Submitted:
27 June 2024
Posted:
28 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Brain Region Related Differences in PAD Isozyme Detection
2.2. Isolation of Citrullinated Target Proteins from Brain Regions and Silver Staining
2.3. Identification of the Brain Region Specific Citrullinomes by LC-MS/MS
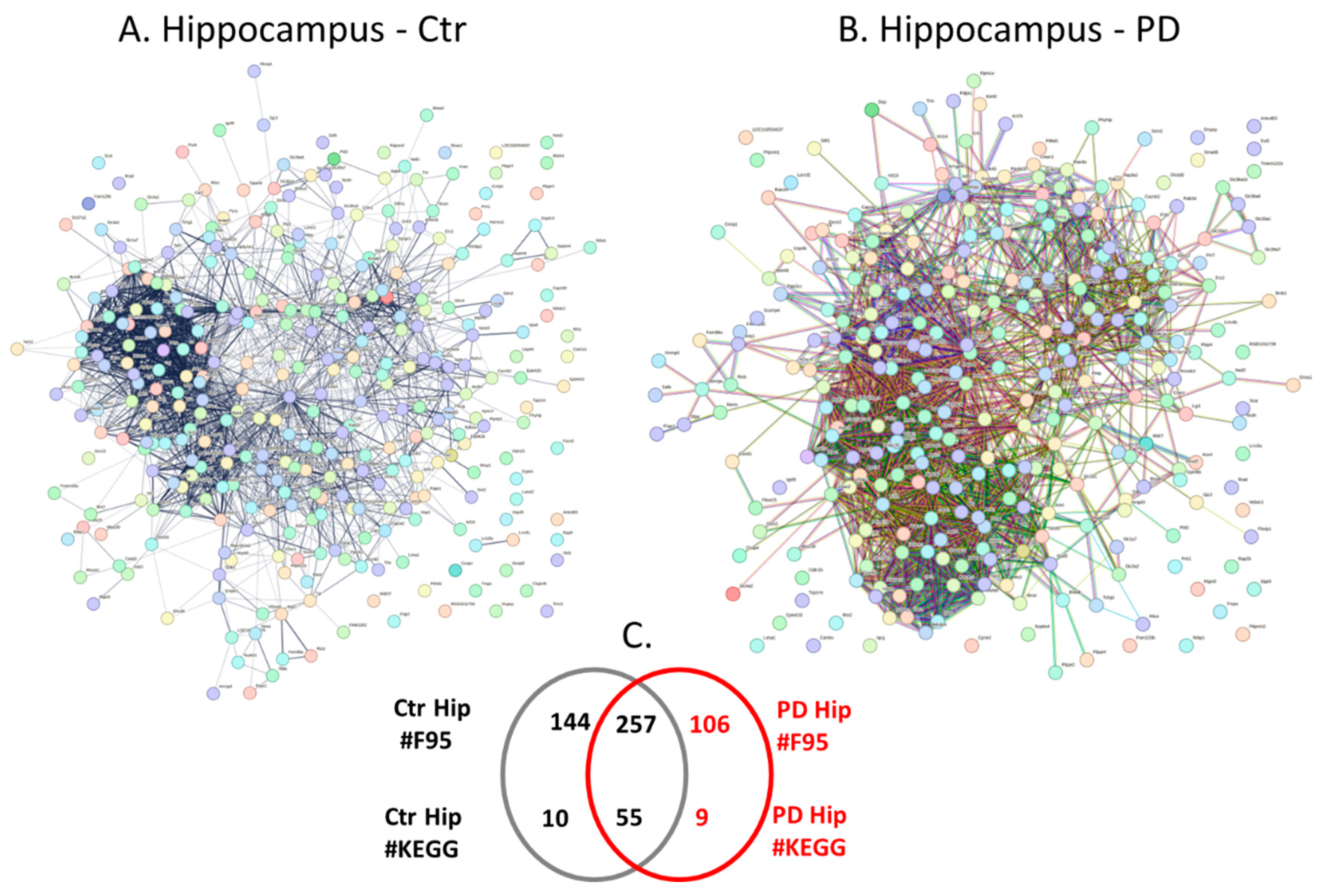
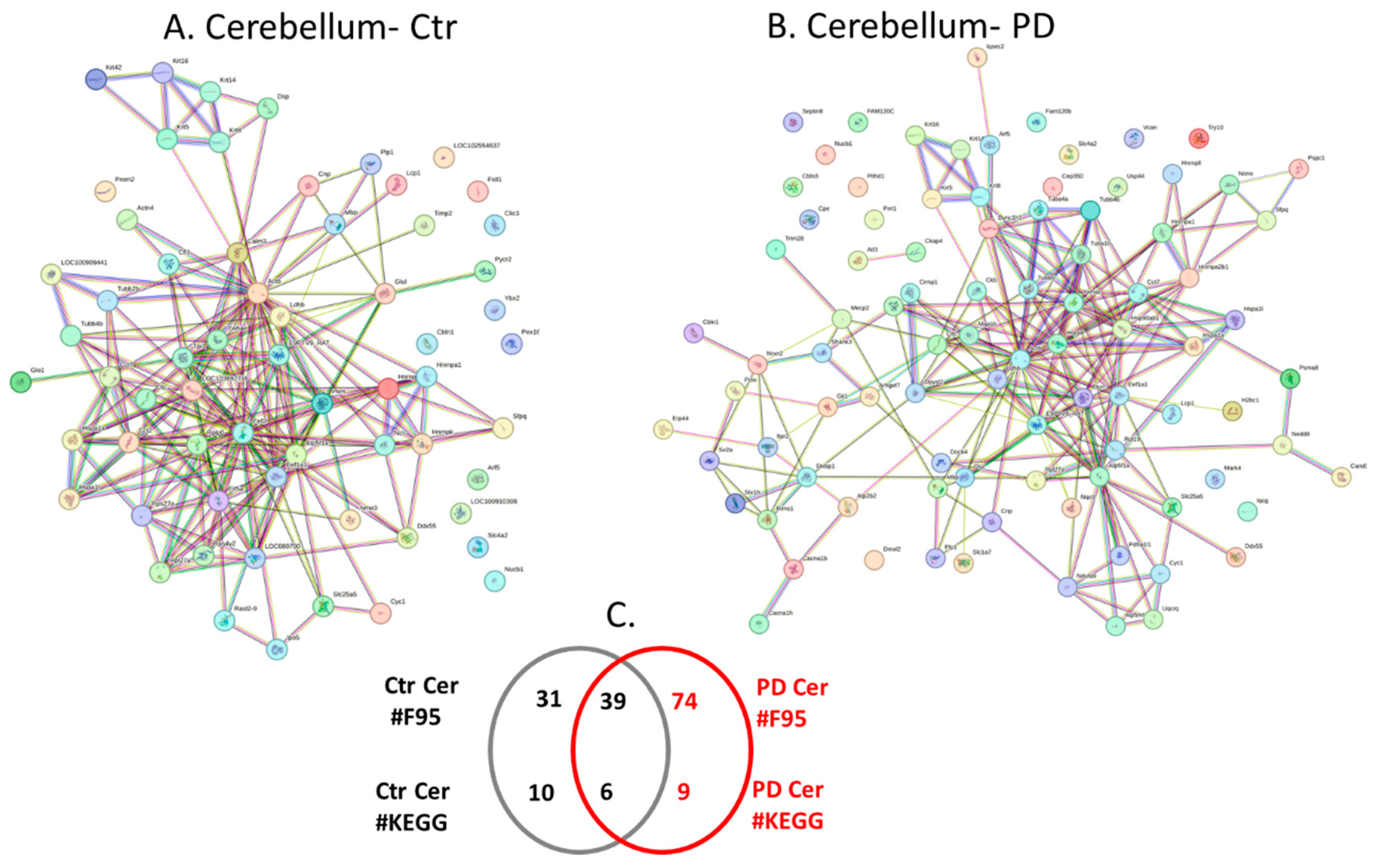
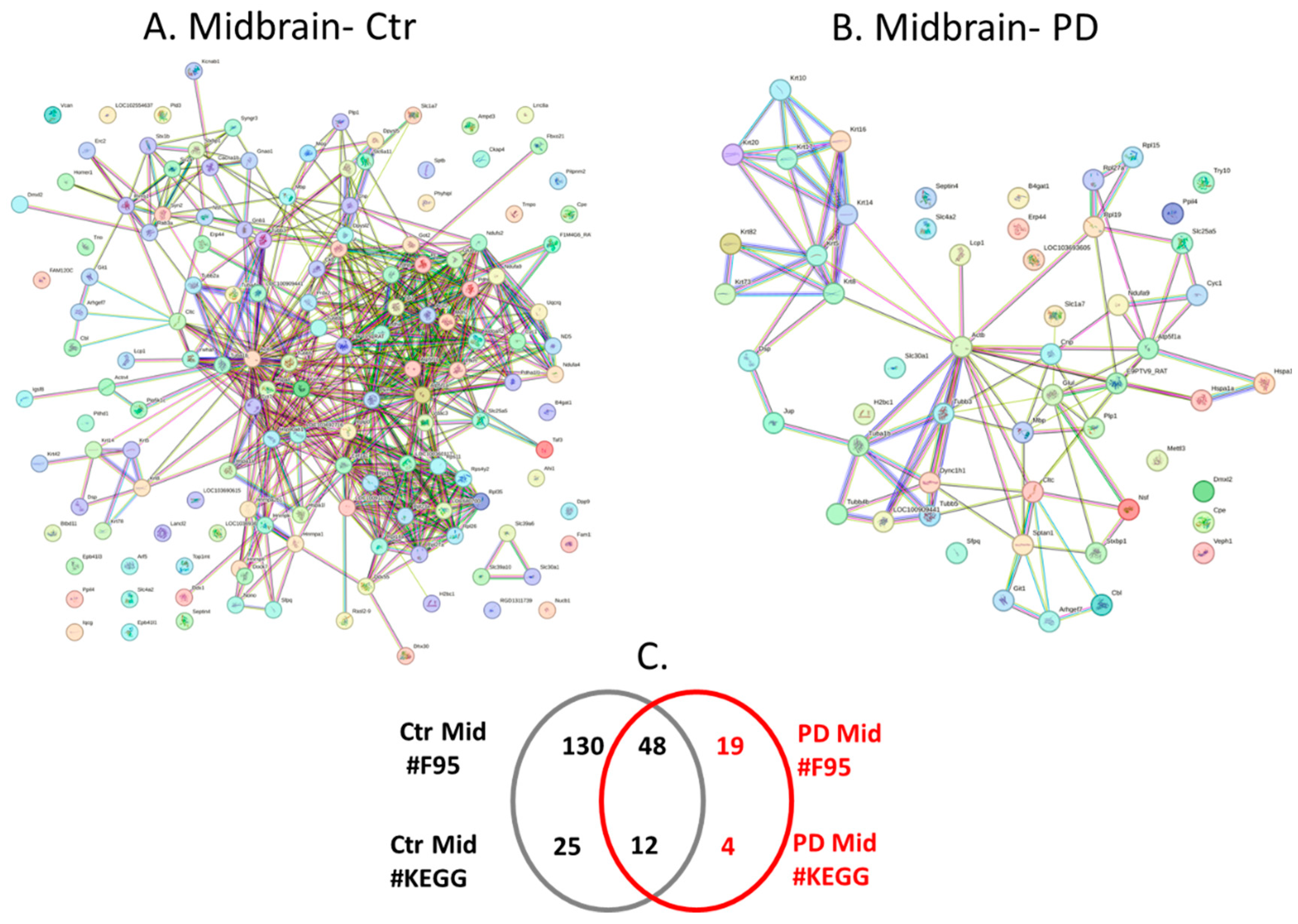
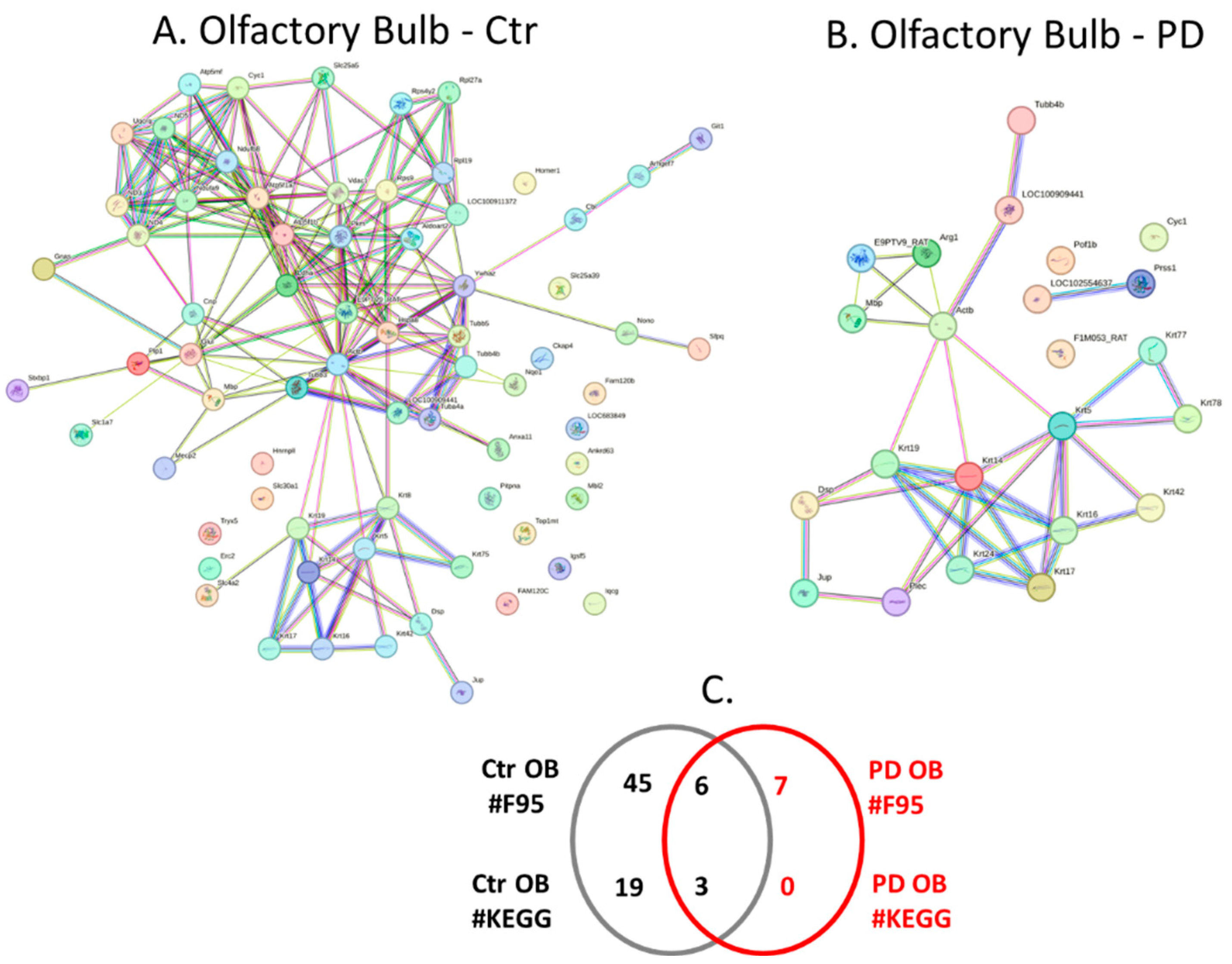
2.4. Protein-Protein Interaction Network Analysis for the Brain Region Specific Citrullinomes in Pre-Motor PD versus Control Brains
3. Discussion
3.1. Brain Region Specific PAD Isozyme Detection
3.2. Brain Region Specific Citrullinomes
3.2.1. Citrullinome KEGG Pathways Shared in Sham/Control and PD Brains
3.2.2. Citrullinome KEGG Pathways in PD Brains Only
3.2.3. Citrullinome KEGG Pathways in Sham/Control Brains Only
3.3. Possible Roles for Citrullination in the Gut-Brain Axis in PD
3.4. Future Prospects for PAD Inhibitors in PD
4. Materials and Methods
4.1. Pre-Motor PD Rat Model
4.2. Protein Isolation from Brain Tissue
4.3. Western Blotting
4.4. Isolation of Citrullinated Proteins from Brain Tissue
4.5. Liquid Chromatography with Tandem Mass Spectrometry (LC-MS/MS)
4.6. Protein-Protein Interaction Network Analysis
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siderowf, A.; Lang, A.E. Premotor Parkinson’s disease: Concepts and definitions. Mov. Disord. 2012, 27, 608. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ciesielski, O.; Biesiekierska, M.; Panthu, B.; Soszyński, M.; Pirola, L.; Balcerczyk, A. Citrullination in the pathology of inflammatory and autoimmune disorders: Recent advances and future perspectives. Cell Mol Life Sci. 2022, 79. [Google Scholar] [CrossRef] [PubMed]
- Nicholas, A.P. Dual immunofluorescence study of citrullinated proteins in Parkinson diseased substantia nigra. Neurosci Lett. 2011, 495, 26. [Google Scholar] [CrossRef] [PubMed]
- Sancandi, M.; Uysal-Onganer, P.; Kraev, I.; Mercer, A.; Lange, S. Protein Deimination Signatures in Plasma and Plasma-EVs and Protein Deimination in the Brain Vasculature in a Rat Model of Pre-Motor Parkinson’s Disease. Int J Mol Sci. 2020, 21. [Google Scholar] [CrossRef] [PubMed]
- Mercer, A.; Jaunmuktane, Z.; Hristova, M.; Lange, S. Differential, Stage Dependent Detection of Peptidylarginine Deiminases and Protein Deimination in Lewy Body Diseases-Findings from a Pilot Study. Int J Mol Sci. 2022, 23. [Google Scholar] [CrossRef]
- Witalison, E.E.; Thompson, P.R.; Hofseth, L.J. Protein arginine deiminases and associated citrullination: Physiological functions and diseases associated with dysregulation. Curr. Drug Targets 2015, 16, 700–710. [Google Scholar] [CrossRef] [PubMed]
- György, B.; Tóth, E.; Tarcsa, E.; Falus, A.; Buzás, E.I. Citrullination: A posttranslational modification in health and disease. Int. J. Biochem. Cell Biol. 2006, 38, 1662–1677. [Google Scholar] [CrossRef] [PubMed]
- Alghamdi, M.; Alasmari, D.; Assiri, A.; Mattar, E.; Aljaddawi, A.A.; Alattas, S.G.; Redwan, E.M. An Overview of the Intrinsic Role of Citrullination in Autoimmune Disorders. J. Immunol. Res. 2019, 2019, 7592851. [Google Scholar] [CrossRef]
- Vossenaar, E.R.; Zendman, A.J.; van Venrooij, W.J.; Pruijn, G.J. PAD, a growing family of citrullinating enzymes: Genes, features and involvement in disease. Bioessays. 2003, 25(11), 1106. [Google Scholar] [CrossRef]
- Magnadóttir, B.; Hayes, P.; Hristova, M.; Bragason, B.T.; Nicholas, A.P.; Dodds, A.W.; Guðmundsdóttir, S.; Lange, S. Post-translational protein deimination in cod (Gadus morhua L.) ontogeny novel roles in tissue remodelling and mucosal immune defences? Dev Comp Immunol. 2018, 87, 157. [Google Scholar] [CrossRef]
- Christophorou, M.A. The virtues and vices of protein citrullination. R Soc Open Sci. 2022, 9. [Google Scholar] [CrossRef]
- Shimada, N.; Handa, S.; Uchida, Y.; Fukuda, M.; Maruyama, N.; Asaga, H.; Choi, E.K.; Lee, J.; Ishigami, A. Developmental and age-related changes of peptidylarginine deiminase 2 in the mouse brain. J. Neurosci. Res. 2010, 88, 798–806. [Google Scholar] [CrossRef]
- Ishigami, A.; Ohsawa, T.; Hiratsuka, M.; Taguchi, H.; Kobayashi, S.; Saito, Y.; Murayama, S.; Asaga, H.; Toda, T.; Kimura, N.; Maruyama, N. Abnormal accumulation of citrullinated proteins catalyzed by peptidylarginine deiminase in hippocampal extracts from patients with Alzheimer’s disease. J. Neurosci. Res. 2005, 80, 120–128. [Google Scholar] [CrossRef]
- Ishigami, A.; Masutomi, H.; Handa, S.; Nakamura, M.; Nakaya, S.; Uchida, Y.; Saito, Y.; Murayama, S.; Jang, B.; Jeon, Y.C.; Choi, E.K.; Kim, Y.S.; Kasahara, Y.; Maruyama, N.; Toda, T. Mass spectrometric identification of citrullination sites and immunohistochemical detection of citrullinated glial fibrillary acidic protein in Alzheimer’s disease brains. J. Neurosci. Res. 2015, 93, 1664–1674. [Google Scholar] [CrossRef]
- Jang, B.; Jin, J.K.; Jeon, Y.C.; Cho, H.J.; Ishigami, A.; Choi, K.C.; Carp, R.I.; Maruyama, N.; Kim, Y.S.; Choi, E.K. Involvement of peptidylarginine deiminase-mediated post-translational citrullination in pathogenesis of sporadic Creutzfeldt-Jakob disease. Acta Neuropathol. 2009, 119, 199–210. [Google Scholar] [CrossRef]
- Jang, B.; Ishigami, A.; Maruyama, N.; Carp, R.I.; Kim, Y.S.; Choi, E.K. Peptidylarginine deiminase and protein citrullination in prion diseases: Strong evidence of neurodegeneration. Prion 2013, 7, 42–46. [Google Scholar] [CrossRef]
- Yusuf, I.O.; Qiao, T.; Parsi, S.; Tilvawala, R.; Thompson, P.R.; Xu, Z. Protein citrullination marks myelin protein aggregation and disease progression in mouse ALS models. Acta Neuropathol Commun. 2022, 10. [Google Scholar] [CrossRef]
- Raijmakers, R.; Vogelzangs, J.; Raats, J.; Panzenbeck, M.; Corby, M.; Jiang, H.; Thibodeau, M.; Haynes, N.; van Venrooij, W.J.; Pruijn, G. J.; Werneburg, B. Experimental autoimmune encephalomyelitis induction in peptidylarginine deiminase 2 knockout mice. J Comp Neurol. 2006, 498, 217. [Google Scholar] [CrossRef]
- van Beers, J.J.; Zendman, A.J.; Raijmakers, R.; Stammen-Vogelzangs, J.; Pruijn, G.J. Peptidylarginine deiminase expression and activity in PAD2 knock-out and PAD4-low mice. Biochimie. 2013, 95, 299. [Google Scholar] [CrossRef]
- Mastronardi, F.G.; Wood, D.D.; Mei, J.; Raijmakers, R.; Tseveleki, V.; Dosch, H.M.; Probert, L.; Casaccia-Bonnefil, P.; Moscarello, M.A. Increased citrullination of histone H3 in multiple sclerosis brain and animal models of demyelination: A role for tumor necrosis factor-induced peptidylarginine deiminase 4 translocation. J. Neurosci. 2006, 26, 11387–11396. [Google Scholar] [CrossRef]
- Faigle, W.; Cruciani, C.; Wolski, W.; Roschitzki, B.; Puthenparampil, M.; Tomas-Ojer, P.; Sellés-Moreno, C.; Zeis, T.; Jelcic, I.; Schaeren-Wiemers, N.; Sospedra, M.; Martin, R. Brain Citrullination Patterns and T Cell Reactivity of Cerebrospinal Fluid-Derived CD4+ T Cells in Multiple Sclerosis. Front. Immunol. 2019, 10, 540. [Google Scholar] [CrossRef]
- Lazarus, R.C.; Buonora, J.E.; Flora, M.N.; Freedy, J.G.; Holstein, G.R.; Martinelli, G.P.; Jacobowitz, D.M.; Mueller, G.P. Protein Citrullination: A Proposed Mechanism for Pathology in Traumatic Brain Injury. Front. Neurol. 2015, 6, 204. [Google Scholar] [CrossRef] [PubMed]
- Lange, S.; Rocha-Ferreira, E.; Thei, L.; Mawjee, P.; Bennett, K.; Thompson, P.R.; Subramanian, V.; Nicholas, A.P.; Peebles, D.; Hristova, M.; et al. Peptidylarginine deiminases: Novel drug targets for prevention of neuronal damage following hypoxic ischemic insult (HI) in neonates. J. Neurochem. 2014, 130, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Lange, S. Peptidylarginine deiminases and extracellular vesicles: Prospective drug targets and biomarkers in central nervous system diseases and repair. Neural. Regen. Res. 2021, 16, 934–938. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.; Liu, L.; Cao, Y.; et al. Inhibition of neutrophil extracellular trap formation ameliorates neuroinflammation and neuronal apoptosis via STING-dependent IRE1α/ASK1/JNK signaling pathway in mice with traumatic brain injury. J Neuroinflammation. 2023, 20. [Google Scholar] [CrossRef] [PubMed]
- Lange, S.; Wray, S.; Devine, M.; Matarin, M.; Hardy, J. Protein Deimination in Protein Misfolding Disorders–Modelled in Human Induced Pluripotent Stem Cells (iPSCs). In Protein Deimination in Human Health and Disease; Nicholas, A.P., Bhattacharya, S.K., Eds.; Springer Science and Business Media: New York, NY, USA, 2017; Volume 2. [Google Scholar]
- Lange, S.; Gögel, S.; Leung, K.Y.; Vernay, B.; Nicholas, A.P.; Causey, C.P.; Thompson, P.R.; Greene, N.D.; Ferretti, P. Protein deiminases: New players in the developmentally regulated loss of neural regenerative ability. Dev. Biol. 2011, 355, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Kosgodage, U.S.; Uysal-Onganer, P.; MacLatchy, A.; Kraev, I.; Chatterton, N.P.; Nicholas, A.P.; Inal, J.M.; Lange, S. Peptidylarginine Deiminases Post-Translationally Deiminate Prohibitin and Modulate Extracellular Vesicle Release and MicroRNAs in Glioblastoma Multiforme. Int. J. Mol. Sci. 2018, 20, 103. [Google Scholar] [CrossRef]
- Uysal-Onganer, P.; MacLatchy, A.; Mahmoud, R.; Kraev, I.; Thompson, P.R.; Inal, J.M.; Lange, S. Peptidylarginine Deiminase Isozyme-Specific PAD2, PAD3 and PAD4 Inhibitors Differentially Modulate Extracellular Vesicle Signatures and Cell Invasion in Two Glioblastoma Multiforme Cell Lines. Int. J. Mol. Sci. 2020, 21, 1495. [Google Scholar] [CrossRef]
- U, K.P.; Subramanian, V.; Nicholas, A.P.; Thompson, P.R.; Ferretti, P. Modulation of calcium-induced cell death in human neural stem cells by the novel peptidylarginine deiminase-AIF pathway. Biochim Biophys Acta. 2014, 1843, 1162. [Google Scholar] [CrossRef]
- Pasquero, S.; Gugliesi, F.; Biolatti, M.; et al. Citrullination profile analysis reveals peptidylarginine deaminase 3 as an HSV-1 target to dampen the activity of candidate antiviral restriction factors. PLoS Pathog. 2023, 19. [Google Scholar] [CrossRef]
- Méchin, M.C.; Takahara, H.; Simon, M. Deimination and peptidylarginine deiminases in skin physiology and diseases. Int. J. Mol. Sci. 2020, 21, 566. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, X.; Zhang, M.; Li, T.; Muth, A.; Thompson, P.R.; Coonrod, S.A.; Zhang, X. Peptidylarginine deiminase 1-catalyzed histone citrullination is essential for early embryo development. Sci. Rep. 2016, 6, 38727. [Google Scholar] [CrossRef]
- Qin, H.; Liu, X.; Li, F.; Miao, L.; Li, T.; Xu, B.; An, X.; Muth, A.; Thompson, P.R.; Coonrod, S.A.; Zhang, X. PAD1 promotes epithelial-mesenchymal transition and metastasis in triple-negative breast cancer cells by regulating MEK1-ERK1/2-MMP2 signaling. Cancer Lett. 2017, 409, 30–41. [Google Scholar] [CrossRef]
- Coassolo, S.; Davidson, G.; Negroni, L.; Gambi, G.; Daujat, S.; Romier, C.; Davidson, I. Citrullination of pyruvate kinase M2 by PADI1 and PADI3 regulates glycolysis and cancer cell proliferation. Nat. Commun. 2021, 12, 1718. [Google Scholar] [CrossRef]
- Williams, J.P.C.; Walport, L.J. PADI6: What we know about the elusive fifth member of the peptidyl arginine deiminase family. Philos Trans R Soc Lond B Biol Sci. 2023, 378. [Google Scholar] [CrossRef]
- Yurttas, P.; Vitale, A.M.; Fitzhenry, R.J.; Cohen-Gould, L.; Wu, W.; Gossen, J.A.; Coonrod, S.A. Role for PADI6 and the cytoplasmic lattices in ribosomal storage in oocytes and translational control in the early mouse embryo. Development 2008, 135, 2627–2636. [Google Scholar] [CrossRef]
- Xu, Y.; Shi, Y.; Fu, J.; Yu, M.; Feng, R.; Sang, Q.; Liang, B.; Chen, B.; Qu, R.; Li, B.; Yan, Z.; Mao, X.; Kuang, Y.; Jin, L.; He, L. , Sun, X.; Wang, L. Mutations in PADI6 Cause Female Infertility Characterized by Early Embryonic Arrest. Am. J. Hum. Genet. 2016, 99, 744–752. [Google Scholar] [CrossRef]
- Inal, J.M.; Hristova, M.; Lange, S. A Pilot Study on Peptidylarginine Deiminases and Protein Deimination in Animal Cancers across Vertebrate Species. Int. J. Mol. Sci. 2022, 23, 8697. [Google Scholar] [CrossRef]
- D’Alessio, S.; Cheng, H.; Eaton, L.; Kraev, I.; Pamenter, M.E.; Lange, S. Acute Hypoxia Alters Extracellular Vesicle Signatures and the Brain Citrullinome of Naked Mole-Rats (Heterocephalus glaber). Int J Mol Sci. 2022, 23. [Google Scholar] [CrossRef] [PubMed]
- Lyu, Y. , Huang, Y., Shi, G., Lei, X., Li, K., Zhou, R., Bai, L., Qin, C. Transcriptome profiling of five brain regions in a 6-hydroxydopamine rat model of Parkinson’s disease. CNS Neurosci Ther. 2021, 27, 1289. [Google Scholar] [CrossRef] [PubMed]
- Vaibhav, K.; Braun, M.; Alverson, K.; Khodadadi, H.; Kutiyanawalla, A.; Ward, A.; Banerjee, C.; Sparks, T.; Malik, A.; Rashid, M.H.; et al. Neutrophil extracellular traps exacerbate neurological deficits after traumatic brain injury. Sci. Adv. 2020, 6, eaax8847. [Google Scholar] [CrossRef]
- Nicholas, A.P.; Lu, L.; Heaven, M.; Kadish, I.; van Groen, T.; Accavitti-Loper, M.A.; Wewering, S.; Kofskey, D.; Gambetti, P.; Brenner, M. Ongoing studies of deimination in neurodegenerative diseases using the F95 antibody. In Protein Deimination in Human Health and Disease; Nicholas, A.P., Bhattacharya, S.K., Eds.; Springer-Verlag: New York, NY, USA, 2014; pp. 257–280. [Google Scholar]
- Petrozziello, T.; Mills, A.N.; Vaine, C.A.; Penney, E.B.; Fernandez-Cerado, C.; Legarda, G.; Velasco-Andrada, M.S.; Acuña, P.J.; Ang, M.A.; Muñoz, E.L.; Diesta, C. C. E.; Macalintal-Canlas, R.; Acuña-Sunshine, G.; Ozelius, L. J.; Sharma, N.; Bragg, D. C.; Sadri-Vakili, G. Neuroinflammation and histone H3 citrullination are increased in X-linked Dystonia Parkinsonism post-mortem prefrontal cortex. Neurobiol. Dis. 2020, 144, 105032. [Google Scholar] [CrossRef]
- Mondal, S.; Thompson, P.R. Protein Arginine Deiminases (PADs): Biochemistry and Chemical Biology of Protein Citrullination. Acc Chem Res. 2019, 52, 818. [Google Scholar] [CrossRef]
- Verma, D.K.; Seo, B.A.; Ghosh, A.; Ma, S.X.; Hernandez-Quijada, K.; Andersen, J.K.; Ko, H.S.; Kim, Y.H. Alpha-Synuclein Preformed Fibrils Induce Cellular Senescence in Parkinson’s Disease Models. Cells. 2021, 10. [Google Scholar] [CrossRef]
- Melis, M.; Pistis, M. Endocannabinoid signaling in midbrain dopamine neurons: More than physiology? Curr. Neuropharmacol. 2007, 5, 268–277. [Google Scholar] [CrossRef]
- Orgado, J.M.; Fernández-Ruiz, J.; Romero, J. The endocannabinoid system in neuropathological states. Int. Rev. Psychiatry 2009, 21, 172–180. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Q.J.; Wang, H.S.; Wang, T.; Liu, J. Genome-wide microarray analysis identifies a potential role for striatal retrograde endocannabinoid signaling in the pathogenesis of experimental L-DOPA-induced dyskinesia. Synapse 2014, 68, 332–343. [Google Scholar] [CrossRef]
- Aymerich, M.S.; Aso, E.; Abellanas, M.A.; Tolon, R.M.; Ramos, J.A.; Ferrer, I.; Romero, J.; Fernández-Ruiz, J. Cannabinoid pharmacology/therapeutics in chronic degenerative disorders affecting the central nervous system. Biochem. Pharmacol. 2018, 157, 67–84. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, S.P.; Bano, S.; Sen, S.; Suchal, K.; Kumar, S.; Nikolajeff, F.; Dey, S.K; Sharma, V. Altered neural cell junctions and ion-channels leading to disrupted neuron communication in Parkinson’s disease. NPJ Parkinsons Dis. 2022, 8. [Google Scholar] [CrossRef]
- Sen, S.; Lagas, S.; Roy, A.; Kumar, H. Cytoskeleton saga: Its regulation in normal physiology and modulation in neurodegenerative disorders. Eur J Pharmacol. 2022, 925. [Google Scholar] [CrossRef]
- Calabresi, P.; Di Filippo, M.; Gallina, A.; Wang, Y.; Stankowski, J.N.; Picconi, B.; Dawson, V.L.; Dawson, T.M. New synaptic and molecular targets for neuroprotection in Parkinson’s disease. Mov Disord. 2013, 28, 51. [Google Scholar] [CrossRef] [PubMed]
- Barber, C.N.; Huganir, R.L.; Raben, D.M. Phosphatidic acid-producing enzymes regulating the synaptic vesicle cycle: Role for PLD? Adv Biol Regul. 2018, 67, 141. [Google Scholar] [CrossRef]
- Beilina, A.; Bonet-Ponce, L.; Kumaran, R.; et al. The Parkinson’s Disease Protein LRRK2 Interacts with the GARP Complex to Promote Retrograde Transport to the trans-Golgi Network. Cell Rep. 2020, 31. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Hwang, B.; Reva, M.; Lee, J.; Lee, B. E.; Lee, Y.; Cho, E. J.; Jeong, M.; Lee, S.E.; Myung, K.; Baik, J.H.; Park, J.H.; Kim, J.I. GABAergic-like dopamine synapses in the brain. Cell Rep. 2023, 42. [Google Scholar] [CrossRef] [PubMed]
- Bohnen, N.I.; Roytman, S.; Kanel, P.; et al. Progression of regional cortical cholinergic denervation in Parkinson’s disease. Brain Commun. 2022, 4, fcac. [Google Scholar] [CrossRef] [PubMed]
- Gcwensa, N.Z.; Russell, D.L.; Cowell, R.M.; Volpicelli-Daley, L.A. Molecular Mechanisms Underlying Synaptic and Axon Degeneration in Parkinson’s Disease. Front Cell Neurosci. 2021, 15. [Google Scholar] [CrossRef]
- de Natale, E.R.; Wilson, H.; Politis, M. Serotonergic imaging in Parkinson’s disease. Prog Brain Res. 2021, 261, 303. [Google Scholar] [CrossRef]
- Richard, I.H. Depression in Parkinson’s Disease. Curr Treat Options Neurol. 2000, 2, 263. [Google Scholar] [CrossRef] [PubMed]
- Weintraub, D.; Aarsland, D.; Chaudhuri, K.R.; Dobkin, R.D.; Leentjens, A.F.; Rodriguez-Violante, M.; Schrag, A. The neuropsychiatry of Parkinson’s disease: Advances and challenges. Lancet Neurol. 2022, 21, 89. [Google Scholar] [CrossRef]
- Reumann, D.; Krauditsch, C.; Novatchkova, M.; Sozzi, E.; Wong, S.N.; Zabolocki, M.; Priouret, M.; Doleschall, B.; Ritzau-Reid, K. I.; Piber, M.; Morassut, I.; Fieseler, C.; Fiorenzano, A.; Stevens, M.M.; Zimmer, M.; Bardy, C.; Parmar, M.; Knoblich, J.A. In vitro modeling of the human dopaminergic system using spatially arranged ventral midbrain-striatum-cortex assembloids. Nat Methods. 2023, 20, 2034. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Yang, Q.; B Joshi, R.; Liu, Y.; Akbar, M.; Song, B. J.; Zhou, S.; Wang, X. Role of Alcohol Drinking in Alzheimer’s Disease, Parkinson’s Disease, and Amyotrophic Lateral Sclerosis. Int J Mol Sci. 2020, 21. [Google Scholar] [CrossRef]
- Jang, Y.; Pletnikova, O.; Troncoso, J.C.; Pantelyat, A.Y.; Dawson, T.M.; Rosenthal, L.S.; Na, C.H. Mass Spectrometry-Based Proteomics Analysis of Human Substantia Nigra From Parkinson’s Disease Patients Identifies Multiple Pathways Potentially Involved in the Disease. Mol Cell Proteomics. 2023, 22. [Google Scholar] [CrossRef]
- Iovino, M.; Messana, T.; Tortora, A.; Giusti, C.; Lisco, G.; Giagulli, V.A.; Guastamacchia, E.; De Pergola, G.; Triggiani, V. Oxytocin Signaling Pathway: From Cell Biology to Clinical Implications. Endocr Metab Immune Disord Drug Targets. 2021, 21, 91. [Google Scholar] [CrossRef]
- Higashida, H.; Hashii, M.; Tanaka, Y.; et al. CD38, CD157, and RAGE as Molecular Determinants for Social Behavior. Cells. 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Lestón Pinilla, L.; Ugun-Klusek, A.; Rutella, S.; De Girolamo, L.A. Hypoxia Signaling in Parkinson’s Disease: There Is Use in Asking “What HIF?”. Biology (Basel). 2021, 10. [Google Scholar] [CrossRef] [PubMed]
- Trist, B.G.; Hare, D.J.; Double, K.L. Oxidative stress in the aging substantia nigra and the etiology of Parkinson’s disease. Aging Cell. 2019, 18. [Google Scholar] [CrossRef] [PubMed]
- Peggion, C.; Barazzuol, L.; Poggio, E.; Calì, T.; Brini, M. Ca2+ signalling: A common language for organelles crosstalk in Parkinson’s disease. Cell Calcium. 2023, 115. [Google Scholar] [CrossRef]
- Barazzuol, L.; Giamogante, F.; Brini, M.; Calì, T. PINK1/Parkin Mediated Mitophagy, Ca2+ Signalling, and ER-Mitochondria Contacts in Parkinson’s Disease. Int J Mol Sci. 2020, 21. [Google Scholar] [CrossRef]
- Virdi, G.S.; Choi, M.L.; Evans, J.R.; et al. Protein aggregation and calcium dysregulation are hallmarks of familial Parkinson’s disease in midbrain dopaminergic neurons. NPJ Parkinsons Dis. 2022, 8. [Google Scholar] [CrossRef]
- Wang, J.; Zhao, J.; Zhao, K.; Wu, S.; Chen, X.; Hu, W. The Role of Calcium and Iron Homeostasis in Parkinson’s Disease. Brain Sci. 2024, 14. [Google Scholar] [CrossRef]
- Wang, Z.L.; Yuan, L.; Li, W.; Li, J.Y. Ferroptosis in Parkinson’s disease: Glia-neuron crosstalk. Trends Mol Med. 2022, 28, 258. [Google Scholar] [CrossRef]
- Mahoney-Sánchez, L.; Bouchaoui, H.; Ayton, S.; Devos, D.; Duce, J.A.; Devedjian, J.C. Ferroptosis and its potential role in the physiopathology of Parkinson’s Disease. Prog Neurobiol. 2021, 196. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.S.; Gao, L.; Han, Z.; et al. Ferroptosis in Parkinson’s disease: Molecular mechanisms and therapeutic potential. Ageing Res Rev. 2023, 91. [Google Scholar] [CrossRef] [PubMed]
- Thapa, K.; Khan, H.; Kanojia, N.; Singh, T.G.; Kaur, A.; Kaur, G. Therapeutic Insights on Ferroptosis in Parkinson’s disease. Eur J Pharmacol. 2022, 930. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Gao, X.; Zhao, L.; Wang, Y.; Zhang, J.; Liu, S. Effects of Cannabidiol on Parkinson’s Disease in a Transgenic Mouse Model by Gut-Brain Metabolic Analysis. Evid Based Complement Alternat Med. 2022, 2022. [Google Scholar] [CrossRef] [PubMed]
- Fusco, C.M.; Desch, K.; Dörrbaum, A.R.; Wang, M.; Staab, A.; Chan, I.C.W.; Vail, E.; Villeri, V.; Langer, J.D.; Schuman, E.M. Neuronal ribosomes exhibit dynamic and context-dependent exchange of ribosomal proteins. Nat Commun. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Tresse, E.; Marturia-Navarro, J.; Sew, W.Q.G.; et al. Mitochondrial DNA damage triggers spread of Parkinson’s disease-like pathology. Mol Psychiatry. 2023, 28, 4902. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Meng, X.; Yang, C.; Fang, D.; Wang, X.; An, J.; Zhang, J.; Wang, L.; Lu, T.; Ruan, H.B.; et al. Brown adipose tissue activation in a rat model of Parkinson’s disease. Am. J. Physiol. Endocrinol. Metab. 2017, 313, E731–E736. [Google Scholar] [CrossRef]
- Naeem, U.; Arshad, A.R.; Jawed, A.; et al. Glycolysis: The Next Big Breakthrough in Parkinson’s Disease. Neurotox Res. 2022, 40, 1707. [Google Scholar] [CrossRef]
- Tang, B.L. Glucose, glycolysis, and neurodegenerative diseases. J Cell Physiol. 2020, 235, 7653. [Google Scholar] [CrossRef] [PubMed]
- Pathania, A.; Garg, P.; Sandhir, R. Impaired mitochondrial functions and energy metabolism in MPTP-induced Parkinson’s disease: Comparison of mice strains and dose regimens. Metab Brain Dis. 2021, 36, 2343. [Google Scholar] [CrossRef] [PubMed]
- Kopp, K.O.; Glotfelty, E.J.; Li, Y.; Greig, N.H. Glucagon-like peptide-1 (GLP-1) receptor agonists and neuroinflammation: Implications for neurodegenerative disease treatment. Pharmacol Res. 2022, 186. [Google Scholar] [CrossRef] [PubMed]
- Jha, S.K.; Jha, N.K.; Kumar, D.; Ambasta, R.K.; Kumar, P. Linking mitochondrial dysfunction, metabolic syndrome and stress signaling in Neurodegeneration. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 1132–1146. [Google Scholar] [CrossRef]
- Limphaibool, N.; Iwanowski, P.; Holstad, M.J.V.; Perkowska, K. Parkinsonism in Inherited Metabolic Disorders: Key Considerations and Major Features. Front. Neurol. 2018, 9, 857. [Google Scholar] [CrossRef] [PubMed]
- Özcan, E.; Çakır, T. Genome-Scale Brain Metabolic Networks as Scaffolds for the Systems Biology of Neurodegenerative Diseases: Mapping Metabolic Alterations. Adv. Neurobiol. 2018, 21, 195–217. [Google Scholar]
- Gonçalves, V.C.; Cuenca-Bermejo, L.; Fernandez-Villalba, E.; Martin-Balbuena, S.; da Silva Fernandes, M.J.; Scorza, C.A.; Herrero, M.T. Heart Matters: Cardiac Dysfunction and Other Autonomic Changes in Parkinson’s Disease. Neuroscientist. 2022, 28, 530. [Google Scholar] [CrossRef] [PubMed]
- Smeyne, R.J.; Noyce, A.J.; Byrne, M.; Savica, R.; Marras, C. Infection and Risk of Parkinson’s Disease. J Parkinsons Dis. 2021, 11, 31. [Google Scholar] [CrossRef] [PubMed]
- Arisan, E.D., Uysal-Onganer. Uysal-Onganer, P., Lange, S. Putative Roles for Peptidylarginine Deiminases in COVID-. Int J Mol Sci. 2020, 21. [Google Scholar] [CrossRef] [PubMed]
- Leta, V.; Boura, I.; van Wamelen, D.J.; Rodriguez-Violante, M.; Antonini, A.; Chaudhuri, K.R. Covid-19 and Parkinson’s disease: Acute clinical implications, long-COVID and post-COVID-19 parkinsonism. Int Rev Neurobiol. 2022, 165, 63. [Google Scholar] [CrossRef]
- Priyanka, *!!! REPLACE !!!*; Seth, P. Insights Into the Role of Mortalin in Alzheimer’s Disease, Parkinson’s Disease, and HIV-1-Associated Neurocognitive Disorders. Front Cell Dev Biol. 2022, 10. [Google Scholar] [CrossRef]
- Olson, K.E.; Bade, A.N.; Namminga, K.L.; Potash, M.J.; Mosley, R.L.; Poluektova, L.Y.; Volsky, D.J.; Gendelman, H.E. Persistent EcoHIV infection induces nigral degeneration in 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-intoxicated mice. J Neurovirol. 2018, 24, 398. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.M.; Dahodwala, N. Sex differences in Parkinson’s disease and other movement disorders. Exp Neurol. 2014, 259, 44. [Google Scholar] [CrossRef] [PubMed]
- Al-Sweidi, S.; Morissette, M.; Bourque, M.; Di Paolo, T. Estrogen receptors and gonadal steroids in vulnerability and protection of dopamine neurons in a mouse model of Parkinson’s disease. Neuropharmacology. 2011, 61, 583. [Google Scholar] [CrossRef] [PubMed]
- Tan, E.K.; Chan, L.L.; Auchus, A.P. Reversible Parkinsonism in systemic lupus erythematosus. J. Neurol. Sci. 2001, 193, 53–57. [Google Scholar] [CrossRef]
- Kivity, S.; Agmon-Levin, N.; Zandman-Goddard, G.; Chapman, J.; Shoenfeld, Y. Neuropsychiatric lupus: A mosaic of clinical presentations. BMC Med. 2015, 13, 43. [Google Scholar] [CrossRef]
- Li, M.; Wan, J.; Xu, Z.; Tang, B. The association between Parkinson’s disease and autoimmune diseases: A systematic review and meta-analysis. Front Immunol. 2023, 14. [Google Scholar] [CrossRef] [PubMed]
- Kalaani, J.; Roche, J.; Hamade, E.; Badran, B.; Jaber, M.; Gaillard, A.; Prestoz, L. Axon guidance molecule expression after cell therapy in a mouse model of Parkinson’s disease. Restor Neurol Neurosci. 2016, 34, 877. [Google Scholar] [CrossRef]
- Lesnick, T.G.; Sorenson, E.J.; Ahlskog, J.E.; Henley, J.R.; Shehadeh, L.; Papapetropoulos, S.; Maraganore, D.M. Beyond Parkinson disease: Amyotrophic lateral sclerosis and the axon guidance pathway. PLoS ONE. 2008, 3. [Google Scholar] [CrossRef]
- Lin, L.; Lesnick, T.G.; Maraganore, D.M.; Isacson, O. Axon guidance and synaptic maintenance: Preclinical markers for neurodegenerative disease and therapeutics. Trends Neurosci. 2009, 32, 142. [Google Scholar] [CrossRef]
- Sullivan, R.; Yau, W.Y.; O’Connor, E.; Houlden, H. Spinocerebellar ataxia: An update. J Neurol. 2019, 266, 533. [Google Scholar] [CrossRef]
- Horvath, J.D.; Casas, M.; Kutchukian, C.; Sánchez, S.C.; Pergande, M.R.; Cologna, S.M.; Simó, S.; Dixon, R.E.; Dickson, E.J. α-Synuclein-dependent increases in PIP5K1γ drive inositol signaling to promote neurotoxicity. Cell Rep. 2023, 42. [Google Scholar] [CrossRef]
- Napolitano, F.; D’Angelo, L.; de Girolamo, P.; Avallone, L.; de Lange, P.; Usiello, A. The Thyroid Hormone-target Gene Rhes a Novel Crossroad for Neurological and Psychiatric Disorders: New Insights from Animal Models. Neuroscience. 2018, 384, 419. [Google Scholar] [CrossRef]
- Li, Y.; Xia, Y.; Yin, S.; Wan, F.; Hu, J.; Kou, L.; Sun, Y.; Wu, J.; Zhou, Q.; Huang, J.; Xiong, N.; Wang, T. Targeting Microglial α-Synuclein/TLRs/NF-kappaB/NLRP3 Inflammasome Axis in Parkinson’s Disease. Front Immunol. 2021, 12. [Google Scholar] [CrossRef]
- Piancone, F.; La Rosa, F.; Marventano, I.; Saresella, M.; Clerici, M. The Role of the Inflammasome in Neurodegenerative Diseases. Molecules. 2021, 26. [Google Scholar] [CrossRef]
- Panicker, N.; Kanthasamy, A.; Kanthasamy, A.G. Fyn amplifies NLRP3 inflammasome signaling in Parkinson’s disease. Aging (Albany NY). 2019, 11, 5871. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhang, B.; Tian, T.; Zhang, B.; Shi, G.; Zhang, C.; Li, G. , Huang, M. Taurine protects dopaminergic neurons in paraquat-induced Parkinson’s disease mouse model through PI3K/Akt signaling pathways. Amino Acids. 2022, 54, 1. [Google Scholar] [CrossRef]
- Deneubourg, C.; Ramm, M.; Smith, L.J.; Baron, O.; Singh, K.; Byrne, S.C.; Duchen, M.R.; Gautel, M.; Eskelinen, E.L.; Fanto, M.; Jungbluth, H. The spectrum of neurodevelopmental, neuromuscular and neurodegenerative disorders due to defective autophagy. Autophagy. 2022, 18, 496. [Google Scholar] [CrossRef]
- Ferrer-Raventós, P.; Beyer, K. Alternative platelet activation pathways and their role in neurodegenerative diseases. Neurobiol Dis. 2021, 159. [Google Scholar] [CrossRef]
- Oliynyk, Z.; Rudyk, M.; Dovbynchuk, T.; Dzubenko, N.; Tolstanova, G.; Skivka, L. Inflammatory hallmarks in 6-OHDA- and LPS-induced Parkinson’s disease in rats. Brain Behav Immun Health. 2023, 30. [Google Scholar] [CrossRef]
- Liu, X.; Chen, J.; Guan, T.; Yao, H.; Zhang, W.; Guan, Z.; Wang, Y. miRNAs and target genes in the blood as biomarkers for the early diagnosis of Parkinson’s disease. BMC Syst Biol. 2019, 13. [Google Scholar] [CrossRef]
- Moya, I.M.; Halder, G. Hippo-YAP/TAZ signalling in organ regeneration and regenerative medicine. Nat Rev Mol Cell Biol. 2019, 20, 211. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.; Hu, Y.; Lan, T.; Guan, K.L.; Luo, T.; Luo, M. The Hippo signalling pathway and its implications in human health and diseases. Signal Transduct Target Ther. 2022, 7. [Google Scholar] [CrossRef] [PubMed]
- Eddin, L.B.; Azimullah, S.; Jha, N.K.; Nagoor Meeran, M.F.; Beiram, R.; Ojha, S. Limonene, a Monoterpene, Mitigates Rotenone-Induced Dopaminergic Neurodegeneration by Modulating Neuroinflammation, Hippo Signaling and Apoptosis in Rats. Int J Mol Sci. 2023, 24. [Google Scholar] [CrossRef] [PubMed]
- Varela, L.; Garcia-Rendueles, M.E.R. Oncogenic Pathways in Neurodegenerative Diseases. Int J Mol Sci. 2022, 23. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Huang, G.; Liu, J.; Ge, J.; Zhang, W.; Mei, Z. An update on the role of Hippo signaling pathway in ischemia-associated central nervous system diseases. Biomed Pharmacother. 2023, 162. [Google Scholar] [CrossRef]
- Xu, D.D.; Li, G.Q.; Wu, Z.S.; Liu, X.Q.; Yang, X.X.; Wang, J.H. Bioinformatics analysis and identification of genes and molecular pathways involved in Parkinson’s disease in patients with mutations in the glucocerebrosidase gene. Neuroreport. 2021, 32, 918. [Google Scholar] [CrossRef] [PubMed]
- Stamou, M.; Grodzki, A.C.; van Oostrum, M.; Wollscheid, B.; Lein, P.J. Fc gamma receptors are expressed in the developing rat brain and activate downstream signaling molecules upon cross-linking with immune complex. J Neuroinflammation 2018, 15. [Google Scholar] [CrossRef]
- Park, Y.H.; Hodges, A.; Risacher, S.L.; Lin, K.; Jang, J.W.; Ahn, S.; Kim, S.; Lovestone, S.; Simmons, A.; Weiner, M.W.; Saykin, A.J.; Nho, K. AddNeuroMed consortium and the Alzheimer’s Disease Neuroimaging Initiative. Dysregulated Fc gamma receptor-mediated phagocytosis pathway in Alzheimer’s disease: Network-based gene expression analysis. Neurobiol Aging 2020, 88, 24. [Google Scholar] [CrossRef] [PubMed]
- Massey, W.J.; Kay, K.E.; Jaramillo, T.C.; et al. Metaorganismal choline metabolism shapes olfactory perception. J Biol Chem. 2023, 299. [Google Scholar] [CrossRef]
- Yang, Y.; Qian, J.; Mei, J.; Zhong, K.; Niu, C. In vivo detection of metabolic changes in the striatum of proteasomal inhibition-induced Parkinson’s disease in rats using proton MR spectroscopy at 9.4 T. Int J Neurosci. 2020, 130, 153. [Google Scholar] [CrossRef]
- Sanami, S.; Shamsabadi, S.; Dayhimi, A.; Pirhayati, M.; Ahmad, S.; Pirhayati, A.; Ajami, M.; Hemati, S.; Shirvani, M.; Alagha, A.; Abbarin, D.; Alizadeh, A.; Pazoki-Toroudi, H. Association between cytomegalovirus infection and neurological disorders: A systematic review. Rev Med Virol. 2024, 34, 34. [Google Scholar] [CrossRef]
- Bu, X.L.; Wang, X.; Xiang, Y.; Shen, L.L.; Wang, Q.H.; Liu, Y.H.; Jiao, S.S.; Wang, Y.R.; Cao, H.Y.; Yi, X.; Liu, C.H.; Deng, B.; Yao, X.Q.; Xu, Z.Q.; Zhou, H.D.; Wang, Y.J. The association between infectious burden and Parkinson’s disease: A case-control study. Parkinsonism Relat Disord. 2015, 21, 877. [Google Scholar] [CrossRef] [PubMed]
- Bhattarai, Y.; Kashyap, P.C. Parkinson’s disease: Are gut microbes involved? Am J Physiol Gastrointest Liver Physiol. 2020, 2020 319, G529. [Google Scholar] [CrossRef]
- Tan, C.; Liu, X.; Chen, J. Microarray Analysis of the Molecular Mechanism Involved in Parkinson’s Disease. Parkinsons Dis. 2018. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.N.; Liu, J.C.; Ju, J.Q.; Wang, Y.; Sun, S.C. LRRK2 regulates actin assembly for spindle migration and mitochondrial function in mouse oocyte meiosis. J Mol Cell Biol. 2022, 2022 14, mjab. [Google Scholar] [CrossRef]
- Esposito, G.; Vitale, A.M.; Leijten, F.P.; Strik, A.M.; Koonen-Reemst, A.M.; Yurttas, P.; Robben, T.J.; Coonrod, S.; Gossen, J.A. Peptidylarginine deiminase (PAD) 6 is essential for oocyte cytoskeletal sheet formation and female fertility. Mol Cell Endocrinol. 2007, 273, 25. [Google Scholar] [CrossRef] [PubMed]
- Horibata, S.; Coonrod, S.A.; Cherrington, B.D. Role for peptidylarginine deiminase enzymes in disease and female reproduction. J Reprod Dev. 2012, 58, 274. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, T.G.; Di Paolo, G. Phospholipase D in brain function and Alzheimer’s disease. Biochim Biophys Acta. 2010, 1801, 799. [Google Scholar] [CrossRef]
- Lindsley, C.W.; Brown, H.A. Phospholipase D as a therapeutic target in brain disorders. Neuropsychopharmacology. 2012, 37, 301. [Google Scholar] [CrossRef]
- Zhang, W.; Zhu, F.; Zhu, J.; Liu, K. Phospholipase D, a Novel Therapeutic Target Contributes to the Pathogenesis of Neurodegenerative and Neuroimmune Diseases. Anal Cell Pathol (Amst). 2024, 2024. [Google Scholar] [CrossRef]
- Bottero, V.; Powers, D.; Yalamanchi, A.; Quinn, J.P.; Potashkin, J.A. Key Disease Mechanisms Linked to Alzheimer’s Disease in the Entorhinal Cortex. Int J Mol Sci. 2021, 22. [Google Scholar] [CrossRef] [PubMed]
- Manfredi-Lozano, M.; Leysen, V.; Adamo, M.; et al. GnRH replacement rescues cognition in Down syndrome. Science. 2022, 377, eabq. [Google Scholar] [CrossRef] [PubMed]
- Quigley, E.B.; DeVore, S.B.; Khan, S.A.; Geisterfer, Z.M.; Rothfuss, H.M.; Sequoia, A.O.; Thompson, P.R.; Gatlin, J.C. , Cherrington, B.D., Navratil, A.M. GnRH Induces Citrullination of the Cytoskeleton in Murine Gonadotrope Cells. Int J Mol Sci. 2024, 25. [Google Scholar] [CrossRef]
- Roy, A.; Al-bataineh, M.M.; Pastor-Soler, N.M. Collecting duct intercalated cell function and regulation. Clin J Am Soc Nephrol. 2015, 10, 305. [Google Scholar] [CrossRef] [PubMed]
- Reaux-Le Goazigo, A.; Bodineau, L.; De Mota, N.; Jeandel, L.; Chartrel, N.; Knauf, C.; Raad, C.; Valet, P.; Llorens-Cortes, C. Apelin and the proopiomelanocortin system: A new regulatory pathway of hypothalamic α-MSH release. Am. J. Physiol. Endocrinol. Metab. 2011, 301, E955–E966. [Google Scholar] [CrossRef]
- Masoumi, J.; Abbasloui, M.; Parvan, R.; Mohammadnejad, D.; Pavon-Djavid, G.; Barzegari, A.; Abdolalizadeh, J. Apelin, a promising target for Alzheimer disease prevention and treatment. Neuropeptides 2018, 70, 76–86. [Google Scholar] [CrossRef]
- Zhu, J.; Gao, W.; Shan, X.; Wang, C.; Wang, H.; Shao, Z.; Dou, S.; Jiang, Y.; Wang, C.; Cheng, B. Apelin-36 mediates neuroprotective effects by regulating oxidative stress, autophagy and apoptosis in MPTP-induced Parkinson’s disease model mice. Brain Res. 2020, 1726, 146493. [Google Scholar] [CrossRef]
- Angelopoulou, E.; Paudel, Y.N.; Bougea, A.; Piperi, C. Impact of the apelin/APJ axis in the pathogenesis of Parkinson’s disease with therapeutic potential. J Neurosci Res. 2021, 99, 2117. [Google Scholar] [CrossRef]
- Doxakis, E. Insights into the multifaceted role of circular RNAs: Implications for Parkinson’s disease pathogenesis and diagnosis. NPJ Parkinsons Dis. 2022, 8. [Google Scholar] [CrossRef]
- Shvetcov, A.; Thomson, S.; Spathos, J.; et al. Blood-Based Transcriptomic Biomarkers Are Predictive of Neurodegeneration Rather Than Alzheimer’s Disease. Int J Mol Sci. 2023, 24. [Google Scholar] [CrossRef]
- Lan, A.P.; Chen, J.; Zhao, Y.; Chai, Z.; Hu, Y. mTOR Signaling in Parkinson’s Disease. Neuromolecular Med. 2017, 19, 1. [Google Scholar] [CrossRef] [PubMed]
- Pena-Leon, V.; Perez-Lois, R.; Seoane, L.M. mTOR Pathway is Involved in Energy Homeostasis Regulation as a Part of the Gut-Brain Axis. Int J Mol Sci. 2020, 21. [Google Scholar] [CrossRef]
- Tan, C.; Ai, J.; Zhu, Y. mTORC1-Dependent Protein and Parkinson’s Disease: A Mendelian Randomization Study. Brain Sci. 2023, 13. [Google Scholar] [CrossRef]
- Khan, M.R.; Yin, X.; Kang, S.U.; et al. Enhanced mTORC1 signaling and protein synthesis in pathologic α-synuclein cellular and animal models of Parkinson’s disease. Sci Transl Med. 2023, 15, eadd. [Google Scholar] [CrossRef] [PubMed]
- Fahmy, A.M.; Boulais, J.; Desjardins, M.; Matheoud, D. Mitochondrial antigen presentation: A mechanism linking Parkinson’s disease to autoimmunity. Curr Opin Immunol. 2019, 58, 31. [Google Scholar] [CrossRef] [PubMed]
- Hobson, B.D.; Sulzer, D. Neuronal Presentation of Antigen and Its Possible Role in Parkinson’s Disease. J Parkinsons Dis. 2022, 12, S137–S. [Google Scholar] [CrossRef]
- Schonhoff, A.M.; Figge, D.A.; Williams, G.P.; et al. Border-associated macrophages mediate the neuroinflammatory response in an alpha-synuclein model of Parkinson disease. Nat Commun. 2023, 14. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Yamada, T. Viral etiology for Parkinson’s disease--a possible role of influenza A virus infection. Jpn J Infect Dis. 1999, 52, 89–98. [Google Scholar] [CrossRef]
- Ghemrawi, R.; Khair, M. Endoplasmic Reticulum Stress and Unfolded Protein Response in Neurodegenerative Diseases. Int J Mol Sci. 2020, 21, Published–2020. [Google Scholar] [CrossRef]
- Mou, Z.; Yuan, Y.H.; Zhang, Z.; Song, L.K.; Chen, N.H. Endoplasmic reticulum stress, an important factor in the development of Parkinson’s disease. Toxicol Lett. 2020, 324, 20. [Google Scholar] [CrossRef]
- Li, P.; Nie, Y.; Yu, J. An Effective Method to Identify Shared Pathways and Common Factors among Neurodegenerative Diseases. PLoS ONE 2015, 10, e0143045. [Google Scholar] [CrossRef]
- Attilio, P.J.; Flora, M.; Kamnaksh, A.; Bradshaw, D.J.; Agoston, D.; Mueller, G.P. The Effects of Blast Exposure on Protein Deimination in the Brain. Oxid. Med. Cell Longev. 2017, 2017, 8398072. [Google Scholar] [CrossRef] [PubMed]
- Gallart-Palau, X.; Lee, B.S.; Adav, S.S.; et al. Gender differences in white matter pathology and mitochondrial dysfunction in Alzheimer’s disease with cerebrovascular disease. Mol Brain. 2016, 9. [Google Scholar] [CrossRef]
- Munoz-Pinto, M.F.; Empadinhas, N.; Cardoso, S.M. The neuromicrobiology of Parkinson’s disease: A unifying theory. Ageing Res Rev. 2021, 70. [Google Scholar] [CrossRef]
- Griffante, G.; Gugliesi, F.; Pasquero, S.; et al. Human cytomegalovirus-induced host protein citrullination is crucial for viral replication. Nat Commun. 2021, 12. [Google Scholar] [CrossRef]
- Casanova, V.; Sousa, F.H.; Shakamuri, P.; et al. Citrullination Alters the Antiviral and Immunomodulatory Activities of the Human Cathelicidin LL-37 During Rhinovirus Infection. Front Immunol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Martens, C.P.; Peetermans, M.; Vanassche, T.; Verhamme, P.; Jacquemin, M.; Martinod, K. Peptidylarginine deiminase 4 and ADAMTS13 activity in Staphylococcusaureus bacteraemia. Philos Trans R Soc Lond B Biol Sci. 2023, 378. [Google Scholar] [CrossRef]
- Sinha, S.; Mittal, S.; Roy, R. Parkinson’s Disease and the COVID-19 Pandemic: A Review Article on the Association between SARS-CoV-2 and α-Synucleinopathy. J Mov Disord. 2021, 14, 184. [Google Scholar] [CrossRef]
- Boura, I.; Qamar, M.A.; Daddoveri, F.; et al. SARS-CoV-2 and Parkinson’s Disease: A Review of Where We Are Now. Biomedicines. 2023, 11. [Google Scholar] [CrossRef]
- Calculli, A.; Bocci, T.; Porcino, M.; et al. Parkinson disease following COVID-19: Report of six cases. Eur J Neurol. 2023, 30, 1272. [Google Scholar] [CrossRef]
- Li, Z.; Lu, G.; Luo, E.; et al. Oral, Nasal, and Gut Microbiota in Parkinson’s Disease. Neuroscience. 2022, 480, 65. [Google Scholar] [CrossRef]
- Lazarini, F.; Roze, E.; Lannuzel, A.; Lledo, P.M. The microbiome-nose-brain axis in health and disease. Trends Neurosci. 2022, 45, 718. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, A.; Pirracchio, L.; Imber, J.C.; et al. Citrullination in periodontium is associated with Porphyromonas gingivalis. Arch Oral Biol. 2020, 114. [Google Scholar] [CrossRef] [PubMed]
- Bennett, J.P. Jr.; Keeney, P.M.; Brohawn, D.G. RNA Sequencing Reveals Small and Variable Contributions of Infectious Agents to Transcriptomes of Postmortem Nervous Tissues From Amyotrophic Lateral Sclerosis, Alzheimer’s Disease and Parkinson’s Disease Subjects, and Increased Expression of Genes From Disease-Activated Microglia. Front Neurosci. 2019, 13. [Google Scholar] [CrossRef]
- Gawron, K.; Bereta, G.; Nowakowska, Z.; et al. Peptidylarginine deiminase from Porphyromonas gingivalis contributes to infection of gingival fibroblasts and induction of prostaglandin E2 -signaling pathway. Mol Oral Microbiol. 2014, 29, 321. [Google Scholar] [CrossRef] [PubMed]
- Emery, D.C.; Davies, M.; Cerajewska, T.L.; et al. High resolution 16S rRNA gene Next Generation Sequencing study of brain areas associated with Alzheimer’s and Parkinson’s disease. Front Aging Neurosci. 2022, 14. [Google Scholar] [CrossRef]
- Cirstea, M.S.; Kliger, D.; MacLellan, A.D.; et al. The Oral and Fecal Microbiota in a Canadian Cohort of Alzheimer’s Disease. J Alzheimers Dis. 2022, 87, 247. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.K.; Wu, Q.L.; Peng, Y.W.; et al. Oral P. gingivalis impairs gut permeability and mediates immune responses associated with neurodegeneration in LRRK2 R1441G mice. J Neuroinflammation. 2020, 17. [Google Scholar] [CrossRef]
- Yay, E.; Yilmaz, M.; Toygar, H.; et al. Parkinson’s disease alters the composition of subgingival microbiome. J Oral Microbiol. 2023, 15. [Google Scholar] [CrossRef]
- Perricone, C.; Ceccarelli, F.; Saccucci, M.; Di Carlo, G.; Bogdanos, D.P.; Lucchetti, R.; Pilloni, A.; Valesini, G.; Polimeni, A.; Conti, F. Porphyromonas gingivalis and rheumatoid arthritis. Curr Opin Rheumatol. 2019, 31, 517. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Kurita-Ochiai, T.; Kobayashi, R.; Suzuki, T.; Ando, T. Activation of the NLRP3 inflammasome in Porphyromonas gingivalis-accelerated atherosclerosis. Pathog Dis. 2015, 73, ftv. [Google Scholar] [CrossRef] [PubMed]
- Blenkinsopp, H.C.; Seidler, K.; Barrow, M. Microbial Imbalance and Intestinal Permeability in the Pathogenesis of Rheumatoid Arthritis: A Mechanism Review with a Focus on Bacterial Translocation, Citrullination, and Probiotic Intervention. J Am Nutr Assoc. 2024, 43, 59. [Google Scholar] [CrossRef] [PubMed]
- Bielecka, E.; Scavenius, C.; Kantyka, T.; Jusko, M.; Mizgalska, D.; Szmigielski, B.; Potempa, B.; Enghild, J.J.; Prossnitz, E.R.; Blom, A.M.; Potempa, J. Peptidyl arginine deiminase from Porphyromonas gingivalis abolishes anaphylatoxin C5a activity. J. Biol. Chem. 2014, 289, 32481–32487. [Google Scholar] [CrossRef]
- Kosgodage, U.S.; Matewele, P.; Mastroianni, G.; Kraev, I.; Brotherton, D.; Awamaria, B.; Nicholas, A.P.; Lange, S.; Inal, J.M. Peptidylarginine Deiminase Inhibitors Reduce Bacterial Membrane Vesicle Release and Sensitize Bacteria to Antibiotic Treatment. Front. Cell Infect. Microbiol. 2019, 9, 227. [Google Scholar] [CrossRef]
- Sancandi, M.; De Caro, C.; Cypaite, N.; Marascio, N.; Avagliano, C.; De Marco, C.; Russo, E.; Constanti, A.; Mercer, A. Effects of a probiotic suspension Symprove™ on a rat early-stage Parkinson’s disease model. Front Aging Neurosci. 2023, 14. [Google Scholar] [CrossRef] [PubMed]
- Adams, B.; Nunes, J.M.; Page, M.J.; Roberts, T.; Carr, J.; Nell, T.A.; Kell, D.B.; Pretorius, E. Parkinson’s Disease: A Systemic Inflammatory Disease Accompanied by Bacterial Inflammagens. Front Aging Neurosci. 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- Olsen, I.; Kell, D.B.; Pretorius, E. Is Porphyromonas gingivalis involved in Parkinson’s disease? Eur J Clin Microbiol Infect Dis. 2020, 39, 2013. [Google Scholar] [CrossRef]
- Li, D.; Ren, T.; Li, H.; Liao, G.; Zhang, X. Porphyromonas gingivalis: A key role in Parkinson’s disease with cognitive impairment? Front Neurol. 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Ramazi, S.; Allahverdi, A.; Zahiri, J. Evaluation of post-translational modifications in histone proteins: A review on histone modification defects in developmental and neurological disorders. J Biosci 2020, 45, 135. [Google Scholar] [CrossRef]
- Antunes, A.S.L.M. Post-translational Modifications in Parkinson’s Disease. Adv Exp Med Biol. 2022, 1382, 1382, 85. [Google Scholar] [CrossRef]
- Zhang, S.; Zhu, R.; Pan, B.; et al. Post-translational modifications of soluble α-synuclein regulate the amplification of pathological α-synuclein. Nat Neurosci. 2023, 26, 213. [Google Scholar] [CrossRef] [PubMed]
- Bogetofte, H.; Ryan, B.J.; Jensen, P.; et al. Post-translational proteomics platform identifies neurite outgrowth impairments in Parkinson’s disease GBA-N370S dopamine neurons. Cell Rep. 2023, 42. [Google Scholar] [CrossRef]
- Kim, S.; Song, Y.K.; Cho, C.S.; Kim, H.J.; Fang, S.; Jo, D.H.; Kim, H. Inhibition of protein arginine deiminase II suppresses retinoblastoma in orthotopic transplantation in mice. Oncol Rep. 2023, 50. [Google Scholar] [CrossRef] [PubMed]
- Sancandi, M.; Schul, E.V.; Economides, G.; Constanti, A.; Mercer, A. Structural Changes Observed in the Piriform Cortex in a Rat Model of Pre-motor Parkinson’s Disease. Front. Cell Neurosci. 2018, 12, 479. [Google Scholar] [CrossRef]
- Nicholas, A.P.; Whitaker, J.N. Preparation of a monoclonal antibody to citrullinated epitopes: Its characterization and some applications to immunohistochemistry in human brain. Glia 2002, 37, 328–336. [Google Scholar] [CrossRef]
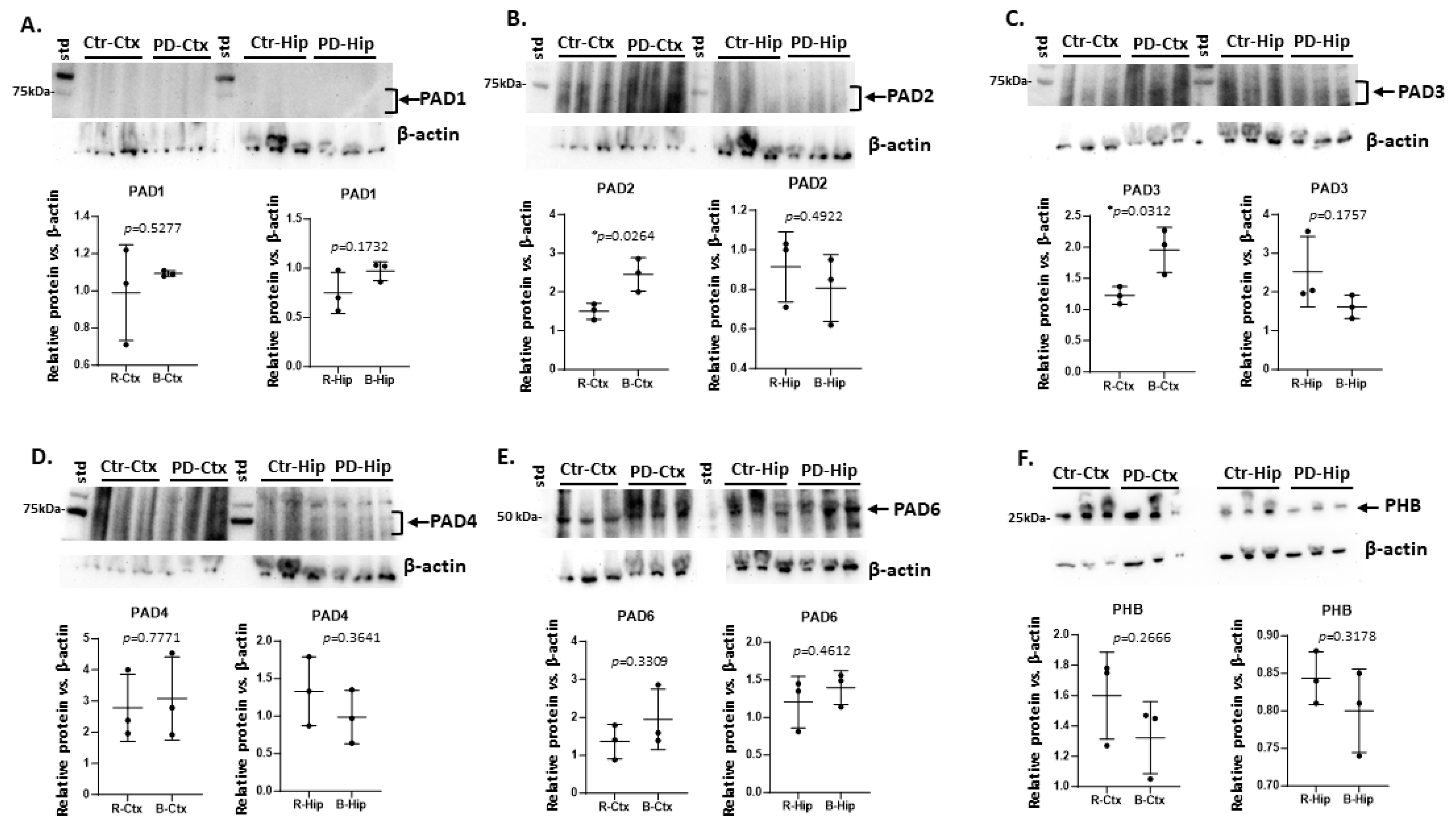
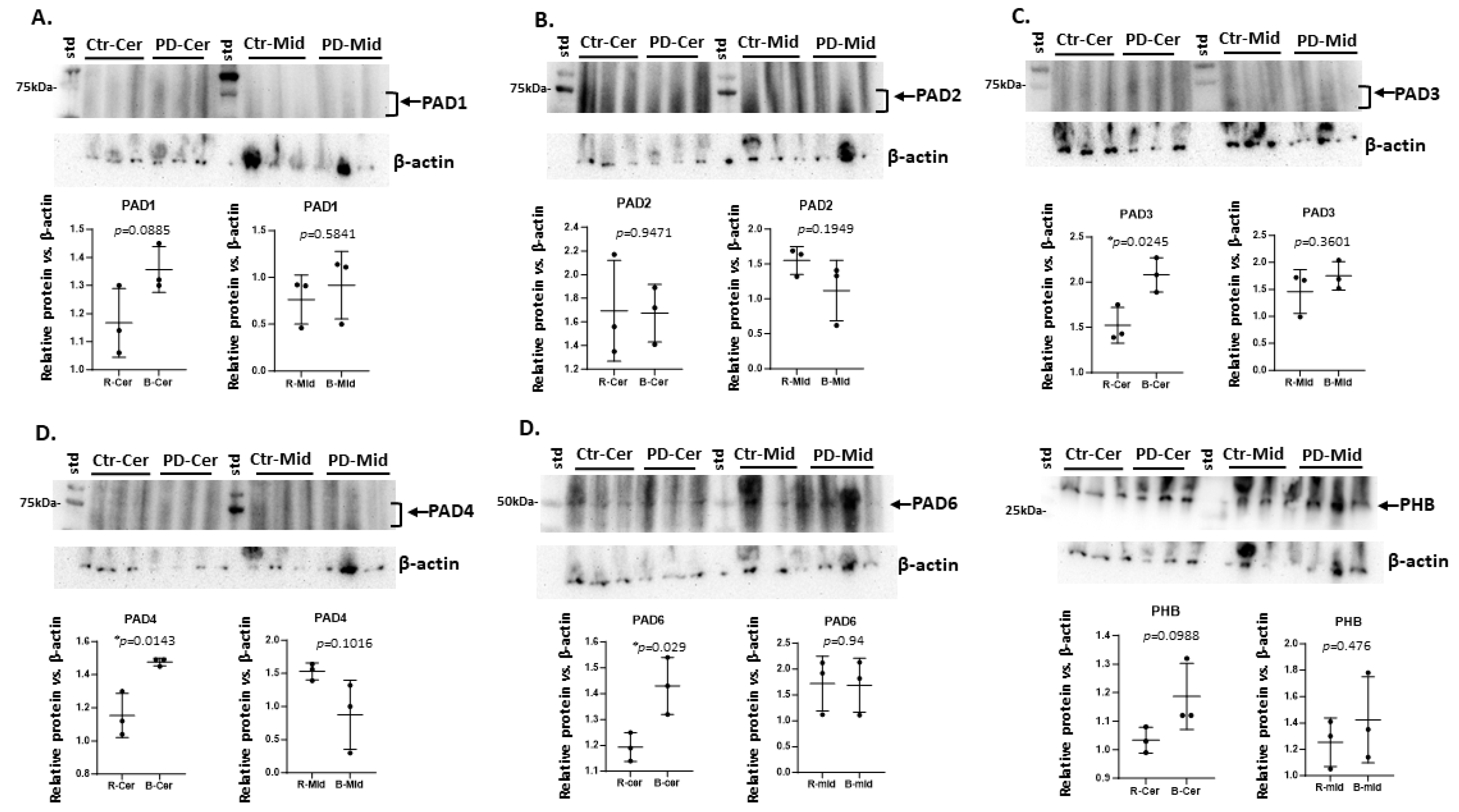
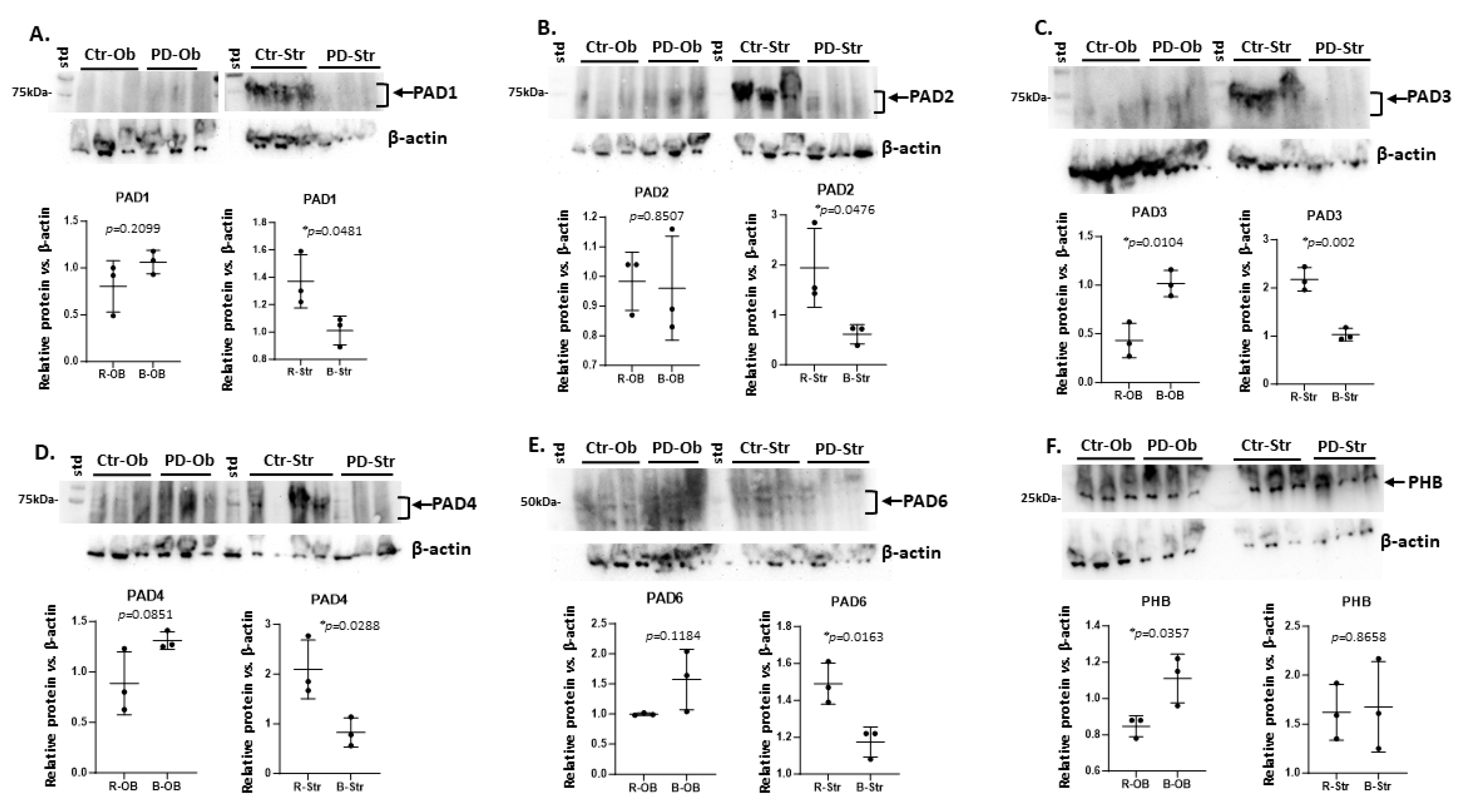
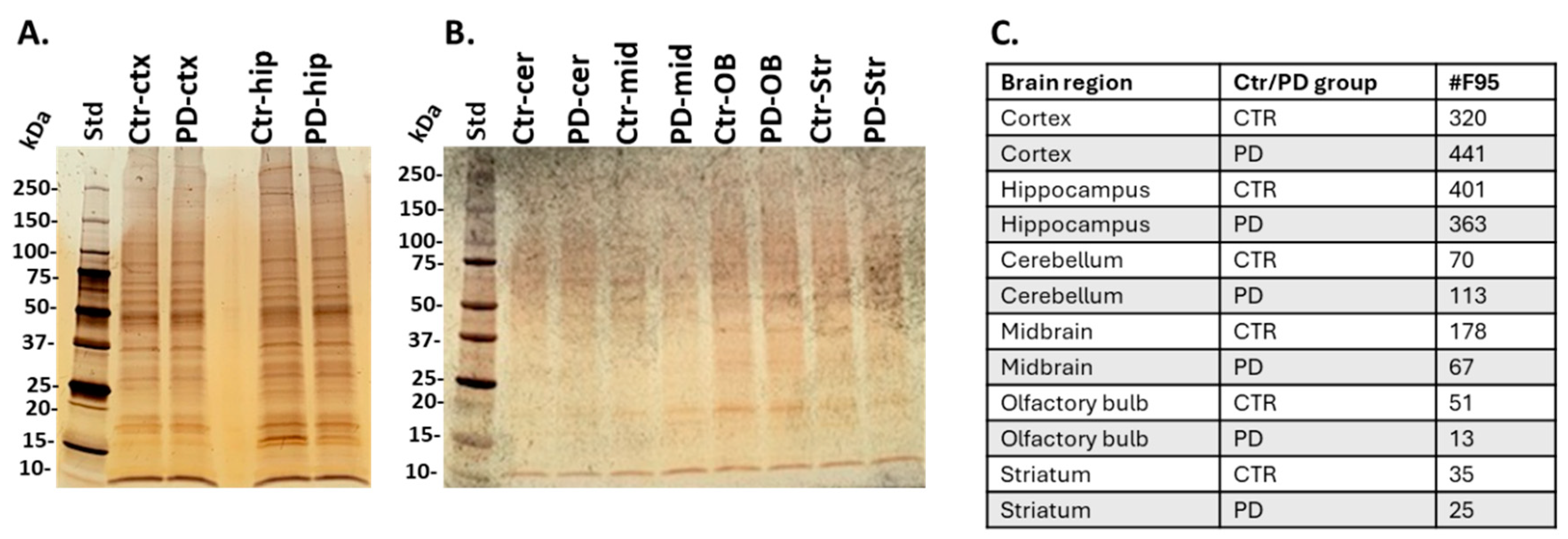

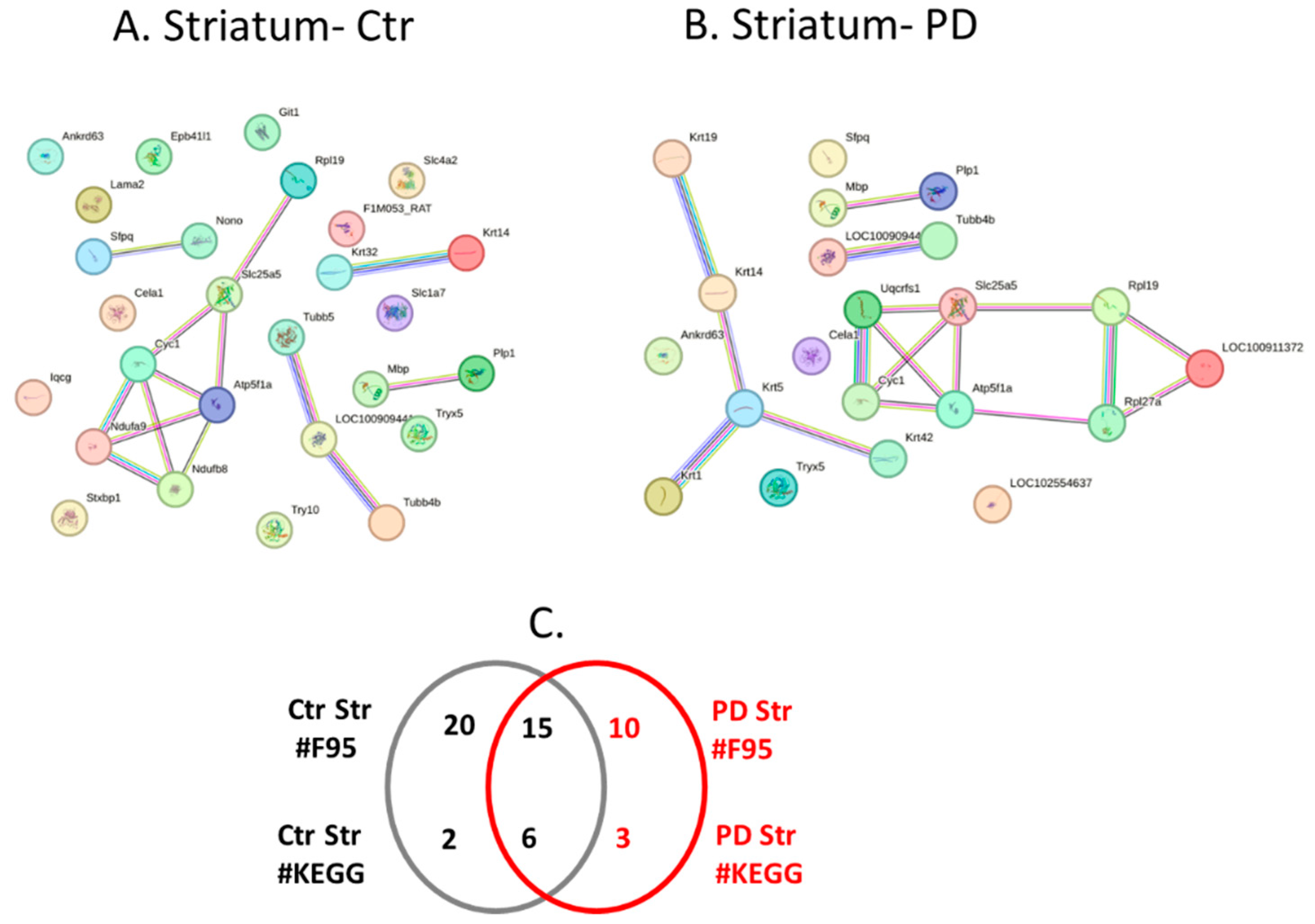
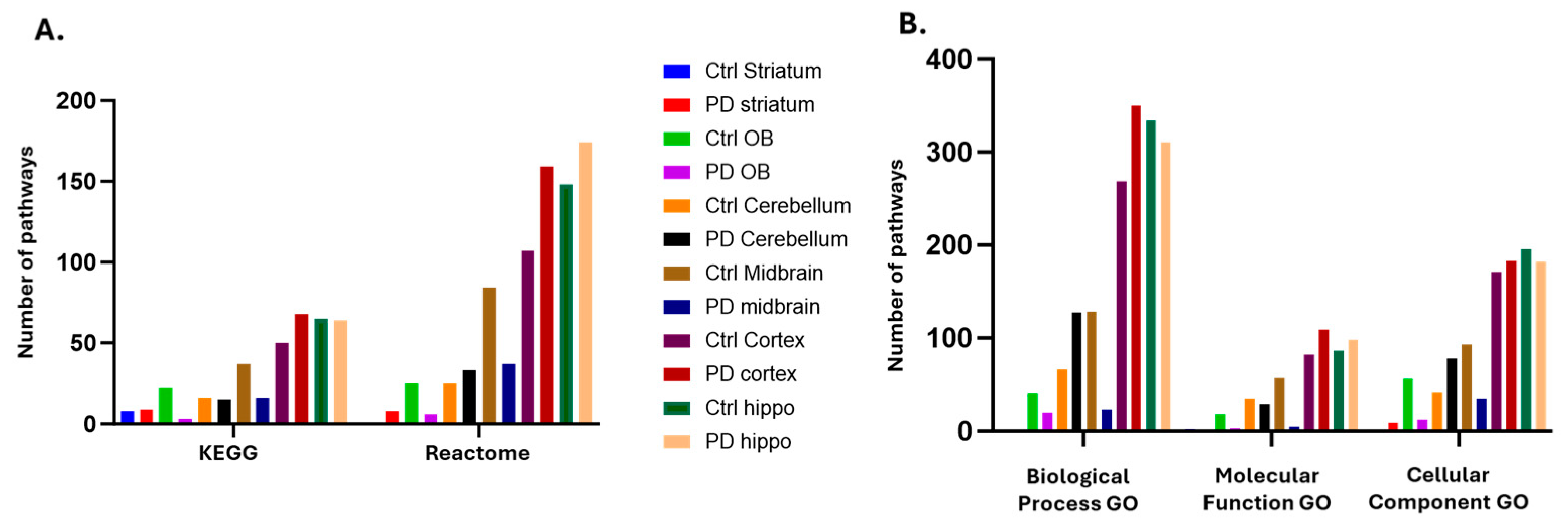
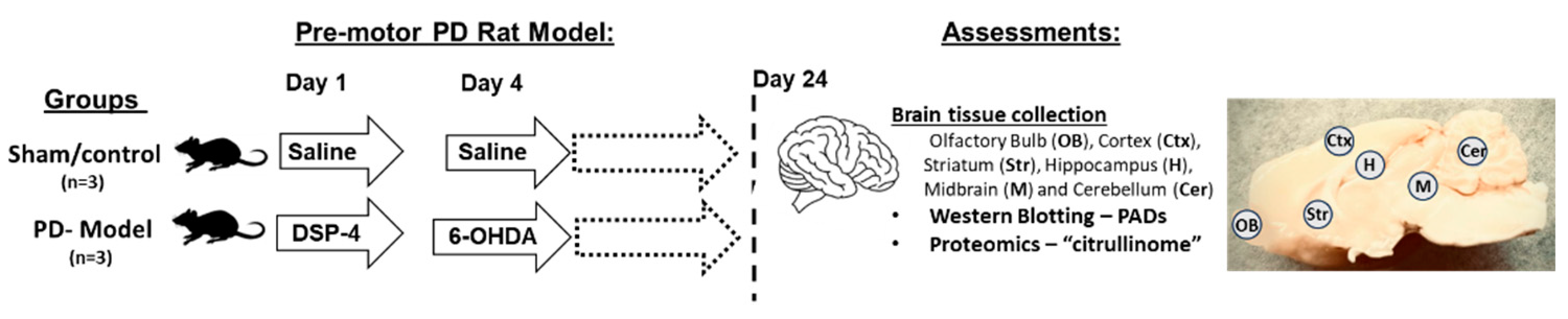
| Brain region | Exp group | PAD1 | PAD2 | PAD3 | PAD4 | PAD6 |
| Cortex | CTR | (+) | ++ | + | +++ | ++ |
| Cortex | PD | (+) | +++ * | ++ * | +++ | +++ |
| Hippocampus | CTR | (+) | + | ++ | ++ | ++ |
| Hippocampus | PD | (+) | (+) | +(+) | +(+) | ++(+) |
| Cerebellum | CTR | + | ++ | + | + | + |
| Cerebellum | PD | + | ++ | ++ * | +(+) * | +(+) * |
| Midbrain | CTR | (+) | ++ | +(+) | +(+) | ++ |
| Midbrain | PD | + | ++ | +(+) | + | ++ |
| Olfactory bulb | CTR | (+) | + | + | +(+) | + |
| Olfactory bulb | PD | + | + | +(+) * | ++ | ++ |
| Striatum | CTR | ++ | +++ | ++ | +++ | + |
| Striatum | PD | + * | + * | (+) * | + * | (+) * |
| KEGG pathway | Ctr CTX | PD CTX | CtrlHIP | PD HIP | Ctr STR |
PD STR | Ctr OB | PD OB | Ctr MB | PD MB | Ctr CER | PD CER |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Oxidative phosphorylation | V | V | V | V | V | V | V | V | V | |||
| Synaptic vesicle cycle | V | V | V | V | V | V | V | |||||
| Ribosome | V | V | V | V | V | V | V | V | ||||
| Parkinson’s disease | V | V | V | V | V | V | V | V | V | V | V | |
| Retrograde endocannabinoid signalling | V | V | V | V | V | V | ||||||
| Huntington disease | V | V | V | V | V | V | V | V | V | V | V | |
| Prion disease | V | V | V | V | V | V | V | V | V | V | V | |
| Gap junction | V | V | V | V | V | V | V | V | ||||
| Non-alcoholic fatty liver disease | V | V | V | V | V | V | V | |||||
| Thermogenesis | V | V | V | V | V | V | V | V | V | V | ||
| Alzheimer’s disease | V | V | V | V | V | V | V | V | V | V | ||
| GABAergic synapse | V | V | V | V | V | |||||||
| Amyotrophic lateral sclerosis | V | V | V | V | V | V | V | V | V | V | ||
| Endocrine and other factor-regulated calcium reabsorption | V | V | ||||||||||
| Glutamatergic synapse | V | V | V | V | V | V | V | |||||
| Phagosome | V | V | V | V | V | V | V | V | ||||
| Cardiac muscle contraction | V | V | V | V | ||||||||
| Biosynthesis of amino acids | V | V | V | V | V | V | V | |||||
| Carbon metabolism | V | V | V | V | V | |||||||
| Oestrogen signalling pathway | V | V | V | V | V | V | V | V | V | V | V | |
| Necroptosis | V | V | V | V | V | |||||||
| Endocytosis | V | V | V | V | V | V | V | |||||
| Metabolic pathways | V | V | V | V | V | V | ||||||
| Cocaine addiction | V | V | ||||||||||
| Insulin secretion | V | V | V | V | ||||||||
| Apoptosis | V | V | V | V | V | V | ||||||
| cGMP-PKG signalling pathway | V | V | V | |||||||||
| Alcoholism | V | V | V | |||||||||
| Pyruvate metabolism | V | V | V | V | V | V | ||||||
| Inositol phosphate metabolism | V | |||||||||||
| Phosphatidylinositol signalling system | V | V | ||||||||||
| Progesterone mediated oocyte maturation | V | |||||||||||
| Axon guidance | V | |||||||||||
| Oocyte meiosis | V | V | ||||||||||
| Cholinergic synapse | V | V | V | V | ||||||||
| Spinocerebellar ataxia | V | |||||||||||
| Phospholipase D signalling pathway | V | |||||||||||
| Rap1 signalling pathway | V | |||||||||||
| 2-Oxocarboxylic acid metabolism | V | V | ||||||||||
| Nitrogen metabolism | V | V | V | V | ||||||||
| Pentose phosphate pathway | V | V | ||||||||||
| Fructose and mannose metabolism | V | V | V | V | ||||||||
| Bacterial invasion of epithelial cells | V | V | V | V | V | |||||||
| Leukocyte transendothelial migration | V | V | ||||||||||
| Morphine addiction | V | V | V | V | ||||||||
| Regulation of actin cytoskeleton | V | V | V | |||||||||
| Citrate cycle (TCA cycle) | V | V | V | V | V | |||||||
| Vasopressin-regulated water reabsorption | V | V | V | |||||||||
| Arginine biosynthesis | V | V | V | V | ||||||||
| Glycolysis/Gluconeogenesis | V | V | V | V | V | V | V | |||||
| Central carbon metabolism in cancer | V | V | V | V | V | |||||||
| SNARE interactions in vesicular transport | V | V | V | |||||||||
| Alanine, aspartate and glutamate metabolism | V | V | V | V | ||||||||
| Cysteine and methionine metabolism | V | V | V | V | ||||||||
| HIF-1 signalling pathway | V | V | V | V | V | |||||||
| Legionellosis | V | V | V | V | V | |||||||
| Salmonella infection | V | V | V | V | ||||||||
| Oxytocin signalling pathway | V | V | V | V | ||||||||
| Tight junction | V | V | V | |||||||||
| mTOR signalling pathway | V | |||||||||||
| Viral carcinogenesis | V | V | V | V | V | |||||||
| Collecting duct acid secretion | V | |||||||||||
| Glyoxylate and dicarboxylate metabolism | V | V | V | |||||||||
| Butanoate metabolism | V | |||||||||||
| Dopaminergic synapse | V | V | V | V | ||||||||
| Calcium signalling pathway | V | V | V | V | ||||||||
| Ferroptosis | V | V | ||||||||||
| Systemic lupus erythematosus | V | V | ||||||||||
| Long-term depression | V | V | ||||||||||
| Hippo signalling pathway | V | |||||||||||
| Staphylococcus aureus infection | V | V | V | V | V | |||||||
| Gastric acid secretion | V | V | V | V | ||||||||
| Arrhythmogenic right ventricular cardiomyopathy | V | V | V | |||||||||
| Influenza A | V | |||||||||||
| Glucagon signalling pathway | V | V | V | V | V | V | V | |||||
| GnRH secretion | V | |||||||||||
| Type II diabetes mellitus | V | V | V | V | ||||||||
| Apelin signalling pathway | V | |||||||||||
| Proteoglycans in cancer | V | V | V | |||||||||
| NOD-like receptor signalling pathway | V | |||||||||||
| Platelet activation | V | |||||||||||
| Cellular senescence | V | V | ||||||||||
| Yersinia infection | V | |||||||||||
| Human immunodeficiency virus 1 infection | V | V | V | V | ||||||||
| Propanoate metabolism | V | V | ||||||||||
| Serotonergic synapse | V | V | V | |||||||||
| Beta-Alanine metabolism | V | |||||||||||
| Thyroid hormone signalling pathway | V | |||||||||||
| Fc gamma R-mediated phagocytosis | V | |||||||||||
| Choline metabolism in cancer | V | |||||||||||
| Spliceosome | V | |||||||||||
| Antigen processing and presentation | V | V | ||||||||||
| Protein processing in endoplasmic reticulum | V | |||||||||||
| Human cytomegalovirus infection | V |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





