Introduction
While there have been reviews on the integration of organs-on-chips and DL algorithms in the past, the rapid developments in both the technology and successful integrations have made it necessary for a more recent review. Additionally, this review focuses exclusively on CNNs and their model variations since their ability to analyze images makes them extremely valuable for streamlining research with the abundant data OoCs create. Following a brief overview of OoCs, CNNs, and types of CNNs, the applications of CNNs in OoCs covered in this article include device parameters, super-resolution, tracking and predicting cell trajectories, image classification, and image segmentation.
Organ-on-a-Chip
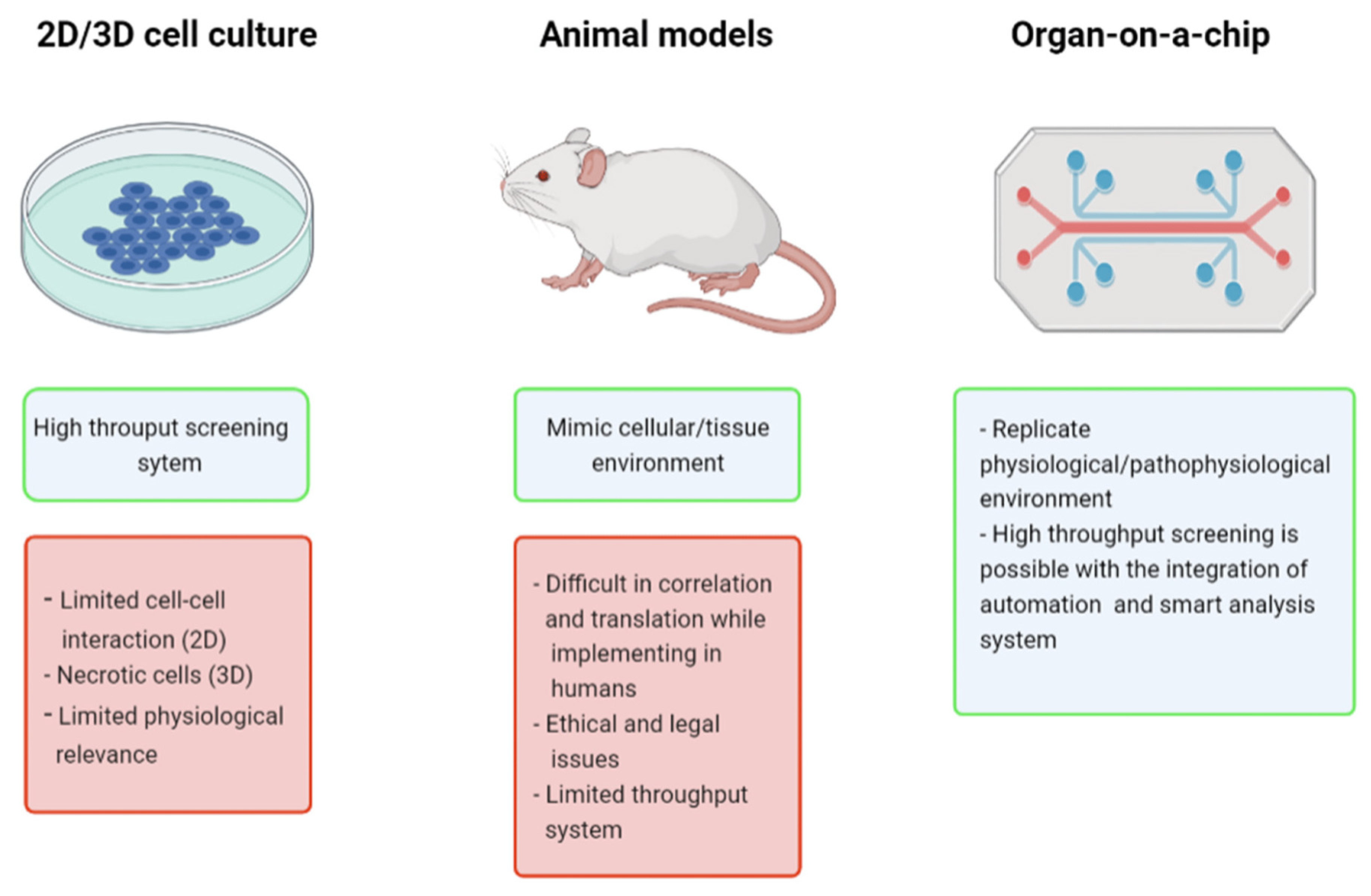
Organ-on-a-chip (OoC) technology is a 3-dimensional (3D) cell culture that utilizes advanced biomaterials (Osório et al., 2021) and microfluidics (Pattanayak et al., 2021) to accurately simulate an organ functioning in the body. By controlling and manipulating fluids, often through a series of valves, pumps, and microchannels containing living cells, researchers can create a highly accurate and controlled microenvironment that mimics the physiology and structure of an organ. With OoCs, researchers can control many more factors compared to traditional cell cultures (
Figure 1) such as chemical concentration gradient, tissue mechanical force, cell spatial configurational culture, multiple-cell coculture, and organ-to-organ interaction (Li et al., 2022). OoCs can precisely test and deliver compounds to the many individual living cells, allowing extremely high throughput and efficient use of resources. Because of their efficiency and quality of data, OoCs are often used in many applications such as life-saving cancer research and drug discovery studies (Leung et al., 2022). With the data provided by the sensors on cancer-on-a-chip platforms, the data accumulation can be the equivalent of around two gigabytes a day. With months upon months of data collection, databases can become tremendously large when researchers are working with OoCs (Fetah et al., 2019). This can present a challenge for researchers who have to examine all of the data to draw valuable conclusions.
Although OoCs are an incredible development for biomedical research, researchers still struggle with the optimization and extensive data analysis involved with this technology. As the field of artificial intelligence (AI) and deep learning (DL) continues to grow, complex algorithms like CNNs can continue to effectively address many of OoCs’ shortcomings.
Convolutional Neural Networks (CNNs)
CNNs are a type of neural network which are commonly used to extract features from images in order to draw certain conclusions. Based on interconnected neurons, CNNs are comprised of three main layers of neurons: the convolutional layer, the pooling layer, and the fully connected layer. Each layer is further overviewed below:
Convolutional Layer
The convolutional layer takes an image as input and applies kernels (a filter) over various sections of the image to produce a feature map to pass on to the next layer (
Figure 2). The most important parameters to consider in this layer are the number of kernels and the size of the kernels. This helps simplify the image to its core features. Typically, early convolutional layers highlight the low-level features of an image such as the gradient, edges, and colors, while later convolutional layers find high-level patterns within the image (Salehi et al., 2023).
Pooling Layer
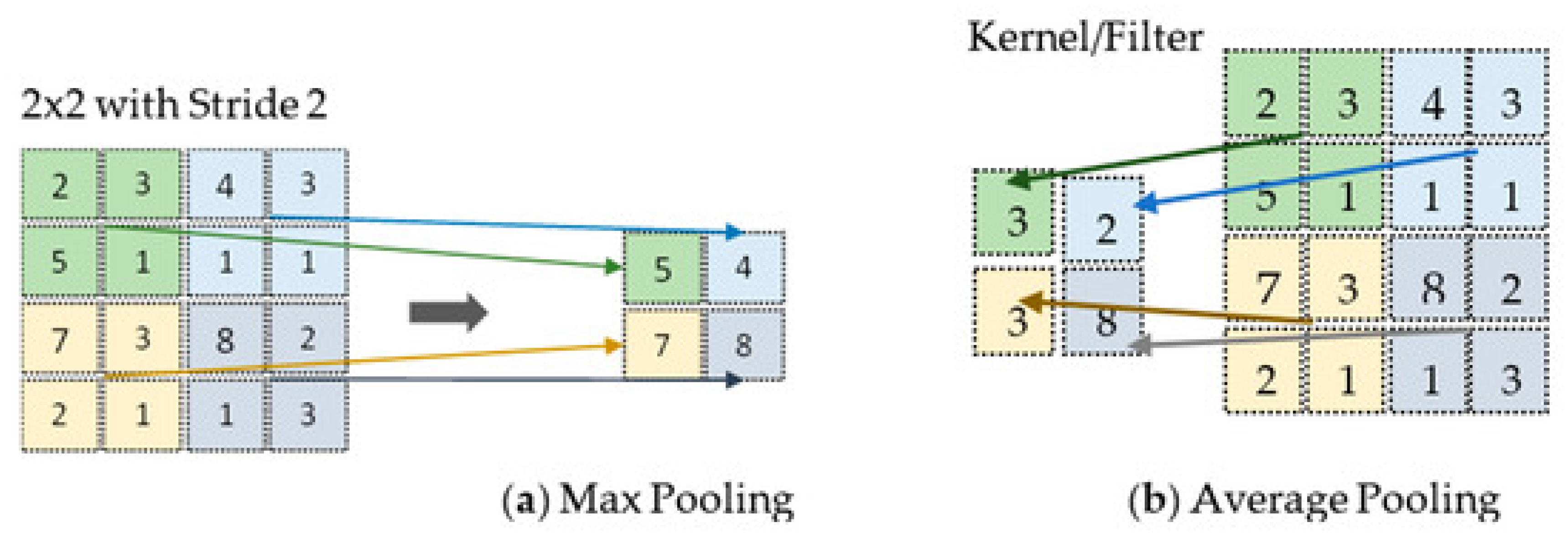
The pooling layer reduces the spatial complexity of the input. It achieves downsampling by dividing the feature map into non-overlapping regions and applying operations like max pooling which takes the highest value within each region, or average pooling which takes the average of the values within a region (
Figure 3). Through this process, pooling enhances translation invariance by emphasizing the features’ presence rather than their exact locations (Salehi et al., 2023).
Fully Connected Layer
In the fully connected layer, the input is flattened into a one-dimensional vector so it can be processed. Each element in the flattened vector connects to every neuron in the fully connected layer. Each neuron performs a weighted sum of its inputs and then performs an activation function like softmax or ReLU which performs a non-linear transformation, allowing the CNN to learn more complex patterns of features. The fully connected layer maps the learned features to the desired output, such as class probabilities for image classification (Salehi et al., 2023).
Types of Convolutional Neural Networks
Advancements in deep learning have allowed the development of more complex CNN architectures that are preferable for performing specific tasks that have many uses in research involving OoCs. There are three main types of CNNs used in OoC applications: Fully Convolutional Networks, Faster R-CNNs, and Mask R-CNNs.
Fully convolutional networks (FCNs) use convolution, pooling, and upscaling without dense/fully-connected layers to reduce the number of parameters and allow accurate pixel-level semantic segmentation in images (
Figure 4) (Long et al., 2015).
Popular FCNs such as U-Net (Ronneberger et al., 2015) are often used with research in the biomedical field due to their proficiency at image segmentation at the cellular level even with a limited data set.
Faster R-CNNs (Ren et al., 2015) use a Regional Proposal Network (a form of FCN) in combination with a Fast R-CNN (Girshick, 2015) for quick real-time object detection, practical for image classification. Based on Faster R-CNNs, Mask R-CNNs introduce a third branch for object masking, making the network capable of performing both object detection as well as semantic segmentation on each of the detected objects (K. He et al., 2017).
Device Parameters
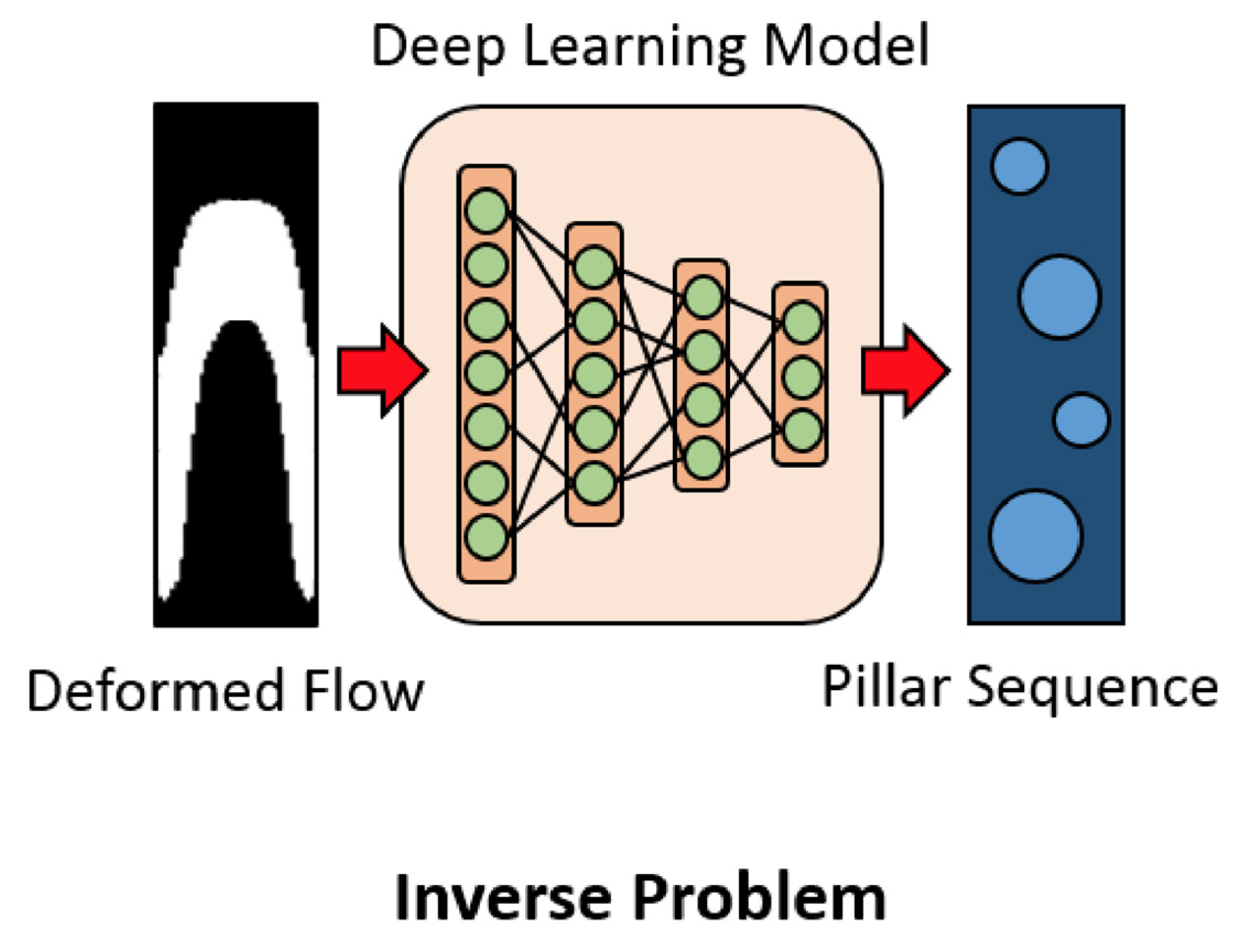
When developing OoCs and microfluidic devices in general, researchers must set and tweak many parameters before and after manufacturing that impact the flow of fluids and conditions within the chip. To expedite the design process, CNNs have been applied to determine the correct design parameters on a chip to get the desired result. For example, CNNs have been applied to microfluidic design for flow sculpting, which passively uses pillars in a microchannel to influence the inertial flow of fluid (Lore et al., 2015). Handcrafting the flows can be tedious and unintuitive, but CNN’s ability to identify patterns and trends makes them extremely useful for determining the pillar placement based on the required flow (
Figure 5) (Lore et al., 2015). Stoecklein et al. (2017) tested incorporating a CNN in the flow sculpting design process by having it predict the pillar placement according to the deformed flow shape in images. This allowed the group to quickly determine the optimal design based on the predictions of the CNN. Lore et al. (2015) compared CNN models to deep neural networks (DNN) and state-of-the-art genetic algorithm (GA) models for determining the organization of pillars to create a desired flow. The CNN model was able to outperform the DNN in accuracy and was only marginally worse than the GA. Nevertheless, the CNN proposed a solution within seconds while the GA computed for hours, meaning that CNNs are preferable for real-time or time-sensitive applications.
Another design parameter CNNs can be applied is micromixing (Lee et al., 2011). Wang et al. (2021) used a CNN to predict the outflow rates and concentration after micromixing. Being able to accurately predict the outflow rates and concentration gives researchers a better idea of how to adjust the micromixer in the microfluidic system.
Super-Resolution
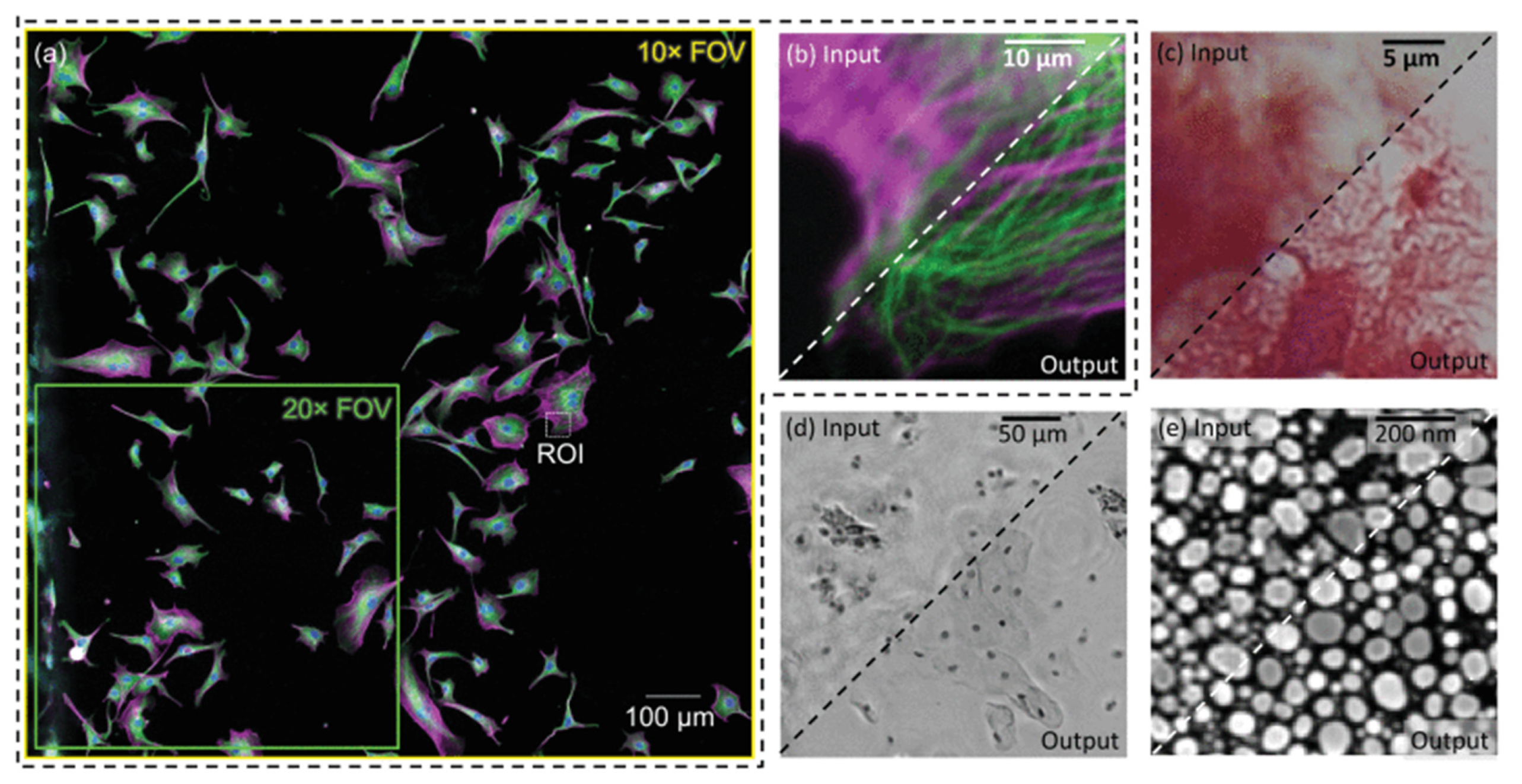
Clinical trials involving OoCs frequently require images and videos at the microscopic level to be analyzed by researchers or DL algorithms to get insight into drug pharmacokinetics and potential side effects on cells. However, there are many physical and financial challenges in capturing high-quality images in OoCs (Siu et al., 2023). To get quality data from OoCs, CNNs can be applied to accurately increase the resolution of images and videos to facilitate analysis. DL algorithms, particularly CNNs, have frequently been used in the past to enhance and reconstruct images obtained through optical microscopy (
Figure 6) (De Haan et al., 2020).
CNNs could similarly now be applied to enhance the resolution of images and videos captured of cells on OoCs which can often be blurry and difficult to interpret (Siu et al., 2023). Cascarano et al. (2021) implemented a recursive version of a Deep Image Prior (DIP) (Ulyanov et al., 2018), an image reconstruction model based on deep convolutional neural networks, to achieve super-resolution with time-lapsed microscopy (TLM) video. After the DIP was able to successfully improve the video quality for synthetic and real experiments, edge detection, cell tracking, and cell localization were noticeably improved .
This successful application demonstrates that utilizing CNNs for super-resolution for OoCs can provide easier, more accurate analysis for researchers and CNNs. It also allows for analysis of complex drug and cell interactions that cannot be seen at lower resolution (Cascarano et al., 2021).
Tracking and Predicting Cell Trajectories
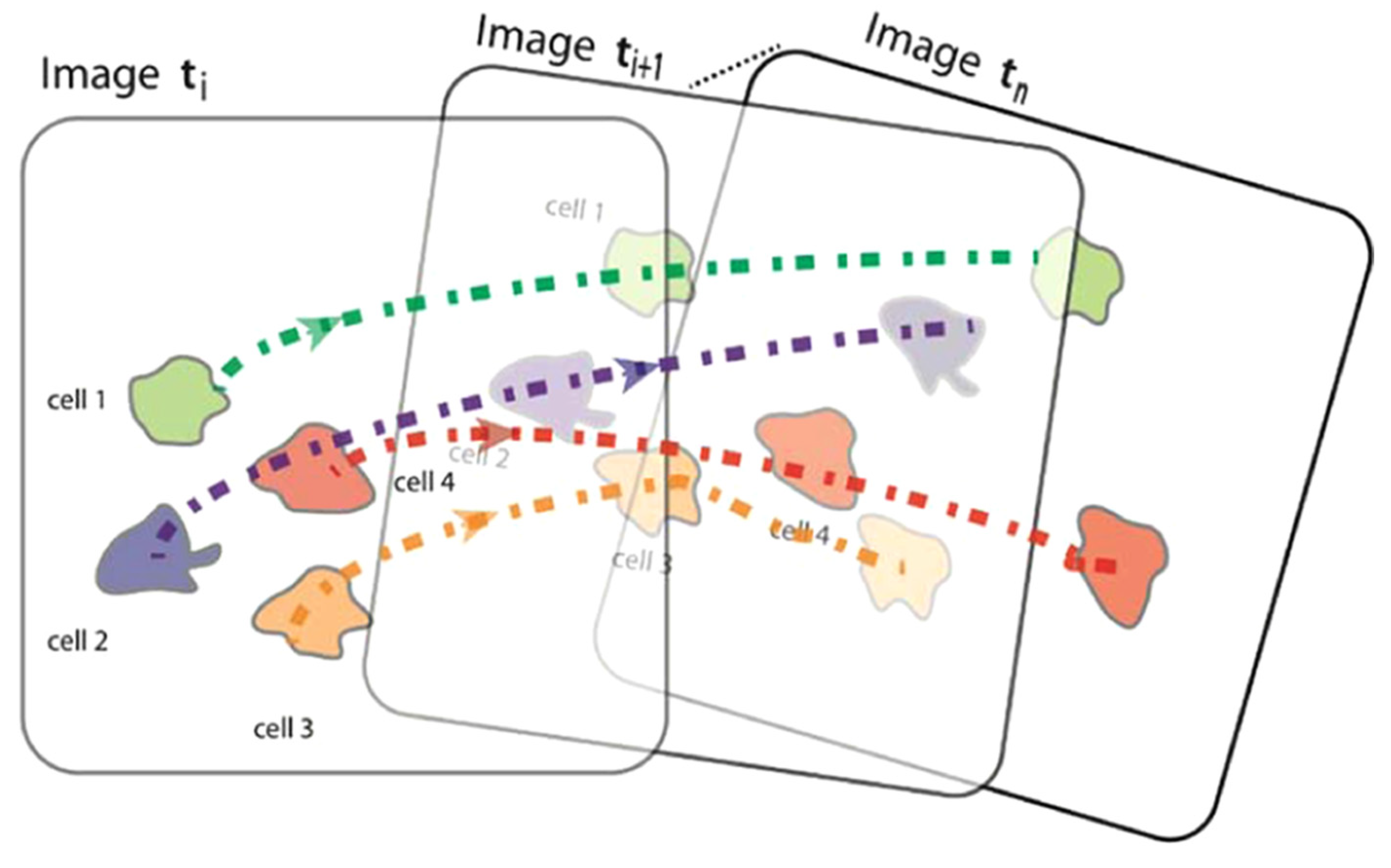
Inside the body, cell motility is important for tissue repair and immune responses (Comes et al., 2021). When doing research with OoCs, sometimes tracking cell trajectories (
Figure 7) is crucial for understanding how cells migrate, interact with their surroundings, or react to drug or chemical administration (Mencattini et al., 2020).
One example where tracking cells is important is cancer research. Mencattini et al. (2020) used a pre-trained CNN called AlexNet on a tumor-on-a-chip with time lapse-microscopy video. They determined that the immunotherapy drug trastuzumab increased cancer-immune cell interactions based on the tracked movements of the cells.
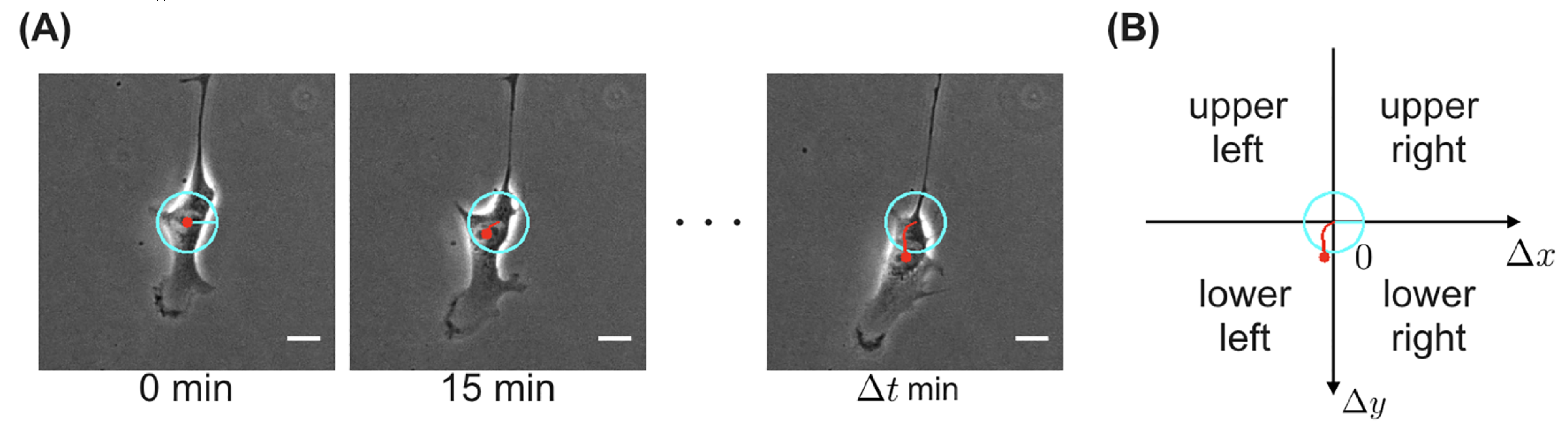
In addition to tracking cell trajectories, predicting cell trajectories can be equally important, especially when working with OoCs that produce a lot of data. One of the drawbacks of OoCs is the need for massive amounts of electronic data storage. This can be challenging for many researchers who have neither the access nor the finances to host massive data servers. Nishimoto et al. (2019) used a CNN to produce the migration direction in the next frame as an output by taking in the previous frame as an input (
Figure 8). After identifying the features of the cell, the CNN was able to accurately predict the direction of trajectory instead of researchers having to film hundreds of hours of time-lapsed video.
Kimmel et al. (2021) used a combination of a recurrent neural network (RNN), capable of analyzing sequential data (Salehinejad et al., 2018), and a deep CNN to predict the trajectories of cells with high accuracy by representing the predicted cell trajectories in the form of multi-channel time series. Due to the high throughput nature of OoCs, cell tracking and predictive movement have extensive utility in research and hold tremendous potential for further application in OoCs.
Image Segmentation
In some cases, researchers working with OoCs might only want to focus on certain sections of the microscopic images taken of the cells on the device. These sections could have special significance in the study or be hard-to-see features. The segmentation of images by CNNs can immediately analyze or streamline the analysis of images by easing the burden of identifying structures, especially when considering the massive image datasets OoCs produce (Siu et al., 2023). After image preprocessing, de Keizer (2019) applied U-Net, a popular FCN model, to phase contrast images of OoCs to segment blood vessels. They also used another CNN to segment the nuclei of the cells from the background. These segmentations are crucial to faster and more robust analysis of data from OoCs. Su et al. (2023) applied two CNNs to a blood-brain barrier-on-a-chip (BBB-oC) in order to monitor the protein secretions made by endothelial cells. One of the CNNs segmented the images into image defects (blurring or distortions) from the background pixels. The other CNN identified, counted, and then segmented out the “On” microwells with the segmented defects removed. The CNNs provided valuable segmentations to the images that helped improve the quality of the data and facilitate analysis.
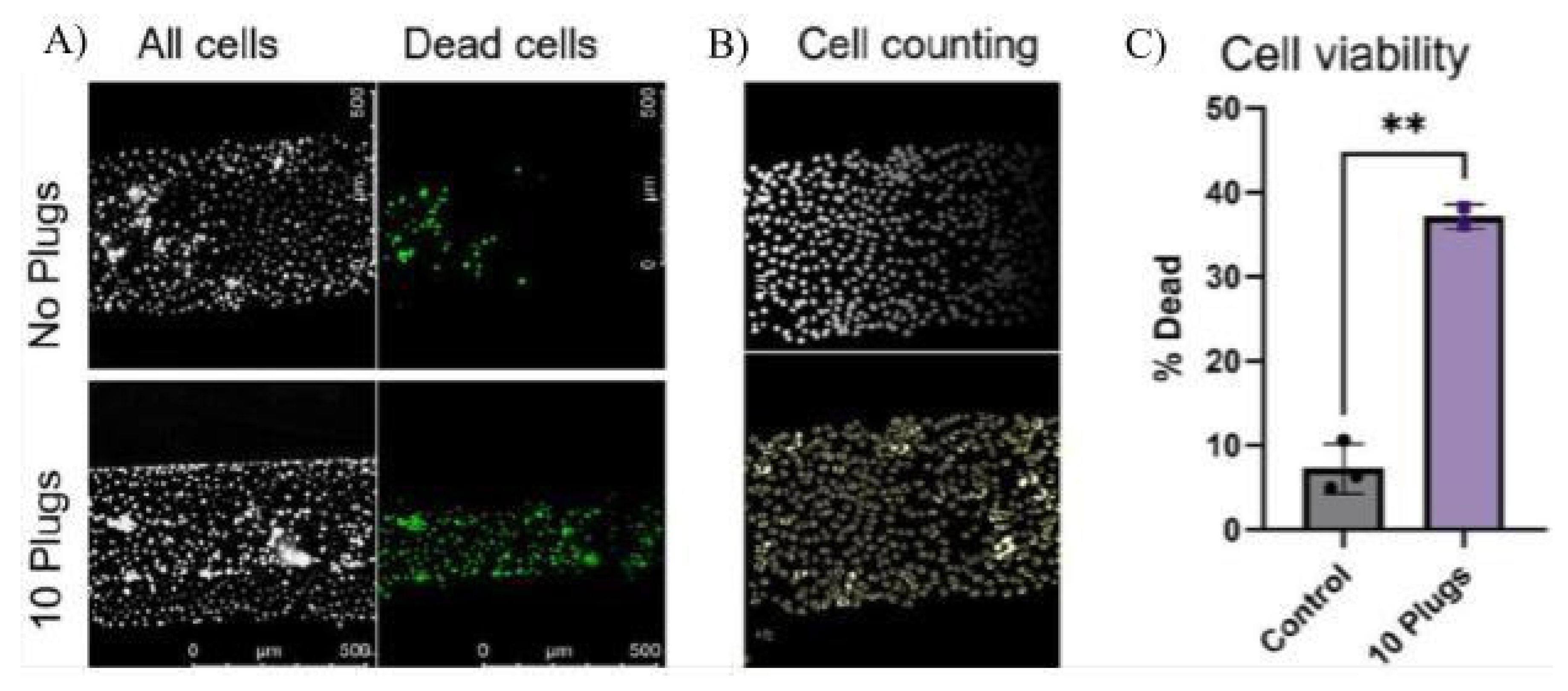
Within the past few years, models based on U-Net, Cellpose (Stringer et al., 2021) and Cellpose2.0 (Carsen Stringer & Marius Pachitariu, 2022), have been especially useful in research involving OoCs because of their ability to segment individual cell bodies. Cellpose2.0 was used in a study by Ehlers et al. (2023) to quantify the number of VE-Cadherin adhesion molecules expressed on an OoC with blood vessels through image segmentation. Observing changes in VE-Cadherin expression is a useful metric in quantifying the inflammatory response, so CNN models like Cellpose2.0 play an important role in quickly analyzing the thousands of images created by experiments on OoCs. Similarly, Viola et al. (2023) produced a plug on an airway-on-a-chip, which is useful in recreating the environment of the lung. They used Cellpose to quantify the number of cells after exposure to the plug (
Figure 9).
When testing for the ability of a new pump-less, recirculating OoC to sustain cells and run experiments, Busek et al. (2023) applied Cellpose2.0 to segment out the nuclei of the cells for analysis.
Image segmentation performed by CNNs has been used to expedite the data examination process, which is especially important when examining large data sets like the ones produced by OoCs. With this technology, analysis becomes even easier and researchers are able to produce concrete quantifiable results much quicker than before.
Image Classification
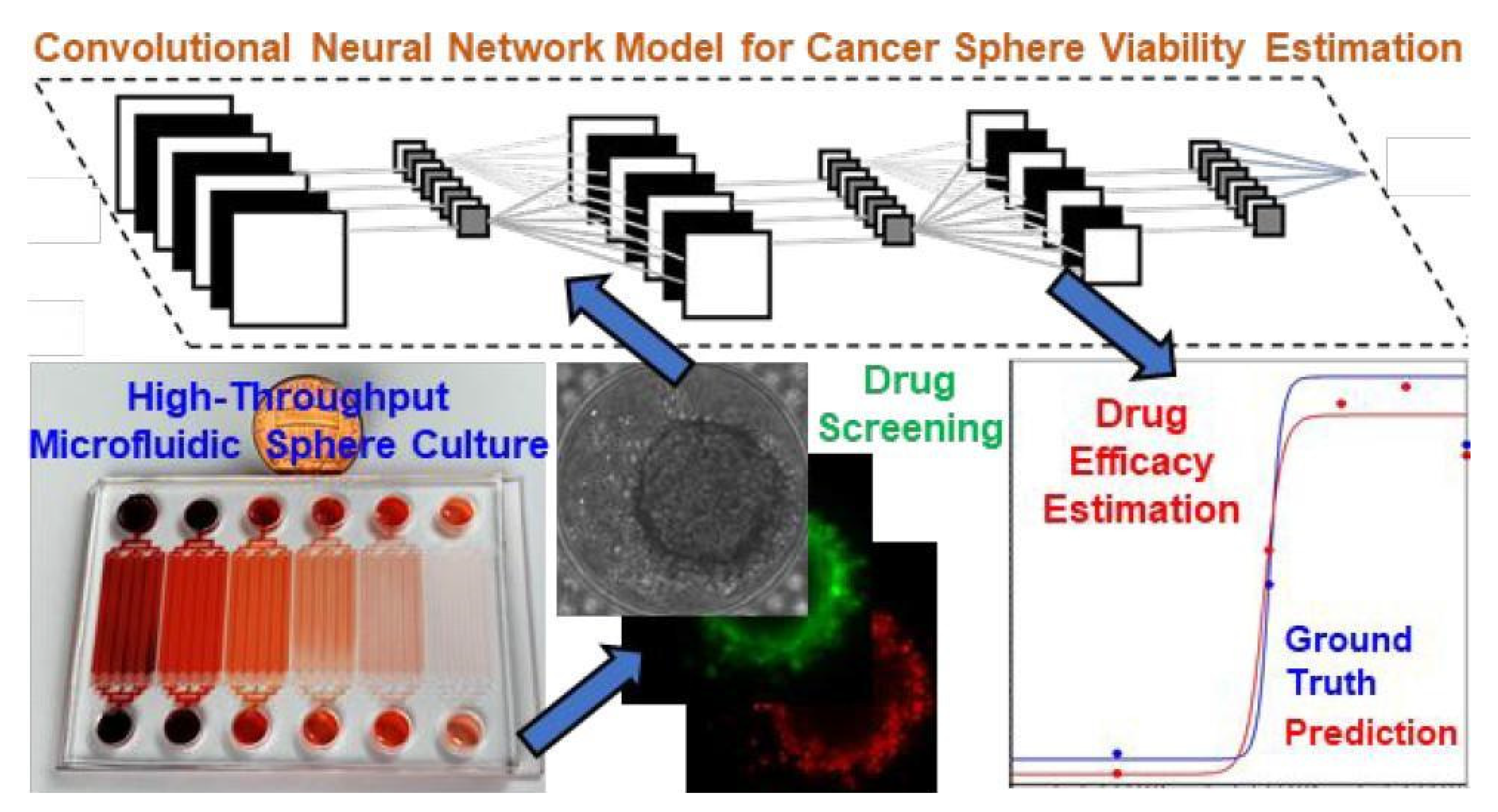
In many experiments, researchers often need to quantify the number of times a certain item or state is present for a variety of reasons, meaning the classification of objects based on images is crucial to many experiments. Instead of a researcher having to manually label every image of a massive data set, CNNs can quickly and accurately identify what type or state of an object is present within an image. Zhang et al. (2019) collected images from a tumor-on-a-chip platform with 1,920 tumor spheres. A CNN was then used to quickly classify the drug concentration each tumor cell received and estimate the viability for each tumor sphere after exposure to a therapeutic drug (
Figure 10). When implemented, CNNs can exponentially hasten cancer drug development analysis involving OoCs through classification.
Jena et al. (2019) propose that studying muscle-on-a-chip devices with CNNs could prove valuable for better understanding human muscle cells and creating a definitive Skeletal Muscle Cell Atlas. Looking at the markers on muscle cells, a CNN trained on OoCs could immediately classify the metabolic state (e.g., aerobic vs anaerobic), contractile type (slow vs fast twitch fibers), and the overall performance of muscle cells from imaged biopsy material.
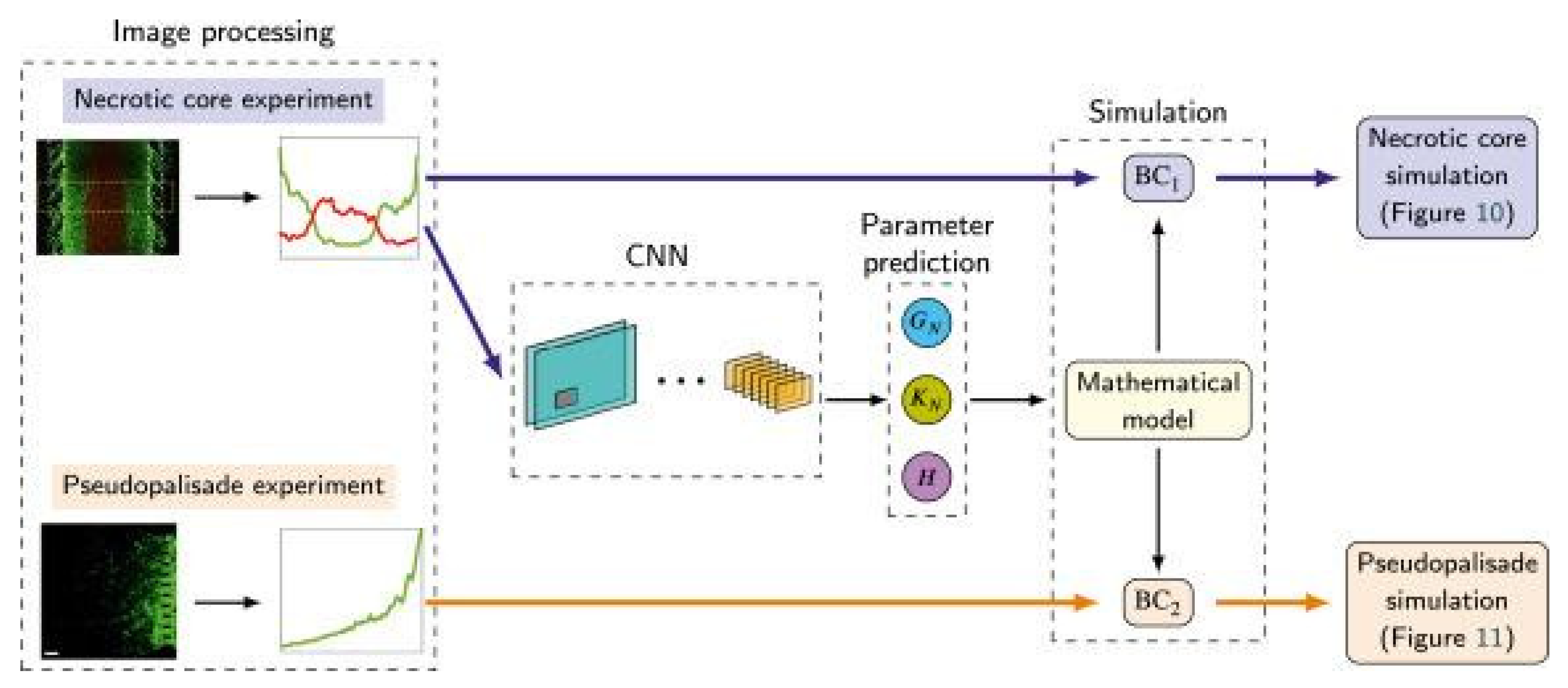
Pérez-Aliacar et al. (2021) developed and trained a CNN from images taken from a glioblastoma-on-a-chip (a type of deadly brain tumor) to classify and predict the parameters that influence the growth of the tumor (
Figure 11). In clinical applications, these factors are important for prognosis and treatment response.
Not only does this publication display that CNNs for classification can aid in clinical applications but also that OoCs can provide the massive datasets required to train powerful CNNs with uses in the medical field.
Opportunities and Challenges
Scalability
Most of the current applications of CNNs involve analysis from only one OoC device. While CNNs have been shown to be successful on that scale, research always would benefit from a larger collection of data to ensure thorough testing and discovery. To produce even higher quality research, CNNs must be able to be applied at a larger scale with multiple organ-on-a-chip devices. As the scale increases, however, the computational load becomes progressively more difficult to accommodate. With limited resources and funding, the optimization of CNNs (Habib & Qureshi, 2022) is crucial to their scalability in OoCs. To address the issue of scalability, customized design of models, compression of model designs (Zhao et al., 2022), and iterative improvements to models over time as deep learning progresses are all actions that could be taken in order to increase computational efficiency.
Accessibility
CNNs are a powerful tool for researchers using OoCs. However, CNNs are extremely complex models that many researchers who specialize in the biomedical field and know little about AI might not be able to implement into their work successfully. To make applying DL more accessible in research, the creation of a user-friendly service for researchers interested in involving AI in their research, providing general and more specific algorithms.
Further development of the AutoML (X. He et al., 2021) field and neural architecture searching (NAS) (Mok et al., 2021) could allow researchers lacking expertise in the ML field to create useful neural networks to incorporate into their research. With a provided data set, NAS can automatically design, test, and tweak parameters to optimize a neural network without the need for researchers to have extensive knowledge of ML. This greatly improves the usability of neural networks to expedite and aid in research.
Human-on-a-Chip
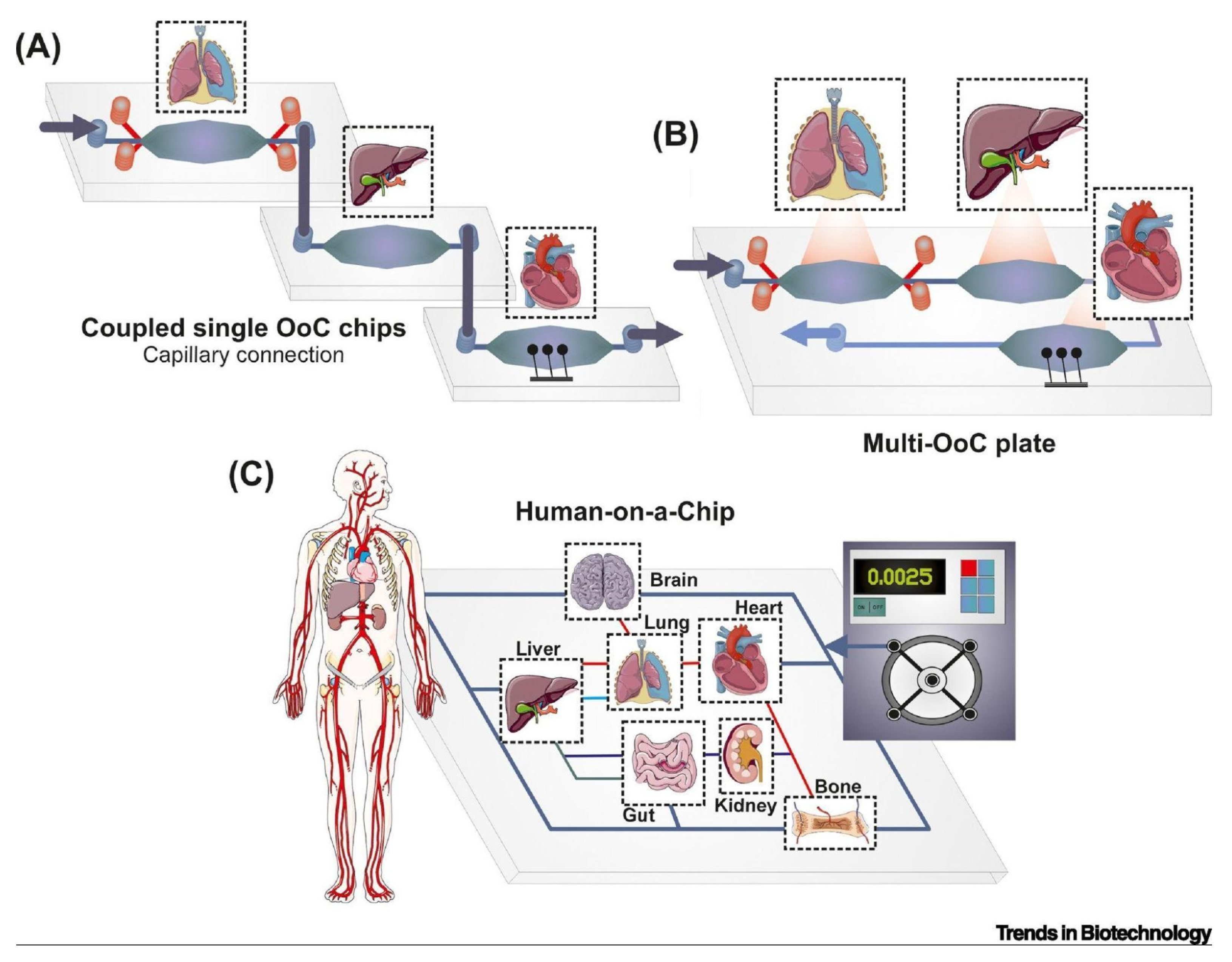
Human-on-a-chip (HoC), an emerging on-a-chip system, devices include several human organs in a hierarchical and physiological pattern on a chip in order to simulate an in vitro human. This platform allows for a comprehensive view of cellular behavior in response to chemical and mechanical cues and could serve as one of the best and most inexpensive clinical models for researchers instead of clinical trial participants. Coupling single OoCs together to create HoC platforms through a capillary connection or a microfluidic motherboard are some of the emerging methods of merging microfluidic cell cultures together (
Figure 12) (Picollet-D’hahan et al., 2021).
However, the coculturing of many different types of cells, which often require different conditions and nutrients, makes this a difficult development in the world of OoCs (Syama & Mohanan, 2021).
Automated Microfluidic Systems
During an experiment, researchers must constantly monitor and adjust the conditions within the device to achieve an exact environment. CNNs could potentially aid researchers during the experimental process by providing continuous monitoring of OoCs based on live images for semi or complete automation. While this is still a developing application, the continued integration of DL models with OoC models paves the way for entirely autonomous OoCs to soon become a reality (Polini & Moroni, 2021; Sabaté Del Río et al., 2023).
Data Limitations and Its Solutions
One limitation of using CNNs in OoCs is the need for a large, labeled data set to train and test the network. Since these CNNs are often trained to do very specific tasks, researchers might have to start the data collection process from nothing. The collection and labeling of this data can be a costly and sometimes time-consuming process. Some solutions exist that can address this data scarcity problem by reducing the need for a large, labeled training data set or artificially increasing it.
Unsupervised and Semi-Supervised Learning
Unsupervised learning (Bengio et al., 2014) feeds only unlabeled data to train the CNN. The CNN then uses techniques such as clustering (Moriya et al., 2018) to learn features from the images on its own. This has had many successful implementations in the field of medical imaging (Chen et al., 2019; Raza & Singh, 2018) and serves as a useful method to completely forgo the need for a labeled data set.
Semi-supervised learning (Zhu, 2005) reduces the dependency on a large, labeled data set by training the network on both unlabeled and labeled data. Different techniques in semi-supervised learning such as active learning (Budd et al., 2021; Kim et al., 2020) and Cross Teaching (Luo et al., 2022) have been used to train CNNs for purposes in medical imaging (Jiao et al., 2022). These articles demonstrate that a successful network can be trained even when data is limited.
Transfer Learning
Transfer learning (Zhuang et al., 2021) can be a useful tool for creating an accurate model without a large, labeled data set. Transfer learning uses a preexisting network that works in a similar domain and provides it with a relatively smaller data set. The model is able to pick up on the features faster than a completely new untrained model because it has a basis on which it can transfer its current skills to a new, but similar, task. Transfer learning can be conducive to using CNNs in OoCs as many existing models specialize in cell image analysis whose skills could be transferred (Salehi et al., 2023).
Data Augmentation
When working with a small data set, CNNs can overfit the training data, leading to poor performance in actual applications with new data. Data augmentation (Shorten & Khoshgoftaar, 2019) can artificially increase the size of the database by performing changes called augmentations to the existing data. This effectively provides “new” data for the CNN to train on.
Automation of Data Labeling
In the coming years, the development of automated data labeling systems could reduce the labor-intensive process of data labeling. Massive unlabeled data sets could be automatically labeled and used for supervised training of CNNs. Desmond et al. (2021) created a semi-automated data labeling system, which could expedite the data labeling process by assisting researchers. Further development of this technology could eventually lead to a more automated data labeling system.
Conclusion
This review first introduced an overview of OoCs and CNNs, highlighting key CNN models frequently applied to OoCs. Then, many current applications of CNNs in OoCs like device parameters, predicting and tracking cell trajectories, super-resolution, image classification, and image segmentation were explained and delved into. Lastly, several challenges and opportunities involving CNNs and OoCs were presented.
Overall, both the field of DL and OoCs are rapidly evolving as these are both emerging technologies. The optimization and development of both technologies have led to many creative, interdisciplinary applications. The creation and recent application of CNNs have allowed for streamlined image analysis through a variety of methods that are widely applicable in research with OoCs. Predicting and tracking cell trajectories, super-resolution, image classification, and image segmentation performed by CNNs minimize the burden of analyzing the inherently high throughput OoCs models. CNNs additionally facilitate the creation of microfluidic models like OoCs because of their ability to recognize patterns and make predictions.
Although CNNs can be a useful tool, challenges such as data limitations, scalability, and accessibility of these models restrict the more extensive use of CNNs with OoCs. These challenges may be daunting, but some plausible solutions continue to become more feasible as the field of AI and OoCs progresses. Possible developments with human-on-a-chip models and automated microfluidics platforms present an exciting future for more productive and efficient research. This review offers valuable insights and hopes to serve as a comprehensive guide for researchers interested in exploring the beneficial applications of CNNs in OoC platforms.
Acknowledgments
The accomplishment of this review paper is greatly attributed to the guidance and encouragement provided by my mentor, Morteza Sarmadi. His feedback played a vital role in enabling me to enhance and improve my research.
References
- Y. Bengio, A. Courville and P. Vincent, “Representation Learning: A Review and New Perspectives,” in IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 35, no. 8, pp. 1798-1828, Aug. 2013. [CrossRef]
- Budd, S., Robinson, E. C., & Kainz, B. (2021). A survey on active learning and human-in-the-loop deep learning for medical image analysis. Medical Image Analysis, 71, 102062. [CrossRef]
- Busek, M., Aizenshtadt, A., Koch, T., Frank, A., Delon, L., Martinez, M. A., Golovin, A., Dumas, C., Stokowiec, J., Gruenzner, S., Melum, E., & Krauss, S. (2023). Pump-less, recirculating organ-on-a-chip (rOoC) platform. Lab on a Chip, 23(4), 591–608. [CrossRef]
- Carsen Stringer & Marius Pachitariu. (2022). Cellpose 2.0: How to train your own model. BioRxiv, 2022.04.01.486764. [CrossRef]
- Cascarano, P., Comes, M. C., Mencattini, A., Parrini, M. C., Piccolomini, E. L., & Martinelli, E. (2021). Recursive Deep Prior Video: A super resolution algorithm for time-lapse microscopy of organ-on-chip experiments. Medical Image Analysis, 72, 102124. [CrossRef]
- Chen, L., Bentley, P., Mori, K., Misawa, K., Fujiwara, M., & Rueckert, D. (2019). Self-supervised learning for medical image analysis using image context restoration. Medical Image Analysis, 58, 101539. [CrossRef]
- Comes, M. C., Filippi, J., Mencattini, A., Corsi, F., Casti, P., De Ninno, A., Di Giuseppe, D., D’Orazio, M., Ghibelli, L., Mattei, F., Schiavoni, G., Businaro, L., Di Natale, C., & Martinelli, E. (2020). Accelerating the experimental responses on cell behaviors: A long-term prediction of cell trajectories using Social Generative Adversarial Network. Scientific Reports, 10(1), 15635. [CrossRef]
- De Haan, K., Rivenson, Y., Wu, Y., & Ozcan, A. (2020). Deep-Learning-Based Image Reconstruction and Enhancement in Optical Microscopy. Proceedings of the IEEE, 108(1), 30–50. [CrossRef]
- de Keizer, C. (2019). Phase Contrast Image Preprocessing and Segmentation of Vascular Networks in Human Organ-on-Chips (Doctoral dissertation, Tilburg University).
- Desmond, M., Duesterwald, E., Brimijoin, K., Brachman, M., & Pan, Q. (2021). Semi-Automated Data Labeling. Proceedings of the NeurIPS 2020 Competition and Demonstration Track, 156–169. https://proceedings.mlr.press/v133/desmond21a.html.
- Ehlers, H., Nicolas, A., Schavemaker, F., Heijmans, J. P. M., Bulst, M., Trietsch, S. J., & Van Den Broek, L. J. (2023). Vascular inflammation on a chip: A scalable platform for trans-endothelial electrical resistance and immune cell migration. Frontiers in Immunology, 14, 1118624. [CrossRef]
- Fetah, K. L., DiPardo, B. J., Kongadzem, E., Tomlinson, J. S., Elzagheid, A., Elmusrati, M., Khademhosseini, A., & Ashammakhi, N. (2019). Cancer Modeling-on-a-Chip with Future Artificial Intelligence Integration. Small, 15(50), 1901985. [CrossRef]
- Girshick, R. (2015). Fast r-cnn. In Proceedings of the IEEE international conference on computer vision (pp. 1440-1448). https://openaccess.thecvf.com/content_iccv_2015/html/Girshick_Fast_R-CNN_ICCV_2015_paper.html.
- Habib, G., & Qureshi, S. (2022). Optimization and acceleration of convolutional neural networks: A survey. Journal of King Saud University - Computer and Information Sciences, 34(7), 4244–4268. [CrossRef]
- Hannah L. Viola, Vishwa Vasani, Kendra Washington, Ji-Hoon Lee, Cauviya Selva, Andrea Li, Carlos J. Llorente, Yoshinobu Murayama, James B. Grotberg, Francesco Romanò, & Shuichi Takayama. (2023). Liquid plug propagation in computer-controlled microfluidic airway-on-a-chip with semi-circular microchannels. BioRxiv, 2023.05.24.542177. [CrossRef]
- He, K., Gkioxari, G., Dollár, P., & Girshick, R. (2017). Mask r-cnn. In Proceedings of the IEEE international conference on computer vision (pp. 2961-2969). https://openaccess.thecvf.com/content_iccv_2017/html/He_Mask_R-CNN_ICCV_2017_paper.html.
- He, X., Zhao, K., & Chu, X. (2021). AutoML: A survey of the state-of-the-art. Knowledge-Based Systems, 212, 106622. [CrossRef]
- Jena, B. P., Gatti, D. L., Arslanturk, S., Pernal, S., & Taatjes, D. J. (2019). Human skeletal muscle cell atlas: Unraveling cellular secrets utilizing ‘muscle-on-a-chip’, differential expansion microscopy, mass spectrometry, nanothermometry and machine learning. Micron, 117, 55–59. [CrossRef]
- Jiao, R., Zhang, Y., Ding, L., Cai, R., & Zhang, J. (2022). Learning with Limited Annotations: A Survey on Deep Semi-Supervised Learning for Medical Image Segmentation. [CrossRef]
- Kim, T., Lee, K. H., Ham, S., Park, B., Lee, S., Hong, D., Kim, G. B., Kyung, Y. S., Kim, C.-S., & Kim, N. (2020). Active learning for accuracy enhancement of semantic segmentation with CNN-corrected label curations: Evaluation on kidney segmentation in abdominal CT. Scientific Reports, 10(1), 366. [CrossRef]
- Kimmel, J. C., Brack, A. S., & Marshall, W. F. (2021). Deep Convolutional and Recurrent Neural Networks for Cell Motility Discrimination and Prediction. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 18(2), 562–574. [CrossRef]
- Koyilot, M. C., Natarajan, P., Hunt, C. R., Sivarajkumar, S., Roy, R., Joglekar, S., Pandita, S., Tong, C. W., Marakkar, S., Subramanian, L., Yadav, S. S., Cherian, A. V., Pandita, T. K., Shameer, K., & Yadav, K. K. (2022). Breakthroughs and Applications of Organ-on-a-Chip Technology. Cells, 11(11), 1828. [CrossRef]
- Lee, C.-Y., Chang, C.-L., Wang, Y.-N., & Fu, L.-M. (2011). Microfluidic Mixing: A Review. International Journal of Molecular Sciences, 12(5), 3263–3287. [CrossRef]
- Leung, C. M., De Haan, P., Ronaldson-Bouchard, K., Kim, G.-A., Ko, J., Rho, H. S., Chen, Z., Habibovic, P., Jeon, N. L., Takayama, S., Shuler, M. L., Vunjak-Novakovic, G., Frey, O., Verpoorte, E., & Toh, Y.-C. (2022). A guide to the organ-on-a-chip. Nature Reviews Methods Primers, 2(1), 33. [CrossRef]
- Li, J., Chen, J., Bai, H., Wang, H., Hao, S., Ding, Y., Peng, B., Zhang, J., Li, L., & Huang, W. (2022). An Overview of Organs-on-Chips Based on Deep Learning. Research, 2022, 2022/9869518. [CrossRef]
- Liu, X., Deng, Z. & Yang, Y. Recent progress in semantic image segmentation. Artif Intell Rev 52, 1089–1106 (2019). [CrossRef]
- J. Long, E. Shelhamer and T. Darrell, “Fully convolutional networks for semantic segmentation,” 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 2015, pp. 3431-3440. [CrossRef]
- Lore, K. G., Stoecklein, D., Davies, M., Ganapathysubramanian, B., & Sarkar, S. (2015). Hierarchical Feature Extraction for Efficient Design of Microfluidic Flow Patterns. Proceedings of the 1st International Workshop on Feature Extraction: Modern Questions and Challenges at NIPS 2015, 213–225. https://proceedings.mlr.press/v44/lore15.html.
- Luo, X., Hu, M., Song, T., Wang, G., & Zhang, S. (2022). Semi-Supervised Medical Image Segmentation via Cross Teaching between CNN and Transformer. Proceedings of The 5th International Conference on Medical Imaging with Deep Learning, 820–833. https://proceedings.mlr.press/v172/luo22b.html.
- Mencattini, A., Di Giuseppe, D., Comes, M. C., Casti, P., Corsi, F., Bertani, F. R., Ghibelli, L., Businaro, L., Di Natale, C., Parrini, M. C., & Martinelli, E. (2020). Discovering the hidden messages within cell trajectories using a deep learning approach for in vitro evaluation of cancer drug treatments. Scientific Reports, 10(1), 7653. [CrossRef]
- Mok, J., Na, B., Choe, H., & Yoon, S. (2021). AdvRush: Searching for Adversarially Robust Neural Architectures. 12322–12332. https://openaccess.thecvf.com/content/ICCV2021/html/Mok_AdvRush_Searching_for_Adversarially_Robust_Neural_Architectures_ICCV_2021_paper.html.
- Moriya, T., Roth, H. R., Nakamura, S., Oda, H., Nagara, K., Oda, M., & Mori, K. (2018). Unsupervised segmentation of 3D medical images based on clustering and deep representation learning. In B. Gimi & A. Krol (Eds.), Medical Imaging 2018: Biomedical Applications in Molecular, Structural, and Functional Imaging (p. 71). SPIE. [CrossRef]
- Nishimoto, S., Tokuoka, Y., Yamada, T. G., Hiroi, N. F., & Funahashi, A. (2019). Predicting the future direction of cell movement with convolutional neural networks. PLOS ONE, 14(9), e0221245. [CrossRef]
- Osório, L. A., Silva, E., & Mackay, R. E. (2021). A Review of Biomaterials and Scaffold Fabrication for Organ-on-a-Chip (OOAC) Systems. Bioengineering, 8(8), Article 8. [CrossRef]
- Pattanayak, P., Singh, S. K., Gulati, M., Vishwas, S., Kapoor, B., Chellappan, D. K., Anand, K., Gupta, G., Jha, N. K., Gupta, P. K., Prasher, P., Dua, K., Dureja, H., Kumar, D., & Kumar, V. (2021). Microfluidic chips: Recent advances, critical strategies in design, applications and future perspectives. Microfluidics and Nanofluidics, 25(12), 99. [CrossRef]
- Pérez-Aliacar, M., Doweidar, M. H., Doblaré, M., & Ayensa-Jiménez, J. (2021). Predicting cell behaviour parameters from glioblastoma on a chip images. A deep learning approach. Computers in Biology and Medicine, 135, 104547. [CrossRef]
- Picollet-D’hahan, N., Zuchowska, A., Lemeunier, I., & Gac, S. L. (2021). Multiorgan-on-a-Chip: A Systemic Approach To Model and Decipher Inter-Organ Communication. Trends in Biotechnology, 39(8), 788–810. [CrossRef]
- Polini, A., & Moroni, L. (2021). The convergence of high-tech emerging technologies into the next stage of organ-on-a-chips. Biomaterials and Biosystems, 1, 100012. [CrossRef]
- Ren, S., He, K., Girshick, R., & Sun, J. (2015). Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. Advances in Neural Information Processing Systems, 28. https://proceedings.neurips.cc/paper_files/paper/2015/hash/14bfa6bb14875e45bba028a21ed38046-Abstract.html.
- Ronneberger, O., Fischer, P., & Brox, T. (2015). U-Net: Convolutional Networks for Biomedical Image Segmentation. In N. Navab, J. Hornegger, W. M. Wells, & A. F. Frangi (Eds.), Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015 (Vol. 9351, pp. 234–241). Springer International Publishing. [CrossRef]
- Sabaté Del Río, J., Ro, J., Yoon, H., Park, T.-E., & Cho, Y.-K. (2023). Integrated technologies for continuous monitoring of organs-on-chips: Current challenges and potential solutions. Biosensors and Bioelectronics, 224, 115057. [CrossRef]
- Salehi, A. W., Khan, S., Gupta, G., Alabduallah, B. I., Almjally, A., Alsolai, H., Siddiqui, T., & Mellit, A. (2023). A Study of CNN and Transfer Learning in Medical Imaging: Advantages, Challenges, Future Scope. Sustainability, 15(7), 5930. [CrossRef]
- Salehinejad, H., Sankar, S., Barfett, J., Colak, E., & Valaee, S. (2018). Recent Advances in Recurrent Neural Networks. arXiv:1801.01078. [CrossRef]
- Shorten, C., & Khoshgoftaar, T. M. (2019). A survey on Image Data Augmentation for Deep Learning. Journal of Big Data, 6(1), 60. [CrossRef]
- Siu, D. M. D., Lee, K. C. M., Chung, B. M. F., Wong, J. S. J., Zheng, G., & Tsia, K. K. (2023). Optofluidic imaging meets deep learning: From merging to emerging. Lab on a Chip, 23(5), 1011–1033. [CrossRef]
- Stoecklein, D., Lore, K. G., Davies, M., Sarkar, S., & Ganapathysubramanian, B. (2017). Deep Learning for Flow Sculpting: Insights into Efficient Learning using Scientific Simulation Data. Scientific Reports, 7(1), 46368. [CrossRef]
- Stringer, C., Wang, T., Michaelos, M., & Pachitariu, M. (2021). Cellpose: A generalist algorithm for cellular segmentation. Nature Methods, 18(1), 100–106. [CrossRef]
- Su, S.-H., Song, Y., Stephens, A., Situ, M., McCloskey, M. C., McGrath, J. L., Andjelkovic, A. V., Singer, B. H., & Kurabayashi, K. (2023). A tissue chip with integrated digital immunosensors: In situ brain endothelial barrier cytokine secretion monitoring. Biosensors and Bioelectronics, 224, 115030. [CrossRef]
- Syama, S., & Mohanan, P. V. (2021). Microfluidic based human-on-a-chip: A revolutionary technology in scientific research. Trends in Food Science & Technology, 110, 711–728. [CrossRef]
- Ulyanov, D., Vedaldi, A., & Lempitsky, V. (2018). Deep Image Prior. 9446–9454. https://openaccess.thecvf.com/content_cvpr_2018/html/Ulyanov_Deep_Image_Prior_CVPR_2018_paper.html.
- Wang, J., Zhang, N., Chen, J., Su, G., Yao, H., Ho, T.-Y., & Sun, L. (2021). Predicting the fluid behavior of random microfluidic mixers using convolutional neural networks. Lab on a Chip, 21(2), 296–309. [CrossRef]
- Zhang, Z., Chen, L., Wang, Y., Zhang, T., Chen, Y.-C., & Yoon, E. (2019). Label-Free Estimation of Therapeutic Efficacy on 3D Cancer Spheres Using Convolutional Neural Network Image Analysis. Analytical Chemistry, 91(21), 14093–14100. [CrossRef]
- Zhao, M., Li, M., Peng, S.-L., & Li, J. (2022). A Novel Deep Learning Model Compression Algorithm. Electronics, 11(7), 1066. [CrossRef]
- Zhu, X. (Jerry). (2005). Semi-Supervised Learning Literature Survey [Technical Report]. University of Wisconsin-Madison Department of Computer Sciences. https://minds.wisconsin.edu/handle/1793/60444.
- Zhuang, F., Qi, Z., Duan, K., Xi, D., Zhu, Y., Zhu, H., Xiong, H., & He, Q. (2021). A Comprehensive Survey on Transfer Learning. Proceedings of the IEEE, 109(1), 43–76. [CrossRef]
Figure 1.
A comparison of different models used in clinical trials. Source image from Koyilot et al., 2022.
Figure 1.
A comparison of different models used in clinical trials. Source image from Koyilot et al., 2022.
Figure 2.
Visual model of the convolutional process. The kernel matrix performs a dot product with the image patch to extract key features. Source image from Salehi et al., 2023.
Figure 2.
Visual model of the convolutional process. The kernel matrix performs a dot product with the image patch to extract key features. Source image from Salehi et al., 2023.
Figure 3.
Visual example of pooling methods. Source image from Salehi et al., 2023.
Figure 3.
Visual example of pooling methods. Source image from Salehi et al., 2023.
Figure 4.
Visual example of semantic segmentation of an image using FCNs. From left to right and top to bottom, the first segmentation shows the object masks. The second segmentation corresponds to the object parts (e.g., body parts, mug parts, table parts). The third segmentation shows parts of the heads (e.g., eyes, mouth, and nose). Source image from Liu et al., 2018.
Figure 4.
Visual example of semantic segmentation of an image using FCNs. From left to right and top to bottom, the first segmentation shows the object masks. The second segmentation corresponds to the object parts (e.g., body parts, mug parts, table parts). The third segmentation shows parts of the heads (e.g., eyes, mouth, and nose). Source image from Liu et al., 2018.
Figure 5.
CNNs can determine the pillar sequence needed to create the chosen deformed flow. Source image from Lore et al., 2015.
Figure 5.
CNNs can determine the pillar sequence needed to create the chosen deformed flow. Source image from Lore et al., 2015.
Figure 6.
Super-resolution and image quality enhancement using deep learning. (a) Fluorescence microscopy image. (b) Zoomed-in region of (a) showing a network perform super-resolution. (c) Super-resolution of a bright-field microscope image. (d) Super-resolution of an image created using digital holographic microscopy. (e) Super-resolution of an SEM image. Source image from De Haan et al., 2020.
Figure 6.
Super-resolution and image quality enhancement using deep learning. (a) Fluorescence microscopy image. (b) Zoomed-in region of (a) showing a network perform super-resolution. (c) Super-resolution of a bright-field microscope image. (d) Super-resolution of an image created using digital holographic microscopy. (e) Super-resolution of an SEM image. Source image from De Haan et al., 2020.
Figure 7.
Visual model of tracking individual cells through video sequences. Source image from reference (Mencattini et al., 2020).
Figure 7.
Visual model of tracking individual cells through video sequences. Source image from reference (Mencattini et al., 2020).
Figure 8.
Visual model of CNN tracking a cell. (A) Time-lapse images of a migrating cell. (B) Calculated net displacement of x and y. Source image from Nishimoto et al., 2019.
Figure 8.
Visual model of CNN tracking a cell. (A) Time-lapse images of a migrating cell. (B) Calculated net displacement of x and y. Source image from Nishimoto et al., 2019.
Figure 9.
Visual data from Viola et al., 2023 (A) Through segmentation of the cells, Cellpose counted the number of alive and dead cells. (B) The Cellpose counted the total number of cells. (C) A graph comparing the percentage of dead cells between the control and experimental groups was created with the data from the Cellpose. Source image from Viola et al., 2023.
Figure 9.
Visual data from Viola et al., 2023 (A) Through segmentation of the cells, Cellpose counted the number of alive and dead cells. (B) The Cellpose counted the total number of cells. (C) A graph comparing the percentage of dead cells between the control and experimental groups was created with the data from the Cellpose. Source image from Viola et al., 2023.
Figure 10.
The inclusion of a CNN to analyze images from a tumor-on-a-chip to quickly determine drug efficacy. Reprinted (adapted) with permission from Zhang et al., 2019. Copyright 2019 American Chemical Society.
Figure 10.
The inclusion of a CNN to analyze images from a tumor-on-a-chip to quickly determine drug efficacy. Reprinted (adapted) with permission from Zhang et al., 2019. Copyright 2019 American Chemical Society.
Figure 11.
Trained a CNN to predict parameters that influence the growth of the tumor based on glioblastoma-on-a-chip images with high accuracy. Source image from Pérez-Aliacar et al., 2021.
Figure 11.
Trained a CNN to predict parameters that influence the growth of the tumor based on glioblastoma-on-a-chip images with high accuracy. Source image from Pérez-Aliacar et al., 2021.
Figure 12.
(a) Visual model of proposed capillary connection and (b) use of a microfluidic mother to create HoCs. (c) Visual model of an HoC with many simulated organs. Source image from Picollet-D’hahan et al., 2021.
Figure 12.
(a) Visual model of proposed capillary connection and (b) use of a microfluidic mother to create HoCs. (c) Visual model of an HoC with many simulated organs. Source image from Picollet-D’hahan et al., 2021.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).