1. Introduction
In recent years, cancer genomic medicine has been implemented in clinical practice, and comprehensive genomic profiling (CGP) using next-generation sequencers (NGS), which can detect a large number of gene sequences in tumor tissue specimens, has become available [
1,
2,
3,
4,
5,
6,
7,
8,
9,
10,
11]. In pancreatic cancer, CGP has made it possible to select treatment options, such as molecularly targeted drugs and immune checkpoint inhibitors, that are suited to the patient, with new treatment options expected in the future [
12,
13,
14,
15,
16,
17,
18,
19,
20,
21]. Therefore, endoscopic ultrasound-guided tissue acquisition (EUS–TA) in unresectable pancreatic cancer requires not only a tissue diagnosis but also tissue collection in anticipation of CGP [
22].
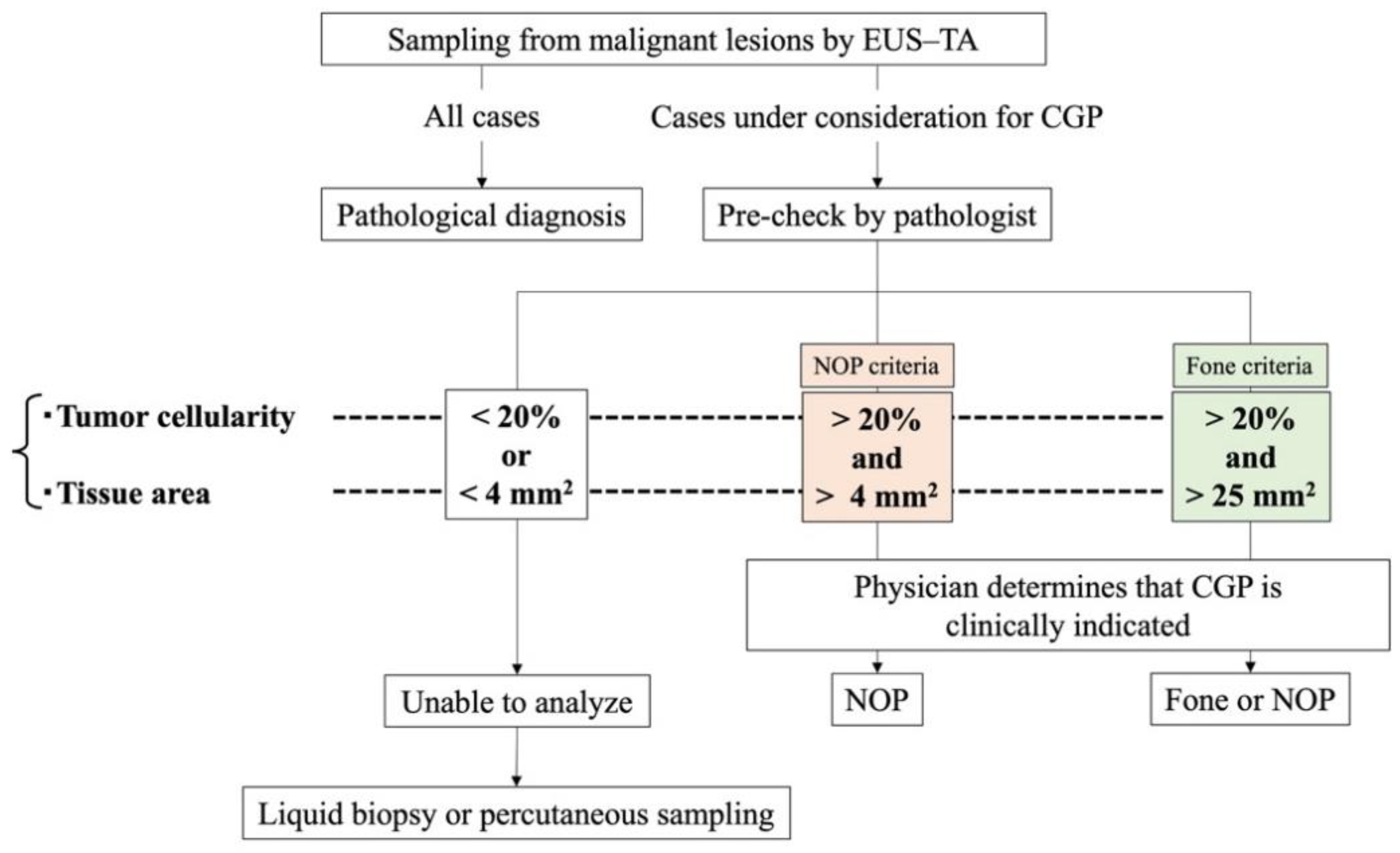
CGP systems currently approved in Japan include the OncoGuide NCC Oncopanel System (NOP; Sysmex Corporation, Hyogo, Japan) and Foundation One CDx Cancer Genome Profile (Fone; Foundation Medicine, Cambridge, MA, USA). The specimen criteria for the NOP system are tumor cellularity ≥ 20% and tissue area ≥ 4 mm
2, and that for the Fone system are tumor cellularity ≥ 20% and tissue area ≥ 25 mm
2 (
Table 1).
In clinical settings, a pathologist evaluates the quality of the tissue specimen obtained by EUS–TA (during pre-check), and only cases that meet the criteria are subjected to CGP analysis (
Figure 1). As pancreatic cancer is representative of low-cellularity tumors with abundant stromal components, the tumor cell content is reported to be 5%–20%; accordingly, a greater amount of adequate tissue for CGP is required [
23].
Two prospective studies have examined the proportion of successful CGP analyses of pancreatic tumor specimens. Carrera et al. reported a success rate of 97% (32/33) for CGP using NGS of cryopreserved tissue samples obtained by fine-needle biopsy (FNB) with a 22G needle [
24]. Hisada et al. analyzed the pre-check results of EUS–TA specimens obtained by EUS–FNB with a 19G needle from a single lesion in patients with unresectable pancreatic cancer with metastasis (UR–M) and found that 63.6% (21/33) of specimens met NOP criteria [
25]. Additionally, several retrospective studies have examined the success rate of genomic analysis of pancreatic cancer specimens obtained using a FNB needle [
24,
26,
27,
28,
29,
30], including two analyses that focused exclusively on invasive pancreatic ductal carcinoma. Elhanafi et al. performed a retrospective study of EUS–TA in 167 patients with pancreatic cancer and found that 70.1% of specimens met NGS analysis criteria [
29]. However, this study utilized a low threshold in the criterion for tumor cellularity (≥ 10%) in the pre-check. Park et al. performed a retrospective study of 190 patients with invasive pancreatic ductal adenocarcinoma and reported an NGS analysis success rate of 57.4% [
30]. However, this study utilized a lower threshold for DNA content (≥ 50 ng) than that required for the NOP system. Thus, the reported percentages of specimens meeting NGS analysis criteria show considerable variation (57.4%–100%), likely because of differences in outcome measures (percentage of successful analyses rather than that of the pre-check), specimen preservation methods, patient population (invasive pancreatic ductal adenocarcinoma with or without other cancer types), and specimen criteria (unspecified or lower criteria than that required for the NOP system).
The relationship between the EUS–TA technique and percentage of successful CGP analyses has also been evaluated. Park et al. reported that the puncture needle diameter (22G or 19G) and tumor site (body-tail), but not needle type (fine needle aspiration or FNB), contributed to a successful CGP analysis of primary pancreatic specimens collected using EUS–TA [
30]. Larson et al. reported that large FNB needle diameter (19G, n = 2; 22G, n = 41; and 25G, n = 2) was associated with a high success rate for meeting NGS analysis criteria (p = 0.05) [
28]. Therefore, when performing EUS–TA with an FNB needle for CGP, a large puncture needle diameter, with a large amount of tissue sampled, is recommended to achieve a high analysis success rate.
However, the optimal puncture target remains controversial. Larson et al. compared the proportion of specimens that met CGP criteria between those obtained by EUS–TA for primary pancreatic lesions and EUS–TA or percutaneous biopsies for metastatic lesions and found a significantly higher success rate for metastatic lesions than for primary lesions, despite the difference in sampling modalities (p = 0.036) [
28]. Ikeda et al. reported that, overall, 39.2% (60/153) of pancreatic specimens obtained with an FNB needle met NOP analysis criteria during pre-check; however, primary lesion specimens (37.1%, 53/143) were significantly less likely than metastatic lesion specimens (70%, 7/10) to meet analysis criteria (p = 0.049).[
31] Nevertheless, data on the proportion of metastatic lesion specimens that meet CGP analysis criteria remain insufficient, and it is not yet clear whether primary or metastatic lesions have a high success rate for CGP analysis of invasive pancreatic adenocarcinoma.
Therefore, we designed a study to clarify the appropriate target for EUS–TA when CGP is planned for pancreatic ductal adenocarcinoma by comparing the success rates for meeting NOP analysis criteria between primary and metastatic lesion specimens obtained during the same EUS–TA session in patients with invasive pancreatic ductal adenocarcinoma. The protocols are described herein.
2. Study Design
2.1. Patients, Ethics, and Results Dissemination
The study has been approved by the National Cancer Center Institutional Review Board (Research No. 2022-168). Protocol version 1.2 was finally approved on the May 23, 2023. A summary of the study, its progress, and main results will be made available in the clinical registry system of the University Hospital Medical Information Network (UMIN ID: 000048966). The results will be reported at an international conference and published in an international peer-reviewed journal. All patients will be briefed on the study, and informed consent will be obtained before enrollment.
2.2. Study Design
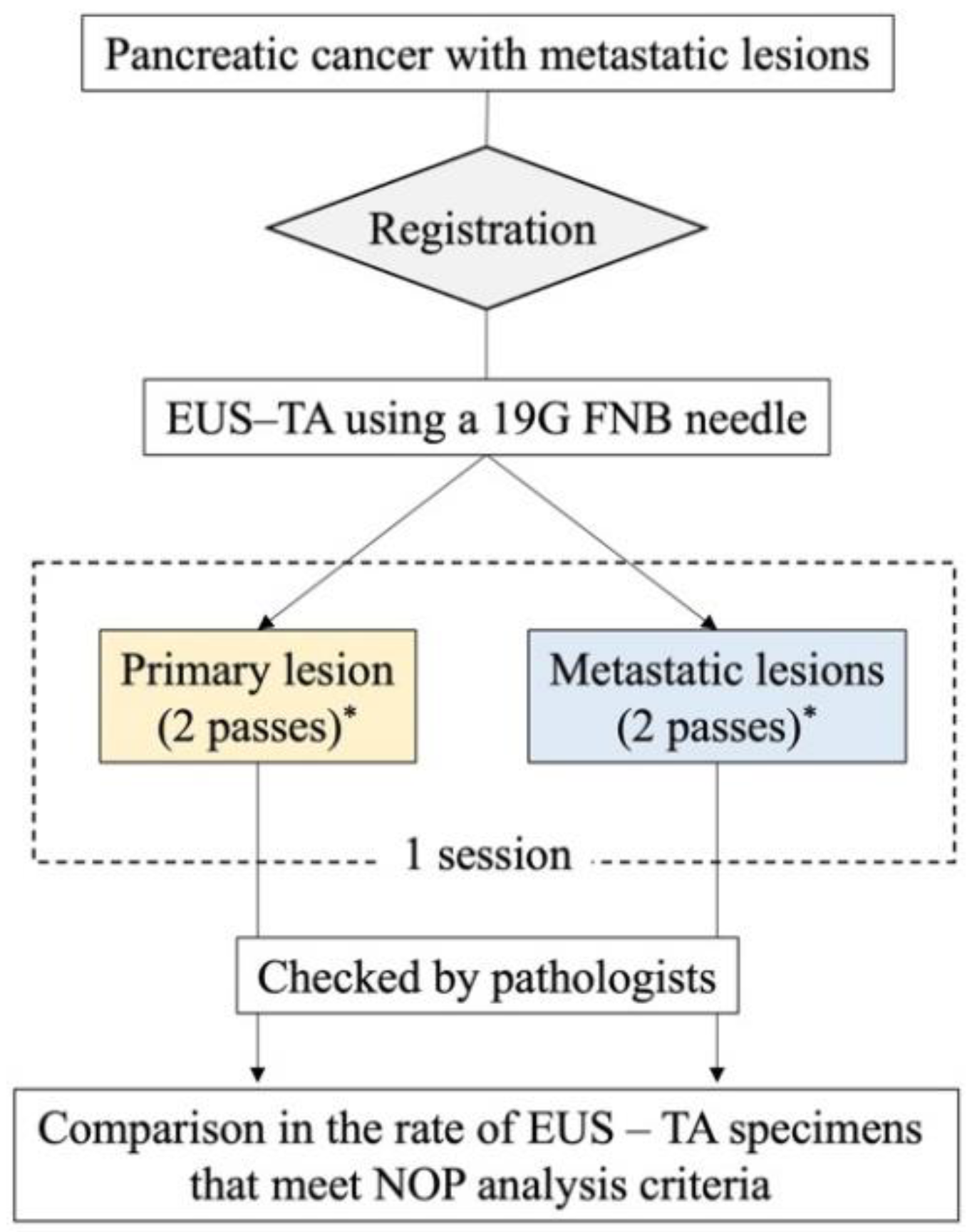
This is an ongoing single-center prospective self-controlled trial. The study flowchart is shown in
Figure 2.
2.3. Endpoints
The primary endpoint is the proportion of EUS–TA specimens obtained from primary and metastatic lesions that meet NOP analysis criteria. Although the NOP product catalog indicates tumor cellularity ≥ 20% and tissue area ≥ 16 mm2 as required, in actual clinical practice, a tissue area of ≥ 4 mm2 provides adequate DNA content (≥ 200 ng) for analysis. Thus, specimens with tumor cellularity ≥ 20% and tissue area ≥ 4 mm2 are considered to meet NOP analysis criteria.
The secondary endpoints are as follows: rate of patients with primary or metastatic lesion specimens that meet NOP analysis criteria; technical success rates of EUS–TA with a TopGain 19G needle in primary and metastatic lesions; technical success rates of EUS–TA (regardless of needle type) in primary and metastatic lesions; adverse event rate, details, and severity; sensitivity, specificity, and accuracy of the histological differentiation of malignant and benign lesions in primary or metastatic lesion specimens; rates of successful NOP analysis of primary and metastatic lesion specimens; rate of patients who receive a drug targeting detected genetic mutations; details of genetic mutations detected on NOP analysis; tumor cellularity and tissue area of all EUS–TA sessions; tumor cellularity and tissue area according to the nCounter system for all EUS–TA sessions; rate of patients who do not meet NOP criteria for both primary and metastatic lesion specimens and underwent liquid biopsy; rate of the necessity of percutaneous biopsy because specimens do not meet NOP analysis criteria for primary or metastatic lesions; and DNA amount in cases analyzed for DNA amount.
2.4. Eligibility Criteria
Eligibility criteria are as follows: suspected invasive pancreatic ductal adenocarcinoma (or a confirmed diagnosis of invasive pancreatic ductal adenocarcinoma); metastatic lesions on pre-procedure imaging or prior EUS examination; aged older than 18 years; Class 0–2 on the Eastern Cooperative Oncology Group Performance Status scale; no gastrointestinal strictures in the pharynx or esophagus on pre-procedural imaging or prior endoscopy examination; pre-procedure imaging or EUS indicating that both primary and metastatic lesions can be safely punctured in EUS–TA with a 19G FNB needle, without interventions on blood vessels, other tumors, or other organs; no prior radiotherapy or chemotherapy for pancreatic cancer; able to withdraw from antithrombotic medication based on guideline criteria [
32]; no specific bleeding risk (prothrombin time-international normalized ratio < 1.5 or platelet count > 50,000/L); and no more than a moderate volume of ascites (i.e., clear accumulation of ascites at the site of the possible puncture route for EUS–TA).
2.5. Exclusion Criteria
The exclusion criteria are as follows: two or more lesions in the pancreas suspected of pancreatic cancer; psychosis or psychiatric symptoms that interfere with daily life and make study participation difficult; and enrollment in the study deemed as inappropriate by a physician.
2.6. Trial Examination Procedures
Participants undergo EUS performed by a physician who has performed at least 100 cases of EUS–TA as a primary physician. Pancreatic primary and metastatic lesions are characterized, and the maximum lesion diameter is measured in B-mode, followed by confirmation that there are no intervening vessels in the puncture route on Doppler mode. EUS–TA is typically performed using a TopGain 19G needle (SonoTip TopGain; Medi-Globe, Achenmuhle, Germany). Negative pressure is applied using suction with a 20-mL syringe or the slow-pull method, and approximately 20 actuations per pass are performed. When a lesion cannot be identified or the puncture line is technically difficult, a change in the puncture needle or physician is considered. If the puncture remains difficult, it is not forced, and the patient is considered as a technical failure. If both primary and metastatic lesions are difficult to puncture, alternative biopsies, such as percutaneous biopsies, are considered.
For both primary and metastatic lesions, obtained specimens are placed in shale, and a portion of the specimen is cut and considered for rapid on-site evaluation (ROSE). However, ROSE is not mandatory. When ROSE is performed, the remaining specimens are not included in this study because of the small number of specimens. If malignant cells are not confirmed by ROSE, changes in the puncture site, target, and needle are considered; however, a diagnosis of malignancy may be made by histological examination in some cases. Therefore, even if malignant cells cannot be confirmed using ROSE, additional puncturing, without changing the site or puncture method, is considered as acceptable. All collected specimens, including residual specimens after ROSE, are individually preserved in formalin bottles for histological evaluation.
The procedure is terminated once two specimens each are obtained from primary and metastatic lesions, after confirming by EUS and endoscopy that there are no findings suggestive of hemorrhage. If bleeding is suspected, endoscopic hemostasis or endovascular treatment will be considered.
Specimens obtained by EUS–TA are thinly sliced after formalin fixation and stained with hematoxylin and eosin. Specimens, excluding those from the ROSE session, are checked to determine whether they meet NOP analysis criteria. The sample volume required for NOP analysis is five 10-μm-thick sections, with tumor cellularity ≥ 20% and total DNA content ≥ 200 ng per slide. A tissue area of approximately 16 mm2 per slide is recommended; however, a tissue area of 4 mm2 or more yields > 200 ng of total DNA. Therefore, we consider specimens with tumor cellularity ≥ 20% and tissue area ≥ 4 mm2 as successfully meeting NOP analysis criteria. The pathological evaluation is performed by two pathologists, and any discrepancies in the measurements are resolved by discussion. In this evaluation, tumor cellularity is evaluated at five levels (>80%, 60%–80%, 40%–60%, 20%–40%, and <20%), and tissue area is evaluated at four levels (≥25 mm2, 16–25 mm2, 4–16 mm2, and <4 mm2).
2.7. Sample Size Determination and Statistical Analysis
The proportion of metastatic lesion specimens that meet NOP analysis criteria is expected to be approximately 70%. The proportion of primary lesions that meet NOP analysis criteria is likely to be lower than the 63.6% reported by Hisada et al. because of the reduced number of passes in the PRIMATE study. In addition, in our previous retrospective study, the proportion of primary tumor specimens obtained by EUS–TA with a 19G needle meeting criteria on pre-check was 53.8%, with a median number of punctures of four. Therefore, the proportion of primary lesion specimens that meet NOP analysis criteria in the present study is expected to be approximately 45%. Accordingly, the minimum sample size required for a Fisher's exact test is 56 patients, assuming a one-sided significance level of 0.05 and a power of 80%. Assuming that approximately 10% of registered patients will experience difficulty in puncturing, a sample size of 61 patients was planned.
The proportion of EUS–TA specimens meeting NOP analysis criteria will be compared between primary and metastatic lesions using Fisher's exact test. Statistical significance is set at p < 0.05.
3. Conclusions
In clinical settings, the first choice for the target of EUS–TA in UR–M pancreatic ductal adenocarcinoma is the pancreatic primary lesion. However, the success rate for meeting CGP analysis criteria is not high in specimens acquired from the primary lesion because one characteristic of pancreatic ductal adenocarcinoma is low cellularity. If this study finds that the percentage of metastatic lesion specimens meeting NOP analysis criteria (trial examination) exceeds the percentage of primary lesion specimens meeting the criteria (standard examination), metastatic lesions should be recommended as the target of EUS–TA when considering CGP in UR–M pancreatic ductal adenocarcinoma. This may allow more patients with UR–M pancreatic ductal adenocarcinoma to benefit from CGP. It may be possible to eliminate the conventional two-step procedure of EUS–TA for the primary lesion for diagnosis and percutaneous liver biopsy of the metastatic lesion for CGP. With their incorporation into future guidelines, the results of this study may change the standard for selecting targets for EUS–TA in pancreatic ductal adenocarcinoma with metastatic lesions.
Author Contributions
Conceptualization, Kotaro Takeshita; Data curation, Yoshikuni Nagashio, Hidenobu Hara and Daiki Agarie; Formal analysis, Daiki Yamashige; Investigation, Hidenobu Hara, Yuki Kawasaki, Tetsuro Takasaki, Shin Yagi, Yuya Hagiwara, Kohei Okamoto, Soma Fukuda, Masaru Kuwada and Yasuhiro Komori; Supervision, Susumu Hijioka and Takuji Okusaka; Writing – original draft, Kotaro Takeshita; Writing – review & editing, Susumu Hijioka, Mao Okada, Yuta Maruki, Chigusa Morizane, Hideki Ueno, Yasushi Yatabe and Takuji Okusaka.
Funding
This research was funded by the National Cancer Center Research and Development Fund, grant number (2022-A-16).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the National Cancer Center Institutional Review Board (Research No. 2022-168).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Kohno, T.; Kato, M.; Kohsaka, S.; Sudo, T.; Tamai, I.; Shiraishi, Y.; Okuma, Y.; Ogasawara, D.; Suzuki, T.; Yoshida, T.; et al. C-CAT: The National Datacenter for Cancer Genomic Medicine in Japan. Cancer Discov. 2022, 12, 2509–2515. [Google Scholar] [CrossRef] [PubMed]
- Aoyagi, Y.; Kano, Y.; Tohyama, K.; Matsudera, S.; Kumaki, Y.; Takahashi, K.; Mitsumura, T.; Harada, Y.; Sato, A.; Nakamura, H.; et al. Clinical utility of comprehensive genomic profiling in Japan: Result of PROFILE-F study. PLoS One. 2022, 17, e0266112. [Google Scholar] [CrossRef] [PubMed]
- Chalmers, Z.R.; Connelly, C.F.; Fabrizio, D.; Gay, L.; Ali, S.M.; Ennis, R.; Schrock, A.; Campbell, B.; Shlien, A.; Chmielecki, J.; et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017, 9, 34. [Google Scholar] [CrossRef] [PubMed]
- Frampton, G.M.; Fichtenholtz, A.; Otto, G.A.; Wang, K.; Downing, S.R.; He, J.; Schnall-Levin, M.; White, J.; Sanford, E.M.; An, P.; et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat. Biotechnol. 2013, 31, 1023–1031. [Google Scholar] [CrossRef] [PubMed]
- Higashigawa, S.; Matsubayashi, H.; Kiyozumi, Y.; Kado, N.; Nishimura, S.; Oishi, T.; Sugino, T.; Fushiki, K.; Shirasu, H.; Yasui, H.; et al. Present status of germline findings in precision medicine for Japanese cancer patients: issues in the current system. Jpn. J. Clin. Oncol. 2022, 52, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Ida, H.; Koyama, T.; Mizuno, T.; Sunami, K.; Kubo, T.; Sudo, K.; Tao, K.; Hirata, M.; Yonemori, K.; Kato, K.; et al. Clinical utility of comprehensive genomic profiling tests for advanced or metastatic solid tumor in clinical practice. Cancer Sci. 2022, 113, 4300–4310. [Google Scholar] [CrossRef]
- Ishikawa, M.; Nakamura, K.; Kawano, R.; Hayashi, H.; Ikeda, T.; Saito, M.; Niida, Y.; Sasaki, J.; Okuda, H.; Ishihara, S.; et al. Clinical and Diagnostic Utility of Genomic Profiling for Digestive Cancers: Real-World Evidence from Japan. Cancers (Basel). 2024, 16. [Google Scholar] [CrossRef]
- Kou, T.; Kanai, M.; Yamamoto, Y.; Kamada, M.; Nakatsui, M.; Sakuma, T.; Mochizuki, H.; Hiroshima, A.; Sugiyama, A.; Nakamura, E.; et al. Clinical sequencing using a next-generation sequencing-based multiplex gene assay in patients with advanced solid tumors. Cancer Sci. 2017, 108, 1440–1446. [Google Scholar] [CrossRef]
- Milbury, C.A.; Creeden, J.; Yip, W.K.; Smith, D.L.; Pattani, V.; Maxwell, K.; Sawchyn, B.; Gjoerup, O.; Meng, W.; Skoletsky, J.; et al. Clinical and analytical validation of FoundationOne(R)CDx, a comprehensive genomic profiling assay for solid tumors. PLoS One. 2022, 17, e0264138. [Google Scholar] [CrossRef]
- Serizawa, M.; Mizuguchi, M.; Urakami, K.; Nagashima, T.; Ohshima, K.; Hatakeyama, K.; Ohnami, S.; Ohnami, S.; Maruyama, K.; Ashizawa, T.; et al. JCGA: the Japanese version of the Cancer Genome Atlas and its contribution to the interpretation of gene alterations detected in clinical cancer genome sequencing. Hum Genome Var. 2021, 8, 38. [Google Scholar] [CrossRef]
- Sunami, K.; Naito, Y.; Aimono, E.; Amano, T.; Ennishi, D.; Kage, H.; Kanai, M.; Komine, K.; Koyama, T.; Maeda, T.; et al. The initial assessment of expert panel performance in core hospitals for cancer genomic medicine in Japan. Int. J. Clin. Oncol. 2021, 26, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Yamai, T.; Ikezawa, K.; Sugimoto, N.; Urabe, M.; Kai, Y.; Takada, R.; Nakabori, T.; Uehara, H.; Kawamura, T.; Kunimasa, K.; et al. Utility of Comprehensive Genomic Profiling Tests for Patients with Incurable Pancreatic Cancer in Clinical Practice. Cancers (Basel). 2023, 15. [Google Scholar] [CrossRef]
- Casolino, R.; Paiella, S.; Azzolina, D.; Beer, P.A.; Corbo, V.; Lorenzoni, G.; Gregori, D.; Golan, T.; Braconi, C.; Froeling, F.E.M.; et al. Homologous Recombination Deficiency in Pancreatic Cancer: A Systematic Review and Prevalence Meta-Analysis. J. Clin. Oncol. 2021, 39, 2617–2631. [Google Scholar] [CrossRef] [PubMed]
- Marabelle, A.; Le, D.T.; Ascierto, P.A.; Di Giacomo, A.M.; De Jesus-Acosta, A.; Delord, J.P.; Geva, R.; Gottfried, M.; Penel, N.; Hansen, A.R.; et al. Efficacy of Pembrolizumab in Patients With Noncolorectal High Microsatellite Instability/Mismatch Repair-Deficient Cancer: Results From the Phase II KEYNOTE-158 Study. J. Clin. Oncol. 2020, 38, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Canto, M.I.; Almario, J.A.; Schulick, R.D.; Yeo, C.J.; Klein, A.; Blackford, A.; Shin, E.J.; Sanyal, A.; Yenokyan, G.; Lennon, A.M.; et al. Risk of Neoplastic Progression in Individuals at High Risk for Pancreatic Cancer Undergoing Long-term Surveillance. Gastroenterology. 2018, 155, 740–751 e742. [Google Scholar] [CrossRef] [PubMed]
- Doebele, R.C.; Drilon, A.; Paz-Ares, L.; Siena, S.; Shaw, A.T.; Farago, A.F.; Blakely, C.M.; Seto, T.; Cho, B.C.; Tosi, D.; et al. Entrectinib in patients with advanced or metastatic NTRK fusion-positive solid tumours: integrated analysis of three phase 1-2 trials. Lancet Oncol. 2020, 21, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Golan, T.; Hammel, P.; Reni, M.; Van Cutsem, E.; Macarulla, T.; Hall, M.J.; Park, J.O.; Hochhauser, D.; Arnold, D.; Oh, D.Y.; et al. Maintenance Olaparib for Germline BRCA-Mutated Metastatic Pancreatic Cancer. N. Engl. J. Med. 2019, 381, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Luchini, C.; Paolino, G.; Mattiolo, P.; Piredda, M.L.; Cavaliere, A.; Gaule, M.; Melisi, D.; Salvia, R.; Malleo, G.; Shin, J.I.; et al. KRAS wild-type pancreatic ductal adenocarcinoma: molecular pathology and therapeutic opportunities. J. Exp. Clin. Cancer Res. 2020, 39, 227. [Google Scholar] [CrossRef] [PubMed]
- Philip, P.A.; Azar, I.; Xiu, J.; Hall, M.J.; Hendifar, A.E.; Lou, E.; Hwang, J.J.; Gong, J.; Feldman, R.; Ellis, M.; et al. Molecular Characterization of KRAS Wild-type Tumors in Patients with Pancreatic Adenocarcinoma. Clin. Cancer Res. 2022, 28, 2704–2714. [Google Scholar] [CrossRef]
- Turpin, A.; Neuzillet, C.; Colle, E.; Dusetti, N.; Nicolle, R.; Cros, J.; de Mestier, L.; Bachet, J.B.; Hammel, P. Therapeutic advances in metastatic pancreatic cancer: a focus on targeted therapies. Ther. Adv. Med. Oncol. 2022, 14, 17588359221118019. [Google Scholar] [CrossRef]
- Wattenberg, M.M.; Asch, D.; Yu, S.; O'Dwyer, P.J.; Domchek, S.M.; Nathanson, K.L.; Rosen, M.A.; Beatty, G.L.; Siegelman, E.S.; Reiss, K.A. Platinum response characteristics of patients with pancreatic ductal adenocarcinoma and a germline BRCA1, BRCA2 or PALB2 mutation. Br. J. Cancer. 2020, 122, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Hijioka, S.; Nagashio, Y.; Maruki, Y.; Kawasaki, Y.; Takeshita, K.; Morizane, C.; Okusaka, T. Endoscopic Ultrasound-Guided Tissue Acquisition of Pancreaticobiliary Cancer Aiming for a Comprehensive Genome Profile. Diagnostics (Basel). 2023, 13. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Electronic address, a.a.d.h.e.; Cancer Genome Atlas Research, N. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell. 2017, 32, 185–203 e113.
- Carrara, S.; Solda, G.; Di Leo, M.; Rahal, D.; Peano, C.; Giunta, M.; Lamonaca, L.; Auriemma, F.; Anderloni, A.; Fugazza, A.; et al. Side-by-side comparison of next-generation sequencing, cytology, and histology in diagnosing locally advanced pancreatic adenocarcinoma. Gastrointest. Endosc. 2021, 93, 597–604 e595. [Google Scholar] [CrossRef] [PubMed]
- Hisada, Y.; Hijioka, S.; Ikeda, G.; Maehara, K.; Hashimoto, T.; Kitamura, H.; Harai, S.; Yoshinari, M.; Kawasaki, Y.; Murashima, Y.; et al. Proportion of unresectable pancreatic cancer specimens obtained by endoscopic ultrasound-guided tissue acquisition meeting the OncoGuide NCC Oncopanel System analysis suitability criteria: a single-arm, phase II clinical trial. J. Gastroenterol. 2022. [CrossRef] [PubMed]
- Young, G.; Wang, K.; He, J.; Otto, G.; Hawryluk, M.; Zwirco, Z.; Brennan, T.; Nahas, M.; Donahue, A.; Yelensky, R.; et al. Clinical next-generation sequencing successfully applied to fine-needle aspirations of pulmonary and pancreatic neoplasms. Cancer Cytopathol. 2013, 121, 688–694. [Google Scholar] [CrossRef] [PubMed]
- Gleeson, F.C.; Kerr, S.E.; Kipp, B.R.; Voss, J.S.; Minot, D.M.; Tu, Z.J.; Henry, M.R.; Graham, R.P.; Vasmatzis, G.; Cheville, J.C.; et al. Targeted next generation sequencing of endoscopic ultrasound acquired cytology from ampullary and pancreatic adenocarcinoma has the potential to aid patient stratification for optimal therapy selection. Oncotarget. 2016, 7, 54526–54536. [Google Scholar] [CrossRef] [PubMed]
- Larson, B.K.; Tuli, R.; Jamil, L.H.; Lo, S.K.; Deng, N.; Hendifar, A.E. Utility of Endoscopic Ultrasound-Guided Biopsy for Next-Generation Sequencing of Pancreatic Exocrine Malignancies. Pancreas. 2018, 47, 990–995. [Google Scholar] [CrossRef]
- Elhanafi, S.; Mahmud, N.; Vergara, N.; Kochman, M.L.; Das, K.K.; Ginsberg, G.G.; Rajala, M.; Chandrasekhara, V. Comparison of endoscopic ultrasound tissue acquisition methods for genomic analysis of pancreatic cancer. J. Gastroenterol. Hepatol. 2019, 34, 907–913. [Google Scholar] [CrossRef]
- Park, J.K.; Lee, J.H.; Noh, D.H.; Park, J.K.; Lee, K.T.; Lee, J.K.; Lee, K.H.; Jang, K.T.; Cho, J. Factors of Endoscopic Ultrasound-Guided Tissue Acquisition for Successful Next-Generation Sequencing in Pancreatic Ductal Adenocarcinoma. Gut Liver. 2020, 14, 387–394. [Google Scholar] [CrossRef]
- Ikeda, G.; Hijioka, S.; Nagashio, Y.; Maruki, Y.; Ohba, A.; Hisada, Y.; Yoshinari, M.; Harai, S.; Kitamura, H.; Koga, T.; et al. Fine-needle biopsies with 19-gauge needle are effective combination modalities of EUS-tissue acquisition for genomic profiling of unresectable pancreatic cancer. Dig. Endosc. 2022.
- Kato, M.; Uedo, N.; Hokimoto, S.; Ieko, M.; Higuchi, K.; Murakami, K.; Fujimoto, K. Guidelines for Gastroenterological Endoscopy in Patients Undergoing Antithrombotic Treatment: 2017 Appendix on Anticoagulants Including Direct Oral Anticoagulants. Dig. Endosc. 2018, 30, 433–440. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).