1. Introduction
Regression analysis is a potent statistical tool
that illuminates the connection between one or more independent variables and a
dependent variable. Essential in data analysis and predictive modeling, it
finds broad application across fields such as economics, finance, healthcare,
and social sciences. However, regression models must meet certain assumptions
to provide reliable and valid results. These assumptions form the foundation of
regression analysis and guide researchers in interpreting the results
accurately. One problematic assumption to avoid is the linear relationship
among the independent variables called multicollinearity, which occurs when two
or more independent variables are correlated, increasing the standard error of
the coefficients. This escalation in standard errors can render the
coefficients of certain independent variables statistically insignificant
despite their potential significance. In essence, multicollinearity distorts
the interpretation of variables by inflating their standard errors [1]. Shrestha [2]
discussed the primary techniques for investigating multicollinearity using
questionnaires for survey data to support customer satisfaction.
Traditional regression techniques often struggle to
handle multicollinearity effectively, leading to biased results and unreliable
predictions. Researchers have developed various methods to mitigate these
challenges, including Liu Regression. Liu Regression is a technique developed
to address multicollinearity in regression analysis. It combines the principles
of Ridge Regression with orthogonalization to effectively mitigate the effects
of multicollinearity. Dawoud et al. [3]
devised a novel modified Liu estimator to employ multicollinearity in a
regression model with a single parameter, incorporating two biasing parameters,
with at least one designed to mitigate this issue. Jahufer [4], on the other hand, employed the Liu estimator
to alleviate the impact of multicollinearity and the influence of specific
observations, devising approximate deletion formulas for identifying
influential points.
Searching for accurate models that can efficiently
handle complex datasets while offering robust predictions is perpetual in
predictive analytics. Among the array of methodologies, the Liu Regression
Model is a game-changer, heralding a new era in predictive modeling. The Liu
Regression Model introduces novel techniques that address the limitations of
traditional regression methods. Unlike conventional approaches that rely solely
on linear relationships between variables, Liu Regression leverages advanced algorithms
to capture non-linear patterns and intricate interactions within the data.
Karlsson et al. [5] introduced a Liu estimator
tailored for the beta regression model with a fixed dispersion parameter,
applicable in various practical scenarios where the correlation level among the
regressors varies.
Liu Regression [6]
involves selecting a Liu estimator to balance the bias-variance trade-off. The
optimal value of the Liu estimator is typically chosen through techniques such
as cross-validation. The Liu estimator, named after its developer, is essential
in managing multicollinearity. It is particularly associated with methodologies
like Ridge Regression with Orthogonalization, often abbreviated as Liu
Regression. Liu [7] enhanced the Liu estimator
within the linear regression model by considering the biasing parameter under
the prediction sum of squares criterion. Yang and Xu [8]
proposed an alternative stochastic restricted Liu estimator for the parameter
vector in a linear regression model, incorporating additional stochastic linear
restrictions. Hubert and Wijekoon [9]
investigated a novel Liu-type biased estimator, termed the stochastic
restricted Liu estimator, and examined its efficiency.
The improvement of the Liu estimator transformed
the multiple regression model to canonical form [10]
to select the biasing parameter called the Liu parameter. The appropriate Liu
parameters have been developed to make minimum mean squares error in the
estimation. Liu [6,7] applied the iterative
method to estimate the Liu parameter as the minimum mean square error in the
smallest of the Liu estimator. Özkale and Kaçiranlar [11]
proposed the new restricted Liu parameter by computing the predicted residual
error sum of squares to determine the biasing parameter. Dawoud et al.[12] proposed a new Liu estimator using the known
mean squares error criterion to handle the multicollinearity problem. Suhail et
al. [13] developed a new method of biasing
parameters to mitigate the multicollinearity data. Lukman et al. [14] introduced a modified Liu estimator to address
multicollinearity issues within the linear regression model.
In this paper, we propose two competing Liu
parameters, following mean squares error and R-squared, to estimate the Liu
estimator via multiple regression model with the multicollinearity problem. We
measure this performance in terms of minimum average of mean absolute
percentage errors for the simulation and real dataset. We also consider the
scale option of independent variables as the center, correlation form, and
standardizes.
The paper is structured as follows: Section 2 presents the multiple regression
estimators and discusses the Liu estimator through the reparameterization of
Liu regression into canonical form, then compared with the OLS estimator. Section 3 generates the independent and
dependent variables to evaluate the performance estimators. Section 4 applies a real dataset to validate
the simulation results. Section 5
discusses the findings, followed by the conclusion in Section 6.
2. The Liu Regression
The multiple regression model is expressed in
matrix form as:
where
is the
column vector of dependent variable, and
is the
independent variable matrix,
is the
multiple regression parameter vector, and
is the
error vector. The following assumptions of error
are made:
,
, and
The efficient parameters (
) in (1) are common estimated to obtain the
ordinary least squares (OLS) estimator in (2) as follows:
The estimation error of
is evaluated by computing
The bias, variance (Var), and mean squares error
(MSE) of the OLS estimator are computed from (3) as follows:
From the above computation, the OLS estimator
presents the unbiased estimator, which reduces the performance in estimating
parameters on the multicollinearity of independent variables.The diagonal
matrix of
is caused the multicollinearity and inflated,
increasing the estimated variance and mean squares error. To overcome this
problems, Liu [6] proposed the Liu esitimator
which provides the better performance than the OLS estimator [11,15]. The Liu estimator based on the
is defined by
where is the Liu parameter in term of the biasing
parameter and is the identity matrix. The OLS form (1) and Liu
estimators from (4) are related to the independent variables that are affected
to the multicollinearity problem because they depend on the OLS estimator.
The estimation error of
is evaluated as the OLS estimator by comparing the
Liu estimator and the parameter of the multiple regression model
The bias [16],
variance (Var), and mean square error (MSE) of the Liu estimator from (5) are
proposed in following:
The Liu estimator is shown as the bias estimator,
and its varaince is greater than that of the OLS estimator when lies on the range of zero to one. Then, Liu [7] developed the shrinkage factor [17] to create the Liu parameter that may lie
outside the range between zero and one. In the following subsection, the
multiple regression model can be transformed into a canonical form to estimate
the OLS and Liu estimators.
2.1. The Reparameterization of Liu Regression
The reparameterization of Liu Regression transforms
a multiple regression model into a canonical form, offering valuable insights
into variable relationships and enhancing predictive accuracy [17]. The optimal Liu parameter is determined by
minimizing the mean squares error. Akdeniz and Kacįranlar [18] introduced a new biased estimator and assessed
its performance against a restricted least squares estimator regarding mean
squares error. The comparison of the Liu estimator’s performance in canonical
form is expressed as follows:
where
,
,
, and
is a diagonal matrix such that
. The OLS estimator of canonical form can be
difined as
Similarly, the Liu estimator [19] can be written as
The bias, variance (Var), and mean square error
(MSE) of the reparameterization of OLS estimator from (7) are expresses as:
The bias, variance (Var), and mean square error
(MSE) of the reparameterization of Liu estimator from (8) are proposed in
following:
The comparison among the OLS and Liu estimator of canonical
form by considering of the variance and MSE.
Given the and , if the is the better estimator than that is if and only if, Recall that
and
Then,
It can observe when .It can conclude that and the Liu estimator outperforms the OLS
estimator.
2.2. Liu Parameter
From the above subsection, we compare the two
estimators. The reparameterization of Liu regression provides the performance
estimator. However, the existing Liu estimator is to select the appropriate Liu
parameter that has been started by Liu [6] and
developed into another model by Suhail et al. [13],
Lukman et al. [14], Abdelwahab et al. [20], and Babar et al. [21].
The optimal Liu parameter is one reason to make the minimum of mean squares
error (MSE) that is excessed to affect the estimation of the Liu estimator of
collinearity on independent variables. However, the trace of a diagonal matrix
of transformation is useful for calculating the optimal Liu parameter. For this
article, we suggest the original Liu parameter, which is proposed by Liu [6], which is defined as the minimum MSE (mm),
optimum (opt), and Cl criterion (cl), respectively following:
,, and.
Furthermore, Liu [7]
improved the Liu parameter in the multiple linear regression under the
approximation of the predicted residual error sum of squares criterion by
calling improved Liu estimator (ILE) as
Özkale and Kaçiranlar [11]
introduced a new two-parameter approach by incorporating the contraction
estimator, encompassing well-known methods such as restricted least squares,
restricted ridge, restricted contraction estimators, and a novel modified,
restricted Liu estimator (RLE). It can be written by
where ,
is the diagonal elements from Liu hat matrix, and is the ith residual at specific value of .
Mallows [22]
discussed the interpretation of Cp-plots by using the display as a basis for
formally selecting a subset-regression model and extending to estimate the Liu
estimator. The Liu parameter is defined to be
where
In this paper, we modify the Liu parameter from
Mallows [22] to introduce the mean squares
error, which is obtained by the mean of sum squares residual (SSR) in the range
between zero and one as follows:
Furthermore, the correlation coefficient often
denoted as R-squared (
), is a critical metric in regression analysis. It
quantifies the proportion of the variance in the dependent variable that can be
predicted from the independent variables. From the significance of R-squared,
we propose the new Liu parameter by computing the correlation coefficient as 1-
which is rewritten by
Scaling options are utilized
to standardize the independent variables and assess their performance via the
Liu estimator. The initial method, introduced by Liu [6],
is centered, standardizing independent variables to have zero mean and unit
variance. The scaled option further standardizes independent variables. Lastly,
the sc option scales independent variables in correlation form, a concept
explored by Belsley [23].
3. Simulation Study
As the previous section’s theoretical comparison
among the Liu estimator, a simulation study covers the Monte Carlo simulation
using the R 4.2.1 programming languages. The objective of the simulation study
is to estimate and compare the Liu parameter to grasp the better performance of
the Liu parameter on the multiple regression model. The independent variables (
) are generated from the multivariate normal
distribution in five, ten, and fifteen independent variables based on Toeplitz
correlation (
) values of 0.1
and 0.9. The multivariate normal distribution based on parameter
means (
) and covariance matrix (
) is simulated as
multicollinearity between independent variables. The probability distribution
is defined by
, where
,
The type of covariance matrix is mentioned in the
Toeplitz correlation model, which implies that closely located independent
variables have a high correlation, and the correlation decreases as independent
variables are farther apart. A matrix with the following pattern characterizes
the relationship:
where the correlation coefficient or level of
multicollinearity is given by 0.1 and
0.9.
The observations on the dependent variable are
obtained from the multiple regression model as
where is generated from the normal distribution to be
mean zero and variance one, the regression coefficients () are defined the constant values.
The performance criterion is used to judge the
performance of different Liu parameters in estimating the Liu estimator.
Evaluated mean absolute percentage error (MAPE) is defined as:
where
is the real dataset and
is the estimated dataset. The average of mean
absolute percentage error of the OLS and eight Liu parameters for five, ten,
and fifteen variables are presented in
Table 1,
Table 2 and
Table 3
according to their correlation coefficient (0.1 and 0.9).
Table 4 presents the Liu parameter values to
estimate the Liu estimator. The average of over 1,000 replications is employed
to approximate the average of mean absolute percentage error. The minimum
average of mean absolute percentage error is shown in bold letters.
Table 1,
Table 2 and
Table 3
describe the simulated average of mean absolute percentage error for two levels
of Toeplitz correlation. In
Table 1,
Table 2 and
Table 3 ,
the smallest value of the MAPE is highlighted in bold letters. The simulation
results show that the modified Liu parameter in terms of R-squared (dR2) has
the smallest values of MAPE, so it outperforms the other methods, especially in
the SC option in Table 3. However, the
dILE, dRLE, and dCp have the weakest performance in all cases. Furthermore, the
MAPE of dmm, dcl, and dopt equals the dR2 in the center and scaled options in Tables 1 and 2. The behavior of sample sizes
can be observed in the sample impact on estimation since the MAPE decreases
when sampling sizes decrease. The MAPE of independent variables is reduced when
the independent variables increase. The Liu parameter of the estimate Liu
estimator is presented in Table 4 and is
varied by sample sizes, independent variables, and the level of correlation.
From Table 4,
the level of the correlation coefficient has a significant effect in computing
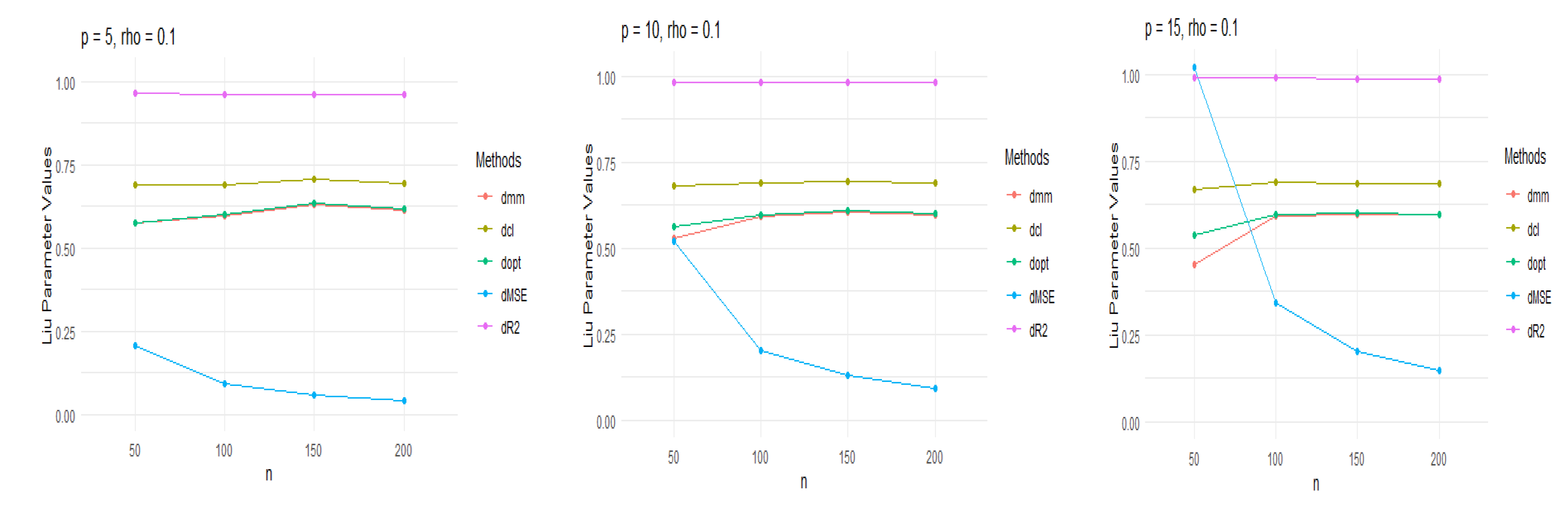
the Liu parameter. The dmm, dcl, and adopt are shown a positive, small
correlation, but the large correlation has exhibited a negative. The dMSE is
stanned from zero to one for small correlation, but the dMSE is more
significant than one for large correlation. The excellent performance in Liu
estimation, dR2, is approximated in the range of zero to one in all cases.
Furthermore, the dILE, dRLE, and dCp have large Liu parameters and show the
lowest performance in Tables 1–3. For a
better understanding, we have plotted the Liu parameter just dmm, dcl, dopt,
dMSE, dR2 for multicollinearity 0.1 and 0.9 in
Figure 1 and
Figure 2, respectively.
4. Application in Actual data
We employed Liu regression to distinguish between
blood donors’ laboratory values and patients’ age using the Hepatitis C
patients dataset sourced from the UCI Machine Learning. This dataset was
retrieved from the
https://archive.ics.uci.edu/ml/datasets/HCV+data. The
dependent variable was the age of patients and independent variables included
Albumin (ALB), Total Protein (PROT), Cholinesterase (CHE), Cholesterol (CHOL),
Alkaline Phosphatase (ALP), Alanine Aminotransferase (ALT), Creatinine (CREA),
Bilirubin (BIL), Aspartate Aminotransferase (AST), and Gamma-Glutamyl
Transferase (GGT). The dataset comprised 589 records displayed the descriptive
statistics about the Hepatitis C dataset in
Table 5.
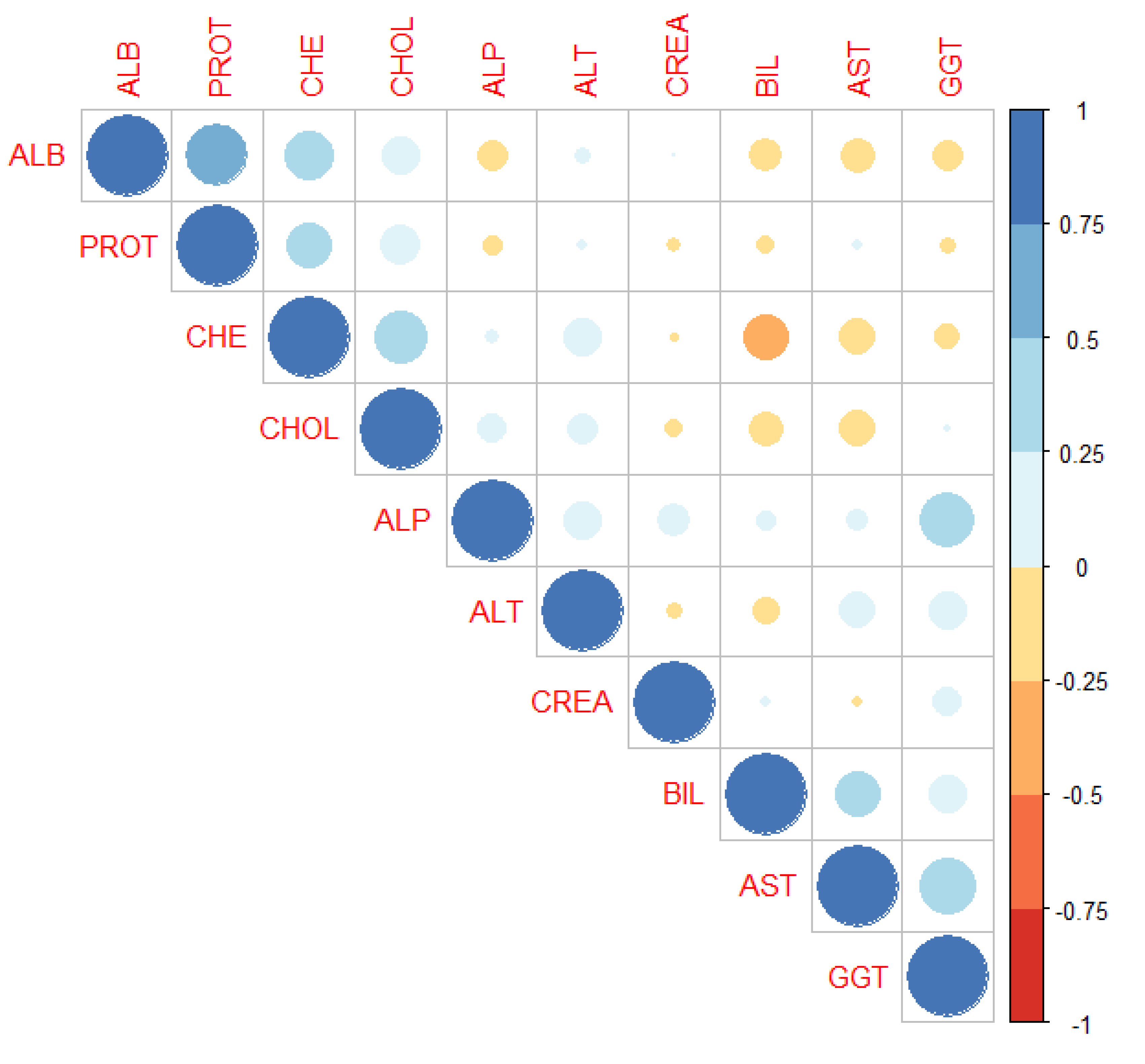
For checking multicollinearity data, Pearson’s
correlation analysis was employed to ascertain any potential relationship among
the ten continuous independent variables. The formula utilized for computing
the correlation between two variables was:
From above formula, the correlation coefficients
for the independent variables are outlined in
Table 6. and
Figure 3.
The null hypothesis stated the no relationship between two variables and the
alternative hypothesis assessed the significance of these relationships. The
t-statistics were evaluated for hypothesis testing of Pearson’s correlation by
with a degree of freedom (df) n-2. Ultimately, a
p-value below 0.05 for the t-statistics signified a rejected null hypothesis
and mean significant relationship between the two variables as demonstrated in Table 6.
Our findings showed that a moderately significant relationship, such as between 0.41-0.6, was observed in most cases. The weak level of significant relationship was evident in some instances, such as between 0.2 and 0.4. Most of the independent variables exhibited a significant relationship, with the exceptions being between Total Protein (PROT) and Alkaline Phosphatase (ALP), Alanine Aminotransferase (ALT), Creatinine (CREA), Bilirubin (BIL), Aspartate Aminotransferase (AST), and Gamma-Glutamyl Transferase (GGT).
The computing Pearson correlation matrix displayed a different color in
Figure 3, derived from
Table 6, utilizes varying shades to enhance clarity. Light shading indicates moderate correlations, while dark shading represents strong correlations. Most independent variables are depicted with moderate and light shadings, suggesting inter-variable correlations or multicollinearity issues. The average of mean absolute percentage error
Table 7was computed using OLS and eight Liu parameters with three scale options by generating 1,000 replications from all dataset. The selection of 50, 100, 150, and 200 sample sizes mirrored those in the simulation data.
Table 8 reveals that modified Liu parameters (dMSE and dR2) exhibited consistent and often superior accuracy prediction across all scenarios. The dCp, dMSE, and dR2 methods notably demonstrated commendable estimation in all sample sizes that better the original method as OLS. Consequently, the Liu parameter adjustment using the dCp, dMSE, and dR2 methods for ten independent variables consistently surpassed expectations and aligned closely with simulation outcomes. Although there were slight discrepancies in estimation when the sample sizes increased, substantial performance enhancements were evident with small sample sizes within the Hepatitis C dataset.
5. Discussion
The simulated results, presented in
Table 1,
Table 2,
Table 3 and
Table 4, revealed that the mean of average percentage error was affected by the number of independent variables and sample sizes. The modified Liu estimator (dR2) exhibited superior performance with all independent variables and all sample sizes, whereas dMSE slightly differed from dR2. However, the average mean of average percentage error for significant independent variables was lower than that for small independent variables. The increase in the correlation coefficient was weak impact estimation in most methods, as indicated by the slight variation in the mean of average percentage error. Moreover, as the sample size increased, the performance estimation of all methods improved consistently.
In the same direction, the real data results in
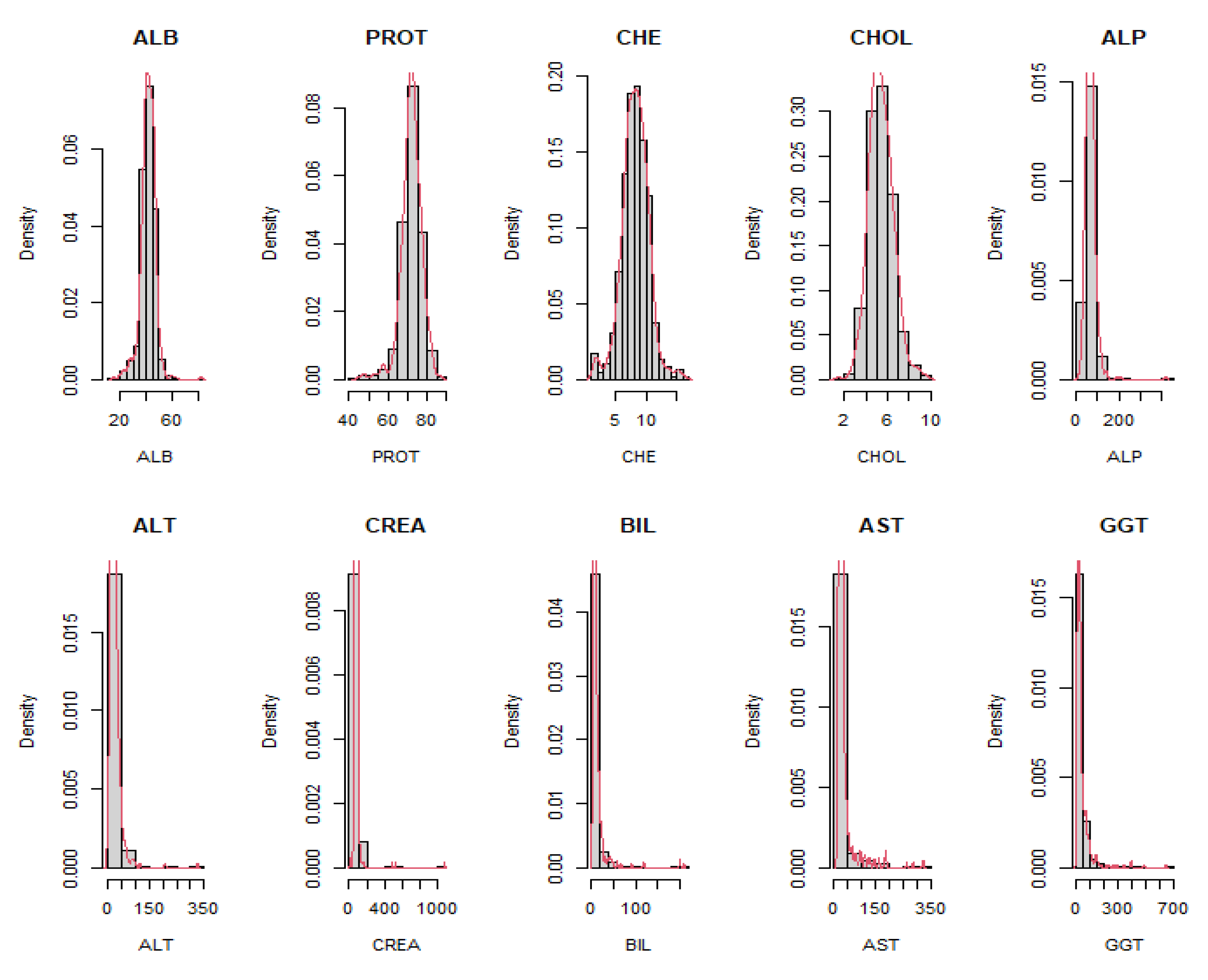
Table 7 showcased that the proposed Liu parameters (dMSE and dR2) achieved the minor mean of average percentage error for datasets with eight independent variables. It was observed that the real data’s independent variables exhibited skewed distributions, as illustrated in
Figure 4, confirmed by the Shapiro-Wilk test [
24], indicating non-normality. So, the dCp effectively estimated large sample sizes using the center option. Notably, the discrepancy between the simulated and real data results emphasized the importance of considering the data source when selecting the Liu parameter.
The proposed Liu parameters (dMSE and dR2) emerged as the most suitable for the Liu estimator. The medical dataset is widely used to predict medical diagnosis enhancement for classification patients. However, the Hepatitis C dataset is a medical dataset used to predict the patient’s age in the multiple regression model with multicollinearity problem among the independent variables. Oladapo et al. [
25] introduced a novel modified Liu Ridge-type estimator for estimating parameters in the general linear model, employing Portland cement data as a case study akin to medical data. Their proposed estimator demonstrates superior performance under certain conditions. Baber et al. [
21] adapted Liu estimators to address multicollinearity issues in linear regression, utilizing tobacco data. They advocate for adopting these new estimators by practitioners facing high to severe multicollinearity among independent variables. Hammond et al. [
26] employed a Liu estimator in inverse Gaussian regression, tackling multicollinearity in chemistry datasets. While considering the Liu estimator in multicollinearity based on multiple regression, the proposed Liu estimator outperforms the other. In summary, we always recommend that the Liu estimator user modify the Liu parameter in high multicollinearity.
6. Conclusions
This paper proposes a Liu parameter to estimate the Liu estimator in a multiple regression model correlated among independent variables, called multicollinearity. The selection of the Liu parameter is investigated and compared to the best performance. According to the simulation studies, the dR2 is always superior in terms of the mean of average percentage error for all levels of correlation, sample sizes, and dependent variables. For application in real data, the dCp, dMSE, and dR2 show the best performance, especially dR2. Moreover, the modified Liu parameter performs better than the OLS method in simulation and real data. The Liu parameter can significantly improve the estimator in terms of the regression model when the independent variables have the multicollinearity problem in low and high correlation. Therefore, the recommendation is to use a Liu parameter in the zero range and one that gives the best estimation. 6. Patents
Acknowledgments
This research is supported by King Mongkut’s Institute of Technology Ladkrabang.
References
- Daoud, J.I. Multicollinearity and regression analysis. J. Phys. Conf. Ser. 2017, 949, 1–7. [Google Scholar] [CrossRef]
- Shrestha, N. Detecting multicollinearity in regression analysis. Am. J. Appl. Math. 2020, 8, 39–42. [Google Scholar] [CrossRef]
- Dawoud, I.; Abonazel, M.R.; Awwad, F.A. Modified Liu estimator to address the multicollinearity problem in regression models: a new biased estimation class. Sci. Afr. 2022, 17, 1–12. [Google Scholar] [CrossRef]
- Jahufer, A. Detecting global influential observations in Liu regression model. Open J. Stat. 2013, 3, 1–7. [Google Scholar] [CrossRef]
- Karlsson, P.; Månsson, K.; Golam Kibria, B.M. A Liu estimator for the beta regression model and its application to chemical data,” J. Chemom. 2020, 24, 2–16. [Google Scholar] [CrossRef]
- Liu, K. A new class of biased estimate in linear regression. Commun. Stat-Theor. M. 1993, 22, 393–402. [Google Scholar]
- Liu, X. -Q. Improved Liu Estimation in a linear regression model. J. Stat. Plan. Inference. 2011, 141, 189–196. [Google Scholar] [CrossRef]
- Yang, H.; Xu, J. An alternative stochastic restricted Liu estimator in linear regression. Stat. Pap. 2009, 50, 639–647. [Google Scholar] [CrossRef]
- Hubert, M.H.; Wijekoon, P. Improvement of the Liu estimator in linear regression model. Stat. Pap. 2006, 47, 471–479. [Google Scholar] [CrossRef]
- Akdeniz, F.; Erol, H. Mean squared error matrix comparison of some biased estimators in linear regression. Commun. Stat-Theor. M. 2003, 32, 2389–2413. [Google Scholar] [CrossRef]
- Özkale, M.R.; Kaçiranlar, S. The restricted and unrestricted two-parameter estimators. Commun. Stat-Theor. M. 2007, 36, 2707–2725. [Google Scholar] [CrossRef]
- Dawoud, I.; Abonazel, M.R.; Awwad, F.A. Modified Liu estimator to address the multicollinearity problem in regression model: A new biased estimation class. Sci. Afr. 2002, 17, 1–12. [Google Scholar] [CrossRef]
- Suhail, M.; Babar, I.; Khan, Y.A.; Imran, M.; Nawaz, Z. Quantile-based estimation of Liu parameter in the linear regression model: Applications to Portland cement and US crime data. Math. Probl. Eng. 2021, 2021, 1–11. [Google Scholar] [CrossRef]
- Lukman, A.F.; Golam Kibria, B.M.; Ayinde, K.; Jegede, S.L. Modified one-parameter Liu estimator for the linear regression model. Mod. Sim. Eng. 2020, 2020, 1–17. [Google Scholar] [CrossRef]
- Lukman, A.F.; Ayinde, K.; Kun, S.S.; Adewuyi, E.T. A Modified new two-parameter estimator in a linear regression model,” Mod. Sim. Eng. 2019, 2019, 1–10. [Google Scholar]
- Filzmoser, P.; Kurnaz, F.S. A robust Liu regression estimator. Commun. Stat. Simul. Comput. 2018, 47, 432–443. [Google Scholar] [CrossRef]
- Druilhet, P.; Mom, A. Shrinkage Structure in Biased Regression. J. Multivar. Anal. 2008, 99, 232–244. [Google Scholar] [CrossRef]
- Akdeniz, F.; Kacįranlar, S. More on the new biased estimator in linear regression. Sankhya: Indian J. Stat., Series B (1960-2002). 2001, 63, 321–325. [Google Scholar]
- Duran, E.R.; Akdeniz, F.; Hu, H. Efficiency of a Liu-type estimator in semiparametric regression models. J. Comput. Appl. Math. 2011, 235, 1418–1428. [Google Scholar] [CrossRef]
- Abdelwahab, M.M.; Abonazel, M.R.; Hammad, A.T.; El-Masry, A.M. Modified two-parameter Liu estimator for addressing multicollinearity in the Poisson regression model. Axioms. 2024, 13, 1–22. [Google Scholar] [CrossRef]
- Babar, I.; Ayed, H.; Chand, S.; Suhail, M.; Khan, Y.A.; Marzouki, R. Modified Liu estimators in the linear regression model: An application to Tobacco data. Plos One. 2021, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Mallows, C.L. Some Comments on Cp. Technometrics. 2012, 42, 87–94. [Google Scholar]
- Belsley, D.A. A Guide to using the collinearity diagnostics. Com. Sci. Eco. Mana. 1991, 4, 33–50. [Google Scholar] [CrossRef]
- Shapiro, S.S.; Wilk, M.P. An analysis of variance test for normality (complete samples). Biometrika. 1965, 52, 591–611. [Google Scholar] [CrossRef]
- Oladapo, O.J.; Owolabi, A.T.; Idowu, J.I.; Ayinde, K. A new modified Liu Ridge-Type estimator for the linear regression model: Simulation and application. Int. J. Clin. Biostat. Biom. 2022, 8, 1–14. [Google Scholar]
- Hammood, N.M.; Jabur, D.M.; Algamal, Z.Y. A Liu estimator in inverse Gaussian regression model with application in chemometrics. Math. Stat. Eng. Appl. 2022, 71, 248–266. [Google Scholar]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).