Submitted:
21 June 2024
Posted:
24 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Animals and Tissues
2.2. Isolation of DNA and RNA
2.3. Real-Time PCR for the Detection of DNA Viruses
2.4. Real-Time Reverse Transcriptase PCR for the Detection of HEV3
2.5. Conventional and Real-Time PCR for the Detection of PERVs
2.6. Western Blot Analysis
3. Results
3.1. Results of the PCR-Based Screening
3.1. Results of the Western Blot-Based Screening
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cooper, D.K.C.; Gaston, R.; Eckhoff, D.; Ladowski, J.; Yamamoto, T.; Wang, L.; Iwase, H.; Hara, H.; Tector, M.; Tector, A.J. Xenotransplantation-the current status and prospects. Br Med Bull. 2018, 125, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Peterson, L.; Yacoub, M.H.; Ayares, D.; Yamada, K.; Eisenson, D.; Griffith, B.P.; Mohiuddin, M.M.; Eyestone, W.; Venter, J.C.; Smolenski, R.T.; Rothblatt, M. Physiological basis for xenotransplantation from genetically modified pigs to humans. Physiol Rev. 2024, 104, 1409–1459. [Google Scholar] [CrossRef]
- Mohiuddin, M.M.; Singh, A.K.; Scobie, L.; Goerlich, C.E.; Grazioli, A.; Saharia, K.; Crossan, C.; Burke, A.; Drachenberg, C.; Oguz, C.; Zhang, T.; Lewis, B.; Hershfeld, A.; Sentz, F.; Tatarov, I.; Mudd, S.; Braileanu, G.; Rice, K.; Paolini, J.F.; Bondensgaard, K.; Vaught, T.; Kuravi, K.; Sorrells, L.; Dandro, A.; Ayares, D.; Lau, C.; Griffith, B.P. Graft dysfunction in compassionate use of genetically engineered pig-to-human cardiac xenotransplantation: A case report. Lancet 2023, 402, 397–410. [Google Scholar] [CrossRef] [PubMed]
- Loupy, A.; Goutaudier, V.; Giarraputo, A.; Mezine, F.; Morgand, E.; Robin, B.; Khalil, K.; Mehta, S.; Keating, B.; Dandro, A.; Certain, A.; Tharaux, P.L.; Narula, N.; Tissier, R.; Giraud, S.; Hauet, T.; Pass, H.I.; Sannier, A.; Wu, M.; Griesemer, A.; Ayares, D.; Tatapudi, V.; Stern, J.; Lefaucheur, C.; Bruneval, P.; Mangiola, M.; Montgomery, R.A. Immune response after pig-to-human kidney xenotransplantation: A multimodal phenotyping study. Lancet 2023, 402, 1158–1169. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, R.A.; Stern, J.M.; Lonze, B.E.; Tatapudi, V.S.; Mangiola, M.; Wu, M.; Weldon, E.; Lawson, N.; Deterville, C.; Dieter, R.A.; Sullivan, B.; Boulton, G.; Parent, B.; Piper, G.; Sommer, P.; Cawthon, S.; Duggan, E.; Ayares, D.; Dandro, A.; Fazio-Kroll, A.; Kokkinaki, M.; Burdorf, L.; Lorber, M.; Boeke, J.D.; Pass, H.; Keating, B.; Griesemer, A.; Ali, N.M.; Mehta, S.A.; Stewart, Z.A. Results of Two Cases of Pig-to-Human Kidney Xenotransplantation. N Engl J Med. 2022, 386, 1889–1898. [Google Scholar] [CrossRef] [PubMed]
- Locke, J.E.; Kumar, V.; Anderson, D.; Porrett, P.M. Normal graft function after pig-to-human kidney xenotransplant. JAMA Surg 2023, 158, 1106–1108. [Google Scholar] [CrossRef] [PubMed]
- Moazami, N.; Stern, J.M.; Khalil, K.; Kim, J.I.; Narula, N.; Mangiola, M.; Weldon, E.P.; Kagermazova, L.; James, L.; Lawson, N.; Piper, G.L.; Sommer, P.M.; Reyentovich, A.; Bamira, D.; Saraon, T.; Kadosh, B.S.; DiVita, M.; Goldberg, R.I.; Hussain, S.T.; Chan, J.; Ngai, J.; Jan, T.; Ali, N.M.; Tatapudi, V.S.; Segev, D.L.; Bisen, S.; Jaffe, I.S.; Piegari, B.; Kowalski, H.; Kokkinaki, M.; Monahan, J.; Sorrells, L.; Burdorf, L.; Boeke, J.D.; Pass, H.; Goparaju, C.; Keating, B.; Ayares, D.; Lorber, M.; Griesemer, A.; Mehta, S.A.; Smith, D.E.; Montgomery, R.A. Pig-to-human heart xenotransplantation in two recently deceased human recipients. Nat Med 2023, 29, 1989–1997. [Google Scholar] [CrossRef]
- First pig liver transplanted into a person lasts for 10 days. Nature 2024, 627, 710–711. Available online: https://www.nature.com/articles/d41586-024-00853-8 (accessed on 13 June 2024). [CrossRef] [PubMed]
- Kidney transplantation in China. Available online: https://www.globaltimes.cn/page/202404/1310234.shtml (accessed on 13 June 2024).
- Transplantation of the second heart in Baltimore. Available online: https://www.nytimes.com/2023/10/31/health/pig-heart-transplant-faucette.html?smid=nytcore-ios-share&referringSource=articleShare (accessed on 13 June 2024).
- Kidney in living individual in Boston STAT. Available online: https://www.statnews.com/2024/03/21/first-xenotransplantation-crispr-gene-edited-pig-kidney-egenesis/?utm_campaign=breaking_news&utm_medium=email&_hsmi=299239261&utm_content=299239261&utm_source=hs_email (accessed on 13 June 2024).
- Available online: https://nyulangone.org/news/first-ever-combined-heart-pump-gene-edited-pig-kidney-transplant-gives-new-hope-patient-terminal-illness (accessed on 8 May 2024).
- Fishman, J.A. Risks of Infectious Disease in Xenotransplantation. N Engl J Med. 2022, 387, 2258–2267. [Google Scholar] [CrossRef]
- Yamada, K.; Tasaki, M.; Sekijima, M.; Wilkinson, R.A.; Villani, V.; Moran, S.G.; Cormack, T.A.; Hanekamp, I.M.; Hawley, R.J.; Arn, J.S.; Fishman, J.A.; Shimizu, A.; Sachs, D.H. Porcine cytomegalovirus infection is associated with early rejection of kidney grafts in a pig to baboon xenotransplantation model. Transplantation. 2014, 98, 411–418.3. [Google Scholar] [CrossRef]
- Sekijima, M.; Waki, S.; Sahara, H.; Tasaki, M.; Wilkinson, R.A.; Villani, V.; Shimatsu, Y.; Nakano, K.; Matsunari, H.; Nagashima, H.; Fishman, J.A.; Shimizu, A.; Yamada, K. Results of life-supporting galactosyltransferase knockout kidneys in cynomolgus monkeys using two different sources of galactosyltransferase knockout Swine. Transplantation. 2014, 98, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Denner, J.; Längin, M.; Reichart, B.; Krüger, L.; Fiebig, U.; Mokelke, M.; Radan, J.; Mayr, T.; Milusev, A.; Luther, F.; Sorvillo, N.; Rieben, R.; Brenner, P.; Walz, C.; Wolf, E.; Roshani, B.; Stahl-Hennig, C.; Abicht, J.M. Impact of porcine cytomegalovirus on long-term orthotopic cardiac xenotransplant survival. Sci Rep. 2020, 10, 17531. [Google Scholar] [CrossRef]
- Denner, J. Xenotransplantation and porcine cytomegalovirus. Xenotransplantation. 2015, 22, 329–335. [Google Scholar] [CrossRef]
- Nellore, A.; Fishman, J.A. Donor-derived infections and infectious risk in xenotransplantation and allotransplantation. Xenotransplantation. 2018, 25, e12423. [Google Scholar] [CrossRef] [PubMed]
- Plotzki, E.; Keller, M.; Ivanusic, D.; Denner, J. A new Western blot assay for the detection of porcine cytomegalovirus (PCMV). J Immunol Methods. 2016, 437, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Noordergraaf, J.; Schucker, A.; Martin, M.; Schuurman, H.J.; Ordway, B.; Cooley, K.; Sheffler, M.; Theis, K.; Armstrong, C.; Klein, L.; Hansen, D.; Olson, M.; Schlechter, L.; Spizzo, T. Pathogen eliminationand prevention within a regulated, Designated Pathogen Free, closed pig herd for long-term breeding and production of xeno-transplantation materials. Xenotransplantation 2018, 25, e12428. [Google Scholar] [CrossRef]
- Denner, J. Sensitive detection systems for infectious agents in xenotransplantation. Xenotransplantation. 2020, 18, e12594. [Google Scholar] [CrossRef] [PubMed]
- Otabi, H.; Miura, H.; Uryu, H.; Kobayashi-Harada, R.; Abe, K.; Nakano, K.; Umeyama, K.; Hasegawa, K.; Tsukahara, T.; Nagashima, H.; Inoue, R. Development of a panel for detection of pathogens in xenotransplantation donor pigs. Xenotransplantation. 2023, 30, e12825. [Google Scholar] [CrossRef]
- Groenendaal, H.; Costard, S.; Ballard, R.; Bienhoff, S.; Challen, D.C.; Dominguez, B.J.; Kern, D.R.; Miller, D.; Noordergraaf, J.; Rudenko, L.; Schuurman, H.J.; Spizzo, T.; Sturos, M.; Zollers, B.; Fishman, J.A. Expert opinion on the identification, risk assessment, and mitigation of microorganisms and parasites relevant to xenotransplantation products from pigs. Xenotransplantation 2023, e12815. [Google Scholar] [CrossRef]
- Krüger, L.; Kristiansen, Y.; Reuber, E.; Möller, L.; Laue, M.; Reimer, C.; Denner, J. A Comprehensive Strategy for Screening for Xenotransplantation-Relevant Viruses in a Second Isolated Population of Gottingen Minipigs. Viruses 2019, 12, 38. [Google Scholar] [CrossRef]
- Denner, J. Zoonosis and xenozoonosis in xenotransplantation: A proposal for a new classification. Zoonoses Public Health. 2023, 70, 578–579. [Google Scholar] [CrossRef] [PubMed]
- Denner, J. Limited availability of methods for the detection of xenotransplantation-relevant viruses in veterinary laboratories. Xenotransplantation 2024, 31, e12851. [Google Scholar] [CrossRef] [PubMed]
- Morozov, V.A.; Ludwig, S.; Ludwig, B.; Rotem, A.; Barkai, U.; Bornstein, S.R.; Denner, J. Islet cell transplantation from Gottingen minipigs to cynomolgus monkeys: Analysis of virus safety. Xenotransplantation. 2016, 23, 320–327. [Google Scholar] [CrossRef]
- Jhelum, H.; Papatsiros, V.; Papakonstantinou, G.; Krabben, L.; Kaufer, B.; Denner, J. Screening for Viruses in Indigenous Greek Black Pigs. Microorganisms 2024, 12, 315. [Google Scholar] [CrossRef] [PubMed]
- Morozov, V.A.; Morozov, A.V.; Rotem, A.; Barkai, U.; Bornstein, S.; Denner, J. Extended Microbiological Characterization of Göttingen Minipigs in the Context of Xenotransplantation: Detection and Vertical Transmission of Hepatitis E Virus. PLoS ONE 2015, 10, e0139893. [Google Scholar] [CrossRef]
- Morozov, V.A.; Plotzki, E.; Rotem, A.; Barkai, U.; Denner, J. Extended microbiological characterization of Göttingen minipigs: Porcine cytomegalovirus and other viruses. Xenotransplantation. 2016, 23, 490–496. [Google Scholar] [CrossRef]
- Heinze, J.; Plotzki, E.; Denner, J. Virus Safety of Xenotransplantation: Prevalence of Porcine Circovirus 2 (PCV2) in Pigs. Ann Virol Res 2016, 2, 1023. [Google Scholar] [CrossRef]
- Krüger, L.; Kristiansen, Y.; Reuber, E.; Möller, L.; Laue, M.; Reimer, C.; Denner, J. A Comprehensive Strategy for Screening for Xenotransplantation-Relevant Viruses in a Second Isolated Population of Gottingen Minipigs. Viruses. 2019, 12, 38. [Google Scholar] [CrossRef]
- Halecker, S.; Metzger, J.; Strube, C.; Krabben, L.; Kaufer, B.; Denner, J. Virological and Parasitological Characterization of Mini-LEWE Minipigs Using Improved Screening Methods and an Overview of Data on Various Minipig Breeds. Microorganisms. 2021, 9, 2617. [Google Scholar] [CrossRef]
- Plotzki, E.; Heinrichs, G.; Kubícková, B.; Ulrich, R.G.; Denner, J. Microbiological characterization of a newly established pig breed, Aachen Minipigs. Xenotransplantation 2016, 23, 159–167. [Google Scholar] [CrossRef]
- Jhelum, H.; Grand, N.; Jacobsen, K.R.; Halecker, S.; Salerno, M.; Prate, R.; Krüger, L.; Kristiansen, Y.; Krabben, L.; Möller, L.; Laue, M.; Kaufer, B.; Kaaber, K.; Denner, J. First virological and pathological study of Gottingen Minipigs with Dippity Pig Syndrome (DPS). PLoS ONE 2023, 18, e0281521. [Google Scholar] [CrossRef] [PubMed]
- Halecker, S.; Papatsiros, V.; Psalla, D.; Krabben, L.; Kaufer, B.; Denner, J. Virological characterization of pigs with erythema multiforme. Microorganisms 2022, 10, 652. [Google Scholar] [CrossRef] [PubMed]
- Jothikumar, N.; Cromeans, T.L.; Robertson, B.H.; Meng, X.J.; Hill, V.R. A broadly reactive one step real-time RT-PCR assay for rapid and sensitive detection of hepatitis E virus. J. Virol. Methods. 2006, 131, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Mueller, N.J.; Barth, R.N.; Yamamoto, S.; Kitamura, H.; Patience, C.; Yamada, K.; Cooper, D.K.; Sachs, D.H.; Kaur, A.; Fishman, J.A. Activation of cytomegalovirus in pig-to-primate organ xenotransplantation. J. Virol. 2002, 76, 4734–4740. [Google Scholar] [CrossRef] [PubMed]
- Chmielewicz, B.; Goltz, M.; Franz, T.; Bauer, C.; Brema, S.; Ellerbrok, H.; Beckmann, S.; Rziha, H.-J.; Lahrmann, K.-H.; Romero, C.; et al. A novel porcine gammaherpesvirus. Virology 2003, 308, 317–329. [Google Scholar] [CrossRef]
- McMahon, K.J.; Minihan, D.; Campion, E.M.; Loughran, S.T.; Allan, G.; McNeilly, F.; Walls, D. Infection of pigs in Ireland with lymphotropic gamma-herpesviruses and relationship to postweaning multisystemic wasting syndrome. Vet. Microbiol. 2006, 116, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Xiao, Y.; Li, X.; Li, S.; Xie, N.; Yan, X.; Li, X.; Zhu, J. Development and application of a quadruplex real-time PCR assay for differential detection of porcine circoviruses (PCV1 to PCV4) in Jiangsu province of China from 2016 to 2020. Transbound. Emerg. Dis. 2021, 68, 1615–1624. [Google Scholar] [CrossRef]
- Palinski, R.; Piñeyro, P.; Shang, P.; Yuan, F.; Guo, R.; Fang, Y.; Byers, E.; Hause, B.M. A Novel Porcine Circovirus Distantly Related to Known Circoviruses Is Associated with Porcine Dermatitis and Nephropathy Syndrome and Reproductive Failure. J. Virol. 2017, 91, e01879. [Google Scholar] [CrossRef]
- Opriessnig, T.; Shen, H.G.; Pal, N.; Ramamoorthy, S.; Huang, Y.W.; Lager, K.M.; Beach, N.M.; Halbur, P.G.; Meng, X. A Live-Attenuated Chimeric Porcine Circovirus Type 2 (PCV2) Vaccine Is Transmitted to Contact Pigs but Is Not Upregulated by Concurrent Infection with Porcine Parvovirus (PPV) and Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) and Is Efficacious in a PCV2b-PRRSV-PPV Challenge Model. Clin. Vaccine Immunol. 2011, 18, 1261–1268. [Google Scholar]
- Duvigneau, J.; Hartl, R.; Groiss, S.; Gemeiner, M. Quantitative simultaneous multiplex real-time PCR for the detection of porcine cytokines. J. Immunol. Methods. 2005, 306, 16–27. [Google Scholar] [CrossRef]
- Kaulitz, D.; Mihica, D.; Adlhoch, C.; Semaan, M.; Denner, J. Improved pig donor screening including newly identified variants of porcine endogenous retrovirus-C (PERV-C). Arch. Virol. 2013, 158, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Wood, J.C.; Quinn, G.; Suling, K.M.; Oldmixon, B.A.; Van Tine, B.A.; Cina, R.; Arn, S.; Huang, C.A.; Scobie, L.; Onions, D.E.; et al. Identification of Exogenous Forms of Human-Tropic Porcine Endogenous Retrovirus in Miniature Swine. J. Virol. 2004, 78, 2494–2501. [Google Scholar] [CrossRef] [PubMed]
- Halecker, S.; Hansen, S.; Krabben, L.; Ebner, F.; Kaufer, B.; Denner, J. How, where and when to screen for porcine cytomegalovirus (PCMV) in donor pigs for xenotransplantation. Sci. Rep. 2022, 12, 21545. [Google Scholar] [CrossRef]
- Chelli, E.; Suffredini, E.; De Santis, P.; De Medici, D.; Di Bella, S.; D’Amato, S.; Gucciardi, F.; Guercio, A.; Ostanello, F.; Perrone, V.; Purpari, G.; Scavia, G.S.; Schembri, P.; Varcasia, B.M.; Di Bartolo, I. Hepatitis E Virus Occurrence in Pigs Slaughtered in Italy. Animals 2021, 11, 277. [Google Scholar] [CrossRef] [PubMed]
- Sooryanarain, H.; Heffron, C.L.; Hill, D.E.; Fredericks, J.; Rosenthal, B.M.; Werre, S.R.; Opriessnig, T.; Meng, X.J. Hepatitis E Virus in Pigs from Slaughterhouses, United States, 2017-2019. Emerg Infect Dis. 2020, 26, 354–357. [Google Scholar] [CrossRef] [PubMed]
- Boxman, I.L.A.; Verhoef, L.; Dop, P.Y.; Vennema, H.; Dirks, R.A.M.; Opsteegh, M. Int J Food Microbiol. 2022, 379, 109830. [CrossRef]
- Jallow, M.M.; Barry, M.A.; Fall, A.; Ndiaye, N.K.; Kiori, D.; Sy, S.; Goudiaby, D.; Niang, M.N.; Fall, G.; Fall, M.; Dia, N. Influenza A Virus in Pigs in Senegal and Risk Assessment of Avian Influenza Virus (AIV) Emergence and Transmission to Human. Microorganisms 2023, 11, 1961. [Google Scholar] [CrossRef] [PubMed]
- Elbers, A.R.; Tielen, M.J.; Cromwijk, W.A.; Hunneman, W.A. Sero-epidemiological screening of pig sera collected at the slaughterhouse to detect herds infected with Aujeszky’s disease virus, porcine influenza virus and Actinobacillus (Haemophilus) pleuropneumoniae in the framework of an integrated quality control (IQC) system. Vet Q. 1990, 12, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Magar, R.; Larochelle, R. Evaluation of the presence of porcine reproductive and respiratory syndrome virus in pig meat and experimental transmission following oral exposure. Can J Vet Res. 2004, 68, 259–266. [Google Scholar]
- Fischer, N.; Gulich, B.; Keßler, B.; Längin, M.; Fishman, J.A.; Wolf, E.; Boller, K.; Tönjes, R.R.; Godehardt, A.W. PCR and peptide based PCMV detection in pig - development and application of a combined testing procedure differentiating newly from latent infected pigs. Xenotransplantation. 2023, 30, e12803. [Google Scholar] [CrossRef]
- Denner, J.; Jhelum, H.; Hansen, S.; Kaufer, B.B. Comparison of methods for the detection of porcine cytomegalovirus/roseolovirus in relation to biosafety monitoring of xenotransplantation products. Xenotransplantation. 2023, 13, e12835. [Google Scholar] [CrossRef]
- Plowright, W.; Edington, N.; Watt, R.G. The behaviour of porcine cytomegalovirus in commercial pig herds. J Hyg (Lond). 1976, 76, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Edington, N. Porcine cytomegalovirus. In Diseases of Swine (eds Straw, B.E. et al.) (Iowa State University Press, 1999).
- Egerer, S.; Fiebig, U.; Kessler, B.; Zakhartchenko, V.; Kurome, M.; Reichart, B.; Kupatt, C.; Klymiuk, N.; Wolf, E.; Denner, J.; Bähr, A. Early weaning completely eliminates porcine cytomegalovirus from a newly established pig donor facility for xenotransplantation. Xenotransplantation. 2018, 25, e12449. [Google Scholar] [CrossRef] [PubMed]
- Mueller, N.J.; Sulling, K.; Gollackner, B.; Yamamoto, S.; Knosalla, C.; Wilkinson, R.A.; Kaur, A.; Sachs, D.H.; Yamada, K.; Cooper, D.K.; Patience, C.; Fishman, J.A. Reduced efficacy of ganciclovir against porcine and baboon cytomegalovirus in pig-to-baboon xenotransplantation. Am J Transplant. 2003, 3, 1057–1064. [Google Scholar] [CrossRef]
- Fryer, J.F.; Griffiths, P.D.; Emery, V.C.; Clark, D.A. Susceptibility of porcine cytomegalovirus to antiviral drugs. J Antimicrob Chemother. 2004, 53, 975–980. [Google Scholar] [CrossRef]
- Denner, J. First transplantation of a pig heart from a multiple gene-modified donor, porcine cytomegalovirus/roseolovirus, and antiviral drugs. Xenotransplantation. 2023, e12800. [Google Scholar] [CrossRef]
- Denner, J. Porcine Lymphotropic Herpesviruses (PLHVs) and Xenotranplantation. Viruses. 2021, 13, 1072. [Google Scholar] [CrossRef]
- Mueller, N.J.; Kuwaki, K.; Knosalla, C.; Dor, F.J.; Gollackner, B.; Wilkinson, R.A.; Arn, S.; Sachs, D.H.; Cooper, D.K.; Fishman, J.A. Early weaning of piglets fails to exclude porcine lymphotropic herpesvirus. Xenotransplantation 2005, 12, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Tucker, A.; McNeill, F.; Meehan, B.; Galbraith, D.; McArdle, P.; Allan, G.; Patience, C. Methods for the exclusion of circoviruses and gammaherpesviruses from pigs. Xenotransplantation 2003, 10, 343–348. [Google Scholar] [CrossRef]
- Denner, J.; Mankertz, A. Porcine Circoviruses and Xenotransplantation. Viruses. 2017, 9, 83. [Google Scholar] [CrossRef]
- Opriessnig, T.; Xiao, C.T.; Mueller, N.J.; Denner, J. Emergence of novel circoviruses in humans and pigs and their possible importance for xenotransplantation and blood transfusions. Xenotransplantation. 2024, 31, e12842. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Wu, J.; Jiang, M.; Xie, Y.; Bu, W.; Liu, C.; Zhang, G.; Luo, M. A Novel Technique for Constructing Infectious Cloning of Type 3 Porcine Circovirus. Front Microbiol. 2020, 11, 1067. [Google Scholar] [CrossRef] [PubMed]
- Yue, W.; Li, Y.; Zhang, X.; He, J.; Ma, H. Prevalence of Porcine Circoviruses in Slaughterhouses in Central Shanxi Province, China. Front Vet Sci. 2022, 9, 820914. [Google Scholar] [CrossRef] [PubMed]
- Krüger, L.; Längin, M.; Reichart, B.; Fiebig, U.; Kristiansen, Y.; Prinz, C.; Kessler, B.; Egerer, S.; Wolf, E.; Abicht, J.M.; Denner, J. Transmission of Porcine Circovirus 3 (PCV3) by xenotransplantation of pig hearts into baboons. Viruses. 2019, 11, 650. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Meng, X.J. Hepatitis E virus: Host tropism and zoonotic infection. Curr Opin Microbiol. 2021, 59, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Denner, J. Xenotransplantation and Hepatitis E virus. Xenotransplantation. 2015, 22, 167–173. [Google Scholar] [CrossRef]
- Takeuchi, Y.; Patience, C.; Magre, S.; Weiss, R.A.; Banerjee, P.T.; Le Tissier, P.; Stoye, J.P. Host range and interference studies of three classes of pig endogenous retrovirus. J Virol. 1998, 72, 9986–9991. [Google Scholar] [CrossRef] [PubMed]
- Denner, J.; Schuurman, H.J. High Prevalence of Recombinant Porcine Endogenous Retroviruses (PERV-A/Cs) in Minipigs: A Review on Origin and Presence. Viruses. 2021, 13, 1869. [Google Scholar] [CrossRef]
- Denner, J. Xenotransplantation - A special case of One Health. One Health. 2017, 3, 17–22. [Google Scholar] [CrossRef]
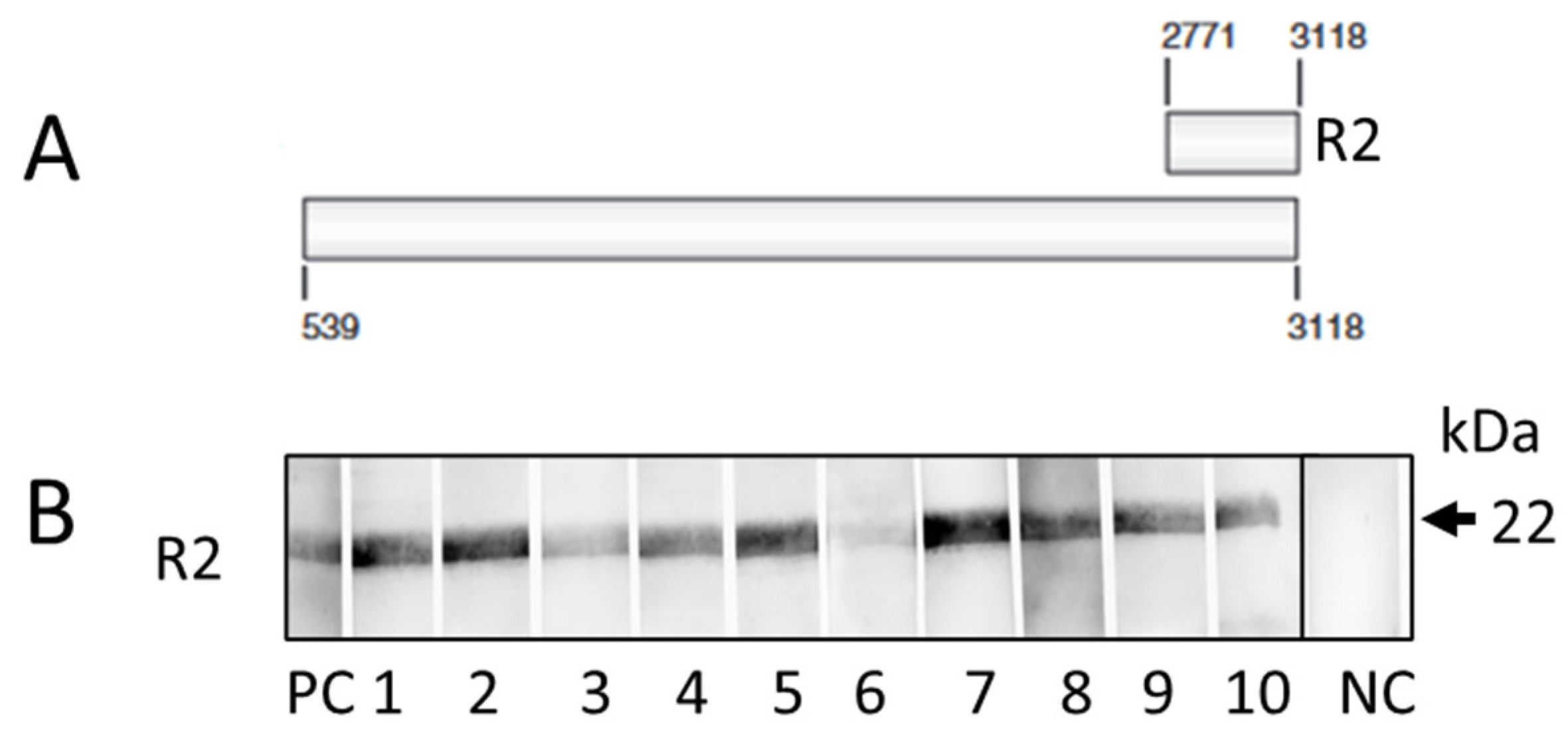
| Animal | Sex |
Weight (kg)* |
PCMV/PRV |
| Western blot | |||
| 1 | male | 110.9 | + |
| 2 | male | 108.1 | + |
| 3 | male | 97.6 | + |
| 4 | female | 97.6 | + |
| 5 | female | 99.1 | + |
| 6 | male | 93.8 | + |
| 7 | female | 108.2 | + |
| 8 | male | 109.3 | + |
| 9 | female | 95.6 | + |
| 10 | female | 94.7 | + |
| Total | 5 and 5 | 10/10 |
| Virus | Primer/Probe | Sequence 5′-3′ | Reference |
| HEV3 | JVHEV3-Fwd | GGT GGT TTC TGG GGT GAC | Jothikumar et al. 2006 [37] |
| JVHEV3-Rev | AGG GGT TGG TTG GAT GAA | ||
| JVHEV3-Probe | 6FAM-TGA TTC TCA GCC CTT CGC-BHQ | ||
| PCMV/PRV | PCMV-Fwd | ACT TCG TCG CAG CTC ATC TGA | Mueller et al. 2002 [38] |
| PCMV-Rev | GTT CTG GGA TTC CGA GGT TG | ||
| PCMV-Probe | 6FAM-CAG GGC GGC GGT CGA GCT C-BHQ | ||
| PLHV-1 | PLHV-1 (1125)-Fwd | CTC ACC TCC AAA TAC AGC GA | Chmielewicz et al. 2003 [39] |
| PLHV-1 (1125)-Rev | GCT TGA ATC GTG TGT TCC ATA G | ||
| PLHV-1 (1125)-Probe | 6FAM-CTG GTC TAC TGA ATC GCC GCT AAC AG-TAMR | ||
| PLHV-2 | PLHV-2 (1155)-Fwd | GTC ACC TGC AAA TAC ACA GG | Chmielewicz et al. 2003 [39] |
| PLHV-2 (1155)-Rev | GGC TTG AAT CGT ATG TTC CAT AT | ||
| PLHV-2 (1155)-Probe | 6FAM-CTG GTC TAC TGA AGC GCT GCC AAT AG-TAMRA | ||
| PLHV-3 | PLHV-3 (210s)-Fwd | AAC AGC GCC AGA AAA AAA GG | McMahon et al. 2006 [40] |
| PLHV-3 (210as)-Rev | GGA AAG GTA GAA GGT GAA CCA TAA AA | ||
| PLHV-3 (210)-Probe | 6-FAM CCA AAG AGG AAA ATC-MGB | ||
| PCV2 | PCV2 (F2020)-Fwd | CTG AGT CTT TTT TAT CAC TTC GTA ATG GT | Chen et al. 2021 [41] |
| PCV2 (F2020)-Rev | ACT GCG TTC GAA AAC AGT ATA TAC GA | ||
| PCV2 (F2020)-Probe | 6FAM-TTA AGT GGG GGG TCT TTA AGA TTA AAT TCT CTG AAT TGT-BHQ2 | ||
| PCV3 | PCV3-Fwd | AGT GCT CCC CAT TGA ACG | Palinski et al. 2017 [42] |
| PCV3-Rev | ACA CAG CCG TTA CTT CAC | ||
| PCV3-Probe | 6FAM-ACC CCA TGG CTC AAC ACA TAT GAC C-BHQ1 | ||
| PCV4 | PCV4 (F2020)-Fwd | ATT ATT AAA CAG ACT TTA TTT GTG TCA TCA CTT | Chen et al. 2021 [41] |
| PCV4 (F2020)-Rev | ACA GGG ATA ATG CGT AGT GAT CAC T | ||
| PCV4 (F2020)-Probe | 6FAM-ATA CTA CAC TTG ATC TTA GCC AAA AGG CTC GTT GA-BHQ1 | ||
| PPV1 | PPV1-Fwd | CAG AAT CAG CAA CCT CAC CA | Opriessnig et al. 2011 [43] |
| PPV1-Rev | GCT GCT GGT GTG TAT GGA AG | ||
| PPV1-Probe | 6FAM-TGC AAG CTT/ZEN/AAT GGT CGC ACT AGA CA-BHQ1 | ||
| pGAPDH | pGAPDH-Fwd | ACA TGG CCT CCA AGG AGT AAG A | Duvigneau et al. 2005 [44] |
| pGAPDH-Rev | GAT CGA GTT GGG GCT GTG ACT | ||
| pGAPDH-Probe | HEX-CCA CCA ACC CCA GCA AGA G-BHQ1 | ||
| PERV-C, PCR1 | PERV-envC-Fwd | GAT TAG AAC TGG AAG CCC CAA GTG CTC T | Kaulitz et al., 2013 [45] |
| PERV-envC-Rev | TCT GAT CCA GAA GTT ATG TTA GAG GAT GGT | ||
| PERV-C, PCR4 | envC.2 for | GATTAGAACTGGAAGCCCCAAGTGCTCT | |
| envC.2 rev | TCTGATCCAGAAGTTATGTTAGAGGATGGT | ||
| PERV-A/C | PERV-A env VRBF-Fwd | CCT ACC AGT TAT AAT CAA TTT AAT TAT GGC | Wood et al. 2004 [46] |
| PERV-C env TMR-Rev | CTC AAA CCA CCC TTG AGT AGT TTC C | ||
| SPLEEN | |||||||||||||
| Animal |
PCMV /PRV |
PLHV-1 | PLHV-2 | PLHV-3 | PPV-1 | PCV2 | PCV3 | PCV4 | PERV-C | PERV-C | PERV-A/C | HEV | |
| Real-time PCR | Real-time PCR | Real-time PCR | Real-time PCR | Real-time PCR | Real-time PCR | Real-time PCR | Real-time PCR | Real-time PCR | PCR1 | PCR4 | PCR | Real-time RT-PCR | |
| 1 | 29.61 | 31.97 | n.d. | 27.41 | n.d. | n.d. | n.d. | n.d. | 24.57 | + | + | - | n.d. |
| 2 | 31.01 | 31.15 | n.d. | 28.24 | n.d. | n.d. | n.d. | n.d. | 21.67 | + | + | - | n.d. |
| 3 | 30.37 | 32.26 | n.d. | 27.77 | n.d. | n.d. | n.d. | n.d. | 26.43 | - | - | - | n.d. |
| 4 | n.d. | 31.21 | n.d. | 29.07 | n.d. | n.d. | 20.09 | n.d. | 25.11 | + | + | - | n.d. |
| 5 | 32.64 | 29.01 | n.d. | 35.62 | n.d. | n.d. | n.d. | n.d. | 25.78 | + | + | - | n.d. |
| 6 | n.d. | 31.75 | n.d. | 35.23 | n.d. | n.d. | n.d. | n.d. | 26.36 | + | + | - | n.d. |
| 7 | 34.10 | 28.56 | n.d. | 31.67 | n.d. | n.d. | n.d. | n.d. | 25.78 | + | + | - | n.d. |
| 8 | n.d. | 30.07 | n.d. | 28.96 | n.d. | n.d. | n.d. | n.d. | 23.49 | + | + | - | n.d. |
| 9 | n.d. | 33.95 | n.d. | 32.73 | n.d. | n.d. | n.d. | n.d. | 29.40 | + | + | - | n.d. |
| 10 | 36.49 | 32.57 | n.d. | 33.81 | n.d. | n.d. | n.d. | n.d. | 27.12 | + | + | - | n.d. |
| Total | 6/10 | 10/10 | 0/10 | 10/10 | 0/10 | 0/10 | 1/10 | 0/10 | 10/10 | 9/10 | 9/10 | 0/10 | 0/10 |
| LIVER | |||||||||||||
| Animal |
PCMV /PRV |
PLHV-1 | PLHV-2 | PLHV-3 | PPV-1 | PCV2 | PCV3 | PCV4 | PERVC | PERV-C | PERV-A/C | HEV | |
| Real-time PCR | Real-time PCR | Real-time PCR | Real-time PCR | Real-time PCR | Real-time PCR | Real-time PCR | Real-time PCR | Real-time PCR | PCR1 | PCR4 | PCR | Real-time RT-PCR | |
| 1 | 36.19 | 31.35 | n.d. | 31.75 | n.d. | n.d. | n.d. | n.d. | 28.66 | + | + | - | n.d. |
| 2 | n.d. | 31.68 | n.d. | 29.91 | n.d. | n.d. | n.d. | n.d. | 22.00 | + | + | - | n.d. |
| 3 | n.d. | 30.79 | n.d. | 33.00 | n.d. | n.d. | n.d. | n.d. | 31.05 | - | - | - | n.d. |
| 4 | n.d. | 30.12 | n.d. | 32.21 | n.d. | n.d. | 25.00 | n.d. | 26.46 | + | + | - | n.d. |
| 5 | n.d. | 27.64 | n.d. | 35.47 | n.d. | n.d. | n.d. | n.d. | 28.90 | + | + | - | n.d. |
| 6 | n.d. | 29.89 | n.d. | 34.92 | n.d. | n.d. | n.d. | n.d. | 26.19 | + | + | - | n.d. |
| 7 | n.d. | 27.46 | n.d. | 34.21 | n.d. | n.d. | n.d. | n.d. | 27.42 | + | + | - | n.d. |
| 8 | n.d. | 29.48 | n.d. | 31.81 | n.d. | n.d. | n.d. | n.d. | 23.05 | + | + | - | n.d. |
| 9 | n.d. | 34.84 | n.d. | 37.02 | n.d. | n.d. | n.d. | n.d. | 31.68 | + | + | - | n.d. |
| 10 | n.d. | 30.68 | n.d. | 35.50 | n.d. | n.d. | n.d. | n.d. | 33.57 | + | + | - | n.d. |
| Total | 1/10 | 10/10 | 010 | 10/10 | 0/10 | 0/10 | 1/10 | 0/10 | 10/10 | 9/10 | 9/10 | o/10 | 0/10 |
| Pig breed |
Virus Detection method |
PCMV/PRV | PLHV-1 | PLHV-2 | PLHV-3 | PPV-1 | PCV1 | PCV2 | PCV3 | PCV4 | PERV-C | PERV-A/C | HEV3 | Reference | ||
|
Facility/ Institution |
Real-time PCR | Western blot |
Real-time PCR | Real-time PCR | Real-time PCR | Real-time PCR | Real-time PCR | Real-time PCR | Real-time PCR | Real-time PCR | Real-time PCR, PCR | PCR | Real-time RT-PCR | Western blot | ||
| Göttingen minipigs | Ellegaard Göttingen Minipigs A/S, Denmark | 12/39 (30%) | 8/67 (12%) | 0/14 (0%) | n.t. | n.t. | n.t. | n.t. | 3/21 (14%) | 0/10 (0%) | n.t. | 28/28 (100%) | 3/13 (23%) |
9/40 (22.5%) | 2/22 (9%) | Morozov et al. [29,30], Plotzki et al. [19], Heinze et al. [31], |
| Göttingen minipigs | University Göttingen, Göttingen, Germany | 0/10 (0%) | n.t. | 2/11 (18%) | 2/11 (18%) | 2/11 (18%) | n.t. | n.t. | 2/10 (20%) | 0/10 (0%) | n.t. | 0/10 (0%) | 0/10 (0%) |
0/10 (0%) | n.t. | Krüger et al. [24] |
| Göttingen minipigs with dippity pig syndrome | Ellegaard Göttingen Minipigs A/S, Denmark, Marshall BioResources, North Rose, New York |
3/7 (42%) |
n.t, | 0/7 (0%) |
0/7 (0%) |
n.t. | n.t. | 3/7 (42%) |
0/7 (0%) |
2/7 (29%) |
0/7 (0%) |
7/7 (100%) |
0/1 (0%) |
0/1 (9%) |
n.t. | Jhelum et al. [35] |
| Aachen minipigs | Aachen Minipig, Heinsberg, Germany |
5/18 (28%) |
n.t. | 0/18 (0%) |
5/18 (28%) |
3/18 (16%) |
n.t. | n.t. | 6/10 (60%) |
n.t. | n.t. | 13/13 (100%) | 2/8 (25%) |
12/18 (67%) |
4/18 (22%) |
Plotzki et al. [33] |
| Mini LEWE | University of Veterinary Medicine Hannover, Germany | 0/10 (0%) |
n.t. | 0/10 (0%) | 0/10 (0%) | 0/10 (0%) | 0/10 (0%) | 0/10 (0%) | 0/10 (0%) | 0/10 (0%) | 0/10 (0%) | 10/10 (110%) |
0/10 (0%) |
0/10 (0%) | n.t. | Halecker et al. [34] |
| Indigenous Greek black pigs | Four farms in Greece | 16/21 (76%) |
11/11 (100%) |
12/21 (57%) |
15/21 (71%) |
21/21 (100%) |
0/21 (0%) |
n.t. | 21/21 (100%) |
6/21 (29%) |
0/21 (0%) |
11/21 (52%) |
0/21 (0%) |
0/21 (0%) |
n.t. | Jhelum et al. [28] |
| Greek pigs with erythema multiforme | Farm in Greece | 0/5 (0%) |
n.t. | 5/5 (100%) |
1/5 (20%) |
4/5 (80%) |
n.t. | 0/5 (0%) |
1/5 (20%) |
1/5 (20%) |
0/5 (0%) |
5/5 (100%) |
0/5 (0%) |
n.t. | n.t. | Halecker et al. [36] |
| German slaughterhouse pigs | Slaughterhouse near Berlin, Germany |
6/10 (60%) |
10/10 (100%) |
10/10 (100%) |
0/10 (0%) |
10/10 (100%) |
0/10 (0%) |
n.t. | 0/10 (0%) |
1/10 (10%) |
0/10 (0%) |
10/10 (100%) |
0/10 (0%) |
0/10 (0%) |
n.t. | This manuscript |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





