1. Introduction
SARS-CoV-2 is closely related to the causal agents of the pandemic severe acute respiratory syndrome coronavirus (SARS) and Middle East respiratory syndrome (MERS), and to endemic viruses associated with mild upper respiratory infection syndromes. Brann et al. (2020) reported that the infection with SARS-CoV-2 is associated with a high rate of disturbances in smell and taste perception.[
1] This olfactory disturbance had a prevalence of over 47% during the first waves of infection although it was less prevalent with the omicron virus ranging between 5.8% to 16.7%.[
2,
3]
The inability to smell (anosmia) and the decreased ability to smell (hyposmia) are more common in the human population than expected with estimations of incidence ranging widely between 3 and 20%.[
4] This wide range depends on the age but other risk factors such as viral infections or upper respiratory infections, head trauma, and neurodegenerative diseases are also involved. The problem of anosmia may not seem significant to many patients simply because many patients may not recognize that they have an olfactory disorder and because the olfactory system has significant “plasticity” represented by a strong interrelationship of the olfactory system and the trigeminal sensory system creating together a uniform flavor experience which may persist after the loss of smell sense.[
5]
The crosstalk of the olfactory and gustatory sensory systems is thought to occur at the level of insular cortex. However, the presence of olfactory signal transduction molecules and olfactory receptors in the fungiform taste papilla cells was documented.[
6]
Olfactory sense is important for the human being not only to localize and appreciate food but also to smell dangerous substances such as gas or fire or even rotten food. In spite of this significance, many smell disorders’ patients reported they were not treated properly in their clinics, visiting at least 2 or more clinics before finding a reasonable health care. Landis et al. (2009) reported that 60% of the patients surveyed received unclear or unsatisfactory information about their olfactory problem and its consequences on their health.[
7]
Respiratory viral infections induce alteration in smell known as post-viral olfactory dysfunction (PVOD) which could be usually temporary (short-term), but in some cases may be persistent (long-term). The recovery process in some patients is taking up to 2 years.[
8] Anosmia is one of the symptoms caused by SARS-CoV-2 which was typically noticeable around the beginning of the infection, and was similar to that observed in most other PVODs.[
9] It is worth mentioning, however, that a comparison of smell and taste disorder rates in unvaccinated patients with COVID-19 and Influenza was made and revealed a higher prevalence of smell and taste disorders in patients with COVID-19 when compared to those with Influenza.[
10] McWilliams et al. (2022) surveyed two-years of follow up of recovery from Covid-19-induced anosmia and showed that among the 267 respondents, 38.2% reported complete recovery, 54.3% partial recovery, and 7.5% reported no recovery at all. Additionally, complete recovery of smell sensation has been observed to be age-related. It was significantly higher in those under 40 years old (45.6% compared to 32.9% in those over 40).[
11] Recent attention has focused on the long-term recovery of smell and taste, alongside the application of objective olfactory assessments. In a prospective multicenter study in Europe, Lechien et al. (2021) documented an objective recovery rate of 85% at 60 days and 95% at 6 months following the onset of olfactory dysfunction.[
12] In a recent meta-analysis involving 18 studies, it was determined that persistent self-reported olfactory and gustatory disorders at 180 days were 4.3% and 2%, respectively.[
13] However, there is currently a lack of long-term (>6 months) outcome data on COVID-19-related olfactory dysfunction in Asians.[
14]
Moreover, olfactory dysfunction was also reported in patients infected with SARS-CoV-2 variants. It has been reported that vaccinated people infected with the omicron variant of SARS-CoV-2 had symptoms shorter and milder than those infected with the delta variant. Anosmia was less common in people infected during the omicron wave than during the delta wave.[
15]
Acute complete loss or impairment of smell sensation caused by acute infection with SARS-CoV-2 was unfamiliar to physicians and attracted huge attention to understand the underlying pathophysiology of this phenomenon. The presumed mechanism was through SARS-CoV-2-induced damage to the olfactory epithelium which is similar to that induced by other viruses such as rhinovirus, coronavirus, parainfluenza virus, or Epstein-Barr virus [
8] although many other mechanisms were proposed and challenged.
2. Hypotheses Explaining Anosmia
Although temporary and persistent olfactory dysfunctions have been observed during SARS-CoV-2 infection, the etiology of both conditions remains unclear. It was initially hypothesized that coronaviruses, and may be other viruses, caused transient loss or impairment in the olfactory neurons function mainly due to host immune responses such as inflammation caused by cytokines or due to down regulation of gene expression of odorant receptors and signaling molecules.[
16,
17] It was proposed that injury to peripheral and central nervous system including olfactory cells occurs as a result of massive activation of cytokines.[
18] Acute anosmia was found to be correlated with the duration of SARS-CoV-2 infection and the associated inflammation in the olfactory neuro-epithelium which may explain the prolonged anosmia in some COVID-19 patients. Experimentally, it was shown that complete olfactory dysfunction induced by SARS-CoV-2 in golden Syrian hamsters lasted as long as the virus persisted in the olfactory epithelium and the olfactory bulb. Indeed, virus and inflammatory mediators’ transcripts were detected in the olfactory mucosa of patients who demonstrated long-term persistence of COVID-19-associated anosmia.[
19] In clinical trials, olfactory training and anti-neuroinflammatory therapy achieved positive results in many patients although 15% of the treated patients did not achieve full recovery of the normal olfactory threshold, and almost 5% had no recovery at all.[
20]
Another hypothesis suggested that olfactory dysfunction resulted from elimination of the support cells (i.e., sustentacular and Bowman’s gland cells) and the deleterious consequences of this elimination on neuronal cells, such as decrease of mucus covering the epithelium, decrease of glucose needed for the function of the cilia, and deciliation or retraction of the olfactory cilia.[
21] Moreover, genes encoding uridine diphosphate glucosyltransferases (like UGT2A1 and UGT2A2) were found to differ in different ethnicities and that was taken as an indication of the involvement of these genes in the variable degrees of anosmia observed during virus infection in different ethnic groups. These enzymes metabolize odorants by glucouronidation in order to clear them from the mucosa, preventing their continuous stimulation of the olfactory receptors (i.e., their saturation) and maintaining the high sensitivity of those receptors.[
22] Furthermore, the time of onset of anosmia was much of importance and created some debate and was taken as a factor pointing to the possible mechanism causing anosmia.[
23,
24] It was argued that since anosmia occurs rapidly and may recover within a week time, this may rule out the probability that anosmia is due to loss of olfactory neurons which require minimally 2-3 weeks to regenerate and resume their function.[
24]
Although several potential mechanisms for COVID-19-induced anosmia have been discussed, the pathophysiological pathways including the receptors and the intracellular signal transduction molecules involved in olfaction and affected by viral infection have not been completely elucidated. Emphasizing the mechanisms of virus-induced anosmia may provide therapeutic strategies especially for patients with persistent post-COVID-syndrome. Accordingly, this review sheds the light on the possible role of various types of receptors expressed on olfactory epithelium and their potential roles in signal transduction in anosmia.
In this review, we focus on anosmia associated with viral infections like SARS-CoV-2, and in particular, we reviewed the literature available on the receptors involved in the smell sense and on the pathways which signals take in order to conceive the smell of an odorant in the brain. The review describes the recent advances that emerged from molecular, physiological, genetic and imaging studies, and highlights many of the remaining questions about post viral olfactory disturbance. We examined the entry pathways of SARS-CoV-2 through sustentacular cells of the olfactory epithelium and connected this to the mechanism of how it disrupts the action potential generation in olfactory sensory neurons. We brought into focus the significance of ion signaling in one of the pathways by which sustentacular cells and olfactory sensory neurons are affecting each other to detect odorants and how their functionality becomes adversely affected in response to SARS-CoV-2 infection-induced anosmia.
3. The Organization of the Olfactory Epithelium
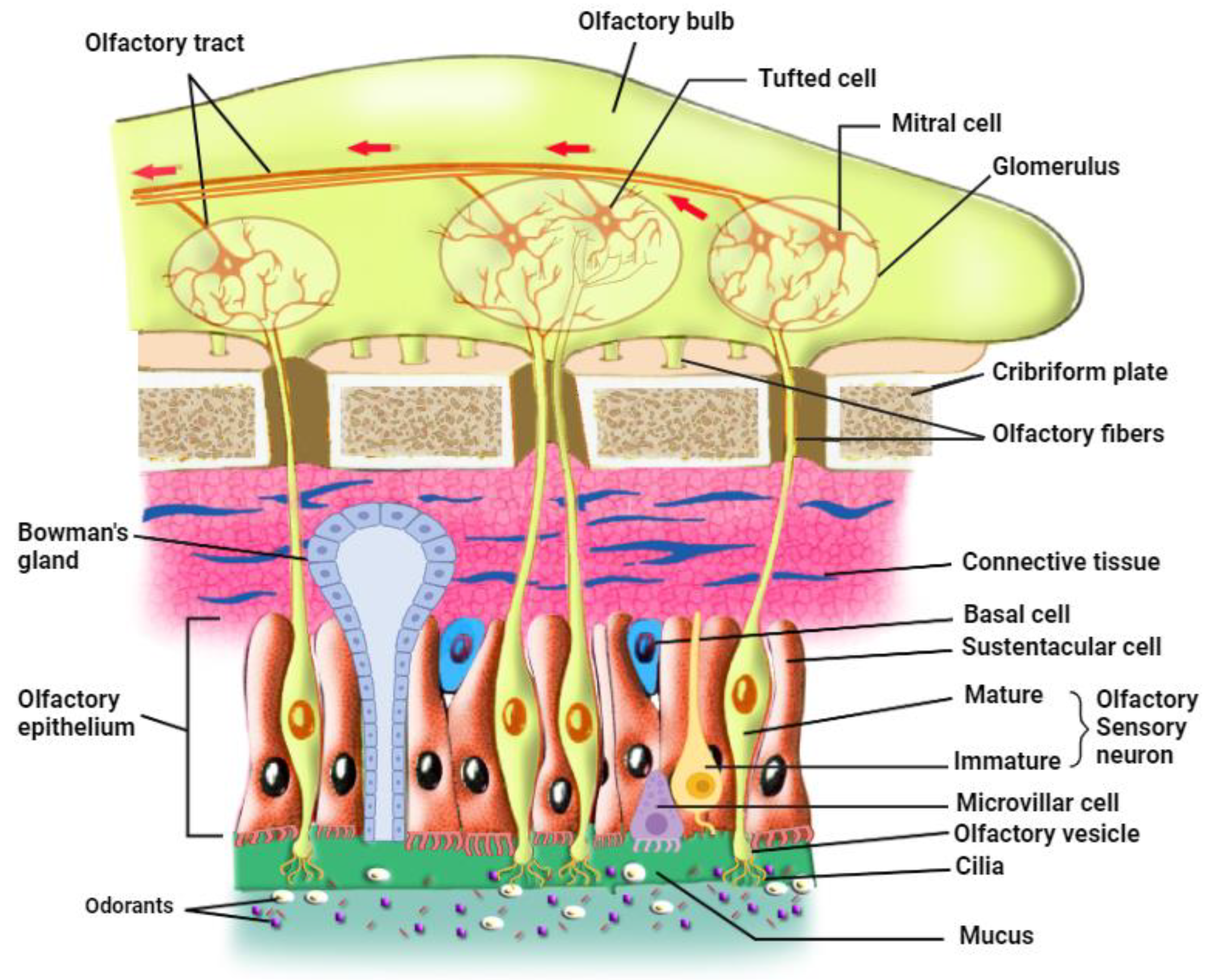
In mammals, the olfactory epithelium (OE) is located in the most dorsal part of the nasal cavity. OE and the underlying lamina propria or connective tissue form together the olfactory mucosa.[
25] There are three principal cell types in the olfactory epithelium: The olfactory sensory neurons (OSNs), also called olfactory receptor neurons, are either immature or mature olfactory sensory neurons (
Figure 1).[
26] OSNs are in direct contact with the inhaled odorants through their cilia that are embedded in the mucus in which odorants dissolve; each OSN extend its dendrite that form multiple, long specialized cilia to get in contact with the outer environment, detecting odorants, pathogens, and infectious agents such as respiratory viruses. On the other hand, axons from OSNs in the OE project to the olfactory bulb (OB), where they merge into glomeruli [
27] and make synapses with mitral and tufted cells that project to make the olfactory tract. Continuously exposed to the respiratory air that contains pathogens and harmful substances, OSNs have a limited life span of 1-2 months and are continually replaced by newborn neurons generated from the basal cells.[
28]
The second cell type is the supporting sustentacular cells (SCs) and the microvillar cells (MVCs) which together make the brush border. The dendrites of OSNs are surrounded by SCs whose nuclei and cell bodies line the external layer of the thick neuro-epithelium that is exposed to the outer environment.[
29,
30] These cell types ensheathe the OSNs and provide them with metabolic support and physical protection. This metabolic support represented by glucose secretion is necessary for the function of the OSNs cilia; when these cilia were deprived of glucose, OSNs deciliate and possibly lose their ability to detect odorants.[
24] Several roles have been assigned to SC cells including secretion, endocytosis, detoxification, communication with the basal cells,[
31] and involvement in the proliferation of the OSNs through purinergic signaling (
see section on purinergic receptors below).[
32] The role of SCs may even be more important than previously thought since they modulate the olfactory thresholds to food odors.[
33] In addition, animal experiments showed that SCs loss may lead to architectural damage to the OE.[
34] Sustentacular cells function as epithelial-like cells when they are involved in secretion, endocytosis, and metabolism of toxicants, but as glia-like cells when they physically and chemically insulate OSNs. They actively phagocytose dead cells, regulate the extracellular ionic environment,[
26,
31] and play a role in the intercellular signaling mechanisms.[
35] Thus, sustentacular cells represent a potential entry door for SARS-CoV-2 into the neuronal sensory system which is in direct connection with the brain. It is important also to note that sustentacular cells and mature and immature OSNs express gap junction subunits (connexin 43). Zhang et al. used immunohistochemical studies, in situ hybridization, and Western blot analysis to determine the pattern of expression of connexin 43 mRNA in mice olfactory epithelium and to verify the presence of connexin protein in nasal tissue. They hypothesized that gap junctions could participate in ion homeostasis in the interstitial fluid that bathes OSNs and sustentacular cells. Furthermore, gap junctions have an important role in the removal of extracellular potassium during periods of high activity of the OSNs. OSNs are electrically coupled to one another or to sustentacular cells at resting membrane potential, and uncoupled during odor stimulation, due either to closure of gap junctions by cAMP or through voltage-dependence of their conductance.[
36] Gap junctions then would lower the noise under resting conditions similar to the role played by cone-cone coupling in the visual information processing.[
37] These results revealed the role of gap junctions in information processing in the olfactory system since they modulate the membrane properties of mature OSNs[
36] and therefore may have a role in the pathophysiological mechanism of anosmia induced by SARS-CoV-2.
The third cell type is the basal cells which constitute the olfactory stem cells (OSCs) which include horizontal basal cells (HBCs) and globose basal cells (GBCs). HBCs are quiescent under normal conditions but upon injury they start dividing vigorously to give the mitotic GBCs and all the other mature cell types of OE.[
38] These stem cells are involved in the renewal of the other cell types of OE upon injury or lesions, but they are not known to have any direct role in odorant detection. An interaction seems to occur between HBCs and SCs; direct loss of SCs was found to activate HBCs which in turn proliferate to substitute the lost cells and restore the olfactory epithelium homeostasis.[
26] More cell types include the, Bowman’s gland cells (BGCs), and the olfactory ensheathing glia. The mucus-secreting Bowman’s gland cells also play important roles in maintaining OE homeostasis and function since they produce not only the mucus that keeps the surface of OE moist and prevents its dryness but also secrete certain proteins that aid in the transport of odorant materials to OSNs.[
1,
25,
39,
40]
4. ACE2 and TMPRSS2 are the Main Entry Receptors for SARS-CoV-2
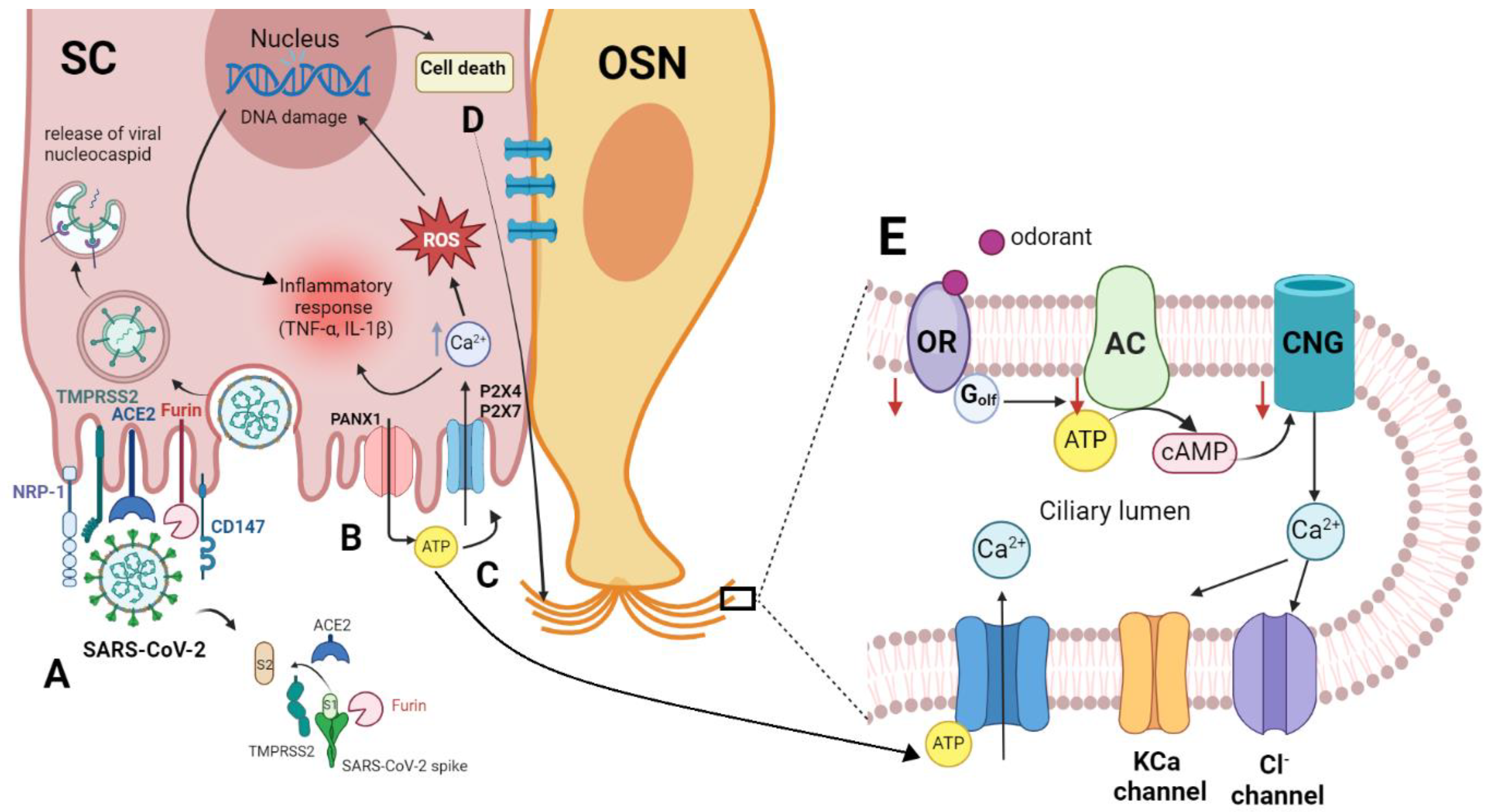
ACE2 receptor has been characterized as the main receptor for entrance of SARS-CoV-2 [
25,
41,
42] as it was previously reported to be the receptor for SARS-CoV-1 and other human respiratory related corona viruses.[
43] In airway epithelia, viral uptake is further facilitated by a priming protease called transmembrane protease serine type2 (TMPRSS2).[
44,
45] Cells with high ACE2 and TMPRSS2 expression have strong virus binding capacity and are particularly susceptible to the infection (
Figure 2). [
41,
42,
46]
Evidence for the role of TMPRSS2 in viral entry was provided by Letko et al. (2020) who demonstrated that the addition of the TMPRSS2 during the course of SARS-CoV infection facilitated entry into cells that exhibit low expression of ACE2 receptors,[
47] and by Chupp et al. (2022) who found that treatment of patients of COVID-19 with camostat methylate, the inhibitor for TMPRSS2, reduced olfactory dysfunction.[
48] Due to the location of SCs in the apical surface of olfactory epithelium, SARS-CoV-2 may first infect these cells, leading to partial or complete breakdown of the olfactory architecture, resulting in a decline in olfaction. These cells have been demonstrated to express a high level of ACE2 and TMPRSS-2 receptors whereas marker staining of immature and mature OSN demonstrated that ACE2 was not found in olfactory neurons.[
3] The presence of these two receptors and their density are important for the initial viral entry, in addition to the viral load. Furin enzymes or the host cell proteases, like TMPRSS2, which cleave the S protein into S1 and S2 can also modify SARS-CoV-1 and SARS-CoV-2 and facilitate the infection.[
49] This cleavage exposes the CendR motif of S1 to the binding pocket at the b1 subdomain of neuropilin (
discussed in the next section below), thus potentiating the viral infectivity.[
50] ACE2 receptors expression are distributed in a gradient from the nasal epithelium (very high) to the alveoli (low).[
51] In addition,
ACE2 gene is expressed in the CNS, lungs and testis.[
46] ACE2 messenger RNA is also predominantly expressed in the bronchi and lung parenchyma as well as in the heart, kidney and gastrointestinal tract. Using human kidney 293T cell line, it was shown that overexpression of ACE2 in vitro efficiently enhanced the replication of SARS-CoV-2 whereas neutralization by ACE2 antibodies inhibited replication of the virus in a dose-dependent manner.[
52] Ge et al. (2021) developed effective therapeutics and vaccines based on the fact that the viral spike (S) glycoprotein of SARS-CoV-2 mediates viral entry and recognition of ACE2 receptor. For example, crystal structural comparisons were used to determine the atomic details of possible engagements of ACE2 receptor in binding the receptor-binding domain (RBD) of the viral spike glycoprotein. Three neutralizing monoclonal antibodies (mAbs) were isolated from SARS-CoV-2 infected individuals which can recognize RBD. Among these three candidates mAbs, P2C-1F11 was the most likely to exert its antiviral activity through functional mimicry of receptor ACE2, and to provide a strong protection against SARS-CoV-2 infection in Ad5-hACE2-sensitized mice.[
53]
The question that remained to be answered was which of these two receptors is more important for virus infectivity. Hou et al. (2020) investigated the relationship between ACE2 receptor expression and SARS-CoV-2 infection by inoculating primary epithelial cultures from several pulmonary regions. They utilized quantitative comparisons of nasal and bronchial airway epithelia obtained as brush samples simultaneously from the same subjects. qPCR results revealed a significantly higher expression of ACE2, but not TMPRSS2, in the nasal than the bronchial tissues. In contrast, the overall expression of TMPRSS2 mRNA was higher in all respiratory tract regions than ACE2.[
54]
5. Neuropilin (NRP1) and Basigin (CD147) are Potential Receptors for Virus Entry
Among the intriguing observations was the ability of SARS-CoV-2 to infect tissues, like the heart or the brain where the density of ACE2 receptors is low or absent. This observation suggested the presence of another facilitating factor(s) for the entry of the virus. Recent evidence indicates that SARS-CoV-2 can use olfactory neurons and other related regions such as the olfactory tubercles and para-olfactory gyri to travel from the periphery into the CNS, and may also enter the brain through the blood-brain barrier (BBB). This transit was demonstrated to occur via neuropilin 1 [
49,
58] or basigin receptors, in addition to cathepsin L (CTSL). In comparison with ACE2 or TMPRSS2, NRP1 and basigin are broadly expressed in the human brain, including the OB. [
9,
59]
Neuropilin-1 (NRP-1) is a one-pass transmembrane receptor that lacks a cytosolic protein kinase domain and is highly expressed in the respiratory and OE. It can enhance the entry of SARS-COV-2 into the brain through the OE. Functioning as a cell surface receptor, it is unclear whether NRP-1 enables viral attachment or enables receptor-mediated endocytosis in the patient’s cells. However, it was shown that NRP-1 has a b1 domain that binds the CendR binding motif of S1 to potentiate infectivity of the virus, and this infectivity was blocked by monoclonal antibodies against the b1 binding pocket of NRP-1.[
49] It was postulated that the b1 subdomain of NRP-1 binds with S1 and makes a strong binding with the cell membrane of the host cell, destabilizing S1/S2 junction, thus bringing quick dissociation of S2 from S1, where S2 then brings membrane fusion of the host and virus and enhances infectivity.[
60,
61] This interaction of S protein with NRP-1 was reported to be disrupted by natural products like esculetin and 3-methylquercetin, giving these natural products a therapeutic potential.[
62] It was found that NRP-1 significantly potentiates SARS-CoV-2 infectivity, which was inhibited by a blocking monoclonal antibody against the extracellular b1b2 domain of NRP-1. analysis of human COVID-19 autopsies revealed SARS-CoV-2 infected NRP-1-positive cells in the olfactory epithelium and bulb as well as in endothelial cells of capillaries and medium vessels.[
58] Further research measured gene expression of NRP-1 using single cell RNA sequencing to examine the detailed expression of NRP-1 in the human brain. Expression of NRP-1 was also assessed by microarray presented as a heat map for six human donors and was detected in the olfactory tubercles and paraolfactory gyri.[
63]
Likewise, Wang et al. discovered CD147 (basigin or emmprin) as a new receptor for SARS-CoV-2 infection. In cells which are reported earlier as resistant to infection with SARS-CoV-2, like BHK-21 cells, the introduction of CD147 alters the virus tropism toward those cells. Moreover, ACE2-deficient T cells can be infected with SARS-CoV-2 pseudovirus, in which CD147 overexpression facilitates the virus infection. CD147 is a transmembrane glycoprotein of the immunoglobulin superfamily, which participates in bacterial and virus infection as well as in tumor development and parasitic invasion.[
55]
The high selectivity of the endothelial cells that form the BBB filters the blood contents available to neurons, and these endothelial cells were found to express high levels of ACE2. But ACE2 receptors are not expressed in the olfactory neurons whereas SARS-CoV-2 was located in the olfactory neurons and other brain parts.[
58] In order to explain the entry of SARS-CoV-2 into olfactory neurons, an alternative pathway for viral entry was suggested. This alternative pathway was NRP1 receptors which are expressed in every cell type in the nasal passages, in the olfactory neurons as well as in the olfactory tract neurons.[
50]
6. Olfactory Receptors Downregulation May be Responsible for Olfactory Dysfunction
Olfactory receptors (ORs) are chemoreceptors expressed in the ciliary membrane of OSNs in the olfactory epithelium.[
64] They are G protein-coupled receptors (GPCRs) that are composed of seven-transmembrane domains. Each OR can detect more than one odorant due to its allosteric recognition. Odorant perception takes place through these ORs on different neurons, inducing intracellular transduction cascades that send signals to the brain via the olfactory bulb.[
65]
SARS-CoV-2 infection of chemosensory cells and the loss of smell is not completely understood. One of the possible pathophysiological mechanisms of smell loss caused by the SARS-CoV-2 virus in COVID-19 patients was suggested to be the downregulation of olfactory receptor proteins within the olfactory sensory neurons.[
9] This assumption was supported by a recent study that showed a significant downregulation of olfactory receptor genes in two types of chemosensory cells: olfactory and taste cells infected with human coronavirus HCoV-OC43 and SARS-CoV-2. Among the OR reported to be downregulated are OR51E1, OR7D4, and TAAR1 (trace amine associated receptor 1).[
18] Similar observations have been reported by Verma et al. (2022) who showed that SARS-CoV-2 infection of sustentacular cells of experimentally infected animals caused inflammation, extensive ciliary damage, and downregulation of OR expression in OSNs, leading to olfactory dysfunction.[
16] In a mouse model, OR genes as well as other molecules involved in the olfactory signal transduction were found to be downregulated[
66] although the same was not found in human patients.[
67] The downregulated proteins include G
olf, CNGA2, ACIII, and ADCY3, another odorant related signaling molecule, which are all important molecules in olfactory signal transduction.[
16]
Another possible mechanism for anosmia associated with viral infection is the induction of apoptosis in olfactory sensory neurons during the early stage of infection. Such mechanism may provide a defensive mechanism that prevents neurovirulent virus invasion of the CNS from the peripherals. This was concluded from a study conducted on female C57BL/6 mice infected with influenza A virus in the nostrils. Using immunohistochemistry and dual immunofluorescent labelling techniques, it was revealed that the expression of Fas ligand molecules was upregulated and the c-Jun N-terminal kinase (JNK/c) signal transduction pathway which is involved in the process of apoptosis was activated in virus-infected OSNs. Furthermore, Iba1-expressing activated microglia/macrophages has a role in the phagocytic activities and helps in clearing apoptotic bodies.[
68] But as we pointed out in the
section hypotheses explaining anosmia above, the time course of the regeneration of the OSNs does not correspond to that of the recovery of smell sense. Recovery of smell sense takes place in about 5-7 days whereas regeneration of OSN after apoptosis may take 2-3weeks,[
24] suggesting that this apoptosis may serve as a feedforward mechanism that prevents the spread of the virus to the brain.
7. Transient Receptor Potential Vanilloid (TRPV1) Trafficking May Lead to Odor Inhibition
TRPV1 is a family of transient receptor potential (TRP) cation channels that are permeable to Ca
2+, and expressed in many tissues including the nervous tissue. Six TRPV channel subtypes (TRPV1-6) have been identified.[
69] TRPV1-4 are localized in OE. Using immunohistochemistry and double staining techniques, it was found that TRPV1 is expressed in various regions of the OE of CBA/J mice. TRPV1 immunofluorescence was detected in the olfactory cilia, basal cells, sustentacular cells and OSNs. TRPVs may perform several functions in the OE since it may participate in olfactory adaptation, olfactory/trigeminal interactions in nasal chemoreception, and in OE homeostasis. It was also suggested to be involved in olfactory transduction and olfactory dysfunction related to sinonasal inflammatory disease.[
70]
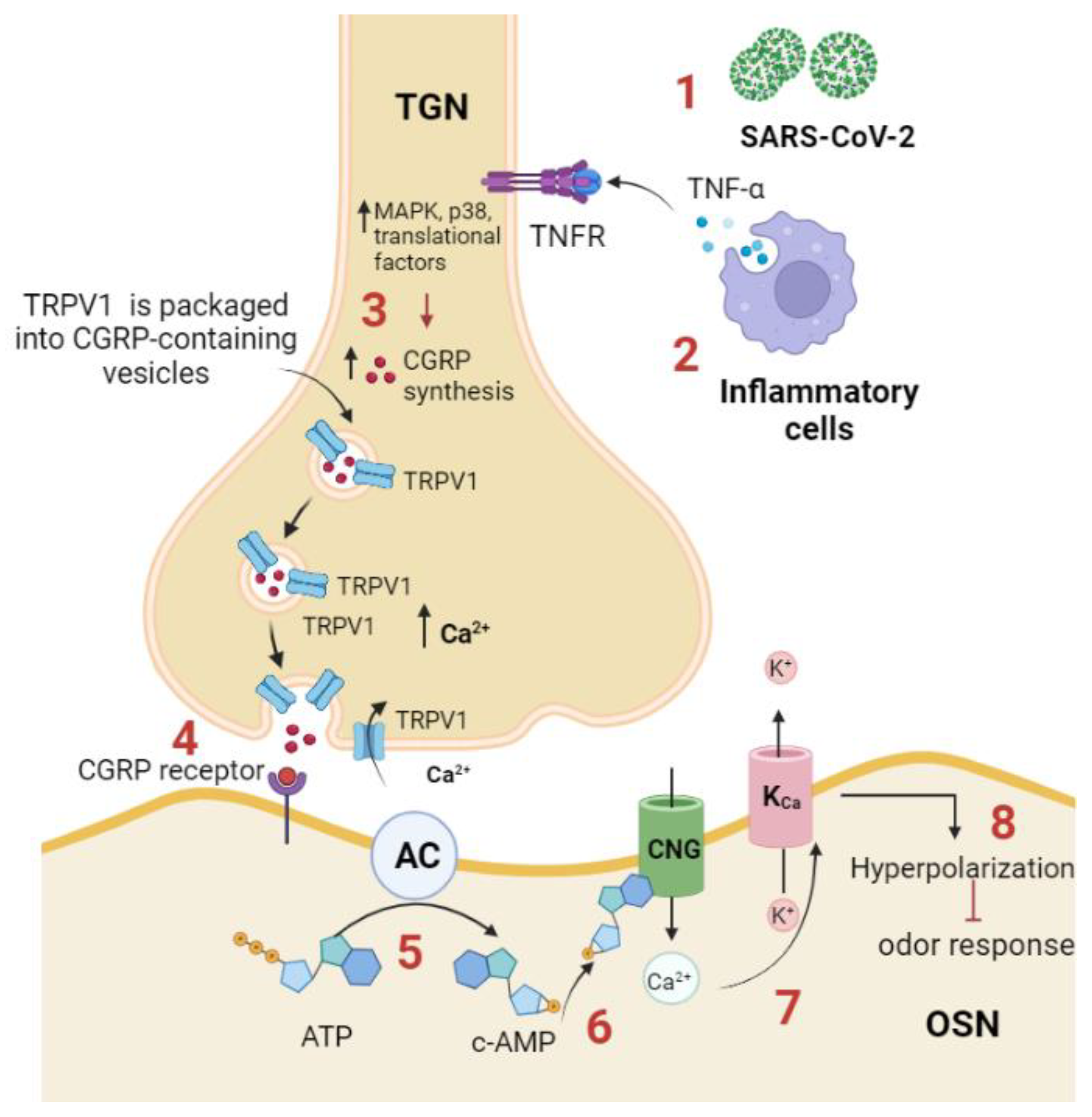
Olfactory dysfunction during infection may be related to the overactivity of trigeminal afferent system that stimulates calcitonin gene-related peptide (CGRP) release and inhibits the detection of the odor by the olfactory receptor.[
71] TRPV1 was found to be expressed on most of the trigeminal afferents innervating the nasal mucosa. During SARS-CoV-2 infection, it was shown that the virus triggers the inflammatory response which is accompanied by an increase in the production of cytokines, specifically TNF-α,[
72] and consequently stimulating trigeminal system in the nasal mucosa (
Figure 3).
TNF-α activates trigeminal ganglion by enhancing the expression of TRPV1 channels and many cytokines including IL-1b and brain-derived neurotrophic factor (BDNF). In response to TNF-α and IL-1b stimulation, activated trigeminal ganglion neurons synthesize and release CGRP.[
73] TNF-α enhances the expression and trafficking of TRPV1 and TRPA1 to the surface of trigeminal neurons and such trafficking is required for CGRP exocytosis.[
74] Experimentally, it was shown that CGRP receptor is expressed on the OSNs and when it binds its ligand, CGRP, it inhibits odor detection by the olfactory receptors. In addition, it was found that cAMP is produced by OSNs in response to CGRP application, presumably through coupling of CGRP receptor with adenylyl cyclase III.[
75] cAMP mediates the inhibitory olfactory transduction pathway by activating a cyclic nucleotide gated (CNG) channel, allowing Ca
2+ permeation. Ca
2+ then activates a Ca
2+-dependent K
+ channel (K
Ca), resulting in a hyperpolarizing receptor potential.[
76]
8. Purinergic Receptors Involvement in Olfactory Signaling (P2X and P2Y Receptors)
Purinergic receptors are expressed on the plasma membrane of neurons, basal, and sustentacular supporting cells of the olfactory epithelium and play an important signal transduction role through binding to the extracellular nucleotides.[
77,
78] Besides the importance of this system in modulating physiological processes like apoptosis, thromboregulation, cell proliferation, platelet aggregation, endothelial vasodilation, and pain, it also maintains the integrity of the olfactory epithelium by inducing proliferation and differentiation of OSNs and regulate olfactory sensation.[
57]
Purinergic receptors can be initially classified into P1 and P2 groups, according to their structural and functional characteristics. P1 receptors are G protein-coupled receptors and are sensitive to a group of signaling molecules including ATP, ADP, AMP and adenosine which are released by all cells as a response to cellular damage caused by the action of pathogens. P1 receptors are subclassified into A1, A2A, A2B, and A3.[
79] On the other hand, ATP, ADP, UTP and UDP are extracellular nucleotides that bind to a specific subtype of purinergic receptors (P2). P2 is further subdivided into two categories: P2X and P2Y receptors. P2X receptor is a ligand-gated ion channel that responds rapidly when bound to its key ligand ATP. It is a nonselective cation channel that allows Na
+, K
+, and Ca
2+ flux. In contrast, P2Y (1, 2, 4, 6, 11-14) receptor is a G protein-coupled receptor that activates a variety of second messengers and intracellular signaling pathways, thus acting at a slower pace than P2X.[
28,
80]
To detect the localization of P2X receptors in OE, Gao et al. used RT-PCR and immunohistochemistry to monitor the expression of these ionotropic purinergic receptors in adult mouse OE. They targeted type III neuron-specific tubulin (TuJ1) using polyclonal rabbit anti-P2X1-3 and monoclonal mouse anti-TuJ1. Their results showed that P2X1-3 receptors were localized at the basal part of OE, including the sustentacular cells, OSNs and basal cells. P2X1 and P2X2 were mainly localized to the cell body of sustentacular cells and basal cells. Strong co-localization of P2X3 with TuJ1 was observed in the olfactory nerve. An immunoreactivity to P2X4-7 receptors was also detected in the OSNs, and to P2X5 receptors in the axons of OSNs only. The localization of P2X receptors in the basal progenitor cells of adult mouse OE may explain the role of purinergic receptors in regulating adult OSN neurogenesis.[
78]
Extracellular ATP participates in the regeneration of the sensory neurons after epithelial damage. The P2X receptor is expressed in the basal cells of olfactory epithelium and blockade of this receptor inhibited the injury-induced proliferation of basal cells.[
78] In addition, it was reported that intranasal ATP and purinergic agonists induced proliferation of the basal cells in the olfactory epithelium, and an opposite effect was observed when purinergic antagonists were used.[
81] The mechanism of ATP-induced olfactory epithelium proliferation through purinergic receptors activation was mediated by growth factors like fibroblast growth factor 2 (FGF2), transforming growth factor alpha (TGFα), and neuropeptide Y (NPY).[
82] Injured OSNs usually release ATP which activates P2X and P2Y receptors and stimulates calcium signaling in microvillar cells and sustentacular cells, thus enhancing the release of NPY. NPY activates Y1 receptors expressed on basal cells, thus stimulating mitogen-activated protein kinase (MAPK), resulting in regeneration of new OSN.[
28]
Anosmia is related, in part, to defect in olfactory neurogenesis in response to various pathogenic infections which stimulate neuro-inflammation and oxidative stress. Many studies indicated that SARS-CoV-2 initially causes neuro-inflammation that leads to damage in the OSNs of the nasal neuroepithelium and spreads through OB to different regions in the brain.[
83] SARS-CoV-2 infection was found to increase the level of extracellular ATP, leading to abnormal activation of purinergic receptors which may cause the hyperactivation of P2X7 receptors that facilitate consequent neuro-inflammatory processes.[
84,
85] Although activation of both P2Y and P2X receptor by ATP caused remarkable decrease in odor responsiveness,[
86] P2X and P2Y receptors exhibited different effects on olfactory transduction.[
87]
Purinergic receptors such as P2Y are GPCRs that may interact with specific subfamilies of olfactory receptors to enhance cell surface expression and modulate odorants responsiveness. The specificity of olfactory receptor (M71) interactions with other GPCRs was examined by Bush et al. (2007). They co-expressed the olfactory receptor (M71) with purinergic receptor (P2Y1R and P2Y2R) in HEK-293 cells and showed that co-expression of the olfactory receptor with purinergic receptors (P2Y1R and P2Y2R) enhanced cell surface expression of the olfactory receptor.[
88] Moreover, activation of P2Y receptors in sustentacular cells of mouse olfactory epithelium was mediated by the phosphorylation of the transcription factor cAMP-response element binding protein (CREB), causing odorant detection. This effect was reduced when a purinergic receptor antagonist like suramin was used.[
35] Nevertheless, cAMP-dependent protein kinase was found to have a role in SARS-CoV-2 infection in Vero E6 cells. The interaction of CREB and CREB-binding protein (CBP) was found necessary for viral replication since the knock down of CREB and CPB with siRNA inhibited the viral infection.[
89] The conclusion to be made is that purinergic receptor, and may be other cAMP-related receptors, are important for assisting olfactory receptors to be localized into the surface membrane, and consequently in their odor responsiveness.
CREB is activated in response to extracellular stimuli by phosphorylation at residue Ser-133.[
90] This activation depends on adenylyl cyclase III (ACIII)-mediated olfactory signaling and upon activation of P2Y purinergic receptors on sustentacular cells. Odorant-induced ATP release from OSNs, leads to long-term activation of CREB in neighboring sustentacular cells via activation of their P2Y receptors and subsequent phosphorylation of CREB. In support of this observation, application of exogenous ATP was found to induce CREB phosphorylation in the nuclei of the sustentacular cells. CREB activation was shown to be purinergic receptor-dependent where the extracellular nucleotides are essential tool in communication between OSNs and sustentacular cells.[
35]
Interestingly, preincubation of OE with SQ22536, an adenylyl cyclase antagonist, inhibited the activation of CREB in mature and immature OSNs, indicating that ATP-mediated activation of CREB in these cells depends completely on ACIII signaling. In sustentacular cell nuclei, this inhibitor was able to reduce but not to abolish the levels of CREB phosphorylation, indicating that ATP-induced phosphorylation of CREB in sustentacular cells is only partially dependent on ACIII signaling.[
35] It is worth mentioning that in the previous section on the role of TRPV1 receptors we reviewed the literature that indicated that cAMP mediates the inhibitory olfactory transduction pathway by activating a cyclic nucleotide gated (CNG) channel, allowing Ca
2+ permeation and activating a Ca
2+-dependent K
+ channel (K
Ca), resulting in a hyperpolarizing receptor potential. This seemed to contradict the role of cAMP/CREB described in this section. This contradiction was reflected in the question raised by Madrid et al. (2005) inquiring how would an OSN generate 2 opposite responses independently of each other if they share a common transduction pathway (i.e., AC/cAMP/CREB/pCREB). These authors speculated that the transduction proteins that participate in each response type form complexes somehow segregated from each other within the cilia, thus allowing them to operate as independent functional units.[
76]
This intercellular communication between olfactory neurons and their supporting counterparts, the sustentacular cells, indicates the importance of sustentacular cells in development of smell sensation. The odorant-mediated activation of OSNs which causes them to fire their action potentials depends on intact sustentacular cells, through induction of CREB phosphorylation, and on extracellular nucleotides that mediate this intercellular communication.[
35] Single-cell RNA sequencing revealed that only sustentacular cells and horizontal stem cells in the olfactory cleft as well as the vascular pericytes in the olfactory bulb co-express ACE2 and TMPRSS2 which explain why the local infection of these cells may cause a block of odor conduction, indicating that general disturbance of the mucosal architecture can obstruct the processing and signal transmission to the brain.[
91] Invasion of sustentacular cells by SARS-CoV-2 is considered a vital factor in disintegrating the architecture of OE, a matter that has transient detrimental effects on smell perception.
In COVID-19, ATP is released by infected cells through pannexin 1 (PANX1) channels as a response to conditions like viral invasion, cell stress, and hypoxia. Once ATP is released, it remains for hours to days in the extracellular environment exerting its pro-inflammatory effects by initiating a cascade that activates purinergic receptors, until it is metabolized. Due to its chemoattractant ability for macrophages, ATP is recognized as a damage-associated molecular pattern that activates P2X and P2Y purinergic receptors.[
56]
It was reported that activation of both P2Y and P2X receptor by exogenous and endogenous ATP caused remarkable decrease in odor responsiveness.[
86] A low level of endogenous ATP was found to increase Ca
2+ and suppress odor sensitivity whereas superfusion of receptor antagonists like suramin and pyridoxal-phosphate-6-azophenyl-2’,4’-disulfonic acid (PPADS) reduced the basal Ca
2+ level and increased sensitivity to odors.[
86] High levels of extracellular ATP result in worsening of the inflammatory responses through PANX1/P2 receptor activation and play an essential role in the acute pulmonary inflammation, edema, and lung dysfunction after ischemia/reperfusion injury. Damaged cells release ATP which activates P2X and P2Y purinergic receptors on the neighboring cells of the olfactory epithelium.[
28] These data indicate that ATP release in the olfactory epithelium after noxious stimuli may serve a physiological role as a neuroprotective mechanism. Moreover, the anosmia associated with COVID-19 infection may serve as a protective mechanism.
9. Interferon Gamma and Interleukins Receptors in Sustentacular Cells
Interferon-gamma (IFN-γ) is a soluble cytokine produced mainly by activated T lymphocyte, macrophages, mucosal epithelial cells and natural killer cells. It modulates antiviral and antibacterial immune responses during infection. IFN-γ mediates cellular responses through its binding to its heterodimeric cell-surface receptor (IFN-γ R) which in turn activates downstream signal transduction pathways and subsequently affects the regulation of gene expression.[
92] Lack of IFN-γ receptor was associated with invasion and persistence of influenza virus A/WSN/33 in the mouse olfactory system[
93] whereas increased IFN-γ expression was correlated with the immune response to sinonasal bacterial biofilms in surgical chronic rhinosinusitis patients.[
94] It has been suggested that IFN-γ mediates the cytokine pathways which may contribute to the pathogenesis of chronic rhinosinusitis-associated anosmia. This assumption has been investigated using electro-olfactography recordings to assess the odorant responses of transgenic mice with enhanced production of IFN-γ by olfactory sustentacular cells. The results showed that chronic IFN-γ expression caused a significant decrease in odorant responsiveness which was explained as due to reduced olfactory receptors expression.[
95] Although there was no inflammatory tissue damage reported in this case, suggesting a direct effect of IFN-γ, it seems that increased levels of IFN- γ and may be other cytokines are involved in COVID-19-induced olfactory disturbance.
As mentioned earlier in this review, inflammation in OE was suggested as one of the possible mechanisms for COVID-19-induced anosmia. Inflammation causes damage in olfactory neurons and activates horizontal basal stem cells-mediated neurogenesis but prolonged inflammation arrests those cells in an undifferentiated state by stimulating ‘‘stemness’’-related transcription factors, leading to olfactory loss. It has been shown that the expression of IFN-γ as well as other cytokines and chemokines were upregulated in the whole olfactory mucosa of the inducible olfactory inflammation mouse model in response to continuous stimulation by tumor necrosis factor (TNF).[
96]
A series of physiopathological mechanisms is related to the disease caused by SARS-CoV-2 infection in which the infection mobilizes a wide variety of biomolecules, mainly proinflammatory cytokines, such as IL-1, IL-6, IL-12, IFN-γ, and TNF-α which preferentially target lung tissue.[
97] Interferon signaling has a crucial role in controlling and regulating disease severity after infection with many viruses, including coronaviruses. SARS-CoV-2 was found to be sensitive to type I and III interferons in human cells in vitro and in mice
in vivo.[
98] These authors infected double knock out C57BL/6J mice for type I and II interferon receptor and wild-type controls with 104 plaque-forming unit of SARS-CoV-2 MA10. Four days post infection, mice displayed much higher congestion scores, which were associated with higher viral titers on 2 and 4 days post infection compared to the wild-type controls. These data suggest that IFNs are important in limiting viral replication and assisting in virus clearance
in vivo. Consistent with these data, lung function abnormalities were more pronounced and prolonged in infected double knock out mice.[
98] Cazzolla et al. investigated the role of IL-6 in smell and taste disorders. A venous blood sample of 67 COVID-19 patients was used to measure IL-6 levels using the chemiluminescence assay. A significant correlation between IL-6 levels and the type of dysfunctions was found. The olfactory and gustatory dysfunctions had a higher score in patients with higher levels of IL-6. Also, the patients with both disorders had even higher levels of IL-6.[
99] Interferons and interleukins levels are inversely proportional to each other. This unique and inappropriate inflammatory response was observed and it has been assumed that increased levels of IL-6 in COVID-19 patients would suppress interferon levels.[
100] The relation of serum IL-6 and IFN-γ concentrations to mortality in the cohort of patients was investigated. It was found that over 60% of peripheral T cells in severely ill COVID-19 patients were unable to produce measurable IFN-γ when stimulated with the potent IFN-γ mitogen phytohemagglutinin. This defect of IFN-γ production was associated with increased levels of IL-6.[
101]
10. Role of Epithelial Sodium Channel (ENaC) in COVID-19 Induced Anosmia
The epithelial sodium channel
α-subunit (ENaC-
α) is a channel protein that is responsible for maintaining the balance of salt and water in the epithelium of many organs including the lungs and olfactory epithelium.[
102,
103] It is also expressed in stem cells as well as in the nervous system such as in brain centers controlling fluid volume or blood pressure, retina, and olfactory bulb.[
104]
Olfactory dysfunction induced by COVID-19 can be linked to the impairment of function.[
105] In sustentacular cells, ENaC helps maintain ion gradients in the liquid layer that surrounds the olfactory epithelium,[
103] a matter that is important to the proper function of OSNs. Impaired ENaC activity in the nasal epithelium is associated with chronic rhinitis, characterized by a continuous runny nose.[
105]
It has been shown that the co-expression of SARS-CoV-1 envelope, SARS-CoV-2 envelope, and SARS-CoV-2 spike protein with ENaC in
Xenopus oocytes, causes a remarkable inhibition in epithelial sodium channel activity.[
106] Furthermore, a study demonstrated that the knockout of ENaC in the olfactory bulb of mice resulted in a decreased number of newly generated neurons and inhibited adult neurogenesis.[
104] Therefore, the reduction in ENaC functionality may impact adult neural stem cell proliferation in the olfactory bulb and, consequently, the olfaction process.
In the human, ENaC-
α is often found in the same types of lung and respiratory tract cells as ACE2.[
102] Anand et al. (2020) performed a systematic expression profiling of the ACE2 and ENaC-
α across 65 published human and mouse single-cell studies including ~1.3 million cells using nferX single cell platform. Their analysis showed a significant overlapping between expression of ENaC-
α and the viral receptor ACE2. SARS-CoV-2 developed a unique S1/S2 cleavage site that is absent in any previous coronavirus sequenced and this site can be cleaved by furin enzymes. On the other hand, ENaC
α-subunit has an identical furin-cleavable peptide. This shows an astonishing imitation by the virus for the sodium channel
α-subunit. This suggests that SARS-CoV-2 may use the same protease (furin) that activates ENaC-
α for the purpose of getting inside human respiratory cells.[
102] In case of COVID-19 infection, this will lead to compromised ENaC activity, compromised fluid reabsorption, and to the lung pathology observed in these patients.
On the other hand, Ozdener et al. (2022) used various molecular techniques to show that TRPV1 is co-localized in cultured adult human fungiform taste cells that express the ENaC δ-subunit, and that modulating TRPV1 activity by high salt media and capsaicin can alter ENaC mRNA expression. Moreover, renin-angiotensin-aldosterone system (RAAS) components function in a complex along with ENaC and TRPV1 expressed in cultured adult human fungiform taste cells. Changes in ACE2 receptor expression can alter the balance between the two major RAAS pathways, ACE1 (Ang II; At
1R) and ACE2 (Ang (1-7); MasR1), leading to changes in ENaC expression and responses to NaCl in salt-sensing human fungiform taste cells.[
107]
11. Physiological Role of Oscillatory Calcium Transients in Sustentacular Cells and Signal Transduction through Olfactory Sensory Neurons
Sustentacular cells express purinergic receptors (P2Y), and they respond to P2R agonists ATP and UTP with an increase in intracellular calcium. This Ca
2+ will be utilized later either to develop intercellular or intracellular Ca
2+ signals. Using confocal calcium imaging of neonatal mouse olfactory epithelium slices, Hegg et al. observed spontaneous intercellular calcium waves and intracellular calcium oscillations in sustentacular cells. This may make sustentacular cells chemically coupled and capable of transferring changes in intracellular calcium between cells through gap junctions.[
31]
When P2Y are stimulated by ATP, diverse intracellular signaling pathways are activated, including the activation of PLC, PKC, and CaM kinase. PLC activation results in the formation of IP3, causing the release of Ca
2+ from IP3-sensitive Ca
2+ stores, thus increasing [Ca
2+]i. [
26,
31,
35] Some P2Y receptors couple to adenylyl cyclase and increase the production of cAMP[
35] with significant sequences as described in purinergic receptors section above.
Dynamic intracellular calcium fluxes in sustentacular cells can serve many different signals in the olfactory epithelium including secretion, proliferation, development of sustentacular cells, and release of chemical signals via calcium-dependent exocytosis. This transient increase in intracellular calcium signal may be translated to an electrical membrane signal through K
+ and Na
+ channels.[
31] In mice sustentacular cells, Ca
2+ waves could serve as a “housekeeping” mechanism to maintain a functional OE. One of the consequences of Ca
2+ increase in these cells is the increased opening of calcium-dependent K channels (K
Ca) for clearance of K
+ to achieve a level of hyperpolarization that is necessary to increase the availability of Na
+ channels and allow sustentacular cells to fire action potentials that may propagate across gap junctions.[
108] These observations point to a key role for sustentacular cells in maintaining a stable intra- and intercellular ionic microenvironment in olfactory epithelium.
In mammals and other vertebrates, once the receptor of OSN binds an odorous molecule, the ligand-bound receptor activates a G protein, and a cascade of events is initiated which activates an adenylyl cyclase (ACIII). The cyclase converts the abundant intracellular molecule ATP into cyclic AMP which binds to the intracellular face of the cyclic nucleotide-gated (CNG) ion channel, enabling it to conduct cations such as Na+ and Ca2+. Inactive OSNs normally maintain a resting voltage across their plasma membrane of about -65 mV. When the CNG channels open, Na+ and Ca2+ ions influx causes the inside of the cell to become less negative. If enough channels are opened long enough, they cause the membrane potential to become about 20 mV less negative; the cell reaches threshold and generates action potentials. The action potentials are then propagated along the axon, which crosses cribriform bone plate into the forebrain where it synapses with second-order neurons in the olfactory bulb.
As much as positive charge entering through the CNG channel, it will be able to activate another ion channel that is permeable to the negatively charged chloride ion. Normally, neuronal Cl
- channels mediate an inhibitory response, as Cl
- ions tend to be distributed in such a way that they will enter the cell through an open channel. Unusually, OSNs have a high intracellular Cl
- concentration maintained via a membrane pump to allow a Cl
- efflux when these channels are activated. This mechanism will add a net positive charge on the membrane that further depolarizes the cell, thus adding to the excitatory response magnitude. Thus, the OSN maintains its own Cl
- voltage, in case the Na
+ gradient in the mucus is insufficient to support a threshold current, and uses it to boost the response,[
109,
110] or when the Na
+ gradient in the mucus has been disturbed as when ENaC-
α function has been compromised when SARS-CoV-2 invades the sustentacular cells.
12. Conclusion
Prolonged olfactory impairment after SARS-CoV-2 infection is a serious problem that requires further investigations to understand the pathology of this deterioration. Significant efforts have been made to explore, at the cellular and molecular levels, the mechanisms of the viral invasion and the signal transduction that leads to olfactory dysfunction. In this regard, the role of the many receptors and channels that may be involved in olfactory dysfunction during COVID-19 was discussed. The sustentacular cells seemed to be the starting site for olfaction dysfunction due to the presence of ACE2 and TMPRSS2 receptors in these cells in higher density than in other types of cells in the olfactory epithelium (e.g., the olfactory sensory neurons which may not have any significant density of those receptors). In addition, the presence of neuropilin-1 in the plasma membrane of SCs seemed to mediate the binding of CendR motif of the S1 protein and enhance the infectivity of the virus. A similar role has been ascribed to basigin (CD147). Moreover, the inflammatory response caused by the viral invasion and the concomitant release of proinflammatory cytokines (i.e., TNF-α) may enhance the expression and trafficking of TRPV1 receptors which may help in the enhanced CGRP synthesis and release, a process that seemed to be involved in odor inhibition. Furthermore, viral infection stimulates Panix1 channels which release ATP extracellularly to bind P2 purinergic receptors that mediate Ca
2+ release and intercellular communication with OSNs through gap junctions, creating Ca
2+ waves in these OSNs. These Ca
2+ waves activate Ca
2+-dependent K
+ channels that mediate a hyperpolarization. Although ATP has an excitatory role on the Ca
2+ responses in non-stimulated OSNs, when co-applied with the odorant it showed an inhibitory role on those responses, indicating its important modulatory role.[
86] This wide and complex spectrum of receptors that mediates the pathophysiology of olfactory dysfunction reflects the many ways by which anosmia can be therapeutically managed.
Additional research is needed to comprehensively understand the molecular processes and the electrophysiological alterations that impact various aspects of olfactory function during viral infections, including the detection threshold, odor discrimination, and odor identification. As new variants of the virus emerge, the occurrence of anosmia may vary, and alterations in their molecular mechanisms would change. Continued investigation is necessary to clarify the pathogenesis and differences in olfactory dysfunction among different variants.