is used as an aggregation layer. The LR algorithm was chosen for this task because its inputs are the four ML models’ probability or the decision function (depending on the type of algorithm).
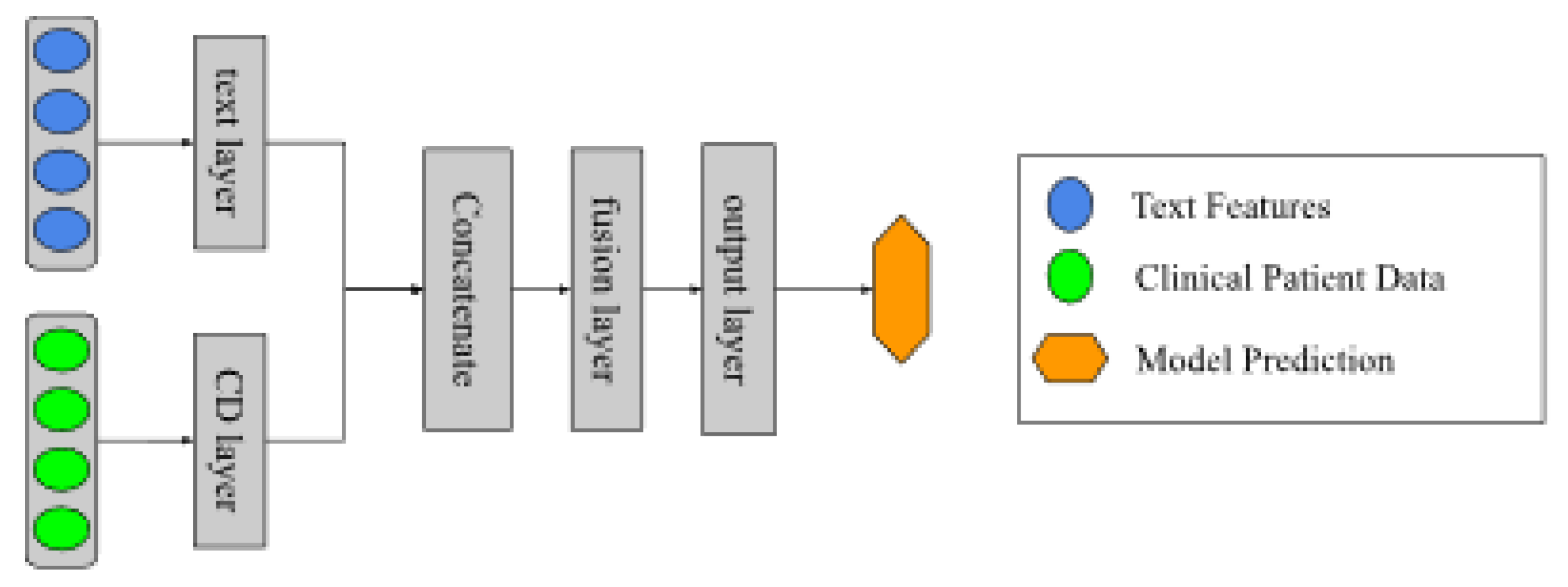
On the other hand, a different model was added from those mentioned above, which consists of a neural network as shown in
Figure 3. This architecture was designed so that two networks would learn simultaneously from the PR and the CD in the learning process. Then, these layers are concatenated, and a new layer oversees the information fusion. The output layer has an output neuron with a sigmoid activation function that does the binary classification.
Stratified
k-fold cross-validation (SKCV) was used to select the best parameters for text representation with TF-IDF, embeddings, and the hyperparameters of the ML models, as shown in
Figure 1. In SKCV, the data were separated into different partitions for training and validation of the approaches, with the objective of being different sets. Stratified means that the divisions preserve the percentage of samples for each class. Three folds were taken because of the database size, maintaining a considerable quantity in the validation set for the evaluation of models correctly. For statistical performance evaluation, the SKCV was repeated ten times, and the mean and standard deviation over the metrics of all repetitions were employed to evaluate and compare the results of the models.
The parameters explored to obtain the TF-IDF measure were the combinations of n-grams and the maximum and minimum values of to include a term in the vocabulary. The number of components for the dimensionality reduction was also explored with the TF-IDF calculated. Both, the calculation of the TF-IDF and the dimensionality reduction, were fitted by employing the training set, and then, applied to the training and testing sets. The methods were fitted with the training set because when predicting the diagnosis of a document, the TF-IDF method does not know the vocabulary within that document. In this way, the test documents simulate adding a new input well.
For the Word2Vec models obtention, the training set was needed to implement the neural networks of the COBW and skip-grams models. In the embedding models, parameters such as the size of the window and the minimum number of times a word appears to be part of the vocabulary were explored. Also, in the pooling step, the maximum and the mean operation were used. For both feature extraction methods, a vector that represents each document with a normalization where a transformation with values into a range between -1 and 1 was applied.
Furthermore, the hyperparameters of the ML algorithms were explored by employing random grid values in a heuristic mode until the best results were found. For models with a small size of hyperparameters, every single option was examined whenever the problem allowed it: LR, ANN, and the SVM kernel models. In the fusion models, the same methodology was also performed. However, this exploration was shorter because of the results for the PR and CD-based models. Also, it is important to note that a combination of the parameters of the text representation and the ML hyperparameters were considered in a coupled mode.
The metrics used to evaluate the results of the SKCV were sensitivity, specificity, and area under the curve (AUC), according to its pertinence in health systems [
49]. These metrics were calculated for each of the testing sets and averaged. Subsequently, ten repetitions were performed to find the average and standard deviation over ten cross-validations using SKCV. The mean and standard deviation of these three metrics allowed us to evaluate the variability of the models. Additionally, due to the unbalanced dataset, the AUC was weighted with the number of samples of each class to have a metric that considers this unbalanced. Finally, the same training and test sets were always used to compare models’ results.
Results and Discussion
Table 3,
Table 4 and
Table 5 show the results of the PR and CD-based isolated models. Each table shows the five algorithms used, and the best model results are shown in bold. In the case of the DC-based models in
Table 3, the best ML algorithm was ANN. However, all models except KNN obtained similar results (see
Table 2 for details of the hyperparameters).
Table 4 and
Table 5 show the models trained using the features extracted from the patient documents. From two used methods used for PR analysis, representation using the TF-IDF approach obtained the best result with an AUC of
,
lower than that presented by the CD-based models.
Table 6 shows the results of the type A data fusion models. As shown in
Figure 1, each of these models receives both sources of information as input. The results are similar to those obtained in
Table 3, which could be explained according to the information provided from CD data compared to patient documents since getting a higher performance with the first source is possible.
As mentioned in the experimental design, four algorithms were used for type B data fusion models (see
Table 7), considering the results of
Table 3,
Table 4 and
Table 5. From
Table 3, the LR and SVM models were taken, which obtained better performance than the other models. ANN model was not considered because it was used in the architecture of
Figure 3. Two models from
Table 4 were taken into account because the TF-IDF method show better results than the embeddings one. Again, the algorithms taken were LR and SVM. There is no AUC metric for the voting classifier because it was not possible to estimate a probability function from the votes.
As mentioned in the experimental design for type II data fusion models, four algorithms were used considering the results of
Table 3,
Table 4 and
Table 5. From
Table 3, the LR and SVM models were taken, which obtained a better performance compared to other models. ANN model was not considered because it was used in the architecture of
Figure 3. Only two models from
Table 4 were taken from
Table 4 and
Table 5 because the TF-IDF method showed better results than the embeddings method. Again, the algorithms taken were LR and SVM. For the voting classifier, and as in the previous scenario, there is no AUC metric because of the lacking possibility to estimate a probability function from the votes.
Comparing the performance of the four proposed schemes, the models based on CD were the ones that obtained the best results in terms of AUC value. Although it was also possible to classify patients by combining the information, the model’s performance remained the same. However, a combination is an option to provide data to the diagnosis system support due to the lack of information unavailable due to different issues, such as the patients avoiding answering questions about HIV status, for example. In these cases, the interview of the patients and the PR created can be used with these tools to decide a patient’s diagnosis.
Despite the result for sensitivity (
) when PR was represented using embeddings, the AUC value was under the CD-based models at
. Comparable results reported an AUC of
and a sensitivity of
with a set of clinical variables similar to this study where an ANN obtained the best result [
15]. In addition, in a similar scenario of limited infrastructure, reported sensitivity values of
[
26]. Furthermore, specificities could have been better, according to
and
values, respectively. It is important to mention that, despite the similarities, the scenarios change with the involved variables and preprocessing stages.
Concerning the data fusion models, type A was better than B, with values of AUC of
and
, respectively. However, the sensitivity for the type B data fusion model reached
, comparable with more available tests based on smear microscopy. In addition, PR should have provided more information when the results were similar for CD-based models (AUC =
) and the best data fusion model (AUC =
), dropping by less than one percentage point. Nevertheless, the best PR-based proposal reached the best sensitivity value of the results with
(see
Table 5). This aspect could be helpful when not all data are available. Health professionals can analyze different proposals for the specific diagnosis scenario and decide with more information than they hold with only CD or PR in an isolated mode.
The use of NLP in the TB diagnosis problem has been reported in terms of detection by employing cough signal analysis and embeddings in [
50], differentiation of TB and Covid-19 from reported cases through text analysis in [
51], and comparing a NLP-based image approach versus natural text generated by radiologists from chest radiographs (CXR) in [
36]. However, works with the present approach based on text from the EMR has not been explored in the same manner as it was developed here. A similar work that employed the fusion of structured and unstructured data for Tuberculosis prediction with results of AUC with mean of
for different models was achieved in [
52]. Nevertheless, this study was carried out with information from 692949 patients and more sophisticated clinical variables such as main complaint hemoptysis, cough, and test erythrocyte sedimentation rate, which is not comparable with the present work. Data fusion between text reports and images of CXR were used in a pre-trained model to establish a report from the available information, reaching a validation accuracy of
. A relevant difference can be found in terms of the number of images with around 3800 images from a known dataset [
53]. Finally, exploratory works where the NLP technique has been used for extracting information on TB from EMR were reported in [
54], but in those cases a classification for supporting the disease diagnosis was not implemented. With this, it is possible to see how the future and incremental work in this field can be improved.
Considering the specific context for the PTB diagnosis, where clinical signs and symptoms are considered as a first sight in the process, the possible patient is acknowledged as a suspect of having the disease. Then, according to the Colombian national protocol, it is necessary to include one of the three tests described in the introduction section. Smear microscopy is developed using sputum to visualize the mycobacterium in the microscopic test. However, due to the sufficient microorganisms that must be in the sample, the sensitivity reached by the test is between 40 and
[
54]. Present results, with proposals based on embeddings and the type B data fusion, obtained sensitivity values of
and
, respectively, could be an option. In this case, the models can accomplish the activities of the health professionals when just smear microscopy is available. Another test with better sensitivity values, as culture in solid or liquid mediums, needs a time interval of two weeks and more sophisticated infrastructure [
55], a scenario that is not always possible in low-income countries. Finally, with the best advantages regarding time and sensitivity results, the molecular test is not an option [
56]. In Colombia, the national health system does not have more than twenty places with quality properties for this test. In addition, the solution is based on the transportation of the necessary samples, which can be compromised by inadequate shipping. Then, the infrastructure for this technology could be improved in developing countries such as Colombia. A tool based on the present proposal can provide information about the patient’s condition to make a proper diagnosis and start treatment timely.
Limitations of the present study are related to the dataset size to propose the models. More samples are better for ML and, in general, AI applications that learn from data. However, the present approach was taken from a real scenario in the Hospital Santa Clara, which represents one of the institutions with the most PTB-treated patients. Unifying the data systems and transferring from different health centers with information previous to confirmed cases continues to be challenging in the same city. Availability of data with the same characteristics is difficult to establish, according to the national protocol for reporting TB-diagnosed patients. Moreover, it is important to mention that, with the advances and use of LLM architectures and its basic unit like the Transformer could be an option that is out of the scope of the present work, where preliminary information can be used. For this, a deeper analysis could be more specific, including the mentioned architectures. Something to pay attention to approaches like that is the lack of a considerable dataset to adjust the model, it will be necessary to increase the size of the data. This depends on the epidemiology aspects of the disease and medical center, which for this case is low but it is the most representative center in Bogota D.C. city.