Submitted:
25 June 2024
Posted:
27 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
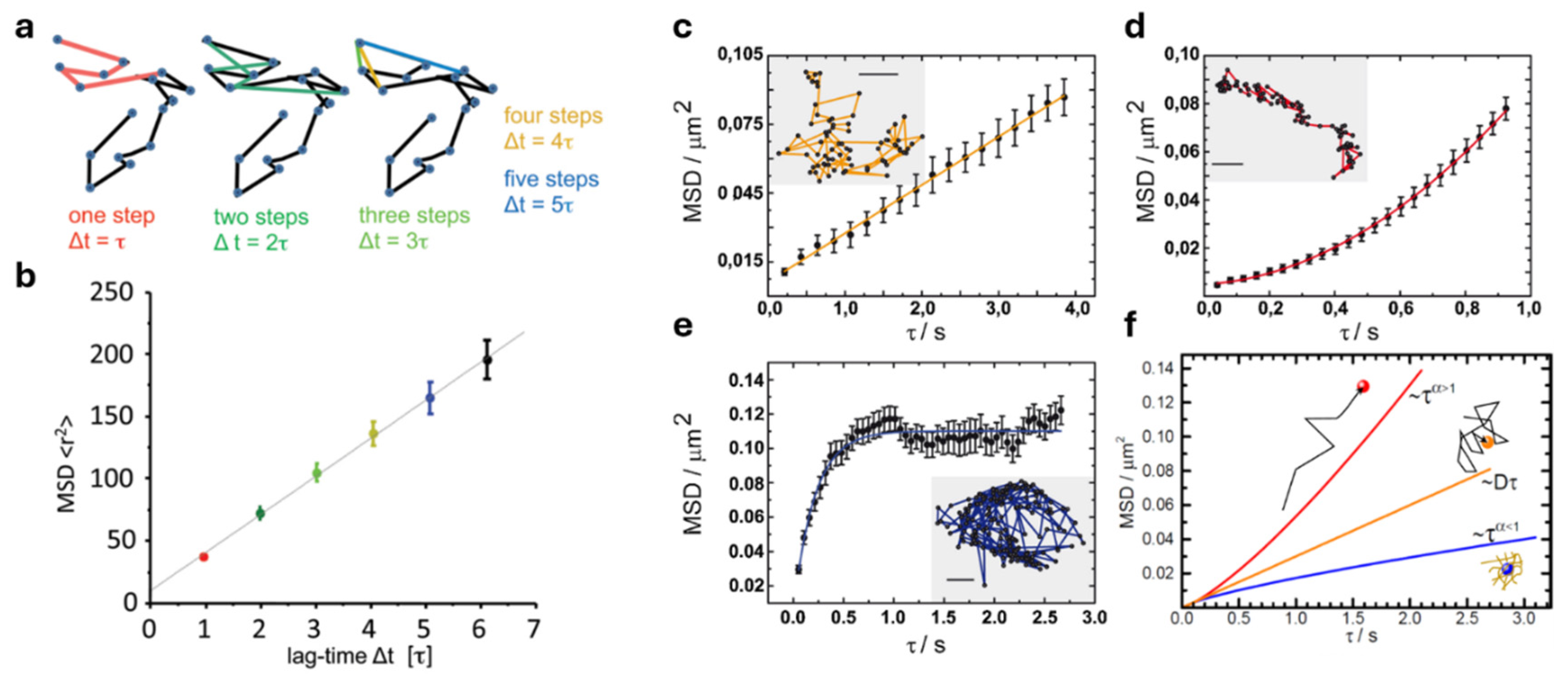
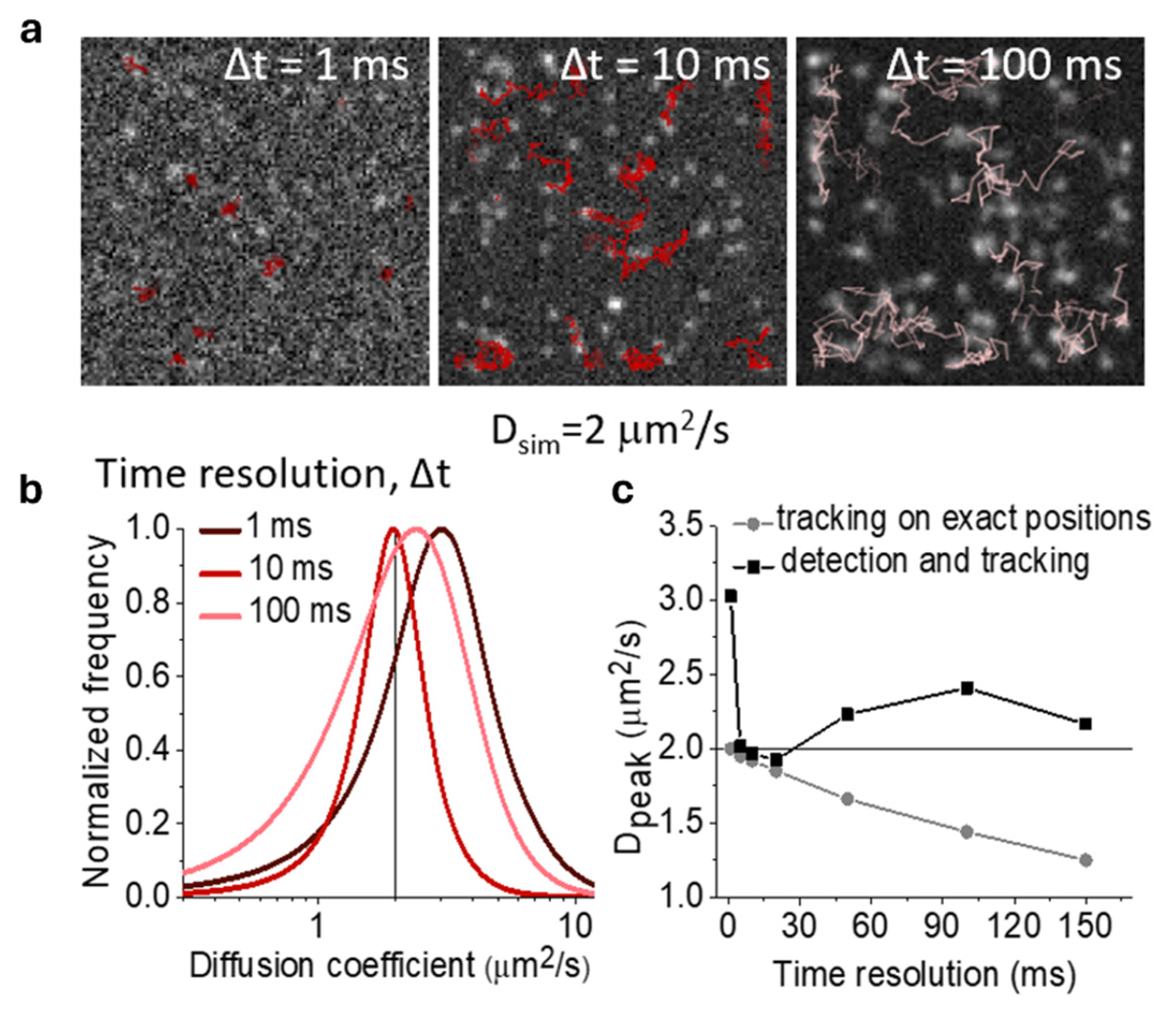
2. Mean Square Displacement (MSD) Analysis
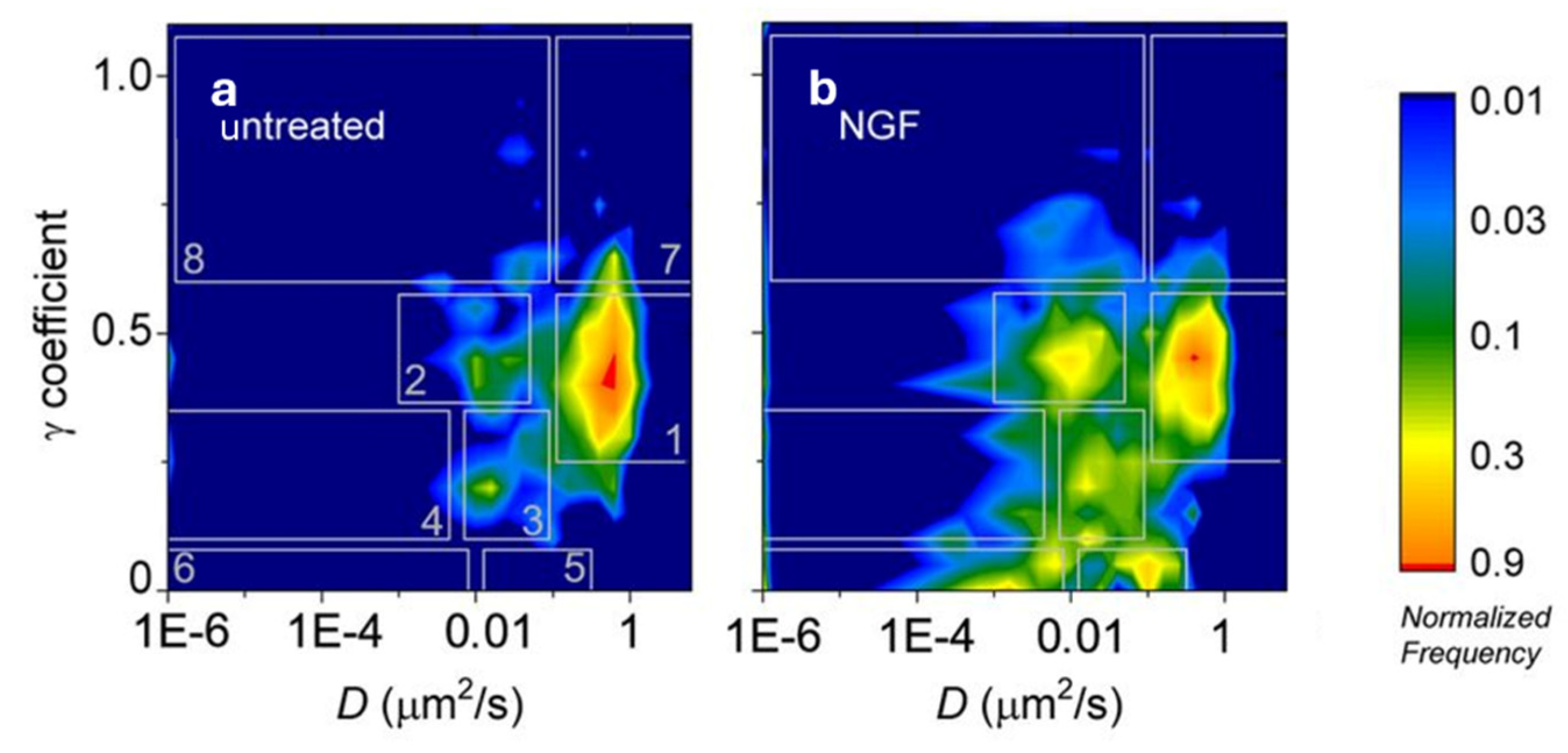
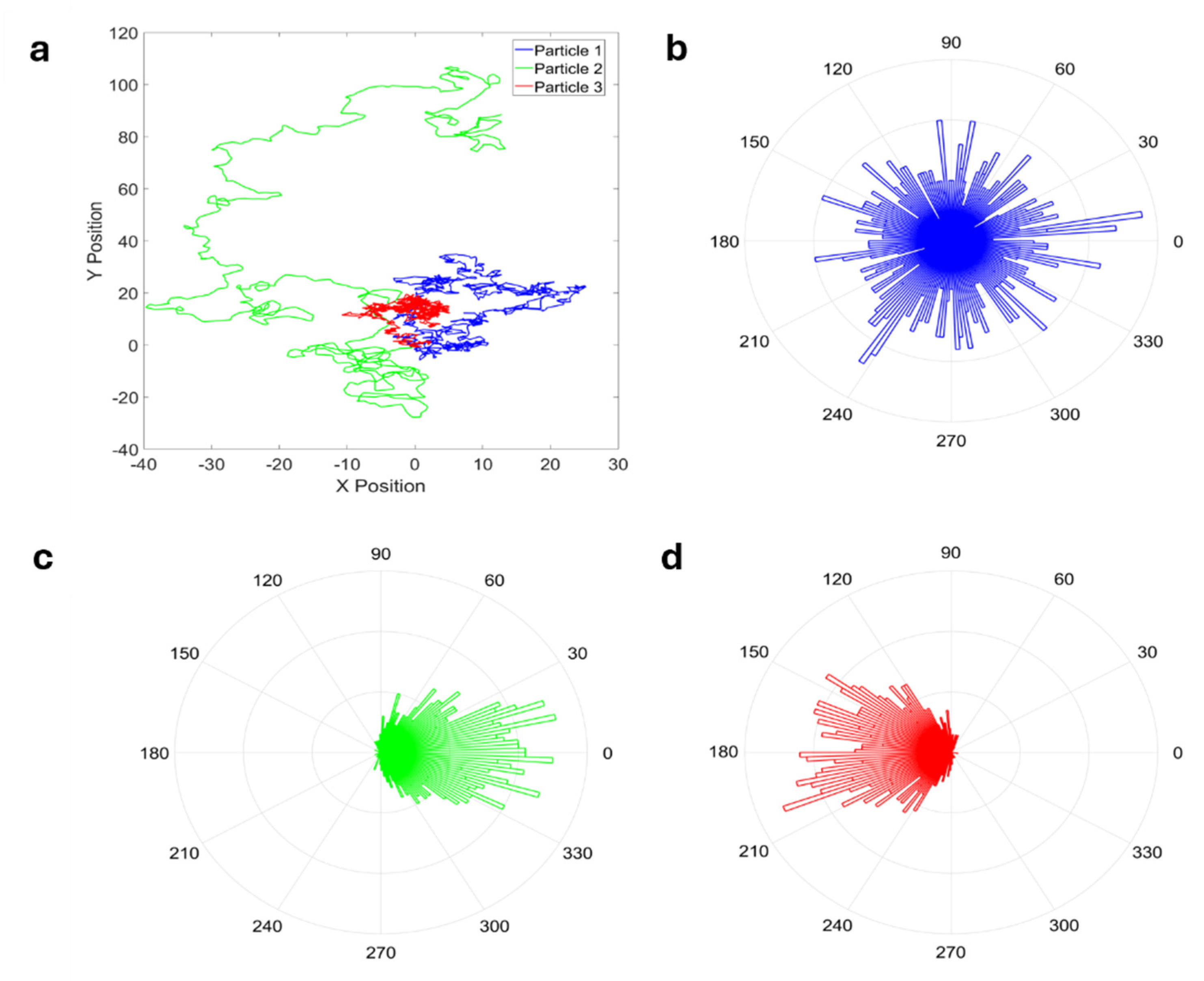
3. Alternatives to MSD Based on Classical Statistics
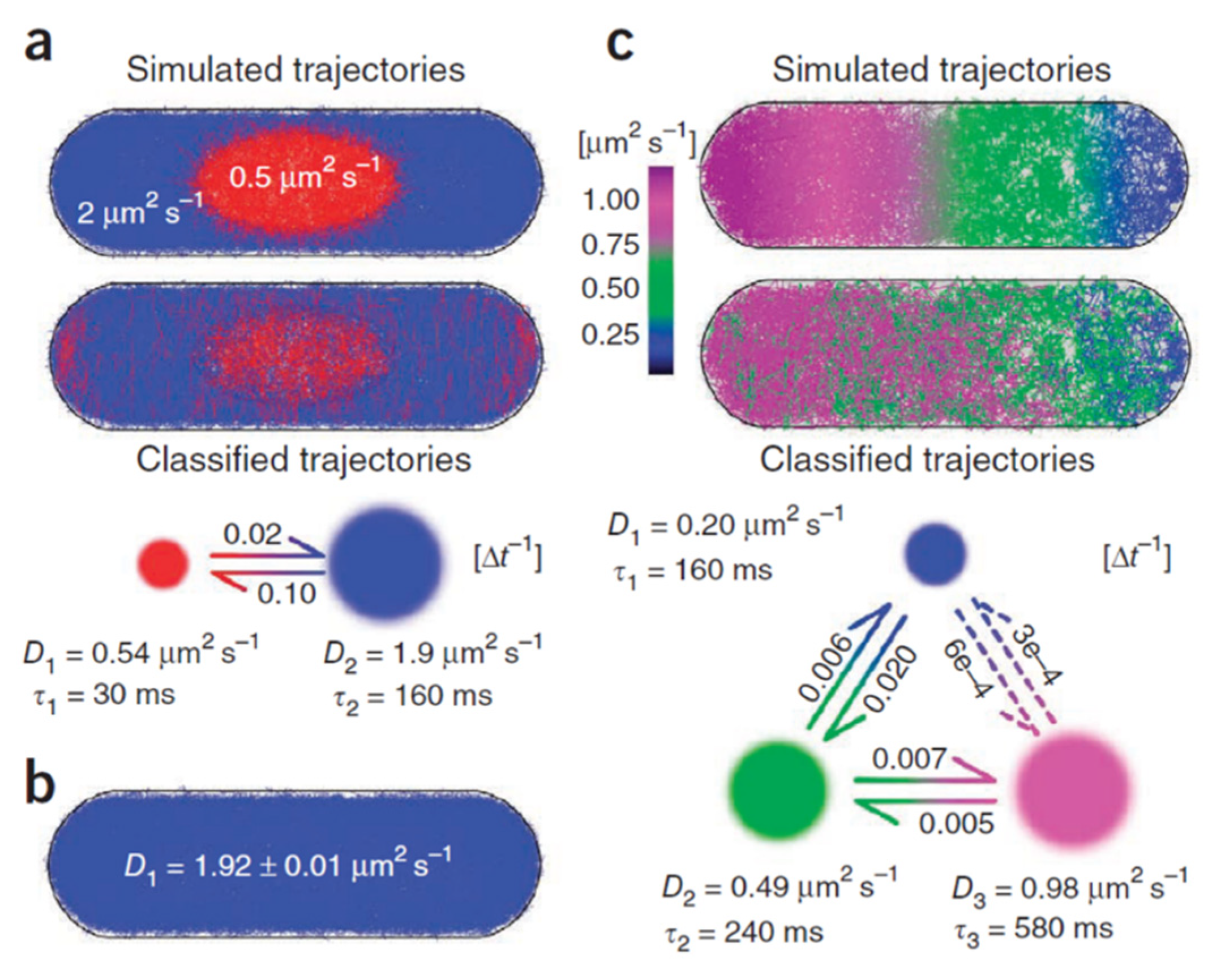
4. Markov Modelling
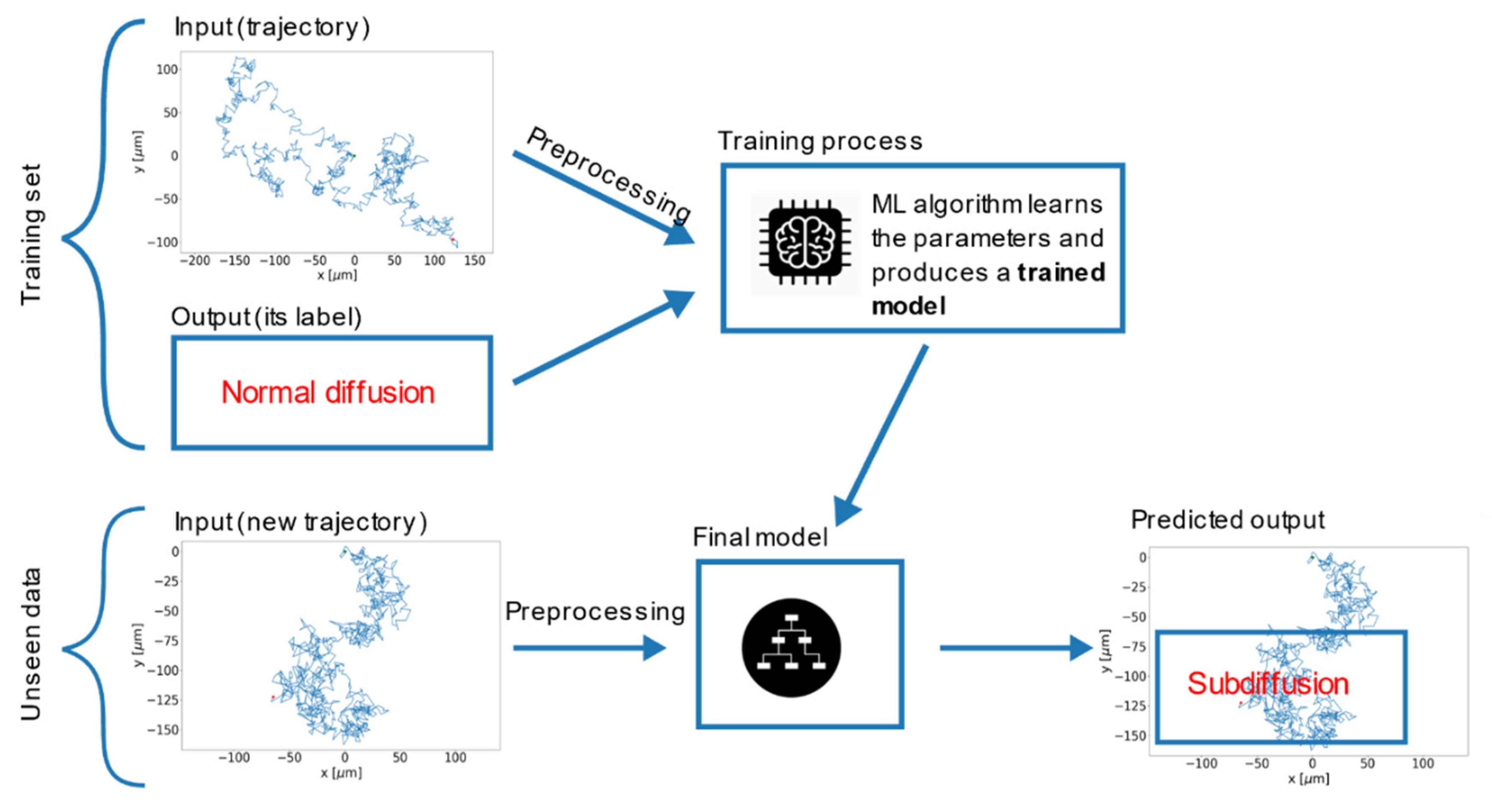
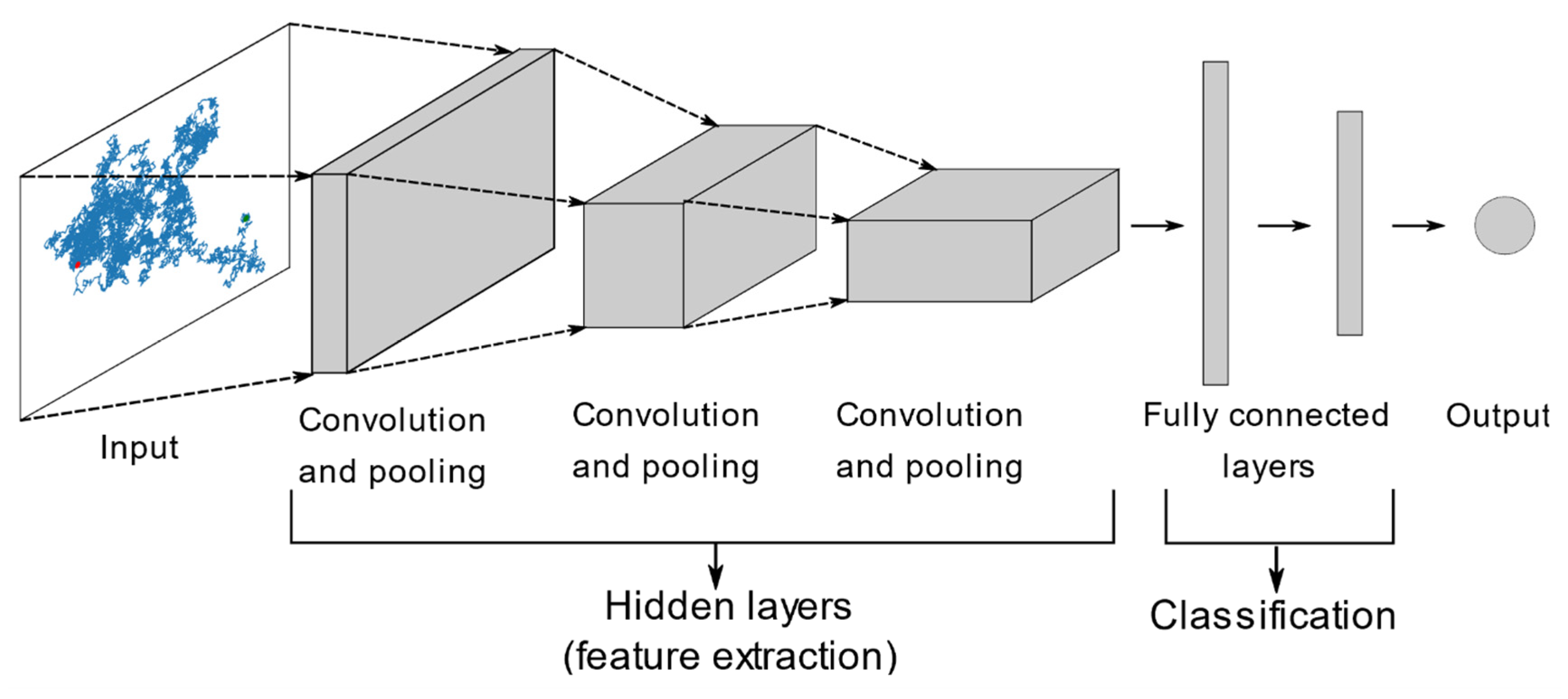
5. Machine Learning Analysis
6. Discussion and Conclusions
Funding
Conflicts of Interest
References
- Wang, Z.; Wang, X.; Zhang, Y.; Xu, W.; Han, X. Principles and Applications of Single Particle Tracking in Cell Research. Small 2021, 17, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, L.; Bonsignore, F.; Gobbo, F.; Amodeo, R.; Calvello, M.; Jacob, A.; Signore, G.; Schirripa Spagnolo, C.; Porciani, D.; Mainardi, M.; et al. Fast-Diffusing P75 NTR Monomers Support Apoptosis and Growth Cone Collapse by Neurotrophin Ligands. Proceedings of the National Academy of Sciences 2019, 116, 21563–21572. [Google Scholar] [CrossRef] [PubMed]
- Ruthardt, N.; Lamb, D.C.; Bräuchle, C. Single-Particle Tracking as a Quantitative Microscopy-Based Approach to Unravel Cell Entry Mechanisms of Viruses and Pharmaceutical Nanoparticles. Molecular Therapy 2011, 19, 1199–1211. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, L.; Luin, S.; Bonsignore, F.; de Nadai, T.; Beltram, F.; Cattaneo, A. Ligand-Induced Dynamics of Neurotrophin Receptors Investigated by Single-Molecule Imaging Approaches. Int J Mol Sci 2015, 16, 1949–1979. [Google Scholar] [CrossRef] [PubMed]
- Schirripa Spagnolo, C.; Moscardini, A.; Amodeo, R.; Beltram, F.; Luin, S. Optimized Two-Color Single-Molecule Tracking of Fast-Diffusing Membrane Receptors. Adv Opt Mater 2023, 2302012. [Google Scholar] [CrossRef]
- Schirripa Spagnolo, C.; Luin, S. Setting up Multicolour TIRF Microscopy down to the Single Molecule Level. Biomol Concepts 2023, 14. [Google Scholar] [CrossRef] [PubMed]
- Schirripa Spagnolo, C.; Luin, S. Choosing the Probe for Single-Molecule Fluorescence Microscopy. Int J Mol Sci 2022, 23, 14949. [Google Scholar] [CrossRef]
- Schirripa Spagnolo, C.; Moscardini, A.; Amodeo, R.; Beltram, F.; Luin, S. Quantitative Determination of Fluorescence Labeling Implemented in Cell Cultures. BMC Biol 2023, 21, 190. [Google Scholar] [CrossRef]
- Manzo, C.; Garcia-Parajo, M.F. A Review of Progress in Single Particle Tracking: From Methods to Biophysical Insights. Reports on Progress in Physics 2015, 78. [Google Scholar] [CrossRef]
- Jaqaman, K.; Loerke, D.; Mettlen, M.; Kuwata, H.; Grinstein, S.; Schmid, S.L.; Danuser, G. Robust Single-Particle Tracking in Live-Cell Time-Lapse Sequences. Nat Methods 2008, 5, 695–702. [Google Scholar] [CrossRef]
- Manzo, C.; Garcia-Parajo, M.F. A Review of Progress in Single Particle Tracking: From Methods to Biophysical Insights. Reports on Progress in Physics 2015, 78. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, X.; Zhang, Y.; Xu, W.; Han, X. Principles and Applications of Single Particle Tracking in Cell Research. Small 2021, 17, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Kepten, E.; Bronshtein, I.; Garini, Y. Improved Estimation of Anomalous Diffusion Exponents in Single-Particle Tracking Experiments. Phys Rev E Stat Nonlin Soft Matter Phys 2013, 87, 052713. [Google Scholar] [CrossRef] [PubMed]
- Kepten, E.; Weron, A.; Sikora, G.; Burnecki, K.; Garini, Y. Guidelines for the Fitting of Anomalous Diffusion Mean Square Displacement Graphs from Single Particle Tracking Experiments. PLoS One 2015, 10, e0117722. [Google Scholar] [CrossRef] [PubMed]
- Gal, N.; Lechtman-Goldstein, D.; Weihs, D. Particle Tracking in Living Cells: A Review of the Mean Square Displacement Method and Beyond. Rheol Acta 2013, 52, 425–443. [Google Scholar] [CrossRef]
- Aaron, J.; Wait, E.; DeSantis, M.; Chew, T.L. Practical Considerations in Particle and Object Tracking and Analysis. Curr Protoc Cell Biol 2019, 83, e88. [Google Scholar] [CrossRef]
- Vestergaard, C.L.; Blainey, P.C.; Flyvbjerg, H. Single-Particle Trajectories Reveal Two-State Diffusion-Kinetics of HOGG1 Proteins on DNA. Nucleic Acids Res 2018, 46, 2446–2458. [Google Scholar] [CrossRef]
- Cairo, C.W.; Das, R.; Albohy, A.; Baca, Q.J.; Pradhan, D.; Morrow, J.S.; Coombs, D.; Golan, D.E. Dynamic Regulation of CD45 Lateral Mobility by the Spectrin-Ankyrin Cytoskeleton of T Cells. Journal of Biological Chemistry 2010, 285, 11392–11401. [Google Scholar] [CrossRef]
- Bewerunge, J.; Ladadwa, I.; Platten, F.; Zunke, C.; Heuer, A.; Egelhaaf, S.U. Time- and Ensemble-Averages in Evolving Systems: The Case of Brownian Particles in Random Potentials. Physical Chemistry Chemical Physics 2016, 18, 18887–18895. [Google Scholar] [CrossRef]
- Kusumi, A.; Sako, Y.; Yamamoto, M. Confined Lateral Diffusion of Membrane Receptors as Studied by Single Particle Tracking (Nanovid Microscopy). Effects of Calcium-Induced Differentiation in Cultured Epithelial Cells. Biophys J 1993, 65, 2021–2040. [Google Scholar] [CrossRef]
- Callegari, A.; Luin, S.; Marchetti, L.; Duci, A.; Cattaneo, A.; Beltram, F. Single Particle Tracking of Acyl Carrier Protein (ACP)-Tagged TrkA Receptors in PC12nnr5 Cells. J Neurosci Methods 2012, 204, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, S.; Sarkar, P.; Preira, P.; Salomé, L.; Chattopadhyay, A. Cholesterol-Dependent Dynamics of the Serotonin1AReceptor Utilizing Single Particle Tracking: Analysis of Diffusion Modes. Journal of Physical Chemistry B 2022, 2022, 6690. [Google Scholar] [CrossRef]
- Triller, A.; Choquet, D. New Concepts in Synaptic Biology Derived from Single-Molecule Imaging. Neuron 2008, 59, 359–374. [Google Scholar] [CrossRef] [PubMed]
- Bannai, H.; Lévi, S.; Schweizer, C.; Dahan, M.; Triller, A. Imaging the Lateral Diffusion of Membrane Molecules with Quantum Dots. Nat Protoc 2007, 1, 2628–2634. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, L.; Callegari, A.; Luin, S.; Signore, G.; Viegi, A.; Beltram, F.; Cattaneo, A. Ligand Signature in the Membrane Dynamics of Single TrkA Receptor Molecules. J Cell Sci 2013, 126, 4445–4456. [Google Scholar] [CrossRef]
- Weiss, M. Resampling Single-Particle Tracking Data Eliminates Localization Errors and Reveals Proper Diffusion Anomalies. Phys Rev E 2019, 100, 042125. [Google Scholar] [CrossRef] [PubMed]
- Metz, M.J.; Pennock, R.L.; Krapf, D.; Hentges, S.T. Temporal Dependence of Shifts in Mu Opioid Receptor Mobility at the Cell Surface after Agonist Binding Observed by Single-Particle Tracking. Sci Rep 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Bronshtein, I.; Kanter, I.; Kepten, E.; Lindner, M.; Berezin, S.; Shav-Tal, Y.; Garini, Y. Exploring Chromatin Organization Mechanisms through Its Dynamic Properties. Nucleus 2016, 7, 27–33. [Google Scholar] [CrossRef]
- Jobin, M.L.; Siddig, S.; Koszegi, Z.; Lanoiselée, Y.; Khayenko, V.; Sungkaworn, T.; Werner, C.; Seier, K.; Misigaiski, C.; Mantovani, G.; et al. Filamin A Organizes Γ-aminobutyric Acid Type B Receptors at the Plasma Membrane. Nature Communications 2023 14:1 2023, 14, 1–14. [Google Scholar] [CrossRef]
- Regner, B.M.; Vučinić, D.; Domnisoru, C.; Bartol, T.M.; Hetzer, M.W.; Tartakovsky, D.M.; Sejnowski, T.J. Anomalous Diffusion of Single Particles in Cytoplasm. Biophys J 2013, 104, 1652. [Google Scholar] [CrossRef]
- Izeddin, I.; Récamier, V.; Bosanac, L.; Cissé, I.I.; Boudarene, L.; Dugast-Darzacq, C.; Proux, F.; Bénichou, O.; Voituriez, R.; Bensaude, O.; et al. Single-Molecule Tracking in Live Cells Reveals Distinct Target-Search Strategies of Transcription Factors in the Nucleus. Elife 2014, 2014. [Google Scholar] [CrossRef]
- Notelaers, K.; Rocha, S.; Paesen, R.; Smisdom, N.; De Clercq, B.; Meier, J.C.; Rigo, J.M.; Hofkens, J.; Ameloot, M. Analysis of A3 GlyR Single Particle Tracking in the Cell Membrane. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 2014, 1843, 544–553. [Google Scholar] [CrossRef] [PubMed]
- Woringer, M.; Izeddin, I.; Favard, C.; Berry, H. Anomalous Subdiffusion in Living Cells: Bridging the Gap Between Experiments and Realistic Models Through Collaborative Challenges. Front Phys 2020, 8, 499989. [Google Scholar] [CrossRef]
- Burov, S.; Jeon, J.H.; Metzler, R.; Barkai, E. Single Particle Tracking in Systems Showing Anomalous Diffusion: The Role of Weak Ergodicity Breaking. Physical Chemistry Chemical Physics 2011, 13, 1800–1812. [Google Scholar] [CrossRef] [PubMed]
- Metzler, R.; Jeon, J.H.; Cherstvy, A.G.; Barkai, E. Anomalous Diffusion Models and Their Properties: Non-Stationarity, Non-Ergodicity, and Ageing at the Centenary of Single Particle Tracking. Physical Chemistry Chemical Physics 2014, 16, 24128–24164. [Google Scholar] [CrossRef]
- Krapf, D. Mechanisms Underlying Anomalous Diffusion in the Plasma Membrane. Curr Top Membr 2015, 75, 167–207. [Google Scholar] [CrossRef] [PubMed]
- Clarke, D.T.; Martin-Fernandez, M.L. A Brief History of Single-Particle Tracking of the Epidermal Growth Factor Receptor. Methods Protoc 2019, 2, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Treppiedi, D.; Jobin, M.L.; Peverelli, E.; Giardino, E.; Sungkaworn, T.; Zabel, U.; Arosio, M.; Spada, A.; Mantovani, G.; Calebiro, D. Single-Molecule Microscopy Reveals Dynamic FLNA Interactions Governing SSTR2 Clustering and Internalization. Endocrinology 2018, 159, 2953–2965. [Google Scholar] [CrossRef] [PubMed]
- Buenaventura, T.; Bitsi, S.; Laughlin, W.E.; Burgoyne, T.; Lyu, Z.; Oqua, A.I.; Norman, H.; McGlone, E.R.; Klymchenko, A.S.; Corrêa, I.R.; et al. Agonist-Induced Membrane Nanodomain Clustering Drives GLP-1 Receptor Responses in Pancreatic Beta Cells. PLoS Biol 2019, 17. [Google Scholar] [CrossRef]
- Drakopoulos, A.; Koszegi, Z.; Lanoiselée, Y.; Hübner, H.; Gmeiner, P.; Calebiro, D.; Decker, M. Investigation of Inactive-State κ Opioid Receptor Homodimerization via Single-Molecule Microscopy Using New Antagonistic Fluorescent Probes. J Med Chem 2020, 63, 3596–3609. [Google Scholar] [CrossRef]
- Savin, T.; Doyle, P.S. Static and Dynamic Errors in Particle Tracking Microrheology. Biophys J 2005, 88, 623–638. [Google Scholar] [CrossRef] [PubMed]
- Deschout, H.; Neyts, K.; Braeckmans, K. The Influence of Movement on the Localization Precision of Sub-Resolution Particles in Fluorescence Microscopy. J Biophotonics 2012, 5, 97–109. [Google Scholar] [CrossRef]
- Berglund, A.J. Statistics of Camera-Based Single-Particle Tracking. Phys Rev E Stat Nonlin Soft Matter Phys 2010, 82. [Google Scholar] [CrossRef] [PubMed]
- Backlund, M.P.; Joyner, R.; Moerner, W.E. Chromosomal Locus Tracking with Proper Accounting of Static and Dynamic Errors. Phys Rev E Stat Nonlin Soft Matter Phys 2015, 91. [Google Scholar] [CrossRef] [PubMed]
- Destainville, N.; Salomé, L. Quantification and Correction of Systematic Errors Due to Detector Time-Averaging in Single-Molecule Tracking Experiments. Biophys J 2006, 90, L17. [Google Scholar] [CrossRef]
- Ernst, D.; Köhler, J. Measuring a Diffusion Coefficient by Single-Particle Tracking: Statistical Analysis of Experimental Mean Squared Displacement Curves. Physical Chemistry Chemical Physics 2012, 15, 845–849. [Google Scholar] [CrossRef] [PubMed]
- Michalet, X. Mean Square Displacement Analysis of Single-Particle Trajectories with Localization Error: Brownian Motion in an Isotropic Medium. Phys Rev E Stat Nonlin Soft Matter Phys 2010, 82, 041914. [Google Scholar] [CrossRef]
- Immune Receptors. 2011, 748. 748. [CrossRef]
- Bayle, V.; Fiche, J.B.; Burny, C.; Platre, M.P.; Nollmann, M.; Martinière, A.; Jaillais, Y. Single-Particle Tracking Photoactivated Localization Microscopy of Membrane Proteins in Living Plant Tissues. Nature Protocols 2021 16:3 2021, 16, 1600–1628. [Google Scholar] [CrossRef]
- Mascalchi, P.; Haanappel, E.; Carayon, K.; Mazères, S.; Salomé, L. Probing the Influence of the Particle in Single Particle Tracking Measurements of Lipid Diffusion. Soft Matter 2012, 8, 4462–4470. [Google Scholar] [CrossRef]
- Shrivastava, S.; Sarkar, P.; Preira, P.; Salomé, L.; Chattopadhyay, A. Role of Actin Cytoskeleton in Dynamics and Function of the Serotonin1A Receptor. Biophys J 2020, 118, 944–956. [Google Scholar] [CrossRef]
- Ritchie, K.; Shan, X.Y.; Kondo, J.; Iwasawa, K.; Fujiwara, T.; Kusumi, A. Detection of Non-Brownian Diffusion in the Cell Membrane in Single Molecule Tracking. Biophys J 2005, 88, 2266–2277. [Google Scholar] [CrossRef]
- Saxton, M.J. Single-Particle Tracking: Effects of Corrals. Biophys J 1995, 69, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Schirripa Spagnolo, C.; Luin, S. Impact of Temporal Resolution in Single Particle Tracking Analysis. Discover Nano 2024, 19, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ewers, H.; Smith, A.E.; Sbalzarini, I.F.; Lilie, H.; Koumoutsakos, P.; Helenius, A. Single-Particle Tracking of Murine Polyoma Virus-like Particles on Live Cells and Artificial Membranes. Proc Natl Acad Sci U S A 2005, 102, 15110–15115. [Google Scholar] [CrossRef] [PubMed]
- Sbalzarini, I.F.; Koumoutsakos, P. Feature Point Tracking and Trajectory Analysis for Video Imaging in Cell Biology. J Struct Biol 2005, 151, 182–195. [Google Scholar] [CrossRef]
- Siebrasse, J.P.; Djuric, I.; Schulze, U.; Schlüter, M.A.; Pavenstädt, H.; Weide, T.; Kubitscheck, U. Trajectories and Single-Particle Tracking Data of Intracellular Vesicles Loaded with Either SNAP-Crb3A or SNAP-Crb3B. Data Brief 2016, 7, 1665–1669. [Google Scholar] [CrossRef] [PubMed]
- Iwao, R.; Yamaguchi, H.; Niimi, T.; Matsuda, Y. Single-Molecule Tracking Measurement of PDMS Layer during Curing Process. Physica A: Statistical Mechanics and its Applications 2021, 565, 125576. [Google Scholar] [CrossRef]
- Iwao, R.; Yamaguchi, H.; Niimi, T.; Matsuda, Y. Single-Molecule Tracking Measurement of PDMS Layer during Curing Process. Physica A: Statistical Mechanics and its Applications 2021, 565, 125576. [Google Scholar] [CrossRef]
- Padmanabhan, P.; Kneynsberg, A.; Cruz, E.; Briner, A.; Götz, J. Single-Molecule Imaging of Tau Reveals How Phosphorylation Affects Its Movement and Confinement in Living Cells. Mol Brain 2024, 17, 1–5. [Google Scholar] [CrossRef]
- Marchetti, L.; Bonsignore, F.; Amodeo, R.; Schirripa Spagnolo, C.; Moscardini, A.; Gobbo, F.; Cattaneo, A.; Beltram, F.; Luin, S. Single Molecule Tracking and Spectroscopy Unveils Molecular Details in Function and Interactions of Membrane Receptors. In Proceedings of the Single Molecule Spectroscopy and Superresolution Imaging XIV; Gregor, I., Erdmann, R., Koberling, F., Eds.; SPIE, March 5 2021; Vol. 11650, p. 20.
- Ewers, H.; Smith, A.E.; Sbalzarini, I.F.; Lilie, H.; Koumoutsakos, P.; Helenius, A. Single-Particle Tracking of Murine Polyoma Virus-like Particles on Live Cells and Artificial Membranes. Proc Natl Acad Sci U S A 2005, 102, 15110–15115. [Google Scholar] [CrossRef]
- Simson, R.; Sheets, E.D.; Jacobson, K. Detection of Temporary Lateral Confinement of Membrane Proteins Using Single-Particle Tracking Analysis. Biophys J 1995, 69, 989–993. [Google Scholar] [CrossRef] [PubMed]
- Meier, J.; Vannier, C.; Sergé, A.; Triller, A.; Choquet, D. Fast and Reversible Trapping of Surface Glycine Receptors by Gephyrin. Nature Neuroscience 2001 4:3 2001, 4, 253–260. [Google Scholar] [CrossRef]
- Mosqueira, A.; Camino, P.A.; Barrantes, F.J. Cholesterol Modulates Acetylcholine Receptor Diffusion by Tuning Confinement Sojourns and Nanocluster Stability. Scientific Reports 2018 8:1 2018, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Sil, P.; Mateos, N.; Nath, S.; Buschow, S.; Manzo, C.; Suzuki, K.G.N.; Fujiwara, T.; Kusumi, A.; Garcia-Parajo, M.F.; Mayor, S. Dynamic Actin-Mediated Nano-Scale Clustering of CD44 Regulates Its Meso-Scale Organization at the Plasma Membrane. Mol Biol Cell 2020, 31, 561–579. [Google Scholar] [CrossRef]
- Fujiwara, T.K.; Tsunoyama, T.A.; Takeuchi, S.; Kalay, Z.; Nagai, Y.; Kalkbrenner, T.; Nemoto, Y.L.; Chen, L.H.; Shibata, A.C.E.; Iwasawa, K.; et al. Ultrafast Single-Molecule Imaging Reveals Focal Adhesion Nano-Architecture and Molecular Dynamics. Journal of Cell Biology 2023, 222. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.G.N.; Fujiwara, T.K.; Edidin, M.; Kusumi, A. Dynamic Recruitment of Phospholipase Cγ at Transiently Immobilized GPI-Anchored Receptor Clusters Induces IP3–Ca2+ Signaling: Single-Molecule Tracking Study 2. J Cell Biol 2007, 177, 731. [Google Scholar] [CrossRef]
- Scheefhals, N.; Westra, M.; MacGillavry, H.D. MGluR5 Is Transiently Confined in Perisynaptic Nanodomains to Shape Synaptic Function. Nature Communications 2023 14:1 2023, 14, 1–20. [Google Scholar] [CrossRef]
- Huet, S.; Karatekin, E.; Tran, V.S.; Fanget, I.; Cribier, S.; Henry, J.P. Analysis of Transient Behavior in Complex Trajectories: Application to Secretory Vesicle Dynamics. Biophys J 2006, 91, 3542–3559. [Google Scholar] [CrossRef]
- Liu, Y.L.; Perillo, E.P.; Liu, C.; Yu, P.; Chou, C.K.; Hung, M.C.; Dunn, A.K.; Yeh, H.C. Segmentation of 3D Trajectories Acquired by TSUNAMI Microscope: An Application to EGFR Trafficking. Biophys J 2016, 111, 2214–2227. [Google Scholar] [CrossRef] [PubMed]
- De Nadai, T.; Marchetti, L.; Di Rienzo, C.; Calvello, M.; Signore, G.; Di Matteo, P.; Gobbo, F.; Turturro, S.; Meucci, S.; Viegi, A.; et al. Precursor and Mature NGF Live Tracking: One versus Many at a Time in the Axons. Sci Rep 2016, 6. [Google Scholar] [CrossRef]
- Convertino, D.; Fabbri, F.; Mishra, N.; Mainardi, M.; Cappello, V.; Testa, G.; Capsoni, S.; Albertazzi, L.; Luin, S.; Marchetti, L.; et al. Graphene Promotes Axon Elongation through Local Stall of Nerve Growth Factor Signaling Endosomes. Nano Lett 2020, 20, 3633–3641. [Google Scholar] [CrossRef] [PubMed]
- Falconieri, A.; De Vincentiis, S.; Cappello, V.; Convertino, D.; Das, R.; Ghignoli, S.; Figoli, S.; Luin, S.; Català-Castro, F.; Marchetti, L.; et al. Axonal Plasticity in Response to Active Forces Generated through Magnetic Nano-Pulling. Cell Rep 2023, 42. [Google Scholar] [CrossRef] [PubMed]
- Durso, W.; Martins, M.; Marchetti, L.; Cremisi, F.; Luin, S.; Cardarelli, F. Lysosome Dynamic Properties during Neuronal Stem Cell Differentiation Studied by Spatiotemporal Fluctuation Spectroscopy and Organelle Tracking. Int J Mol Sci 2020, 21. [Google Scholar] [CrossRef] [PubMed]
- Vega, A.R.; Freeman, S.A.; Grinstein, S.; Jaqaman, K. Multistep Track Segmentation and Motion Classification for Transient Mobility Analysis. Biophys J 2018, 114, 1018–1025. [Google Scholar] [CrossRef] [PubMed]
- Monnier, N.; Barry, Z.; Park, H.Y.; Su, K.C.; Katz, Z.; English, B.P.; Dey, A.; Pan, K.; Cheeseman, I.M.; Singer, R.H.; et al. Inferring Transient Particle Transport Dynamics in Live Cells. Nat Methods 2015, 12, 838–840. [Google Scholar] [CrossRef]
- Burov, S.; Ali Tabei, S.M.; Huynh, T.; Murrell, M.P.; Philipson, L.H.; Rice, S.A.; Gardel, M.L.; Scherer, N.F.; Dinner, A.R. Distribution of Directional Change as a Signature of Complex Dynamics. Proc Natl Acad Sci U S A 2013, 110, 19689–19694. [Google Scholar] [CrossRef] [PubMed]
- Harrison, A.W.; Kenwright, D.A.; Waigh, T.A.; Woodman, P.G.; Allan, V.J. Modes of Correlated Angular Motion in Live Cells across Three Distinct Time Scales. Phys Biol 2013, 10. [Google Scholar] [CrossRef] [PubMed]
- Pierobon, P.; Achouri, S.; Courty, S.; Dunn, A.R.; Spudich, J.A.; Dahan, M.; Cappello, G. Velocity, Processivity, and Individual Steps of Single Myosin V Molecules in Live Cells. Biophys J 2009, 96, 4268. [Google Scholar] [CrossRef] [PubMed]
- Lakadamyali, M.; Rust, M.J.; Babcock, H.P.; Zhuang, X. Visualizing Infection of Individual Influenza Viruses. Proc Natl Acad Sci U S A 2003, 100, 9280–9285. [Google Scholar] [CrossRef]
- Tejedor, V.; Bénichou, O.; Voituriez, R.; Jungmann, R.; Simmel, F.; Selhuber-Unkel, C.; Oddershede, L.B.; Metzler, R. Quantitative Analysis of Single Particle Trajectories: Mean Maximal Excursion Method. Biophys J 2010, 98, 1364–1372. [Google Scholar] [CrossRef]
- Meroz, Y.; Sokolov, I.M. A Toolbox for Determining Subdiffusive Mechanisms. Phys Rep 2015, 573, 1–29. [Google Scholar] [CrossRef]
- Condamin, S.; Tejedor, V.; Voituriez, R.; Bénichou, O.; Klafter, J. Probing Microscopic Origins of Confined Subdiffusion by First-Passage Observables. Proc Natl Acad Sci U S A 2008, 105, 5675–5680. [Google Scholar] [CrossRef]
- Magdziarz, M.; Klafter, J. Detecting Origins of Subdiffusion: P-Variation Test for Confined Systems. [CrossRef]
- Magdziarz, M.; Weron, A.; Burnecki, K.; Klafter, J. Fractional Brownian Motion versus the Continuous-Time Random Walk: A Simple Test for Subdiffusive Dynamics. Phys Rev Lett 2009, 103, 180602. [Google Scholar] [CrossRef] [PubMed]
- Brodin, A.; Turiv, T.; Nazarenko, V. Anomalous Diffusion: Single Particle Trajectory Analysis. Ukrainian Journal of Physics 2014, 59, 775. [Google Scholar] [CrossRef]
- Weber, S.C.; Thompson, M.A.; Moerner, W.E.; Spakowitz, A.J.; Theriot, J.A. Analytical Tools To Distinguish the Effects of Localization Error, Confinement, and Medium Elasticity on the Velocity Autocorrelation Function. Biophys J 2012, 102, 2443–2450. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.H.; Metzler, R. Fractional Brownian Motion and Motion Governed by the Fractional Langevin Equation in Confined Geometries. Phys Rev E Stat Nonlin Soft Matter Phys 2010, 81. [Google Scholar] [CrossRef]
- Metzler, R.; Jeon, J.H.; Cherstvy, A.G. Non-Brownian Diffusion in Lipid Membranes: Experiments and Simulations. Biochimica et Biophysica Acta (BBA) - Biomembranes 2016, 1858, 2451–2467. [Google Scholar] [CrossRef] [PubMed]
- Sposini, V.; Krapf, D.; Marinari, E.; Sunyer, R.; Ritort, F.; Taheri, F.; Selhuber-Unkel, C.; Benelli, R.; Weiss, M.; Metzler, R.; et al. Towards a Robust Criterion of Anomalous Diffusion. Communications Physics 2022 5:1 2022, 5, 1–10. [Google Scholar] [CrossRef]
- Fox, Z.R.; Barkai, E.; Krapf, D. Aging Power Spectrum of Membrane Protein Transport and Other Subordinated Random Walks. Nature Communications 2021 12:1 2021, 12, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Krapf, D.; Lukat, N.; Marinari, E.; Metzler, R.; Oshanin, G.; Selhuber-Unkel, C.; Squarcini, A.; Stadler, L.; Weiss, M.; Xu, X. Spectral Content of a Single Non-Brownian Trajectory. Phys Rev X 2019, 9, 011019. [Google Scholar] [CrossRef]
- Zhao, X.B.; Zhang, X.; Guo, W. Diffusion of Active Brownian Particles under Quenched Disorder. PLoS One 2024, 19. [Google Scholar] [CrossRef]
- Mardoukhi, Y.; Jeon, J.H.; Metzler, R. Geometry Controlled Anomalous Diffusion in Random Fractal Geometries: Looking beyond the Infinite Cluster. Physical Chemistry Chemical Physics 2015, 17, 30134–30147. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.H.; Metzler, R. Analysis of Short Subdiffusive Time Series: Scatter of the Time-Averaged Mean-Squared Displacement. J Phys A Math Theor 2010, 43, 252001. [Google Scholar] [CrossRef]
- Weron, A.; Janczura, J.; Boryczka, E.; Sungkaworn, T.; Calebiro, D. Statistical Testing Approach for Fractional Anomalous Diffusion Classification. Phys Rev E 2019, 99, 042149. [Google Scholar] [CrossRef] [PubMed]
- Das, R.; Cairo, C.W.; Coombs, D. A Hidden Markov Model for Single Particle Tracks Quantifies Dynamic Interactions between LFA-1 and the Actin Cytoskeleton. PLoS Comput Biol 2009, 5, e1000556. [Google Scholar] [CrossRef]
- Slator, P.J.; Cairo, C.W.; Burroughs, N.J. Detection of Diffusion Heterogeneity in Single Particle Tracking Trajectories Using a Hidden Markov Model with Measurement Noise Propagation. PLoS One 2015, 10, e0140759. [Google Scholar] [CrossRef] [PubMed]
- Chung, I.; Akita, R.; Vandlen, R.; Toomre, D.; Schlessinger, J.; Mellman, I. Spatial Control of EGF Receptor Activation by Reversible Dimerization on Living Cells. Nature 2010 464:7289 2010, 464, 783–787. [Google Scholar] [CrossRef]
- Slator, P.J.; Burroughs, N.J. A Hidden Markov Model for Detecting Confinement in Single-Particle Tracking Trajectories. Biophys J 2018, 115, 1741–1754. [Google Scholar] [CrossRef]
- Röding, M.; Guo, M.; Weitz, D.A.; Rudemo, M.; Särkkä, A. Identifying Directional Persistence in Intracellular Particle Motion Using Hidden Markov Models. Math Biosci 2014, 248, 140–145. [Google Scholar] [CrossRef]
- Persson, F.; Lindén, M.; Unoson, C.; Elf, J. Extracting Intracellular Diffusive States and Transition Rates from Single-Molecule Tracking Data. Nat Methods 2013, 10, 265–269. [Google Scholar] [CrossRef]
- VbSPT Download | SourceForge. Available online: https://sourceforge.net/projects/vbspt/ (accessed on 30 May 2024).
- Sungkaworn, T.; Jobin, M.-L.; Burnecki, K.; Weron, A.; Lohse, M.J.; Calebiro, D. Single-Molecule Imaging Reveals Receptor–G Protein Interactions at Cell Surface Hot Spots. Nature 2017, 550. [Google Scholar] [CrossRef] [PubMed]
- Gormal, R.S.; Padmanabhan, P.; Kasula, R.; Bademosi, A.T.; Coakley, S.; Giacomotto, J.; Blum, A.; Joensuu, M.; Wallis, T.P.; Lo, H.P.; et al. Modular Transient Nanoclustering of Activated Β2-Adrenergic Receptors Revealed by Single-Molecule Tracking of Conformation-Specific Nanobodies. Proc Natl Acad Sci U S A 2020, 117, 30476–30487. [Google Scholar] [CrossRef] [PubMed]
- Falcao, R.C.; Coombs, D. Diffusion Analysis of Single Particle Trajectories in a Bayesian Nonparametrics Framework. Phys Biol 2020, 17. [Google Scholar] [CrossRef] [PubMed]
- GitHub - Rcardim/IHMMSPT: Falcao, Rebeca Cardim, and Daniel Coombs. “Diffusion Analysis of Single Particle Trajectories in a Bayesian Nonparametrics Framework.” BioRxiv (2019): 704049. Available online: https://github.com/rcardim/iHMMSPT (accessed on 25 May 2024).
- Biau, G.; Scornet, E. A Random Forest Guided Tour. Test 2016, 25, 197–227. [Google Scholar] [CrossRef]
- Wagner, T.; Kroll, A.; Haramagatti, C.R.; Lipinski, H.G.; Wiemann, M. Classification and Segmentation of Nanoparticle Diffusion Trajectories in Cellular Micro Environments. PLoS One 2017, 12, e0170165. [Google Scholar] [CrossRef] [PubMed]
- GitHub - Thorstenwagner/Ij-Trajectory-Classifier: This Plugin Segments and Classify Diffusion Trajectories. Available online: https://github.com/thorstenwagner/ij-trajectory-classifier (accessed on 20 May 2024).
- Muñoz-Gil, G.; Garcia-March, M.A.; Manzo, C.; Martín-Guerrero, J.D.; Lewenstein, M. Single Trajectory Characterization via Machine Learning. New J Phys 2020, 22, 013010. [Google Scholar] [CrossRef]
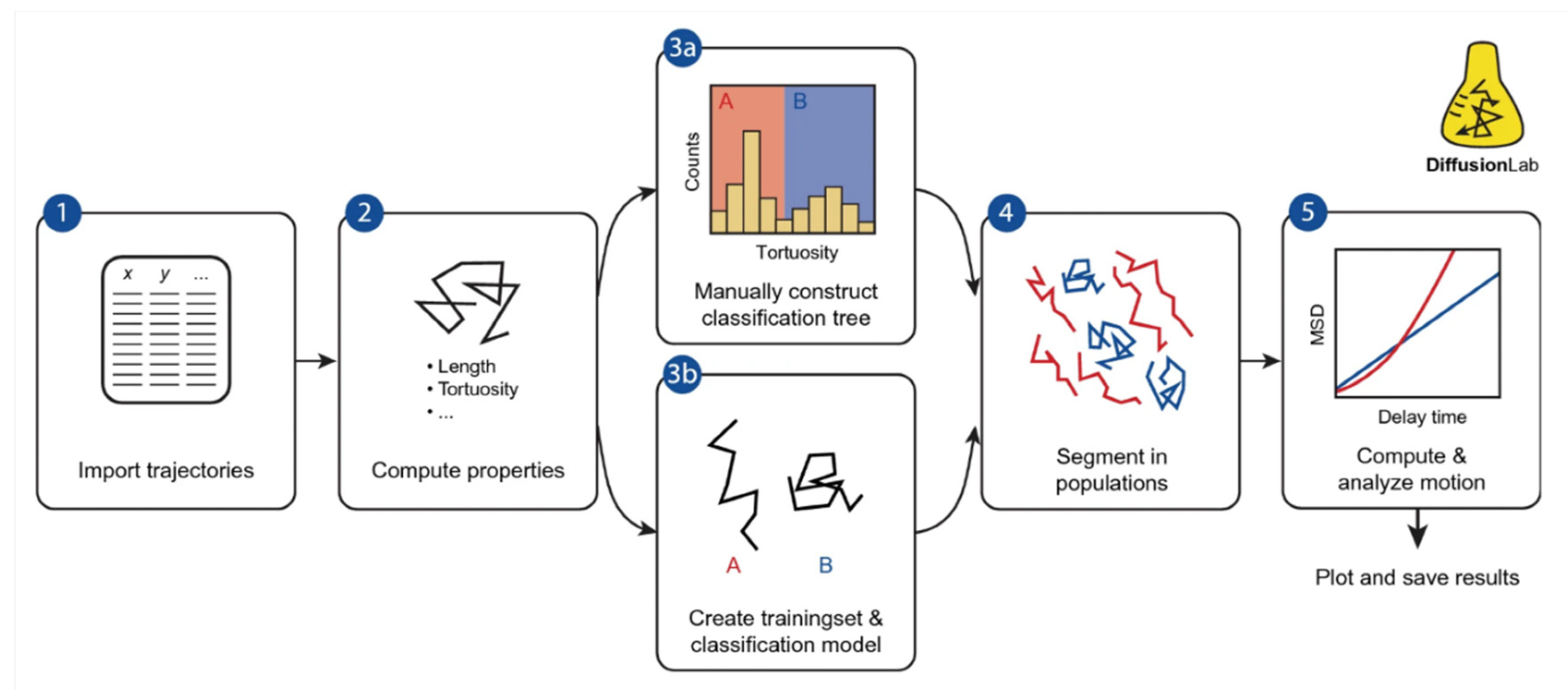
- Maris, J.J.E.; Rabouw, F.T.; Weckhuysen, B.M.; Meirer, F. Classification-Based Motion Analysis of Single-Molecule Trajectories Using DiffusionLab. Sci Rep 2022, 12, 1–10. [Google Scholar] [CrossRef]
- Welcome to DiffusionLab’s Documentation! — DiffusionLab Documentation. Available online: https://diffusionlab.readthedocs.io/en/latest/# (accessed on 22 May 2024).
- GitHub - ErikMaris/DiffusionLab: Single-Molecule Trajectory Analysis. Available online: https://github.com/ErikMaris/DiffusionLab (accessed on 22 May 2024).
- Jaqaman, K.; Loerke, D.; Mettlen, M.; Kuwata, H.; Grinstein, S.; Schmid, S.L.; Danuser, G. Robust Single-Particle Tracking in Live-Cell Time-Lapse Sequences. Nat Methods 2008, 5, 695–702. [Google Scholar] [CrossRef]
- Tinevez, J.Y.; Perry, N.; Schindelin, J.; Hoopes, G.M.; Reynolds, G.D.; Laplantine, E.; Bednarek, S.Y.; Shorte, S.L.; Eliceiri, K.W. TrackMate: An Open and Extensible Platform for Single-Particle Tracking. Methods 2017, 115, 80–90. [Google Scholar] [CrossRef]
- Pinholt, H.D.; Bohr, S.S.R.; Iversen, J.F.; Boomsma, W.; Hatzakis, N.S. Single-Particle Diffusional Fingerprinting: A Machine-Learning Framework for Quantitative Analysis of Heterogeneous Diffusion. Proc Natl Acad Sci U S A 2021, 118, e2104624118. [Google Scholar] [CrossRef]
- Wythoff, B.J. Backpropagation Neural Networks: A Tutorial. Chemometrics and Intelligent Laboratory Systems 1993, 18, 115–155. [Google Scholar] [CrossRef]
- Dosset, P.; Rassam, P.; Fernandez, L.; Espenel, C.; Rubinstein, E.; Margeat, E.; Milhiet, P.E. Automatic Detection of Diffusion Modes within Biological Membranes Using Back-Propagation Neural Network. BMC Bioinformatics 2016, 17, 1–12. [Google Scholar] [CrossRef]
- Kowalek, P.; Loch-Olszewska, H.; Szwabiński, J. Classification of Diffusion Modes in Single-Particle Tracking Data: Feature-Based versus Deep-Learning Approach. Phys Rev E 2019, 100, 032410. [Google Scholar] [CrossRef]
- Gu, J.; Wang, Z.; Kuen, J.; Ma, L.; Shahroudy, A.; Shuai, B.; Liu, T.; Wang, X.; Wang, G.; Cai, J.; et al. Recent Advances in Convolutional Neural Networks. Pattern Recognit 2018, 77, 354–377. [Google Scholar] [CrossRef]
- Sadouk, L. CNN Approaches for Time Series Classification. Time Series Analysis - Data, Methods, and Applications, 2019. [Google Scholar] [CrossRef]
- Gamboa, J.C.B. Deep Learning for Time-Series Analysis. 2017.
- Janczura, J.; Kowalek, P.; Loch-Olszewska, H.; Szwabiński, J.; Weron, A. Classification of Particle Trajectories in Living Cells: Machine Learning versus Statistical Testing Hypothesis for Fractional Anomalous Diffusion. Phys Rev E 2020, 102, 032402. [Google Scholar] [CrossRef]
- Granik, N.; Weiss, L.E.; Nehme, E.; Levin, M.; Chein, M.; Perlson, E.; Roichman, Y.; Shechtman, Y. Single-Particle Diffusion Characterization by Deep Learning. Biophys J 2019, 117, 185–192. [Google Scholar] [CrossRef]
- Muñoz-Gil, G.; Volpe, G.; Garcia-March, M.A.; Aghion, E.; Argun, A.; Hong, C.B.; Bland, T.; Bo, S.; Conejero, J.A.; Firbas, N.; et al. Objective Comparison of Methods to Decode Anomalous Diffusion. Nature Communications 2021 12:1 2021, 12, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Seckler, H.; Szwabiński, J.; Metzler, R. Machine-Learning Solutions for the Analysis of Single-Particle Diffusion Trajectories. Journal of Physical Chemistry Letters 2023, 14, 7910–7923. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Yao, Q.; Huang, Z.; Manzo, C. Extreme Learning Machine for the Characterization of Anomalous Diffusion from Single Trajectories (AnDi-ELM). J Phys A Math Theor 2021, 54, 334002. [Google Scholar] [CrossRef]
- Huang, G.B.; Zhu, Q.Y.; Siew, C.K. Extreme Learning Machine: Theory and Applications. Neurocomputing 2006, 70, 489–501. [Google Scholar] [CrossRef]
- Wang, J.; Lu, S.; Wang, S.H.; Zhang, Y.D. A Review on Extreme Learning Machine. Multimedia Tools and Applications 2021 81:29 2021, 81, 41611–41660. [Google Scholar] [CrossRef]
- Li, D.; Yao, Q.; Huang, Z. WaveNet-Based Deep Neural Networks for the Characterization of Anomalous Diffusion (WADNet). J Phys A Math Theor 2021, 54, 404003. [Google Scholar] [CrossRef]
- Verdier, H.; Duval, M.; Laurent, F.; Cassé, A.; Vestergaard, C.L.; Masson, J.B. Learning Physical Properties of Anomalous Random Walks Using Graph Neural Networks. J Phys A Math Theor 2021, 54, 234001. [Google Scholar] [CrossRef]
- Muñoz-Gil, G.; Bachimanchi, H.; Pineda, J.; Midtvedt, B.; Lewenstein, M.; Metzler, R.; Krapf, D.; Volpe, G.; Manzo, C. Quantitative Evaluation of Methods to Analyze Motion Changes in Single-Particle Experiments. 2023. [CrossRef]
- Challenge 2024 - AnDi Challenge. Available online: http://andi-challenge.org/challenge-2024/ (accessed on 20 May 2024).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




